Abstract
Natural killer (NK) cells are a population of lymphocytes that function in both immune defense and reproduction. Diversifying NK cell phenotype and function are interactions between NK cell receptors and major histocompatibility complex (MHC) class I ligands. As a consequence of strong and variable selection these ligand-receptor systems are polymorphic, rapidly evolving, and considerably species-specific. Counterparts to the human system of HLA class I ligands and killer cell immunoglobulin-like receptors (KIR) are present only in apes and Old World monkeys. HLA-C, the dominant ligand for human KIR and the only polymorphic HLA class I expressed by trophoblast, is further restricted to humans and great apes. Even then, the human system appears qualitatively different from that of chimpanzees, in that it has evolved a genetic balance between particular groups of receptors and ligands that favor reproductive success and other groups of receptors and ligands that have been correlated with disordered placentation. Human populations that have survived successive episodes of epidemic disease and population bottlenecks maintain a breadth of diversity for KIR and HLA class I, implying that loss of such diversity disfavors long-term survival of a human population.
Keywords: HLA, natural killer cells, balancing selection, evolution, immunity, reproduction
Immunogenetics was a term coined originally to describe the use of antibodies as tools for distinguishing the alternative alleles of genes that influence the structure of proteins and carbohydrates on blood cell surfaces [1]. As it developed, immunogenetics adapted and expanded to include all manner of studies that examine the natural, inherited variation of immune-system genes. Consequently, immunogenetics focuses on the less conventional genes without a dominant wild-type having ‘optimal’ function, but are represented instead by a variety of ‘sub-optimal’ forms having complementary functions maintained by balancing selection. For such genes, the members of a population exhibit different allele combinations; making it necessary to consider gene function, not only in the context of the individual, but also of the species and its constituent populations.
Perhaps the most obvious example of balancing selection is the maintenance of both chromosomes X and Y in human populations, reflecting the synergistic functions of men and women in reproduction. While many women appreciate that presence of a Y chromosome is not necessary for the development and survival of a healthy human individual, the presence in a population of XY males, as well as XX females, is essential for creating the next generation and for long-term species survival. Given this centrality of balancing selection in human reproduction, it is hardly surprising that certain molecular and cellular processes of reproduction are also subject to balancing selection and the compromises such selection implies. In particular, formation and function of the placenta involves a nine-month co-operation between the cells of two individuals who share one haploid genome, but differ at the other.
Pregnancy, Transplantation and Immunological Alloreactivity
The unusual physiological situation in pregnancy, where a genetically disparate fetus is implanted within a mother’s womb, was famously compared to that of a foreign organ graft. Ever since Peter Medawar’s influential ruminations on the subject some sixty years ago [2], immunologists have questioned and disputed why the pregnant mother’s immune system does not reject the genetically histoincompatible baby she carries. Less contentious are the positive data showing that pregnant women do indeed eject the babies they carry, but only after an appropriate nine-month period of gestation. It is only then, with the physical trauma of childbirth, that the mother’s immune system is stimulated to make antibodies against those protein products of the child’s paternally-inherited genes that differ in sequence from their maternal counterparts. Antibodies raised against such intra-species (allogeneic) differences are called alloantibodies, their target antigens, alloantigens.
The abundance and strength of alloantibodies can increase with each successive baby a mother conceives by the same father, but this progression has no apparent deleterious effects on reproductive success. Sera donated by multiparous women were a crucial element to the discovery and characterization of the human major histocompatibility complex (MHC), a ~5 megabase region on the short arm of human chromosome 6 that contains the most highly polymorphic genes in the human genome. These genes are also the principal cause of acute graft rejection following organ transplantion and of acute graft-versus-host disease (GVHD) following bone-marrow transplantation. The human MHC was named the human leukocyte antigen (HLA) complex because the highly polymorphic alloantigens it encodes are expressed by the white cells of the blood (leukocytes), but not the red cells (erythrocytes), whose A/B/O, rhesus and other alloantigens had been a previous focus of investigation for several HLA pioneers [3].
Acute graft rejection and acute GVHD are diseases mediated by T-lymphocytes (commonly called T cells) that are stimulated by HLA differences between the transplant donor and the transplant recipient. Acute rejection of an organ transplant is caused by recipient T cells, whereas graft-versus-host disease is caused by donor-derived T cells in the bone-marrow graft that attack and damage the recipient’s tissues, particularly the skin and the intestinal epithelium. Despite the non-physiological circumstances of their occurrence, the T-cell response in these transplantation syndromes reflects the natural physiological function of highly polymorphic HLA molecules. Namely the presentation of peptide antigens to αβ T-cell receptors, engagements necessary for generating protective T-cell mediated immune responses against infection and cancer, as well as a spectrum of undesirable autoimmune and hypersensitivity diseases.
Extra-villous Trophoblast expresses MHC class I but not MHC class II
Polymorphic HLA molecules are of two types that present peptide antigens to functionally distinctive classes of T cell: HLA class I (HLA-A, -B and -C) present antigens to CD8 T cells, whereas HLA class II (HLA-DP, -DQ and -DR) present antigens to CD4 T cells. Of these two structural variations on a common ancestral theme, the class II molecules have the more specialized function as evidenced by their restricted expression to professional antigen-presenting cells (dendritic cells, macrophages, monocytes and B cells) and by their dedicated function as ligands for the αβ TCR of CD4 T cells. In contrast, class I molecules are expressed on a wide range of the body’s cells where they serve as ligands for receptors of the lymphocytes known as natural killer (NK) cells, as well as for the αβ TCR of CD8 T cells. These contrasting characteristics, along with several other lines of circumstantial evidence, are all consistent with an evolutionary model in which class I is closer to the ancestral form of the MHC molecule and MHC class II is more recently derived.
Until recently, all species investigated for the presence of MHC molecules were found either to have or lack both MHC class I and II. Breaking this tie were the results obtained from sequencing the genome of Atlantic cod. This bony fish genome was found to be enriched for MHC class I genes but lacking in genes for MHC class II and also for auxiliary proteins that are dedicated to the MHC class II pathway of antigen presentation [4]. Such species-wide immunodeficiency is consistent with MHC class II being the more specialized and dispensable form of MHC molecule, with MHC class I being the more plastic, but less dispensable, form. In human pregnancy, the only fetal cells that both contact the maternal circulation and have the capacity to stimulate maternal lymphocyte responses against HLA antigenic differences inherited from the father are the extravillous trophoblasts (EVT) of the placenta. Like Atlantic cod, EVT express a variety of forms of HLA class I but do not express HLA class II at all [5, 6]. Consequently, this review will concentrate on MHC class I.
The co-ordinated emergence of HLA-C and NK cells in immunological research
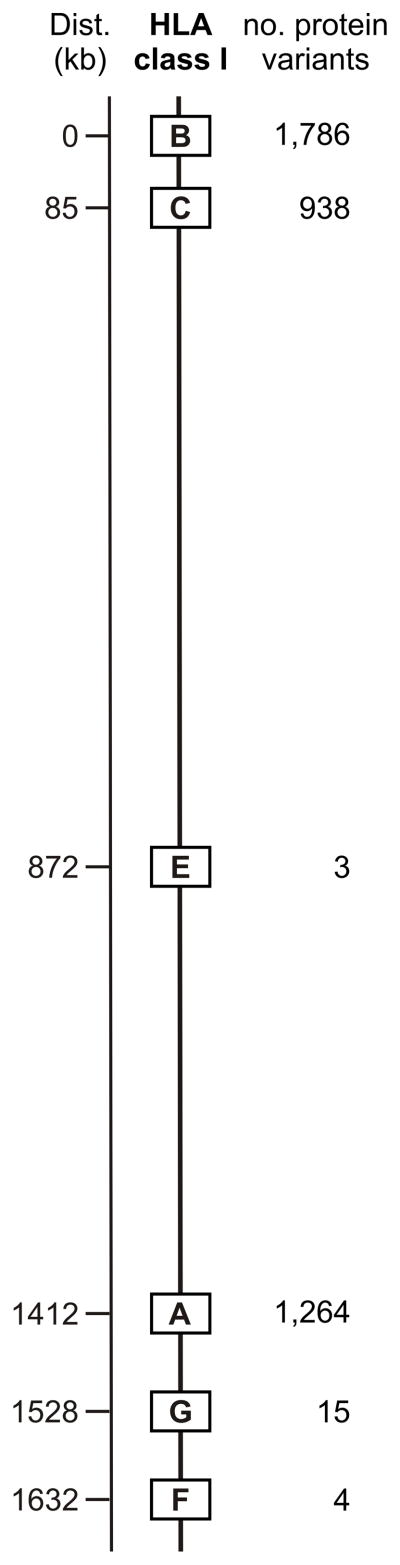
The HLA region contains six functional class I genes (HLA-A,-B,-C, -E, -F, and -G,) all of which are present on all HLA haplotypes (that is, they are fixed genes) (Figure 1). HLA-E, -F, and –G are conserved in the human species and resisted discovery until the late 1980s, when systematic genomic analysis was first used to characterize the class I genes and pseudogenes of the HLA complex [7, 8]. Being highly polymorphic, the HLA-A, -B, and –C genes were susceptible to detection by the alloantisera obtained from multiparous women. More variable and immunogenic than HLA-C, the HLA-A and HLA-B antigens were first uncovered. Throughout the 1960s an ever-increasing number of HLA-A and –B antigens were serologically detected, defined and named [9], a progression facilitated by the initiation of the International Histocompatibility Workshops, a series of HLA-based immunogenetics meetings that continues to the present day [10] (Figure 1). The 16th International Histocompatibility Workshop and Conference will be held at Liverpool, England in 2012 [11].
Figure 1.
Not until the 1970s did evidence for HLA-C antigens as a third highly polymorphic locus begin to emerge. The discoverer of HLA-C was Erik Thorsby, then a young Norwegian clinician scientist working for his PhD, who studied and collaborated with Flemming Kissmeyer-Nielsen in Aarhus, Denmark [12] before returning to make his name in Oslo [13]. (Although now officially retired from being Professor of Immunology at the University and Chairman of the Institute of Immunology at the Rikshospitalet, Thorsby remains internationally very active in HLA immunogenetics.) Despite the strength of Thorsby’s data and his advocacy for the locus, HLA-C remained a cinderella throughout the 1970s and 1980s, overshadowed by its sister loci: HLA-A and –B. That was an era when the presentation of peptides to killer CD8 T cells was considered the exclusive function of polymorphic HLA class I. Pushing HLA-C slowly towards the limelight were observations in the 1990s that HLA-C influenced the alloreactive response of the natural killer (NK) cell [14, 15], a type of lymphocyte that, in common with HLA-C, had been in the shadow of two meretricious siblings, the gene-rearranging B cells and T cells. Compounding interest in HLA-C, among leukemia patients receiving a bone-marrow transplant from an HLA haploidentical family member (a contrived clinical situation resembling that of physiological pregnancy) certain HLA-C mismatches stimulated alloreactive NK cell responses that correlated with protection from leukemic relapse [16, 17].
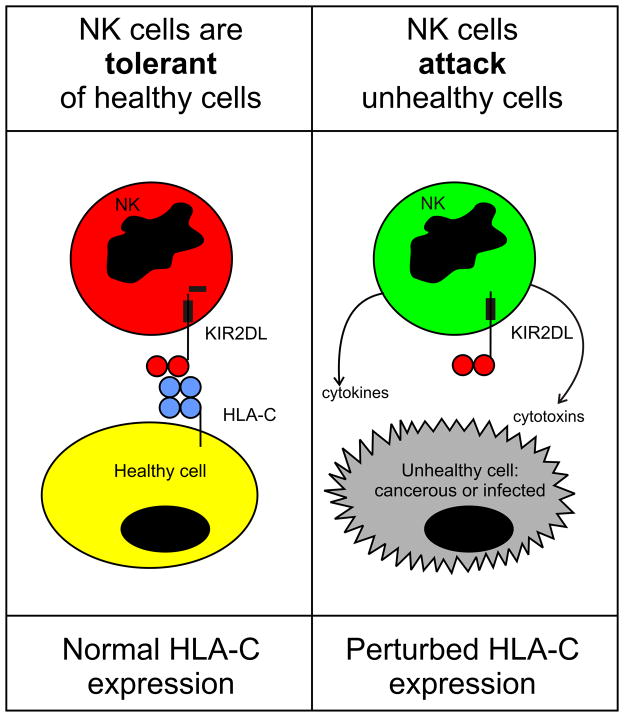
NK cells, long visualized by morphologists as the large granular lymphocytes of healthy blood [18], are distinguished by functional activities, notably antibody-dependent cellular cytotoxicity (ADCC), that were first observed by immunologists in the 1960s [19] and increasingly became chosen as a topic for investigation in the 1970s [20–22]. NK cells were often encountered as the annoying source of a high, ‘non-specific’, background killing in cytotoxic assays of T-cell function. Unlike T cells, NK cells did not require several days of stimulation in culture in order to become competent at killing target cells. They were named natural killer cells because when isolated from blood or tissue they had a natural ability to kill certain tumor target cell lines. A further, and critical, defining characteristic of NK cells was their propensity to kill cells having abnormally low levels of HLA class I on their surfaces: the extent of killing being inversely correlated with the amount of cell-surface MHC class I [23–25]. Because NK cells responded to the loss of self MHC class I (a frequent consequence of viral infection and malignant transformation) Karre and colleagues envisaged NK cells as recognizing ‘missing self’ [26]. The molecular basis for this correlation was found to lie with inhibitory NK cell receptors that recognize MHC class I and prevent NK cell attack (Figure 2). Three types of human inhibitory NK cell receptor recognize different features of HLA class I. A highly conserved interaction is that between HLA-E and CD94:NKG2A [27]. Also conserved is LILRB1, which binds a determinant (epitope) carried by a wide-range of HLA class I forms. However, LILRB1 has strongest avidity for HLA-G [28], which among healthy human cells is expressed only by extra-villous trophoblast [29]. Binding to polymorphic determinants of HLA-A, B and C are the killer cell immunoglobulin-like receptors (KIR) that contribute to the immunogenetics of human placentation [30].
Figure 2.
Evolution of HLA class I recognition by Killer cell immunoglobulin-like receptors
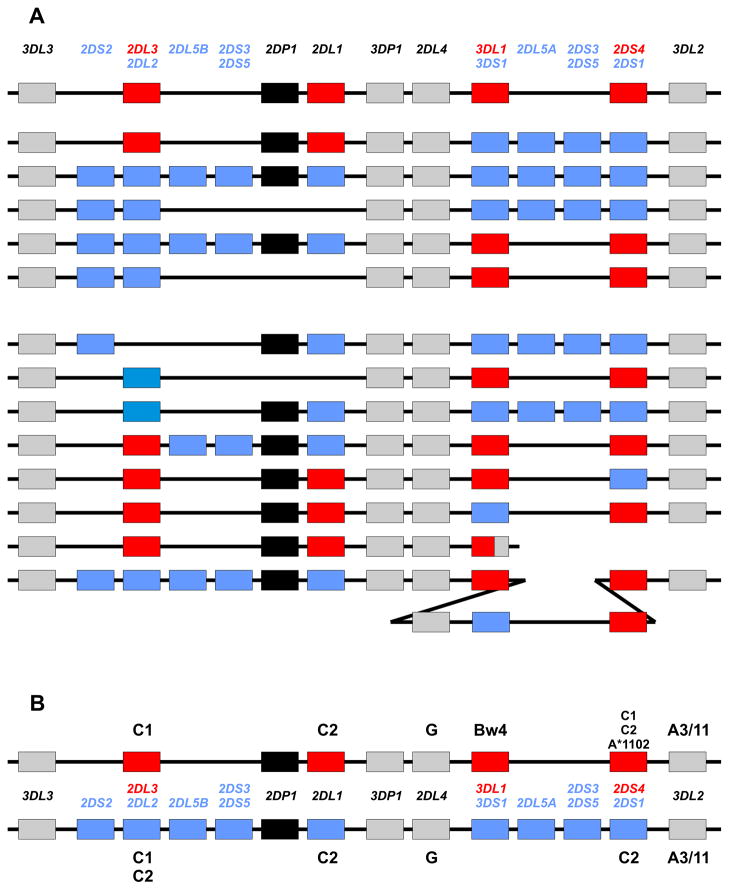
Human killer cell immunoglobulin-like receptors (KIR) are encoded by a variable and polymorphic family of up to 15 genes in the leukocyte-receptor complex on human chromosome 19 [31]. Population studies demonstrate that variation in KIR gene-content, combined with extensive polymorphism for some of the genes (Table I), for example KIR3DL1/S1 [32, 33], make human KIR variability comparable to that of HLA. Beside the genes encoding inhibitory KIR anticipated from missing-self recognition, the locus was found to harbor an unanticipated cohort of genes encoding activating KIR. According to their content of inhibitory and activating KIR genes, distinctive group A and group B KIR haplotype groups were defined [34] and refined [35] (Figure 3). From species comparisons, the KIR gene family is now clearly seen to be highly diverse, rapidly evolving [36, 37], extensively species-specific, and to have originated from one KIR gene in an ancestral simian primate [38]. The vast majority of mammalian species, including prosimians and rodents, do not have NK cell receptors corresponding to the human KIR. Whereas placental reproduction evolved >160 million years ago [39], the system of KIR that is now central to the immunogenetic modulation of placentation in humans and other simian primates, although the latter needs incisive investigation, is from an evolutionary perspective recently elaborated during the last ~85 million years [40].
Table I.
KIR genes, number of encoded proteins, and HLA ligands. Number of encoded proteins from http://www.ebi.ac.uk/ipd/kir/stats.html accessed Oct. 2011.
| KIR | No. protein variants | HLA class I ligands |
|---|---|---|
| 2DL1 | 24 | C2 |
| 2DL2 | 11 | C1, C2 |
| 2DL3 | 17 | C1 |
| 2DL4 | 22 | G? |
| 2DL5A | 5 | - |
| 2DL5B | 11 | - |
| 2DS1 | 7 | C2 |
| 2DS2 | 8 | - |
| 2DS3 | 5 | - |
| 2DS4 | 13 | A11, some C |
| 2DS5 | 11 | - |
| 3DL1 | 58 | Bw4 |
| 3DS1 | 12 | - |
| 3DL2 | 61 | A3/11 |
| 3DL3 | 55 | - |
| 3DP1 | 0 | O |
| 2DP1 | 0 | O |
Figure 3.
Targets for KIR recognition are limited to just four HLA class I epitopes (A3/11, Bw4, C1 and C2) that centre on the helix of the α1 domain and involve residues 78–83. These four epitopes appear to be mutually exclusive variations on a structural theme, in that no HLA class I molecule can carry more than one of them [41]. Thus individual HLA-A, B, C allotypes carry either one of these epitopes or none of them (Figure 4). Carrying the A3/11 epitope is a small minority of HLA-A allotypes, whilst the Bw4 epitope is carried by a larger minority of HLA-A and HLA-B allotypes, such that ~50% of HLA haplotypes furnish a Bw4 ligand, another example of balancing selection [32]. Consequently the majority of HLA-A and HLA-B allotypes do not function as ligands for KIR, meaning they can give dedicated service to the demands of T-cell immunity. In contrast every HLA-C allotype has either the C1 or C2 epitope and functions as a KIR ligand. This is but one of several lines of evidence indicating that HLA-C evolved specifically to provide KIR ligands. Consistent with this proposition, all human individuals have at least C1 or C2, whereas a substantial fraction of individuals lack both A3/11 and Bw4.
Figure 4.
HLA-B*73 and HLA-B*46 are two unusual HLA-B allotypes that carry the C1 epitope and are ligands for C1-specific KIR [41, 42]. Consistent with the epitope exclusion principle, neither HLA-B*73 nor HLA-B*46 carries the Bw4 epitope, a feature of around one third of HLA-B allotypes. HLA-B*73 has an exceptionally divergent sequence, a very strong linkage-disequilibrium with HLA-C*15:05, and is most common in western Asia. These properties are consistent with HLA-B*73 having entered the modern human population as a consequence of meeting and mating with archaic humans who were already living in western Asia during the migration out of Africa some 65 thousand years ago [43]. Neanderthal and Denisovan HLA genotypes suggest this phenomenon of adaptive introgression of archaic HLA class I alleles has had a major influence on the HLA system of today’s European, Asian and Melanesian populations [43]. HLA-B*46 is restricted to south-east Asia where frequencies of up to 25% suggest it has been strongly selected. Unlike HLA-B*73:01 the sequence of HLA-B*46 is not divergent and could have arisen de novo by recombination and mutation in the ‘modern’ human population of south-east Asia [44]. However, it is also possible that the emergence of HLA-B*46 was a consequence of adaptive introgression. For both HLA-B*73 and HLA-B*46 their shared functional property, which may have conferred their selective advantage over other HLA-B allotypes, is the capacity of their C1 epitopes to influence NK cell biology.
In comparisons of human HLA class I to MHC class I of other primate species, we will use the terms MHC-A, MHC-B, and MHC-C to describe the orthologs of HLA-A, HLA-B, and HLA-C, respectively. Such comparison shows that MHC-C emerged more recently than MHC-A or -B, being present only in great apes and humans. By contrast, counterparts of HLA-A and HLA-B are also present in Old World monkeys [38, 45], and in considerable abundance [46, 47]. MHC-C likely evolved from an MHC-B like ancestor, which diverged under natural selection to become a superior ligand for KIR [44, 48]. The focus for this divergence was the α1 helix [49], a major site of contact between KIR and HLA-C [50]. Corresponding with this specialization of MHC-C is the change from using lineage II KIR with three Ig-like domains (D0, D1, and D2) to recognize epitopes of MHC-A and MHC-B to the use of lineage III KIR with two Ig-like domains (D1 and D2) to recognize epitopes of MHC-C. Although the human lineage III KIR do not have a D0 domain, their genes preserve an exon 3 which once encoded this domain, but became inactivated and in various different ways [51]. In contrast to the human situation, some of the chimpanzee lineage III KIR that recognize C1 or C2 retain a D0 domain. Our experiments to remove these D0 domains by mutagenesis had no detectable effect on the avidity and specificity for MHC-C [52].
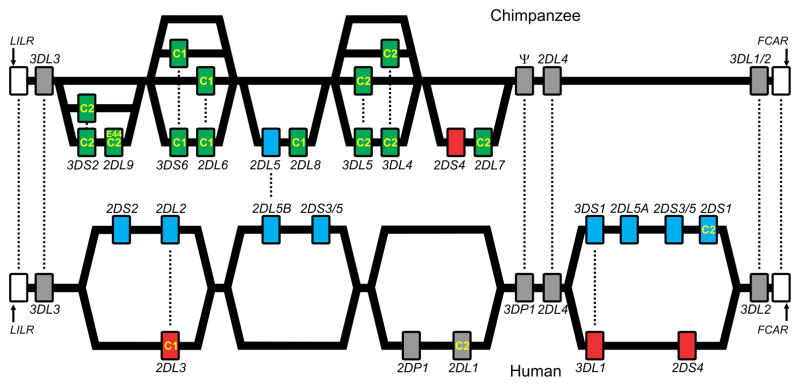
The MHC-B like ancestor of MHC-C is predicted to have carried the C1 epitope [44], consistent with all orangutan MHC-C allotypes carrying C1 [53]. Evolution of C2 from C1, by replacement of asparagine 80 with lysine, occurred in the common ancestor of human, chimpanzee and gorilla, after separation from the orangutan line. Comparison of the human and chimpanzee KIR/MHC systems revealed substantial differences. In humans the C1 and C2 epitopes are almost completely restricted to HLA-C, but in chimpanzee the C1 epitope is also carried by around one quarter of Patr-B allotypes (the chimpanzee ortholog to HLA-B). Although both species have a similar number of KIR genes, with a majority being lineage III KIR that recognize C1 or C2, the chimpanzee receptors are functionally more potent than their human counterparts [44, 54] (Figure 5). Notably, several of the human lineage III activating KIR (KIR2DS2, 2DS3 and 2DS5) have been under selection to reduce or lose their capacity to recognize HLA class I, as is also the case for lineage II KIR3DS1. These attenuated KIR are characteristic of the human group B KIR haplotypes and define their main differences from the group A haplotypes. Overall the chimpanzee KIR haplotypes are closer to human A haplotypes, whereas the B haplotypes are uniquely human (Figure 5). In addition to these qualitative differences, the alleles of other KIR that are associated with B haplotypes tend to be weaker than those associated with A haplotypes. For example, KIR2DL1*004, associated with B haplotypes, is a weaker receptor than KIR2DL1*003 associated with A haplotypes [55, 56]. This comparison indicates that the progressive evolution in hominoids of a system of potent MHC-C receptors, reaches its peak in the chimpanzee and has reversed in human evolution, where balance and compromise between potent A haplotypes and attenuated B haplotypes is now evident.
Figure 5.
Uterine NK cells and placentation
During embryo implantation and placentation, fetal extra-villous trophoblast (EVT) invades the uterine decidua and the inner myometrium with consequent remodeling of the spiral arteries into wider vessels that can better supply the placenta with blood. Among pregnancies there is natural variation in the depth and extent of invasion, which at the extremes of the normal distribution is correlated with a variety of pregnancy complications or disorders. Implicated in control of these processes are the distinctive uterine NK cells (uNK) that populate the decidua and which have phenotype and functional properties that distinguish them from peripheral blood NK cells. EVT, the only HLA class I expressing fetal cells that make direct contact with the maternal circulation and can interact directly with uNK, have a unique combination of HLA class I proteins on the cell surface: HLA-C, HLA-E and HLA-G. All three are ligands for NK cell receptors, but it is the interactions between HLA-C and lineage III KIR that have the greatest immunogenetic potential to vary the co-operation between EVT and uNK and ultimately the course of pregnancy. Consistent with this analysis, uNK cell populations have a bias, not seen for peripheral blood NK cells, towards expression of the lineage III KIR that recognize HLA-C [57, 58].
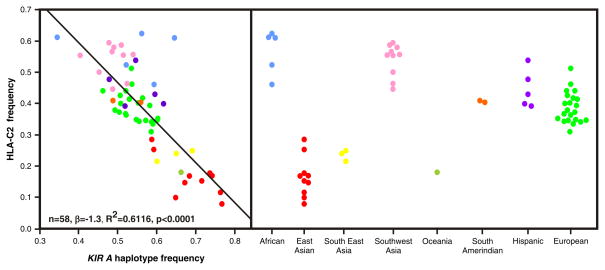
In a series of epidemiological studies [59–63], Moffett and colleagues have shown that susceptibility to several disorders of implantation (pre-eclampsia, recurrent abortion, and fetal growth restriction) is associated with the same genetic combination of a mother who is homozygous for group A KIR haplotypes and a baby who carries the C2 epitope, particularly if the C2 is paternally inherited and absent from the mother. The negative impact of this combination is reflected in the relative frequencies of the KIR A haplotype and the C2 epitope in human populations worldwide (Figure 6). They are inversely correlated, pointing to the strong selective pressure exerted by the consequences of implantation disorders: recurrent abortion represents a failure to reproduce, pre-eclampsia can kill both mother and child, and fetal growth deficit produces less competitive offspring. Although the inverse correlation appears to be universal the frequencies of group A KIR haplotype and C2 epitopes varies between populations. At the extremes are the Japanese, who have high KIR A and low C2 frequencies, and aboriginal Australians who have high KIR B and high C2 frequencies.
Figure 6.
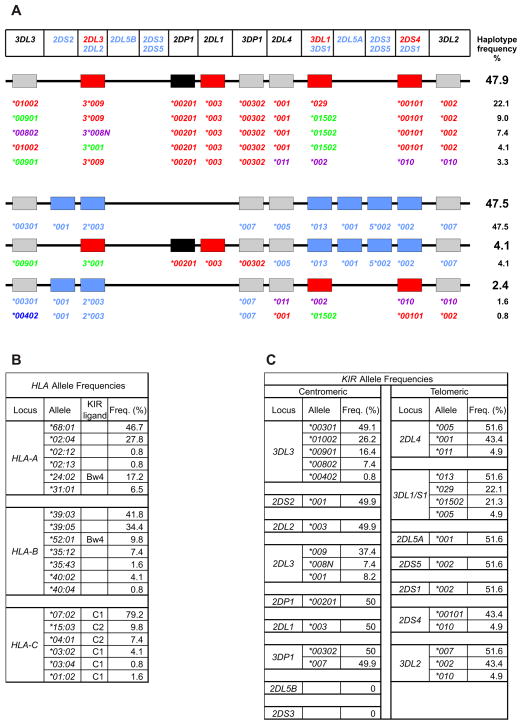
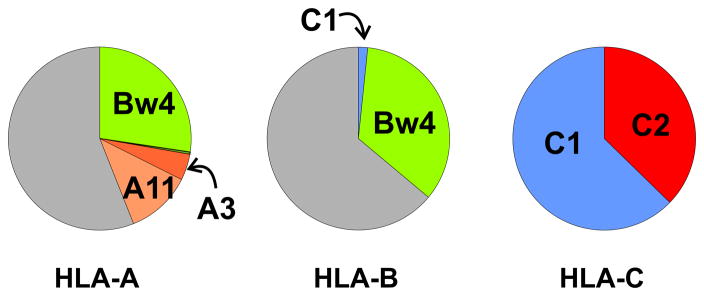
The principal NK cell receptors that recognize the C2 epitope are inhibitory KIR2DL1 and activating KIR2DS1. KIR2DL1 is a fixed gene of KIR A haplotypes, while KIR2DS1 is only present on KIR B haplotypes. Thus implantation disorders are associated with C2 on EVT interacting with KIR2DL1 on uNK cells to cause delivery of a strong inhibitory signal to the NK cells. This suggests a model in which inhibition of the NK cells causes insufficient invasion of the decidua by the EVT and insufficient remodelling of the spiral arteries. As implantation disorders are associated with KIR A homozygosity, a maternal KIR B haplotype can provide dominant protection against implantation disorder. Whereas all KIR A haplotypes have a fixed gene content, the B haplotypes exhibit gene content variability (Figure 3). Some B haplotypes have inhibitory KIR2DL1 in combination with activating KIR2DS1. The presence of KIR2DS1 reduces the likelihood of C2-mediated pregnancy disorder, indicating that interaction of KIR2DS1 with C2 on EVT transduces activating signals to NK cells that serve to counterbalance the inhibitory signals generated by C2 interactions with KIR2DL1 [59, 61]. Another type of KIR B haplotype, which has KIR2DS1 in the absence of KIR2DL1, might be expected to favor reproductive success. That this B haplotype has an unusually high frequency of 47% in Yucpa Amerindians (Figure 7A), a population that has survived successive disease epidemics and population bottlenecks while retaining HLA (Figure 7B) and KIR (Figure 7C) diversity, is consistent with this thesis [64]. It will be important to dissect out the influence of this 2DL1-negative B haplotype in future epidemiological studies of pregnancy disorders. Such analysis will, however, require improvements in the typing and assignment of KIR haplotypes combined with larger study cohorts than has been achieved to date.
Figure 7.
In addition to the presence or absence of the KIR2DL1 and KIR2DS1 genes on KIR haplotypes, a further dimension to the variation is provided by allelic polymorphism, particularly for KIR2DL1. In many populations the common allotype, KIR2DL1*003, is both strong and highly specific. In contrast, KIR2DL1*004, that is associated with B haplotypes, has one substitution in the transmembrane that weakens the signal transduction [55] and others in the extracellular D2 domain that reduces the strength of its binding to C2 (unpublished data). Thus the presence of this weaker form of KIR2DL1 on some B haplotypes may also contribute to the protection they offer against implantation disorders. We are currently comparing the strength and specificity of the ~20 different KIR2DL1 variants that have been defined to date. Polymorphism in the diverse set of HLA-C allotypes that carry the C2 epitope is also likely to be a factor that further diversifies the interaction of KIR2DL1 and KIR2DS1 with C2. The quantitative binding assay that we have used to assess the strength and specificity of KIR for HLA class I shows there is reproducible variation in the binding of the same KIR2DL1 allotype to different C2 bearing HLA-C [42]. We hypothesize, that such differences arise from the different spectra of peptides that are bound to each HLA-C allotype and the extent to which they are permissive for KIR2DL1 binding.
Concluding remarks
Although Medawar’s analysis of immunological aspects to the feto-maternal relationship remains widely cited by reproductive immunologists [65, 66], his assessment of MHC polymorphism and challenge to the emerging field of immunogenetics has proved less of a touchstone. At the time of Medawar’s discussion of ‘Some immunological and endocrinological problems raised by the evolution of viviparity in vertebrates’ [2], nothing was known of the human HLA system. In contrast, for inbred laboratory mice considerable knowledge of an extensive system of MHC class I alloantigens had been acquired through using sera obtained from inter-strain immunizations and serological assays of red-cell agglutination (mouse MHC class I being detectable on red cells), as well as genetic analysis of experimental transplantation [1, 3]. The complexity of these immunogenetic data, combined with their lack of any plausible physiological foundation, induced a bafflement in all but the most broad-minded of non experts that cannot be exaggerated, and continued for another twenty years, well into the 1970s. This is aptly captured by the following two quotes from Medawar [2]:
“It is an interesting fact that the only known consequences of this antigenic diversity are wholly harmful. By ‘antigenic diversity’ I must be understood to mean antigenic diversity as such. If chemistry were a less backward science, all the differences that we are now obliged to recognize as antigenic differences would be chemically defined, and the agglutination tube would be dispossessed of its functions by the ordinary test-tube.”
“Although there are no factual grounds for supposing that antigenic diversity is anything but an unfortunate consequence of constitutional differences between the individuals of a species, yet one is under some obligation to rack one’s brains for evidences of any good it might conceivably do. Only thus can antigenic polymorphism be made genetically respectable”
Although papers and lectures on MHC polymorphism are still reliably capable of baffling their readers and listeners, the 58 years of research since Medawar’s challenge has without doubt given the antigenic diversity of highly polymorphic MHC class I molecules, chemical, genetic, and functional respectability. One proof of this pudding being that ‘antigenic diversity of MHC’ is a phrase no longer used. Essential to this achievement has been the comparison, genotypically, of very many humans with very few mice, members of two species who differ significantly, not only in the nature of their MHC class I molecules, but also in the methods and philosophy investigators have used to study them [67, 68]. Further proof is the individuality that HLA and KIR impose on human immune systems and the reliability with which these polymorphisms are most strongly associated with an extraordinarily wide range of human disease. Following in the twentieth century footsteps of HLA a series of international KIR workshops was initiated in this twenty-first century [69].
Acknowledgments
This research was supported by NIH grants R01 AI17892, R01 AI24258, R01 AI31168, and P01 CA111412 (Project 1) to P.P.
Footnotes
Conflict of Interest
The authors have no conflict of interest to declare.
References
- 1.Snell GD. Immunogenetics: Retrospect and prospect. Immunogenetics. 1974;1(1):1–3. [Google Scholar]
- 2.Medawar PB. Some immunological and endocrinological problems raised by the evolution of viviparity in vertebrates. Symp Soc Exp Biol. 1953;7:320–338. [Google Scholar]
- 3.Thorsby E. A short history of HLA. Tissue Antigens. 2009;74(2):101–16. doi: 10.1111/j.1399-0039.2009.01291.x. [DOI] [PubMed] [Google Scholar]
- 4.Star B, Nederbragt AJ, Jentoft S, Grimholt U, Malmstrom M, Gregers TF, Rounge TB, Paulsen J, Solbakken MH, Sharma A, Wetten OF, Lanzen A, Winer R, Knight J, Vogel JH, Aken B, Andersen O, Lagesen K, Tooming-Klunderud A, Edvardsen RB, Tina KG, Espelund M, Nepal C, Previti C, Karlsen BO, Moum T, Skage M, Berg PR, Gjoen T, Kuhl H, Thorsen J, Malde K, Reinhardt R, Du L, Johansen SD, Searle S, Lien S, Nilsen F, Jonassen I, Omholt SW, Stenseth NC, Jakobsen KS. The genome sequence of Atlantic cod reveals a unique immune system. Nature. 2011;477(7363):207–10. doi: 10.1038/nature10342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Apps R, Murphy SP, Fernando R, Gardner L, Ahad T, Moffett A. Human leucocyte antigen (HLA) expression of primary trophoblast cells and placental cell lines, determined using single antigen beads to characterize allotype specificities of anti-HLA antibodies. Immunology. 2009;127(1):26–39. doi: 10.1111/j.1365-2567.2008.03019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hunt JS. Stranger in a strange land. Immunol Rev. 2006;213:36–47. doi: 10.1111/j.1600-065X.2006.00436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geraghty DE, Koller BH, Hansen JA, Orr HT. The HLA class I gene family includes at least six genes and twelve pseudogenes and gene fragments. J Immunol. 1992;149(6):1934–46. [PubMed] [Google Scholar]
- 8.Shimizu Y, Geraghty DE, Koller BH, Orr HT, DeMars R. Transfer and expression of three cloned human non-HLA-A,B,C class I major histocompatibility complex genes in mutant lymphoblastoid cells. Proc Natl Acad Sci U S A. 1988;85(1):227–31. doi: 10.1073/pnas.85.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marsh SG, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA, Fernandez-Vina M, Geraghty DE, Holdsworth R, Hurley CK, Lau M, Lee KW, Mach B, Maiers M, Mayr WR, Muller CR, Parham P, Petersdorf EW, Sasazuki T, Strominger JL, Svejgaard A, Terasaki PI, Tiercy JM, Trowsdale J. Nomenclature for factors of the HLA system, 2010. Tissue Antigens. 2010;75(4):291–455. doi: 10.1111/j.1399-0039.2010.01466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.http://ihwg.org/about/history.html
- 11.http://www.16ihiw.org/index.html
- 12.Thorsby E, Sandberg L, Lindholm A, Kissmeyer-Nielsen F. The HL-A system: evidence of a third sub-locus. Scand J Haematol. 1970;7(3):195–200. doi: 10.1111/j.1600-0609.1970.tb01887.x. [DOI] [PubMed] [Google Scholar]
- 13.Solheim BG, Bratlie A, Sandberg L, Staub-Nielsen L, Thorsby E. Further evidence of a third HL-A locus. Tissue Antigens. 1973;3(6):439–53. doi: 10.1111/j.1399-0039.1973.tb00514.x. [DOI] [PubMed] [Google Scholar]
- 14.Colonna M, Borsellino G, Falco M, Ferrara GB, Strominger JL. HLA-C is the inhibitory ligand that determines dominant resistance to lysis by NK1- and NK2-specific natural killer cells. Proc Natl Acad Sci U S A. 1993;90(24):12000–4. doi: 10.1073/pnas.90.24.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colonna M, Spies T, Strominger JL, Ciccone E, Moretta A, Moretta L, Pende D, Viale O. Alloantigen recognition by two human natural killer cell clones is associated with HLA-C or a closely linked gene. Proc Natl Acad Sci U S A. 1992;89(17):7983–5. doi: 10.1073/pnas.89.17.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruggeri L, Capanni M, Casucci M, Volpi I, Tosti A, Perruccio K, Urbani E, Negrin RS, Martelli MF, Velardi A. Role of natural killer cell alloreactivity in HLA-mismatched hematopoietic stem cell transplantation. Blood. 1999;94(1):333–9. [PubMed] [Google Scholar]
- 17.Ruggeri L, Capanni M, Urbani E, Perruccio K, Shlomchik WD, Tosti A, Posati S, Rogaia D, Frassoni F, Aversa F, Martelli MF, Velardi A. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science. 2002;295(5562):2097–100. doi: 10.1126/science.1068440. [DOI] [PubMed] [Google Scholar]
- 18.Timonen T, Ortaldo JR, Herberman RB. Characteristics of human large granular lymphocytes and relationship to natural killer and K cells. J Exp Med. 1981;153(3):569–82. doi: 10.1084/jem.153.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacLennan IC, Loewi G. Effect of specific antibody to target cells on their specific and non-specific interactions with lymphocytes. Nature. 1968;219(5158):1069–70. doi: 10.1038/2191069a0. [DOI] [PubMed] [Google Scholar]
- 20.Herberman RB, Holden HT. Natural cell-mediated immunity. Adv Cancer Res. 1978;27:305–77. doi: 10.1016/s0065-230x(08)60936-7. [DOI] [PubMed] [Google Scholar]
- 21.Kiessling R, Haller O. Natural killer cells in the mouse: an alternative immune surveillance mechanism? Contemp Top Immunobiol. 1978;8:171–201. doi: 10.1007/978-1-4684-0922-2_6. [DOI] [PubMed] [Google Scholar]
- 22.Roder JC, Karre K, Kiessling R. Natural killer cells. Prog Allergy. 1981;28:66–159. [PubMed] [Google Scholar]
- 23.Karre K, Ljunggren HG, Piontek G, Kiessling R. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature. 1986;319(6055):675–8. doi: 10.1038/319675a0. [DOI] [PubMed] [Google Scholar]
- 24.Storkus WJ, Alexander J, Payne JA, Dawson JR, Cresswell P. Reversal of natural killing susceptibility in target cells expressing transfected class I HLA genes. Proc Natl Acad Sci U S A. 1989;86(7):2361–4. doi: 10.1073/pnas.86.7.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Storkus WJ, Howell DN, Salter RD, Dawson JR, Cresswell P. NK susceptibility varies inversely with target cell class I HLA antigen expression. J Immunol. 1987;138(6):1657–9. [PubMed] [Google Scholar]
- 26.Ljunggren HG, Karre K. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol Today. 1990;11(7):237–44. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- 27.Sullivan LC, Clements CS, Rossjohn J, Brooks AG. The major histocompatibility complex class Ib molecule HLA-E at the interface between innate and adaptive immunity. Tissue Antigens. 2008;72(5):415–24. doi: 10.1111/j.1399-0039.2008.01138.x. [DOI] [PubMed] [Google Scholar]
- 28.Shiroishi M, Tsumoto K, Amano K, Shirakihara Y, Colonna M, Braud VM, Allan DS, Makadzange A, Rowland-Jones S, Willcox B, Jones EY, van der Merwe PA, Kumagai I, Maenaka K. Human inhibitory receptors Ig-like transcript 2 (ILT2) and ILT4 compete with CD8 for MHC class I binding and bind preferentially to HLA-G. Proc Natl Acad Sci U S A. 2003;100(15):8856–61. doi: 10.1073/pnas.1431057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kovats S, Main EK, Librach C, Stubblebine M, Fisher SJ, DeMars R. A class I antigen, HLA-G, expressed in human trophoblasts. Science. 1990;248(4952):220–3. doi: 10.1126/science.2326636. [DOI] [PubMed] [Google Scholar]
- 30.Moffett A, Loke C. Immunology of placentation in eutherian mammals. Nat Rev Immunol. 2006;6(8):584–94. doi: 10.1038/nri1897. [DOI] [PubMed] [Google Scholar]
- 31.Barrow AD, Trowsdale J. The extended human leukocyte receptor complex: diverse ways of modulating immune responses. Immunol Rev. 2008;224:98–123. doi: 10.1111/j.1600-065X.2008.00653.x. [DOI] [PubMed] [Google Scholar]
- 32.Norman PJ, Abi-Rached L, Gendzekhadze K, Korbel D, Gleimer M, Rowley D, Bruno D, Carrington CV, Chandanayingyong D, Chang YH, Crespi C, Saruhan-Direskeneli G, Fraser PA, Hameed K, Kamkamidze G, Koram KA, Layrisse Z, Matamoros N, Mila J, Park MH, Pitchappan RM, Ramdath DD, Shiau MY, Stephens HA, Struik S, Verity DH, Vaughan RW, Tyan D, Davis RW, Riley EM, Ronaghi M, Parham P. Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nat Genet. 2007;39(9):1092–9. doi: 10.1038/ng2111. [DOI] [PubMed] [Google Scholar]
- 33.Parham P, Norman PJ, Abi-Rached L, Guethlein LA. Variable NK cell receptors exemplified by human KIR3DL1/S1. J Immunol. 2011;187(1):11–9. doi: 10.4049/jimmunol.0902332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, Corliss B, Tyan D, Lanier LL, Parham P. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7(6):753–63. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- 35.Pyo CW, Guethlein LA, Vu Q, Wang R, Abi-Rached L, Norman PJ, Marsh SG, Miller JS, Parham P, Geraghty DE. Different patterns of evolution in the centromeric and telomeric regions of group a and B haplotypes of the human killer cell Ig-like receptor locus. PLoS One. 2010;5(12):e15115. doi: 10.1371/journal.pone.0015115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khakoo SI, Rajalingam R, Shum BP, Weidenbach K, Flodin L, Muir DG, Canavez F, Cooper SL, Valiante NM, Lanier LL, Parham P. Rapid evolution of NK cell receptor systems demonstrated by comparison of chimpanzees and humans. Immunity. 2000;12(6):687–98. doi: 10.1016/s1074-7613(00)80219-8. [DOI] [PubMed] [Google Scholar]
- 37.Vilches C, Parham P. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol. 2002;20:217–51. doi: 10.1146/annurev.immunol.20.092501.134942. [DOI] [PubMed] [Google Scholar]
- 38.Parham P, Abi-Rached L, Matevosyan L, Moesta AK, Norman PJ, Older Aguilar AM, Guethlein LA. Primate-Specific Regulation of Natural Killer Cells. J Med Primatol. 2010;39(4):194–212. doi: 10.1111/j.1600-0684.2010.00432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo ZX, Yuan CX, Meng QJ, Ji Q. A Jurassic eutherian mammal and divergence of marsupials and placentals. Nature. 2011;476(7361):442–5. doi: 10.1038/nature10291. [DOI] [PubMed] [Google Scholar]
- 40.Abi-Rached L, Kuhl H, Roos C, ten Hallers B, Zhu B, Carbone L, de Jong PJ, Mootnick AR, Knaust F, Reinhardt R, Parham P, Walter L. A small, variable, and irregular killer cell Ig-like receptor locus accompanies the absence of MHC-C and MHC-G in gibbons. J Immunol. 2010;184(3):1379–91. doi: 10.4049/jimmunol.0903016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Older Aguilar AM, Guethlein LA, Adams EJ, Abi-Rached L, Moesta AK, Parham P. Coevolution of killer cell Ig-like receptors with HLA-C to become the major variable regulators of human NK cells. J Immunol. 2010;185(7):4238–51. doi: 10.4049/jimmunol.1001494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moesta AK, Norman PJ, Yawata M, Yawata N, Gleimer M, Parham P. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J Immunol. 2008;180(6):3969–79. doi: 10.4049/jimmunol.180.6.3969. [DOI] [PubMed] [Google Scholar]
- 43.Abi-Rached L, Jobin MJ, Kulkarni S, McWhinnie A, Dalva K, Gragert L, Babrzadeh F, Gharizadeh B, Luo M, Plummer FA, Kimani J, Carrington M, Middleton D, Rajalingam R, Beksac M, Marsh SG, Maiers M, Guethlein LA, Tavoularis S, Little AM, Green RE, Norman PJ, Parham P. The shaping of modern human immune systems by multiregional admixture with archaic humans. Science. 2011;334(6052):89–94. doi: 10.1126/science.1209202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abi-Rached L, Moesta AK, Rajalingam R, Guethlein LA, Parham P. Human-specific evolution and adaptation led to major qualitative differences in the variable receptors of human and chimpanzee natural killer cells. PLoS Genet. 2010;6(11):e1001192. doi: 10.1371/journal.pgen.1001192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adams EJ, Parham P. Species-specific evolution of MHC class I genes in the higher primates. Immunol Rev. 2001;183:41–64. doi: 10.1034/j.1600-065x.2001.1830104.x. [DOI] [PubMed] [Google Scholar]
- 46.Daza-Vamenta R, Glusman G, Rowen L, Guthrie B, Geraghty DE. Genetic divergence of the rhesus macaque major histocompatibility complex. Genome Res. 2004;14(8):1501–15. doi: 10.1101/gr.2134504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kulski JK, Anzai T, Shiina T, Inoko H. Rhesus macaque class I duplicon structures, organization, and evolution within the alpha block of the major histocompatibility complex. Mol Biol Evol. 2004;21(11):2079–91. doi: 10.1093/molbev/msh216. [DOI] [PubMed] [Google Scholar]
- 48.Older Aguilar AM, Guethlein LA, Hermes M, Walter L, Parham P. Rhesus macaque KIR bind human MHC class I with broad specificity and recognize HLA-C more effectively than HLA-A and HLA-B. Immunogenetics. 2011;63(9):577–85. doi: 10.1007/s00251-011-0535-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zemmour J, Parham P. Distinctive polymorphism at the HLA-C locus: implications for the expression of HLA-C. J Exp Med. 1992;176(4):937–50. doi: 10.1084/jem.176.4.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boyington JC, Sun PD. A structural perspective on MHC class I recognition by killer cell immunoglobulin-like receptors. Mol Immunol. 2002;38(14):1007–21. doi: 10.1016/s0161-5890(02)00030-5. [DOI] [PubMed] [Google Scholar]
- 51.Vilches C, Pando MJ, Parham P. Genes encoding human killer-cell Ig-like receptors with D1 and D2 extracellular domains all contain untranslated pseudoexons encoding a third Ig-like domain. Immunogenetics. 2000;51(8–9):639–46. doi: 10.1007/s002510000184. [DOI] [PubMed] [Google Scholar]
- 52.Moesta AK, Abi-Rached L, Norman PJ, Parham P. Chimpanzees use more varied receptors and ligands than humans for inhibitory killer cell Ig-like receptor recognition of the MHC-C1 and MHC-C2 epitopes. J Immunol. 2009;182(6):3628–37. doi: 10.4049/jimmunol.0803401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guethlein LA, Flodin LR, Adams EJ, Parham P. NK Cell Receptors of the Orangutan (Pongo pygmaeus): A Pivotal Species for Tracking the Coevolution of Killer Cell Ig-Like Receptors with MHC-C. J Immunol. 2002;169(1):220–9. doi: 10.4049/jimmunol.169.1.220. [DOI] [PubMed] [Google Scholar]
- 54.Moesta AK, Graef T, Abi-Rached L, Older Aguilar AM, Guethlein LA, Parham P. Humans differ from other hominids in lacking an activating NK cell receptor that recognizes the C1 epitope of MHC class I. J Immunol. 2010;185(7):4233–7. doi: 10.4049/jimmunol.1001951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bari R, Bell T, Leung WH, Vong QP, Chan WK, Das Gupta N, Holladay M, Rooney B, Leung W. Significant functional heterogeneity among KIR2DL1 alleles and a pivotal role of arginine 245. Blood. 2009;114(25):5182–90. doi: 10.1182/blood-2009-07-231977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yawata M, Yawata N, Draghi M, Partheniou F, Little AM, Parham P. MHC class I-specific inhibitory receptors and their ligands structure diverse human NK-cell repertoires toward a balance of missing self-response. Blood. 2008;112(6):2369–80. doi: 10.1182/blood-2008-03-143727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Male V, Sharkey A, Masters L, Kennedy PR, Farrell LE, Moffett A. The effect of pregnancy on the uterine NK cell KIR repertoire. Eur J Immunol. 2011;41(10):3017–27. doi: 10.1002/eji.201141445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sharkey AM, Gardner L, Hiby S, Farrell L, Apps R, Masters L, Goodridge J, Lathbury L, Stewart CA, Verma S, Moffett A. Killer Ig-like receptor expression in uterine NK cells is biased toward recognition of HLA-C and alters with gestational age. J Immunol. 2008;181(1):39–46. doi: 10.4049/jimmunol.181.1.39. [DOI] [PubMed] [Google Scholar]
- 59.Chazara O, Xiong S, Moffett A. Maternal KIR and fetal HLA-C: a fine balance. J Leukoc Biol. 2011;90(4):703–16. doi: 10.1189/jlb.0511227. [DOI] [PubMed] [Google Scholar]
- 60.Colucci F, Boulenouar S, Kieckbusch J, Moffett A. How does variability of immune system genes affect placentation? Placenta. 2011;32(8):539–45. doi: 10.1016/j.placenta.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hiby SE, Apps R, Sharkey AM, Farrell LE, Gardner L, Mulder A, Claas FH, Walker JJ, Redman CW, Morgan L, Tower C, Regan L, Moore GE, Carrington M, Moffett A. Maternal activating KIRs protect against human reproductive failure mediated by fetal HLA-C2. J Clin Invest. 2010;120(11):4102–10. doi: 10.1172/JCI43998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hiby SE, Regan L, Lo W, Farrell L, Carrington M, Moffett A. Association of maternal killer-cell immunoglobulin-like receptors and parental HLA-C genotypes with recurrent miscarriage. Hum Reprod. 2008;23(4):972–6. doi: 10.1093/humrep/den011. [DOI] [PubMed] [Google Scholar]
- 63.Hiby SE, Walker JJ, O’Shaughnessy KM, Redman CW, Carrington M, Trowsdale J, Moffett A. Combinations of maternal KIR and fetal HLA-C genes influence the risk of preeclampsia and reproductive success. J Exp Med. 2004;200(8):957–65. doi: 10.1084/jem.20041214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gendzekhadze K, Norman PJ, Abi-Rached L, Graef T, Moesta AK, Layrisse Z, Parham P. Co-evolution of KIR2DL3 with HLA-C in a human population retaining minimal essential diversity of KIR and HLA class I ligands. Proc Natl Acad Sci U S A. 2009;106(44):18692–7. doi: 10.1073/pnas.0906051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Billington WD. The immunological problem of pregnancy: 50 years with the hope of progress. A tribute to Peter Medawar. J Reprod Immunol. 2003;60(1):1–11. doi: 10.1016/s0165-0378(03)00083-4. [DOI] [PubMed] [Google Scholar]
- 66.Manyonda IT. The Immunology of Human Reproduction. London: Taylor & Francis; 2006. [Google Scholar]
- 67.Klein J. Biology of the mouse histocompatibility-2 complex. New York: Springer-Verlag; 1975. [Google Scholar]
- 68.Thorsby E. A tentative new model for the organization of the mouse H-2 histocompatibility system: two segregant series of antigens. Eur J Immunol. 1971;1(1):57–9. doi: 10.1002/eji.1830010112. [DOI] [PubMed] [Google Scholar]
- 69.Malmberg KJ, Michaelsson J, Parham P, Ljunggren HG. Killer cell immunoglobulin-like workshop: insights into evolution, genetics, function and translation. Immunity. 2011 doi: 10.1016/j.immuni.2011.11.007. In Press. [DOI] [PubMed] [Google Scholar]
- 70.Robinson J, Mistry K, McWilliam H, Lopez R, Marsh SG. IPD--the Immuno Polymorphism Database. Nucleic Acids Res. 2010;38(Database issue):D863–9. doi: 10.1093/nar/gkp879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martin MP, Single RM, Wilson MJ, Trowsdale J, Carrington M. KIR haplotypes defined by segregation analysis in 59 Centre d’Etude Polymorphisme Humain (CEPH) families. Immunogenetics. 2008;60(12):767–74. doi: 10.1007/s00251-008-0334-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Middleton D, Meenagh A, Gourraud PA. KIR haplotype content at the allele level in 77 Northern Irish families. Immunogenetics. 2007;59(2):145–58. doi: 10.1007/s00251-006-0181-7. [DOI] [PubMed] [Google Scholar]
- 73.Norman PJ, Cook MA, Carey BS, Carrington CV, Verity DH, Hameed K, Ramdath DD, Chandanayingyong D, Leppert M, Stephens HA, Vaughan RW. SNP haplotypes and allele frequencies show evidence for disruptive and balancing selection in the human leukocyte receptor complex. Immunogenetics. 2004;56(4):225–37. doi: 10.1007/s00251-004-0674-1. [DOI] [PubMed] [Google Scholar]
- 74.Gonzalez-Galarza FF, Christmas S, Middleton D, Jones AR. Allele frequency net: a database and online repository for immune gene frequencies in worldwide populations. Nucleic Acids Res. 2011;39(Database issue):D913–9. doi: 10.1093/nar/gkq1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Parham P, Norman PJ, Abi-Rached L, Guethein LA. Human-specific evolution of killer cell immunoglobulin-like receptor recognition of major histocompatibility complex class I molecules. Philos Trans R Soc Lond B Biol Sci. 2011 doi: 10.1098/rstb.2011.0266. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]