Abstract
In a prospective study conducted by laboratory technologists in a diagnostic laboratory in Cape Town, South Africa, a semi-automated phage-based antibiotic susceptibility assay was implemented and the performance of the luciferase reporter mycobacteriophage (LRP) system for susceptibility testing of clinical Mycobacterium tuberculosis complex (MTC) isolates against rifampin and isoniazid was evaluated. Two hundred consecutive clinical MGIT cultures of MTC species were included in this study. Antibiotic susceptibility assays were setup manually for the LRP and BACTEC radiometric systems and read in a plate luminometer and the BACTEC 460 instrument, respectively. Discrepant susceptibility results were resolved by the conventional agar proportion method. Of the 200 secondary cultures prepared for this study, 9 (4.5%) were lost to contamination (LRP 4, BACTEC 1, both 4). All of the remaining 191 cultures underwent susceptibility testing by both methods and the overall agreement between the LRP and BACTEC was 98.4% (rifampin 100%; isoniazid 96.9%). Of the 6 discrepant cultures tested by the agar proportion method, 2 gave results in agreement with the LRP. The sensitivity of the LRP for detection of drug-resistant isolates was 100% for both rifampin (n=9) and isoniazid (n=12). The median turnaround time for susceptibility testing was 2 days with the LRP and 9 days with BACTEC. In conclusion, the semi-automated LRP-based assay offers a rapid and practical approach for accurate susceptibility testing of Mycobacterium tuberculosis cultures in diagnostic laboratories with limited financial resources but with competent technologists.
Keywords: Mycobacterium tuberculosis, drug susceptibility testing, luciferase reporter mycobacteriophages, drug resistance
Introduction
Mycobacterium tuberculosis (Mtb) is amongst the leading infectious causes of death worldwide. 1 The latest surveillance report showed that drug-resistant isolates are ubiquitous worldwide and in the Western Cape Province of South Africa, resistance rates were 5.6% and 1.9% for isoniazid and rifampin, respectively, and 1.9% for multidrug resistance. 2 Drug resistance is a serious global threat as number of studies have shown that in patients with drug-resistant isolates, treatment with standard anti-tuberculosis regimens results in significantly higher rates of treatment failure and death. 3-6 Today it is generally accepted that effective management of patients with tuberculosis requires a combination of laboratory susceptibility testing and individualized anti-tuberculosis regimens based on the susceptibility profile of each patient's isolate. 7 Although this approach is feasible in resource-rich countries, susceptibility testing is frequently not performed in resource-poor settings due to financial constraints. Clinical laboratories in the latter countries rely on microscopy and culture isolation for diagnosing and treating tuberculosis. This approach however does not reveal any information on the susceptibility profile of the isolates and could result in treatment failure and emergence of more resistant isolates. Thus, a rapid and affordable assay for antibiotic susceptibility testing of Mtb would be of great value in resource-poor countries.
We have previously reported on the development and evaluation of the luciferase reporter mycobacteriophages (LRP) for susceptibility testing of clinical Mtb cultures. 8-11 It has been shown that the LRP provide rapid and accurate susceptibility results when compared to the BACTEC radiometric and conventional methods. The goals of this study were (i) to evaluate the feasibility and performance of a semi-automated LRP-based assay for antibiotic susceptibility testing of clinical isolates against isoniazid and rifampin and (ii) to determine whether if this assay can be performed by laboratory technologists in a routine hospital laboratory in Cape Town, South Africa.
Material and Methods
Clinical isolates
From April to November of 2003, 200 consecutive M. tuberculosis complex cultures, isolated from patient specimens in the laboratory of clinical microbiology at the Groote Schuur Hospital, Cape Town, South Africa, were included in this study. Specimens consisted of 51 sputa, 1 nasopharyngeal aspirate, 3 tracheal aspirates, 12 bronchoalveolar lavages, 31 pleural fluids, 6 spinal fluids, 13 biopsies, 11 urines, 36 gastric washes/aspirates, 16 fine needle aspirates, 3 ascitic fluids, 9 swabs, and 8 unspecified body fluids.
Luciferase reporter phage (LRP)
Reporter phage phAE142 was used in this study. 12 The phage was propagated and titrated on lawns of M. smegmatis mc2 4502 in the hospital laboratory as previously described. 8 High titer phage stocks were stored at 4°C for several months.
Antibiotics
Lyophilized antibiotics (Becton Dickinson) were dissolved in sterile water to make 20× stock concentrations of isoniazid (INH) at 4 μg/ml and rifampin (RIF) at 40 μg/ml and stored at −70°C.
Culture isolation and identification
Specimens were processed according to standard procedures 13 and inoculated into MGIT tubes for cultivation in the MGIT-960 system according to the manufacturer's standard procedure (Becton Dickinson Diagnostic Instrument Systems). The MGIT instrument read tubes hourly and positive cultures were flagged and confirmed by acid-fast microscopy and ruled out for contamination by subculture on blood agar. Contaminant-free cultures were identified with a PCR assay as previously reported 14 and M. tuberculosis isolates were advanced for susceptibility testing.
Susceptibility testing
(i) LRP
Secondary cultures were created by adding 1 ml of MGIT culture to 1 ml of Middlebrook 7H9 broth (Difco) supplemented with 1% (vol/vol) glycerol, and 10% (v/v) ADC (albumin, dextrose, catalase). Secondary cultures were checked for contamination on blood agar. The turbidity of each culture was adjusted to ≤1.5 McFarland when necessary. For each isolate, 5 μl of sterile water and 5 μl of 20× INH (4 μg/ml) and RIF (40 μg/ml) was added to separate wells of a 96-well plate, each containing 95 μl of secondary culture. The plate was sealed with sealing membrane and incubated at 37°C. At 40 h post incubation, sealing membrane was pulled back and 10 μl of phage was added to each well. Following infection for 3 h, the sealing membrane was removed and luciferase activity in each well was quantified in a MicroLumatPlus LB96V Microplate Luminometer (Berthold; 1-s pause, 5-s integration) in a biosafety cabinet, and the susceptibility profiles were calculated according to the following formula [(rluantibiotic)/(rlucontrol)] ×100 where >15% signifies drug resistance. When luciferase activity of the control well (bacteria without antibiotics) was <200 rlu, the results were not interpreted and the secondary culture was incubated to allow for additional growth. All cultures with resistant results were confirmed with a second LRP susceptibility test and only considered resistant when the two tests were in agreement.
(ii) Radiometric
Susceptibility testing was performed according to the manufacturer's instructions. 15 Briefly, 250 μl of each MGIT culture was aseptically inoculated into a 12B vial and incubated at 37°C. Vials were analyzed with the BACTEC 460 instrument and at a growth index of ≥ 500, 0.1 ml of test organism was added to B12 vials containing recommended antibiotic concentrations (RIF 2 μg/ml, INH 0.1 μg/ml). A vial containing a 1:100 dilution of the test organism was inoculated as a control. Vials were analyzed according to the manufacturer's protocols and susceptibility profiles were determined.
(iii) Agar Proportion
Was performed according to standard recommendations. 13 Middlebrook 7H11 slants with RIF (1 μg/ml), INH (0.2 μg/ml), or no antibiotics were inoculated with 0.1 ml of 1:10 dilution of MGIT cultures with a reading of >300. After 14 days of incubation, the colonies were enumerated. Slants with RIF or INH showing ≥1% growth compared to the control slant were considered resistant.
MIC Determination
Was performed using the radiometric method as described for susceptibility testing. Serial two fold dilutions of INH and RIF were prepared and 0.1 ml was added to 12B medium to obtain test concentrations of 0.8, 0.4, 0.2, 0.1, 0.05, 0.025 μg/ml for INH and 640, 320, 160, 80, 40, and 20 μg/ml for RIF. Test organism was added and vials were analyzed in the BACTEC 460 instrument according to the manufacturer's instructions (Becton Dickinson Diagnostic Instrument Systems).
Statistical analysis
The sensitivity, specificity, and accuracy of the susceptibility testing were calculated. Differences in proportions were evaluated by chi-square test.
Results and Discussion
Antibiotic susceptibility testing of M. tuberculosis remains financially challenging in clinical laboratories in countries with limited resources. Although automated susceptibility systems are rapid and easy to use, they are expensive. The aim of this study was to implement and evaluate the performance of a semi-automated phage-based assay for detection of resistance to isoniazid and rifampin in a hospital laboratory in Cape Town, South Africa. Furthermore, unlike the prior studies with the reporter phages, this study was performed by laboratory technologists in its entirety and thus also tested the feasibility of using this assay in settings that are financially constrained but populated with suitably trained and competent technologists.
Two hundred consecutive MGIT cultures with M. tuberculosis complex (MTC) growth, as identified by a PCR assay, were included in this study. Of the 200 secondary cultures prepared for susceptibility testing with the LRP and BACTEC radiometric, 9 (4.5%) were lost to contamination and included 4 LRP, 1 BACTEC, and 4 of both. Contamination of secondary cultures occurred despite steps put in place to detect contaminated primary and secondary cultures. The addition of an antimicrobial cocktail to secondary cultures may further reduce the contamination rates. However, whether this occurs without interfering with the accuracy of susceptibility results remains to be determined.
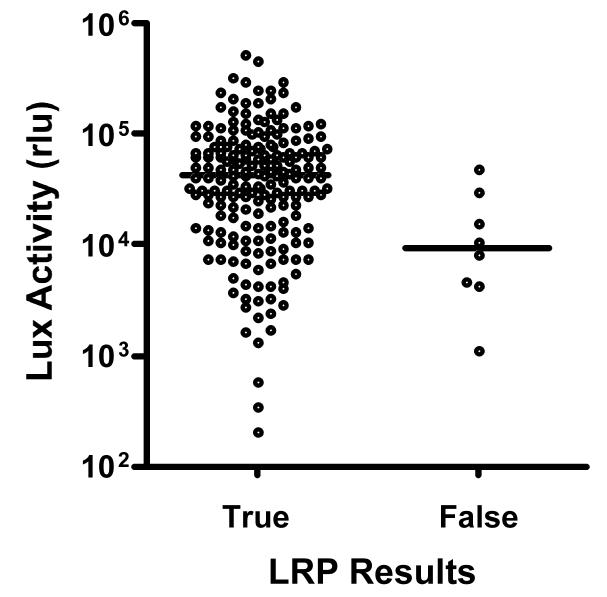
All of the remaining 191 contaminant-free secondary cultures were tested for susceptibility to rifampin and isoniazid by the LRP and BACTEC radiometric methods. The overall agreement between the LRP and BACTEC was 98.4% (rifampin 100%; isoniazid 96.9%) (Table 1). Six discrepant results were obtained with isoniazid (2 BACTEC-resistant LRP-susceptible and 4 BACTEC-susceptible LRP-resistant). Upon testing the discrepant cultures with the agar proportion method, the 2 BACTEC-resistant LRP-susceptible isolates gave results in agreement with the LRP. These isolates had borderline MIC values according to BACTEC (0.1 and 0.8 μg/ml), which may explain the difference in results we obtained with BACTEC compared to LRP and the agar proportion. For the other 4 discrepant cultures (BACTEC-susceptible LRP-resistant), results obtained with the agar proportion method were in agreement with BACTEC and the MIC values were all within the sensitive range (0.05 μg/ml). Retesting of these isolates from newly-grown cultures gave LRP results that were in agreement with BACTEC. To determine if there was a correlation between the discrepant results and the size of the inoculum used in the LRP susceptibility assays, we compared inoculum size between the cultures with true and false isoniazid results. As shown in Figure 1, compared to cultures with true results, the average inoculum size (as a measure of luciferase activity) 16 of the 4 cultures with false results was 4.1 fold lower. Raising the inoculum threshold to 10000 rlu would have prevented 3 of the 4 false isoniazid-resistant results. This increase in specificity would have come at a cost of prolonged turnaround times for 34 (18%) of the remaining 187 isolates. However, 53% of these represented confirmatory tests and given that mycobacteria sediment in standing MGIT tubes, it is possible that an improved sampling technique could have prevented the insufficient inoculum size obtained from these cultures.
Table 1.
Susceptibility of M. tuberculosis complex isolates to isoniazid and rifampin as determined by LRP and BACTEC radiometric system.
| Drug * | Total No. tested |
No. of strains with following results # |
|||
|---|---|---|---|---|---|
| Both S | BACTEC S, LRP R |
BACTEC R, LRP S |
Both R | ||
| RIF | 191 | 182 | 9 | ||
| INH | 191 | 173 | 4 | 2Ω | 12 |
Drug concentrations (in μg/ml) included: RIF, 2 (both systems); INH, 0.2 (LRP) and 0.1(BACTEC)
S, susceptible; R, resistant
agar proportion results were in agreement LRP for these isolates
Figure 1.
Inoculum size for cultures with true and false isoniazid results. The size of inoculum used in the LRP-based susceptibility assay, as a measure of luciferase (Lux) activity, is plotted for cultures with true and false isoniazid results. Cultures were tested in duplicates. The mean rlu value for each group is shown with a horizontal bar.
The sensitivity and specificity of the LRP for detection of drug-resistant isolates were both 100% for rifampin and 100% and 97.7% for isoniazid, respectively. There was no statistically significant difference between LRP and BACTEC for isoniazid susceptibility results (P = 0.40). Although the positive predictive valued of INH resistance was 75%, 92% of INH-resistant cultures would have been correctly reported by raising the inoculum threshold to 10000 rlu. Consistent with a prior study, 9 the median turnaround time for susceptibility testing was 2 days with the LRP and 9 days with BACTEC. Furthermore, none of the South African isolates were found to be resistant to the phage.
As with other rapid methods, the financial cost of LRP-based susceptibility testing includes the cost of an instrument (luminometer). However, unlike the other methods, the reagent costs for the LRP assay are lower and calculated to be ∼0.4 US dollars per isolate for testing with isoniazid and rifampin. Otherwise, the level of competency of technologists and the requirement for other laboratory instruments such as biosafety cabinet and centrifuges are similar between the LRPs and other methods.
In summary, the semi-automated LRP assay produced rapid and highly accurate antibiotic susceptibility results when operated by laboratory technologists in a hospital laboratory in Cape Town, South Africa. This study demonstrates that the semi-automated LRP assay is a suitable alternative for high-volume laboratories with limited financial resources.
Acknowledgements
This work was supported by grants from National Institutes of Health K08 AI061105 (NB), R01 AI26170 and AI43268 (WRJ), and Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Raviglione MC. The TB epidemic from 1992 to 2002. Tuberculosis. 2003;83:4–14. doi: 10.1016/s1472-9792(02)00071-9. [DOI] [PubMed] [Google Scholar]
- 2.WHO/IUATLD Global Project on Anti-Tuberculosis Drug Resistance Surveillance. Anti-tuberculosis drug resistance in the world. Report 3: prevalence and trends. 2004 [Google Scholar]
- 3.DeRiemer K, Garcia-Garcia L, Bobadilla-del-Valle M, Palacios-Martinez M, Martinez-Gamboa A, Small PM, Sifuentes-Osornio J, Ponce-de-Leon A. Does DOTS work in populations with drug-resistant tuberculosis? Lancet. 2005;365:1239–45. doi: 10.1016/S0140-6736(05)74812-1. [DOI] [PubMed] [Google Scholar]
- 4.Espinal MA, Kim SJ, Suarez PG, Kam KM, Khomenko AG, Migliori GB, Baez J, Kochi A, Dye C, Raviglione MC. Standard short-course chemotherapy for drug-resistant tuberculosis: treatment outcomes in 6 countries. Jama. 2000;283:2537–45. doi: 10.1001/jama.283.19.2537. [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Garcia ML, Ponce de Leon A, Jimenez-Corona ME, Jimenez-Corona A, Palacios-Martinez M, Balandrano-Campos S, Ferreyra-Reyes L, Juarez-Sandino L, Sifuentes-Osornio J, Olivera-Diaz H. Clinical consequences and transmissibility of drug-resistant tuberculosis in southern Mexico. Arch Intern Med. 2000;160:630–6. doi: 10.1001/archinte.160.5.630. others. [DOI] [PubMed] [Google Scholar]
- 6.Nachega JB, Chaisson RE. Tuberculosis drug resistance: a global threat. Clin Infect Dis. 2003;36:S24–30. doi: 10.1086/344657. [DOI] [PubMed] [Google Scholar]
- 7.Kim JY, Mukherjee JS, Rich ML, Mate K, Bayona J, Becerra MC. From multidrug-resistant tuberculosis to DOTS expansion and beyond: making the most of a paradigm shift. Tuberculosis. 2003;83:59–65. doi: 10.1016/s1472-9792(02)00078-1. [DOI] [PubMed] [Google Scholar]
- 8.Banaiee N, Bobadilla-Del-Valle M, Bardarov S, Jr., Riska PF, Small PM, Ponce-De-Leon A, Jacobs WR, Jr., Hatfull GF, Sifuentes-Osornio J. Luciferase reporter mycobacteriophages for detection, identification, and antibiotic susceptibility testing of Mycobacterium tuberculosis in Mexico. J Clin Microbiol. 2001;39:3883–8. doi: 10.1128/JCM.39.11.3883-3888.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banaiee N, Bobadilla-del-Valle M, Riska PF, Bardarov S, Jr., Small PM, Ponce-de-Leon A, Jacobs WR, Jr., Hatfull GF, Sifuentes-Osornio J. Rapid identification and susceptibility testing of Mycobacterium tuberculosis from MGIT cultures with luciferase reporter mycobacteriophages. J Med Microbiol. 2003;52:557–61. doi: 10.1099/jmm.0.05149-0. [DOI] [PubMed] [Google Scholar]
- 10.Hazbon MH, Guarin N, Ferro BE, Rodriguez AL, Labrada LA, Tovar R, Riska PF, Jacobs WR., Jr. Photographic and luminometric detection of luciferase reporter phages for drug susceptibility testing of clinical Mycobacterium tuberculosis isolates. J Clin Microbiol. 2003;41:4865–9. doi: 10.1128/JCM.41.10.4865-4869.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacobs WR, Jr., Barletta RG, Udani R, Chan J, Kalkut G, Sosne G, Kieser T, Sarkis GJ, Hatfull GF, Bloom BR. Rapid assessment of drug susceptibilities of Mycobacterium tuberculosis by means of luciferase reporter phages. Science. 1993;260:819–22. doi: 10.1126/science.8484123. [DOI] [PubMed] [Google Scholar]
- 12.Bardarov S, Jr., Dou H, Eisenach K, Banaiee N, Ya S, Chan J, Jacobs WR, Jr., Riska PF. Detection and drug-susceptibility testing of M. tuberculosis from sputum samples using luciferase reporter phage: comparison with the Mycobacteria Growth Indicator Tube (MGIT) system. Diagn Microbiol Infect Dis. 2003;45:53–61. doi: 10.1016/s0732-8893(02)00478-9. [DOI] [PubMed] [Google Scholar]
- 13.Kent PT, Kubica GP. Public health mycobacteriology: a guide for the level III laboratory. U.S. Department of Health, Education, and Welfare, Centers for Disease Control and Prevention; Atlanta, Ga.: 1995. pp. 159–184. [Google Scholar]
- 14.De Wit D, Steyn L, Shoemaker S, Sogin M. Direct detection of Mycobacterium tuberculosis in clinical specimens by DNA amplification. J Clin Microbiol. 1990;28:2437–41. doi: 10.1128/jcm.28.11.2437-2441.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siddiqi SH. BACTEC 460 TB System. Product and procedure manual, revision D. Becton Dickinson Microbiology Systems; Sparks, Md: 1995. [Google Scholar]
- 16.Carriere C, Riska PF, Zimhony O, Kriakov J, Bardarov S, Burns J, Chan J, Jacobs WR., Jr. Conditionally replicating luciferase reporter phages: improved sensitivity for rapid detection and assessment of drug susceptibility of Mycobacterium tuberculosis. J Clin Microbiol. 1997;35:3232–9. doi: 10.1128/jcm.35.12.3232-3239.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]