Imaging mass spectrometry has been used to provide information on the spatial distribution of lipids,[1,2] peptides[3] and proteins,[4] as well as pharmaceuticals[5] and metabolites[6,7] directly from biological tissue sections. Matrix-assisted laser desorption/ionization (MALDI) imaging mass spectrometry has been successful in investigating protein changes corresponding to several diseases, including Alzheimer’s,[8] Parkinson’s,[9,10] and cancer.[11-13] Secondary ion mass spectrometry (SIMS) has been used to map the distribution of cancer agents[14] and lipids[15] with high sensitivity and spatial resolution, although its mass range is limited by extensive molecular fragmentation.[16] Desorption electrospray ionization (DESI), an ambient ionization method, shows promise in the imaging of living tissue without sample preparation,[17,18] but its spatial resolution is currently limited.[19] Common to all surface imaging methods is the ablation from the front side, also referred to as reflection geometry, with the laser beam, cluster beam or liquid stream at an angle to the plane of the surface. Pairing the sensitivity and specificity of mass spectrometry (MS) with imaging techniques has the potential to provide information on the chemical composition of surfaces at the subcellular dimension, but necessitates the development of methods combining high-throughput analysis and high sensitivity with high spatial resolution.
One problem in vacuum MALDI imaging in reflection geometry mode is the loss of mass resolution as well as mass accuracy that accompanies high laser fluence. For this reason, the sample is often ablated near the threshold for ion observation and frequently over 100 laser shots are acquired and their mass spectra summed for each pixel of the image.[20-22] The time required to obtain an image is dependent on several parameters, including laser repetition rate, the desired spatial resolution of the sample, and the speed of data processing.[20] As the spatial resolution increases, so does the time required to image the respective surface. Reasonable speed of analysis in reflection geometry requires the use of high repetition lasers, capable of analyzing multiple pixels per second.[23] Lasers operating in the kHz region significantly reduce analysis time,[20,24,25] but at a significantly higher laser cost.
An alternative configuration is to align the laser in transmission geometry relative to the sample and mass spectrometer orifice. In transmission geometry, laser irradiation occurs from the back of the sample through a transparent sample holder. The first demonstration of tissue analysis in transmission geometry mode has been reported.[26] Transmission geometry had previously been demonstrated in both vacuum[27-30] and atmospheric pressure[31] (AP) MALDI for standard samples.
Laserspray ionization (LSI) operates at AP in either transmission[26] or reflection geometry[32] in the absence of an applied voltage and, as recently shown, the laser is not necessary for ionization.[33] Because ionization occurs inside the transfer capillary,[34,35] any means, including laser ablation, of transferring matrix/analyte to the ion entrance orifice produces ions.[33] Higher temperatures of typically 300°C using 2,5-dihydroxyacetophenone (2,5-DHAP) on Velos LTQ and Orbitrap mass spectrometers have been reported to enhance the abundance of highly charged peptides and protein ions in the positive mode.[32-37] Transmission geometry allows the sample to be placed very near the ion entrance orifice and here the tissue sample was held approximately 1 mm from the mass spectrometer vacuum entrance thus allowing more efficient sampling of the ablated matrix/analyte clusters. A 337 nm nitrogen laser (Newport Corporation, Irvine, CA, USA; VSL-337ND-S) beam aligned at 180° relative to the entrance of the mass spectrometer and focused using a 102 mm focal length lens (CVI Melles Griot, Albuquerque, NM, USA) was used to ablate the sample.
Because LSI produces electrospray ionization (ESI)-like multiply charged ions, newer fragmentation methods can be employed to characterize materials. For example, nearly complete sequence coverage of ubiquitin, a regulatory protein, was obtained by electron transfer dissociation (ETD) using a single laser shot in transmission geometry.[35] ETD, in a single LSI acquisition, was also used to sequence and identify directly from mouse brain tissue the doubly charged peptide of m/z 917.5 [molecular weight (MW) 1833 Da] as a myelin basic protein N-terminal fragment.[37] High MW proteins have also been observed directly from tissue as multiply charged ions using transmission geometry LSI.[37] Multiply charged ions are also beneficial in imaging when combined with ion mobility spectrometry (IMS)-MS because they promote separation of components in mixtures,[38] including isomers.[39]
Another advantage of transmission geometry imaging is that the laser is fired only once at each position because a high laser fluence is used to ablate through the tissue/matrix. This is possible because ionization is at AP, which provides ample collisional cooling to occur before mass analysis.[26,36,37] Each pixel of the image represents a single laser shot producing a full mass spectrum. This offers the potential for rapid analysis[36] and imaging using inexpensive nitrogen lasers. A proof of concept experiment produced a crude image of peptides in mouse brain tissue in 30 min acquisition time using LSI in transmission geometry.[37]
Because the laser travels through the sample in transmission geometry mode, ideally the entire volume of tissue impacted by the laser beam is ablated and ionized in a single pulse. The quality of the obtained image is highly dependent on sample preparation and the thickness of the sample. In the previous transmission geometry images, 2,5-DHAP matrix was applied using the dried-droplet method,[40] applying nanoliters of solution.[37] Solvent-based matrix deposition is inappropriate for high spatial resolution imaging, as solvents cause the delocalization of components in the tissue. Solvents also cause problems related to reproducibility, as ‘hot spots’ can arise from uneven matrix application and crystallization. Because each pixel represents one shot, homogenous matrix coverage is required for shot-to-shot reproducibility. Attempts at spray-applying the matrix solution yielded low abundance signals in LSI operated in transmission geometry.[37]
Solvent-free sample preparation was developed a decade ago,[41,42] and has recently found new applications related to tissue analysis and imaging.[26,43-46] The first reported analysis of tissue in transmission geometry using LSI employed both solvent-free and solvent-based sample preparation of the matrix on the tissue.[26] Tissue imaging was not reported in this study due to limited reproducibility. Another limitation in obtaining high spatial resolution is sensitivity. In a previous report, a strategy was developed in which the glass slides were pre-coated with LSI matrix and a tissue section placed on top of the thin matrix coating.[37] This enhanced the ion abundance but caused significant larger tissue areas to ablate so that arguably no enhancement of ionization efficiency was achieved.[37] Large ablated areas are often required to observe sufficient ion current to obtain the necessary reproducible ion abundance for imaging.
Here, we present the first imaging experiment using transmission geometry. This also represents the first report of LSI producing negative ions. In this proof of principle study, the instrument acquisition time for a mouse brain section was approximately 1 h using single laser shot ablations of ~20 μm achieved by solvent-free sample preparation using automated solvent-free matrix deposition on the tissue section.
Mouse brain tissue was obtained from C57 Bl/6 mice, 20 weeks old, that were euthanized with CO2 gas and transcardially perfused with ice-cold 1× phosphate-buffered saline (PBS, 150 mM NaCl, 100 mM NaH2PO4, pH 7.4) for 5 min to remove red blood cells. The brains were frozen at −22°C and sliced into 10 μm sections in sequence using a Leica CM1850 cryostat (Leica Microsystems Inc., Bannockburn, IL, USA). The tissue sections were placed onto prechilled microscopy glass slides that were briefly warmed with a finger from behind to allow sections to relax and attach. Care was taken to avoid water condensation by storing at −20°C and transporting under dry ice the tissue-mounted glass slides in an airtight, desiccant-containing box until use. 2,5-DHAP LSI matrix was pre-ground in a 5 mL glass vial containing 1.3 mm chrome beads for 30 min at 15 Hz in a TissueLyser II ball mill device (QIAGEN, Valencia, CA, USA) similar to previous procedures.[45] Briefly, the pre-ground matrix was placed in the top compartment of the TissueBox with approximately 30 1.4 mm stainless steel beads. The microscopy slide mounted with mouse brain tissue was separated from the beads and matrix by 5 μm mesh. The TissueBox was placed in the TissueLyser II for 2 min at 25 Hz. Matrix application occurs as the pre-ground matrix is pushed through the mesh by the movement of the TissueBox in the TissueLyser II ball mill device. A 2 min grind time was required to obtain homogenous matrix coverage using 2,5-DHAP. At times greater than 2 min, the layer of applied matrix was too thick to obtain usable MS signals.
LSI-MS and MS/MS experiments were performed on a Thermo Fisher Velos LTQ (Thermo Fisher Scientific, Bremen, Germany) linear ion trap mass spectrometer. As described previously the source housing is removed to allow free access to the ion entrance orifice. The glass microscope slide mounted with mouse brain tissue covered with 2,5-DHAP was attached to a computer-controlled xy-stage (Newmark Systems, Mission Viejo, CA, USA) and moved through the laser beam in transmission geometry mode. The lanes were at 100 μm intervals, and each lane was 10.8 mm long. The instrument was operated in negative ion mode measuring a mass range from m/z 600 to 1000. The laser fluence per pulse was in the range of 0.5–1.0 J cm−2. Tissue images were created using BioMAP 3.7.5.6 (Novartis Institutes for BioMedical Research, Basel, Switzerland). Thermo XCalibur .RAW files were converted into .IMG files using customized software.
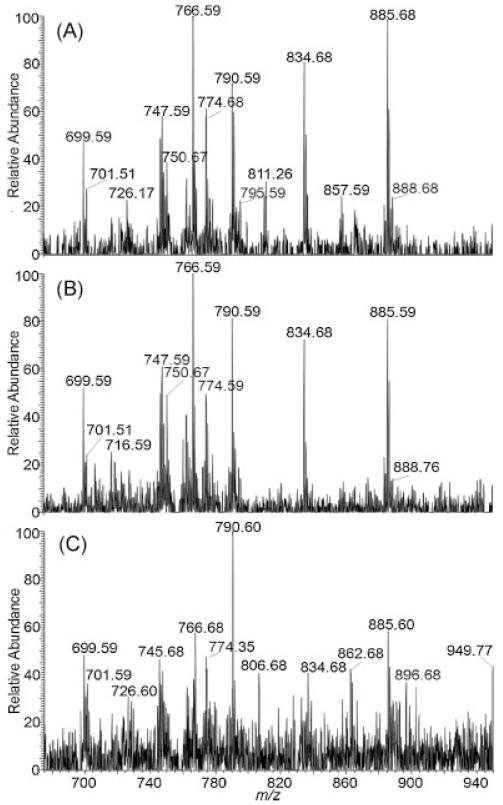
The AP-to-vacuum ion transfer capillary was heated to 450°C. In positive ion mode, 2,5-DHAP reduces the required desolvation temperature to as low as 150°C using solvent-based sample preparation methods.[39] Solvent-free sample preparation uses higher temperatures, as high as 275°C, to produce LSI ions of peptides with high abundance in the positive ion mode.[47] As shown in Fig. 1, thermal requirements are notably high in negative mode measurements using solvent-free sample preparation. At temperatures below 400°C, there was a significant decrease in ion abundance and shot-to-shot reproducibility. Furthermore, the key to solvent-free sample preparation appears to be the ability to minimize the sample crystallinity and compactness but provide intimate contact with analyte material.[48] Using the TissueBox for solvent-free surface coverage, depending on the mesh size used, here 5 μm, produces small matrix particles which have high velocity when using the ball mill at a high frequency. These small projectiles hitting the tissue section may allow sufficient surface penetration so that the spot-to-spot reproducibility is enhanced while keeping the integrity of the tissue relatively undisturbed.
Figure 1.
Transmission geometry negative ion LSI mass spectra of 10 μm thick mouse brain tissue sections using 2,5-DHAP as a matrix and solvent-free sample preparation at (A) 450°C, (B) 400°C, and (C) 350°C ion transfer capillary temperatures. Each spectrum represents a single laser pulse. Mass spectra from similar mouse brain regions are displayed for comparison.
The matrices 2,5-dihydroxybenzoic acid (DHB) and 2,4,6-trihydroxyacetophenone (THAP) were tested, and yielded insufficient negative ion abundance under all experimental conditions evaluated, including variation of temperature and degree of matrix coverage. 2,5-DHAP provided the best sensitivity and reproducibility in negative ion mode measurements and it was thus used exclusively in the following imaging studies.
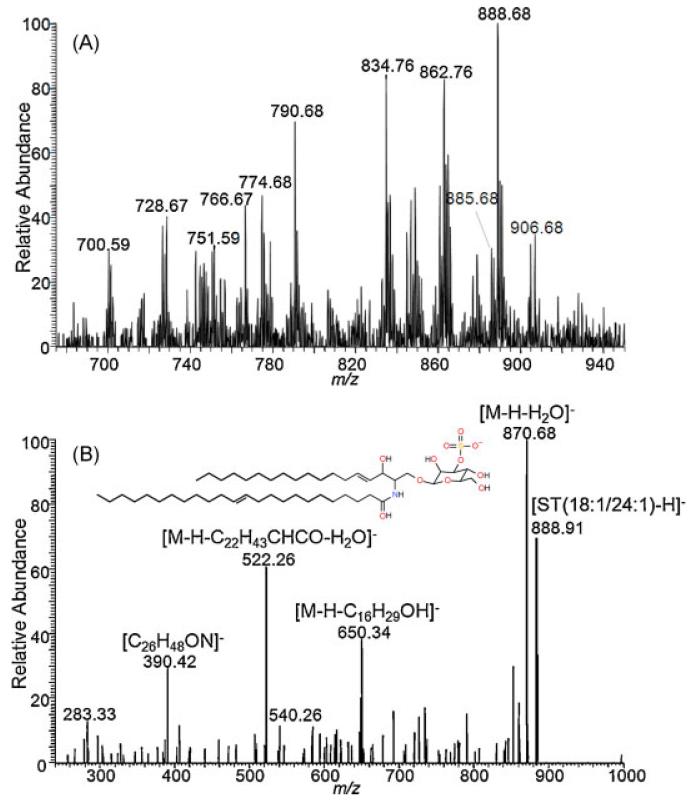
Many polar phospholipids and sphingolipids are preferentially ionized in negative ion mode, including phosphatidylserines, (PS), phosphatidyinisitols (PI), and sulfatides (ST). Several of the detected lipids (Figs. 1 and 2(A)) are similar to those previously reported in negative ion mode.[49,50] LSI MS/MS using collision-induced dissociation (CID) with a collision energy of 40 eV and a precursor ion selection window of 1.0 m/z unit identified the lipid at m/z 888.7 as ST (3-O-sulfogalactosylceramide) 18:1,24:1 (Fig. 2(B)), which is present in the myelin sheath.[51] Differences in intensities and observed lipid species are probably related to solvent-free sample preparation. The observation of different lipids in solvent-free and solvent-based conditions is consistent with previous investigations.[45,46]
Figure 2.
Transmission geometry negative ion LSI mass spectra of 10 μm thick mouse brain tissue sections using 2,5-DHAP as a matrix and solvent-free sample preparation at 450°C ion transfer capillary temperatures. (A) Full mass spectrum. (B) MS/MS spectrum using CID of m/z 888.7 indicating the chemical composition of phosphatidyl sulfatides (ST) 18:1/24:1. The width of the parent selection window is 1.0. Collision energy of 40 eV was used.
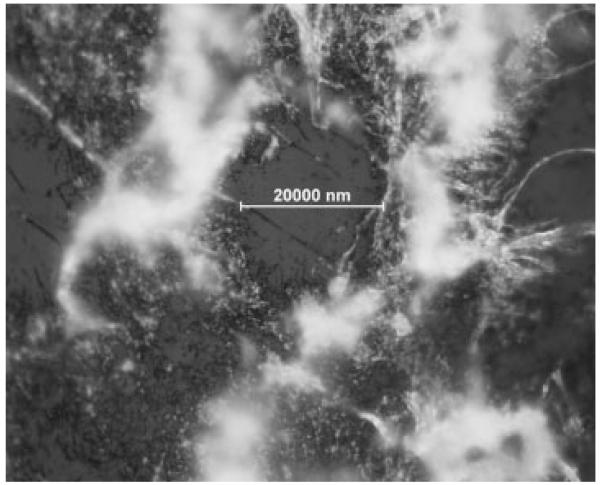
While efforts to reduce the time of analysis for MS imaging have focused on increased laser speed, here an image of mouse brain tissue was obtained in approximately 1 h with a laser operating at approximately 12 Hz. Each row required 33 s of acquisition time, and 93 rows spaced at 100 μm were required to completely image a single section of mouse brain. Laser ablated areas of ca. 20 μm were obtained (Fig. 3). Previous LSI imaging experiments using transmission geometry on delipified mouse brain tissue prepared with 2,5-DHAP and solvent-based sample preparation deposition reported ablated diameters of ca. 15 μm.[37]
Figure 3.
Optical microscopy of mouse brain tissue after LSI imaging in transmission geometry. Laser ablated dimensions are approximately 20 μm.
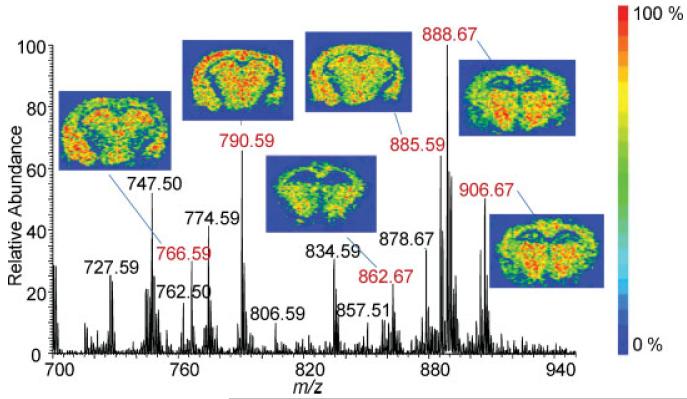
Single laser shots were sufficient to differentiate the distribution of lipids appearing at m/z 888.7, 885.6, and 834.5. Using LSI in negative ion mode and the laser aligned in transmission geometry, sufficient ion abundances were obtained from each laser shot to form an image of lipids from mouse brain tissue (Fig. 4), indicating that inexpensive nitrogen lasers may provide a speed of analysis similar to high repetition rate lasers.
Figure 4.
LSI mass spectrum and ion images of [M–H]− lipids from a 10 μm mouse brain tissue section. The tissue section was covered with 2,5-DHAP solvent-free and the acquisition was obtained with the laser aligned in transmission geometry and with ion transfer capillary temperatures of 450°C in the negative ionization mode. For example m/z 888.7, the crescent-shaped figure on the left side is the external capsule/alveolus, which is rich in myelin. The intensely labeled structure in the center corresponds to the thalamus. Other [M-H]− ion images are of m/z 766.59, 790.59, 862.67, 885.59 and 906.67.
Current work includes optimizing sample preparation to further increase ion abundances and shot-to-shot reproducibility. Previous studies[31] have highlighted the importance of thin matrix coverage for transmission geometry. Application of a thin matrix film that allows the laser to penetrate the complete sample layer while still providing enough matrix coverage for proper ionization of the sample is a challenge that we are addressing. Enhancements are expected with the ability to produce smaller sized and higher velocity crystals. Further, IMS will be incorporated to eliminate washing procedures that delipify tissue and enhance the detection of proteins,[52] allowing gas-phase separation of lipids and proteins and tissue analysis independent of solvent.[38,39,53,54]
In conclusion, the first example of transmission geometry tissue imaging using solvent-free sample preparation is reported. The laser-ablated area of ~20 μm is similar to that obtained with solvent-based sample preparation;[37] however, with solvent-free sample preparation, delocalization of the tissue components is eliminated.[26] The area of ablation reflects the spatial resolution and, combined with the known thickness of the tissue, transmission geometry allows a straightforward determination of the tissue volume that is analyzed, here ~4000 μm3. The ability to achieve the ablation of adjacent holes of ca. 20 μm diameter suggests that transmission geometry capable of subcellular spatial resolution is within reach. More sophisticated laser optics will be needed to achieve this goal. If observed, sensitivity issues at improved spatial resolutions might be overcome, as shown here, by further optimizing the ion transfer capillary temperatures and matrix deposition. Transmission geometry LSI imaging using an inexpensive nitrogen laser allows tissue images to be obtained in a timeframe comparable with high repetition rate (>1 kHz) lasers. Transmission geometry imaging mass spectrometry is potentially useful for many applications including (for example) surface analysis of synthetic materials such as polymers.
Acknowledgements
ST is grateful for having been inspired by Professor J. Michael Walker (1950–2008), Indiana University. Authors are thankful to Professor Charles N. McEwen and Andrew Harron (University of the Sciences in Philadelphia) for providing their help with stitching the mass spectral data to images. Financial support is acknowledged from Wayne State University (Summer Fellowships to ALR and CBL; Start-up Funds to ST), and NSF CAREER 0955975, ASMS Research Award financially supported by Waters Corporation, and DuPont Company Young Investigator Award (to ST), as well as DA021696 and DA011322 (to KM).
REFERENCES
- [1].Vidová V, Pól J, Volný M, Novák P, Havlíček V, Wiedmer SK, Holopainen JM. Visualizing spatial lipid distribution in porcine lens by MALDI imaging high-resolution mass spectrometry. J. Lipid Res. 2010;51:2295. doi: 10.1194/jlr.M005488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ridenour WB, Kliman M, McLean JA, Caprioli RM. Structural characterization of phospholipids and peptides directly from tissue sections by MALDI traveling-wave ion mobility-mass spectrometry. Anal. Chem. 2010;82:1881. doi: 10.1021/ac9026115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Tennessen JA, Woodhams DC, Chaurand P, Reinert LK, Billheimer D, Shyr Y, Caprioli RM, Blouin MS, Rollins-Smith LA. Variations in the expressed antimicrobial peptide repertoire of northern leopard frog (Rana pipiens) populations suggest intraspecies differences in resistance to pathogens. Dev. Comp. Immunol. 2009;33:1247. doi: 10.1016/j.dci.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].van Remoortere A, van Zeijl RJM, van den Oever N, Franck J, Longuespée R, Wisztorski M, Salzet M. MALDI imaging and profiling MS of higher mass proteins from tissue. J. Am. Soc. Mass Spectrom. 2010;21:1922. doi: 10.1016/j.jasms.2010.07.011. [DOI] [PubMed] [Google Scholar]
- [5].Hsieh Y, Chen J, Korfmacher WA. Mapping pharmaceuticals in tissues using MALDI imaging mass spectrometry. J. Pharmacol. Toxicol. Methods. 2007;55:193. doi: 10.1016/j.vascn.2006.06.004. [DOI] [PubMed] [Google Scholar]
- [6].Cornett DS, Frapier SL, Caprioli RM. MALDI-FTICR imaging mass spectrometry of drugs and metabolites in tissue. Anal. Chem. 2008;80:5648. doi: 10.1021/ac800617s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wiseman JM, Ifa DR, Zhu Y, Kissinger CB, Manicke NE, Kissinger PT, Cooks RG. Desorption electrospray ionization mass spectrometry: Imaging drugs and metabolites in tissues. Proc. Natl. Acad. Sci. USA. 2008;105:18120. doi: 10.1073/pnas.0801066105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Rohner TC, Staab D, Stoeckli M. MALDI mass spectrometric imaging of biological tissue sections. Mech. Ageing Dev. 2004;126:177. doi: 10.1016/j.mad.2004.09.032. [DOI] [PubMed] [Google Scholar]
- [9].Pierson J, Norris JL, Aerni H-R, Svenningsson P, Caprioli RM, Andrén PE. Molecular profiling of experimental Parkinson’s disease: direct analysis of peptides and proteins on brain tissue sections by MALDI mass spectrometry. J. Proteome Res. 2004;3:289. doi: 10.1021/pr0499747. [DOI] [PubMed] [Google Scholar]
- [10].Stauber J, Lemaire R, Franck J, Bonnel D, Croix D, Day R, Wisztorski M, Fournier I, Salzet M. MALDI imaging of formalin-fixed paraffin-embedded tissues: application to model animals of Parkinson disease for biomarker hunting. J. Proteome Res. 2008;7:969. doi: 10.1021/pr070464x. [DOI] [PubMed] [Google Scholar]
- [11].Chaurand P, Schwartz SA, Caprioli RM. Assessing protein patterns in disease using imaging mass spectrometry. J. Proteome Res. 2004;3:245. doi: 10.1021/pr0341282. [DOI] [PubMed] [Google Scholar]
- [12].Lemaire R, Menguellet SA, Stauber J, Marchaudon V, Lucot J, Collinet P, Farine M, Vinatier D, Day R, Ducoroy P, Salzet M, Fournier I. Specific MALDI imaging and profiling for biomarker hunting and validation: fragment of the 11S proteasome activator complex, reg alpha fragment, is a new potential ovary cancer biomarker. J. Proteome Res. 2007;6:4127. doi: 10.1021/pr0702722. [DOI] [PubMed] [Google Scholar]
- [13].Rauser S, Marquardt C, Balluff B, Deininger S, Albers C, Belau E, Hartmer R, Suckau D, Specht K, Ebert MP, Schmitt M, Aubele M, Höfler H, Walch A. Classification of HER2 receptor status in breast cancer tissues by MALDI imaging mass spectrometry. J. Proteome Res. 2010;9:1854. doi: 10.1021/pr901008d. [DOI] [PubMed] [Google Scholar]
- [14].Chandra S, Lorey DR. SIMS ion microscopy in cancer research: sinlgle cell isotopic imaging for chemical composition, cytotoxicity and cell cycle recognition. Mol. Biol. Cell. 2001;12:141S. [PubMed] [Google Scholar]
- [15].Jones EA, Lockyer NP, Vickerman JC. Mass spectral analysis and imaging of tissue by ToF-SIMS – The role of buckminsterfullerene, C60+, primary ions. Int. J. Mass Spectrom. 2007;260:146. [Google Scholar]
- [16].Amstalden ER, van Hove D. F. Smith, Heeren RMA. A concise review of mass spectrometry imaging. J. Chromatogr. A. 2010;1217:3946. doi: 10.1016/j.chroma.2010.01.033. [DOI] [PubMed] [Google Scholar]
- [17].Takáts Z, Wiseman JM, Gologan B, Cooks RG. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science. 2004;306:471. doi: 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- [18].Cooks RG, Ouyang Z, Takáts Z, Wiseman JM. Ambient mass spectrometry. Science. 2006;311:1566. doi: 10.1126/science.1119426. [DOI] [PubMed] [Google Scholar]
- [19].Takáts Z, Wiseman JM, Cooks RG. Ambient mass spectrometry using desorption electrospray ionization (DESI): instrumentation, mechanisms and applications in forensics, chemistry, and biology. J. Mass Spectrom. 2005;40:1261. doi: 10.1002/jms.922. [DOI] [PubMed] [Google Scholar]
- [20].Chaurand P, Schwartz SA, Caprioli RM. MALDI imaging MS allows simultaneous mapping of hundreds of peptides and proteins in thin tissue sections with a lateral resolution of ~30–50 μm. Anal. Chem. 2004;76:86A. [Google Scholar]
- [21].Chaurand P, Schriver KE, Caprioli RM. Instrument design and characterization for high resolution MALDI-MS imaging of tissue sections. J. Mass Spectrom. 2007;42:476. doi: 10.1002/jms.1180. [DOI] [PubMed] [Google Scholar]
- [22].Caldwell RL, Caprioli RM. Tissue profiling by mass spectrometry: a review of methodology and applications. Mol. Cell. Proteomics. 2005;4:394. doi: 10.1074/mcp.R500006-MCP200. [DOI] [PubMed] [Google Scholar]
- [23].Becker M, Deninger S, Holle A, Hoehndorf J, Schürenberg M, Pineau C. High definition MALDI imaging: new analytical capabilities for protein biomarker discovery from tissue. J. Biomol. Technol. 2010;21:S33. [Google Scholar]
- [24].Trim PJ, Djidja M, Atkinson SJ, Oakes K, Cole LM, Anderson DMG, Hart PJ, Francese S, Clench MR. Introduction of a 20kHz Nd:YVO4 laser into a hybrid quadrupole time-of-flight mass spectrometer for MAL-DI-MS imaging. Anal. Bioanal. Chem. 2010;397:3409. doi: 10.1007/s00216-010-3874-6. [DOI] [PubMed] [Google Scholar]
- [25].Brown J, Murray P, Claude E, Kenny D. 20 μm resolution MALDI imaging of lipid distribution in tissue using lasers between 1kHz and 10kHz; Proc. 58th ASMS Conf. Mass Spectrometry and Allied Topics; Salt Lake City, Utah. May 27, 2010. [Google Scholar]
- [26].Trimpin S, Herath TN, Inutan ED, Cernat SA, Miller JB, Mackie K, Walker JM. Field-free transmission geometry atmospheric pressure matrix-assisted laser desorption/ionization for rapid analysis of unadulterated tissue samples. Rapid Commun. Mass Spectrom. 2009;23:3023. doi: 10.1002/rcm.4213. [DOI] [PubMed] [Google Scholar]
- [27].Vertes A, Balazs L, Gijbels R. Matrix-assisted laser desorption of peptides in transmission geometry. Rapid Commun. Mass Spectrom. 1990;4:263. [Google Scholar]
- [28].Ehring H, Costa C, Demirev PA, Sundqvist BUR. Photochemical versus thermal mechanisms in matrix-assisted laser desorption/ionization probed by back side desorption. Rapid Commun. Mass Spectrom. 1996;10:821. [Google Scholar]
- [29].Schürenberg M, Schulz T, Dreisewered K, Hillenkamp F. Matrix-assisted laser desorption/ionization in transmission geometry: instrumental implementation and mechanistic implications. Rapid Commun. Mass Spectrom. 1996;10:1873. [Google Scholar]
- [30].Perez J, Petzold CJ, Watkins MA, Vaughn WE, Kenttamaa HI. Laser desorption in transmission geometry inside a Fourier-transform ion cyclotron resonance mass spectrometer. J. Am. Soc. Mass Spectrom. 1999;10:1105. doi: 10.1016/S1044-0305(99)00084-7. [DOI] [PubMed] [Google Scholar]
- [31].Galicia MC, Vertes A, Callahan JH. Atmospheric pressure matrix-assisted laser desorption/ionization in transmission geometry. Anal. Chem. 2002;74:1891. doi: 10.1021/ac011098i. [DOI] [PubMed] [Google Scholar]
- [32].McEwen CN, Larsen BS, Trimpin S. Laserspray ionization on a commercial atmospheric pressure-MALDI mass spectrometer ion source: selecting singly or multiply charged ions. Anal. Chem. 2010;82:4998. doi: 10.1021/ac1006624. [DOI] [PubMed] [Google Scholar]
- [33].McEwen CN, Pagnotti V, Inutan ED, Trimpin S. New paradigm in ionization: multiply charged ion formation from a solid matrix without a laser or voltage. Anal. Chem. 2010;82:9164. doi: 10.1021/ac102339y. [DOI] [PubMed] [Google Scholar]
- [34].McEwen CN, Trimpin S. An alternative ionization paradigm for atmospheric pressure mass spectrometry: flying elephants from Trojan horses. Int. J. Mass Spectrom. 2010 DOI: 10.1016/j.ijms.2010.05.020. [Google Scholar]
- [35].Trimpin S, Inutan ED, Herath TN, McEwen CN. Laserspray ionization – a new AP-MALDI method for producing highly charged gas-phase ions of peptides and proteins directly from solid solutions. Mol. Cell. Proteomics. 2010;9:362. doi: 10.1074/mcp.M900527-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Richards AL, Marshall DD, Inutan ED, McEwen CN, Trimpin S. High-throughput analysis of peptides and proteins by laserspray ionization mass spectrometry. Rapid Commun. Mass Spectrom. 2011;25:247. doi: 10.1002/rcm.4841. [DOI] [PubMed] [Google Scholar]
- [37].Inutan ED, Richards AL, Wager-Miller J, Mackie K, McEwen CN, Trimpin S. A new method for protein analysis directly from tissue at atmospheric pressure and with ultra-high mass resolution and electron transfer dissociation sequencing. Mol. Cell. Proteomics. 2010 doi: 10.1074/mcp.M110.000760. DOI: 10.1074/mcp.M110.000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Inutan ED, Trimpin S. Laserspray ionization (LSI) ion mobility spectrometry (IMS) mass spectrometry (MS) J. Am. Soc. Mass Spectrom. 2010;21:1260. doi: 10.1016/j.jasms.2010.03.039. [DOI] [PubMed] [Google Scholar]
- [39].Inutan ED, Trimpin S. Laserspray ionization-ion mobility spectrometry-mass spectrometry: baseline separation of isomeric amyloids without the use of solvents desorbed and ionized directly from a surface. J. Proteome Res. 2010;9:6077. doi: 10.1021/pr1005923. [DOI] [PubMed] [Google Scholar]
- [40].Karas M, Hillenkamp F. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal. Chem. 1988;60:2299. doi: 10.1021/ac00171a028. [DOI] [PubMed] [Google Scholar]
- [41].Skelton R, Dubois F, Zenobi R. A MALDI sample preparation method suitable for insoluble polymers. Anal. Chem. 2000;72:1707. doi: 10.1021/ac991181u. [DOI] [PubMed] [Google Scholar]
- [42].Trimpin S, Rouhanipour A, Az R, Räder HL, Müllen K. New aspects in matrix-assisted laser desorption/ionization time-of-flight mass spectrometry: a universal solvent-free sample preparation. Rapid Commun. Mass Spectrom. 2001;15:1364. doi: 10.1002/rcm.372. [DOI] [PubMed] [Google Scholar]
- [43].Hankin JA, Barkley RM, Murphy RC. Sublimation as a method of matrix application for mass spectrometric imaging. J. Am. Soc. Mass Spectrom. 2007;18:1646. doi: 10.1016/j.jasms.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Puolitaival SM, Burnum KE, Cornett DS, Caprioli RM. Solvent-free matrix dry-coating for MALDI imaging of phospholipids. J. Am. Soc. Mass Spectrom. 2008;19:882. doi: 10.1016/j.jasms.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Trimpin S, Herath TN, Inutan ED, Wager-Miller J, Kowalski P, Claude E, Walker JM, Mackie K. Automated solvent-free matrix deposition for tissue imaging by mass spectrometry. Anal. Chem. 2010;82:359. doi: 10.1021/ac902065u. [DOI] [PubMed] [Google Scholar]
- [46].Goodwin RJ, MacIntyre L, Watson DG, Scullion SP, Pitt AR. A solvent-free matrix application method for matrix-assisted laser desorption/ionization imaging of small molecules. Rapid Commun. Mass Spectrom. 2010;24:1682. doi: 10.1002/rcm.4567. [DOI] [PubMed] [Google Scholar]
- [47].Wang B, Lietz CL, Inutan ED, Leach S, Trimpin S. Laserspray ionization ion mobility spectrometry mass spectrometry: A total solvent-free analysis approach at atmospheric pressure. Anal. Chem. 2011 doi: 10.1021/ac2000576. submitted. [DOI] [PubMed] [Google Scholar]
- [48].Trimpin S, Räder HJ, Müllen K. Experiments on theoretical principles of matrix-assisted laser desorption/ionization mass spectrometry part I preorganization. Int. J. Mass Spectrom. 2006;253:13. [Google Scholar]
- [49].Wiseman JM, Ifa DR, Song Q, Cooks RG. Tissue imaging at atmospheric pressure using desorption electrospray ionization (DESI) mass spectrometry. Angew. Chem. Int. Ed. 2006;45:7188. doi: 10.1002/anie.200602449. [DOI] [PubMed] [Google Scholar]
- [50].Eberlin LS, Ifa DR, Wu C, Cooks RG. Three-dimensional vizualization of mouse brain by lipid analysis using ambient ionization mass spectrometry. Angew. Chem. 2010;122:885. doi: 10.1002/anie.200906283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Eckhardt M. The role and metabolism of sulfatide in the nervous system. Mol. Neurobiol. 2008;37:93. doi: 10.1007/s12035-008-8022-3. [DOI] [PubMed] [Google Scholar]
- [52].Seeley EH, Oppenheimer SR, Mi D, Chaurand P, Caprioli RM. Enhancement of protein sensitivity for MALDI imaging mass spectrometry after chemical treatment of tissue sections. J. Am. Soc. Mass Spectrom. 2008;19:1069. doi: 10.1016/j.jasms.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Trimpin S. A perspective on MALDI alternatives - total solvent-free analysis and electron transfer dissociation of highly charged ions by laserspray ionization. J. Mass Spectrom. 2010;45:471. doi: 10.1002/jms.1737. [DOI] [PubMed] [Google Scholar]
- [54].Inutan ED, Wang B, Trimpin S. Commercial intermediate pressure MALDI ion mobility spectrometry mass spectrometer capable of producing highly charged laserspray ionization ions. Anal. Chem. 2011;83:678. doi: 10.1021/ac102779e. [DOI] [PubMed] [Google Scholar]