Abstract
Although the current obesity epidemic is of environmental origin, there is substantial genetic variation in individual response to an obesogenic environment. In this study, we perform a genome-wide scan for quantitative trait loci (QTLs) affecting obesity per se, or an obese response to a high-fat diet in mice from the LG/J by SM/J Advanced Intercross (AI) Line (Wustl:LG,SM-G16). A total of 1,002 animals from 78 F16 full sibships were weaned at 3 weeks of age and half of each litter placed on high- and low-fat diets. Animals remained on the diet until 20 weeks of age when they were necropsied and the weights of the reproductive, kidney, mesenteric, and inguinal fat depots were recorded. Effects on these phenotypes, along with total fat depot weight and carcass weight at necropsy, were mapped across the genome using 1,402 autosomal single-nucleotide polymorphism (SNP) markers. Haplotypes were reconstructed and additive, dominance, and imprinting genotype scores were derived every 1 cM along the F16 map. Analysis was performed using a mixed model with additive, dominance, and imprinting genotype scores, their interactions with sex, diet, and with sex-by-diet as fixed effects and with family and its interaction with sex, diet, and sex-by-diet as random effects. We discovered 95 trait-specific QTLs mapping to 40 locations. Most QTLs had additive effects with dominance and imprinting effects occurring at two-thirds of the loci. Nearly every locus interacted with sex and/ or diet in important ways demonstrating that gene effects are primarily context dependent, changing depending on sex and/or diet.
Introduction
The increase in obesity prevalence over the past decades is attributed to an increasing energy imbalance, whereby an individual consumes more energy than is expended (1). Swinburn et al. (2) suggest that most of the increase in body weight has been due to increased energy intake. Even though the increase in obesity in human populations is environmental in origin, significant genetic variation in obesity levels remains (3). Some individuals fed a high-fat diet will become obese, whereas others will remain relatively lean. Part of this variation in response to an obesogenic diet is genetic. Genetic variations interacting with the environment are critical for understanding the genetics of obesity.
Human genome-wide association studies have identified several obesity loci segregating in human populations (4). However, these loci jointly account for only a small portion of the total genetic variance in obesity (5). Several reasons are given for the low levels of genetic variation accounted for by quantitative trait loci (QTLs) in genome-wide association studies, including: low statistical power; unrecognized population structure; the potential contributions of rare alleles; the burden of excessive controls for multiple comparisons; and unrecognized context dependence of gene effects. Context dependency can be with respect to other genes, sex, or environment. Although some human population studies have successfully examined context dependence (e.g. 6,7), many suffer from a lack of power and/or poor measures of the relevant environments.
In addition to investigating the usual genetic effects of QTL and their interactions with environment, we also examine the role that genomic imprinting plays in adult obesity. Genomic imprinting occurs when the effect of an allele differs depending on whether it is inherited from the mother or the father. This is most often discussed in terms of paternal or maternal expression, where the heterozygote expresses the same phenotype as the homozygote that inherited the same paternal or maternal allele, respectively. With genomic imprinting, reciprocal heterozygotes (LS and SL) are genetically identical but phenotypically distinct because they only express one of the two alleles, either that provided by the father or that provided by the mother. A variety of epigenetic mechanisms have been described as leading to genomic imprinting effects, including DNA methylation, histone modification, and noncoding RNAs (8). Genomic imprinting effects on adult obesity have been associated with certain human genetic syndromes, such as Prader–Willi and Angelman syndromes (9) and has been observed in human (10–12) and murine (13–15) mapping studies.
We have utilized the intercross of LG/J and SM/J mouse inbred strains as a model system for investigating genetic variation in obesity and response to a high-fat diet. We originally chose this cross because of the obvious differences in growth and adult body size between the strains (16,17). LG/J has the highest and SM/J the lowest average body mass among the strains listed in the Mouse Phenome Database (phenome.jax. org). Further investigations (18,19) showed that the strains were also distinct in terms of obesity and diabetes-related traits and that their intercross segregates substantial genetic variation in these traits (20,21). As in human populations, variation in complex traits in this cross is due to the segregation of many genes of small effect interacting with each other and with the environment. However, unlike in human populations, mouse gene mapping studies have more power by including: experimental control of population structure; variants that are common by experimental design; and strictly controlled and measured environments.
QTL mapping in the LGXSM Recombinant Inbred strains (22) indicates that many genomic regions are involved in interactions with dietary environment. Here, we present a fine-scale QTL map of obesity and high-fat diet obese response in the LG/J by SM/J Advanced Intercross (AI) Line F16 population (Wustl:LG,SM-G16). Previous quantitative genetic analysis of this population (21) showed that obesity was moderately heritable and that there were substantial gene-by-sex and gene-by-diet interactions. Genetic effects on obesity under the high- and low-fat diets only share 20% of their genetic variation in common, indicating substantial differences in the genetic basis for variation in obesity on different diets.
We have also documented genomic imprinting effects on body size, growth, and body composition in the AI Line (15,23,24). We found that genomic imprinting effects can vary depending on genotypes at other loci (25) and on sex (26). In addition to the usual parental imprinting patterns (paternal or maternal expression), we observed various forms of dominance imprinting (15,24). Unlike earlier obesity mapping studies of this cross (22,27,28), we report variation in dietary fat intake and consequent gene-by-environment interactions, measurement of four different fat depots, not just the reproductive fat depot, an assessment of genomic imprinting effects, and a much finer scale mapping of QTLs than possible in earlier generations.
Methods and Procedures
The mice used in this study were from the F16 generation of the LG/J by SM/J AI Line (Wustl:LG,SM-G16), referred to from now on as the AI Line. We first crossed 10 male SM/J mice with 10 female LG/J mice obtained from the Jackson Laboratories (Bar Harbor, ME). The one-way cross has several consequences for our studies: there is no mitochondrial variation because all animals carry the LG/J mitochondrion; there is no Y chromosome variation as all males carry the SM/J Y chromosome; and the X chromosome complement in the population is 67:33 (LG/J:SM/J) instead of 50:50 requiring modifications of the mapping analysis in AI Line populations. Here we consider only the autosomes.
Starting with the F2 hybrids, the AI Line (29) has been managed as a pseudorandomly mated population: (i) brother–sister mating is not allowed; and (ii) we chose one male and one female from each family as breeders for the next generation, when possible, rather than choosing randomly from the population as a whole. Eliminating variation in familial contributions to the next generation doubles the effective population size of a colony relative to its census size and is the most effective, simple measure that can be taken to reduce the rate of inbreeding in a closed colony (30). The average number of breeding pairs used in AI Line production is 75, for a census size of 150 and an effective size of ~300. Earlier mapping studies of obesity per se in the AI Line include mapping in the F2 and F3 generations (27,31) and in the F10 generation (28). Animals in the earlier studies were fed a standard chow diet (PicoLab Rodent Chow 20, cat. no. 5053) and only the reproductive fat depot was measured.
There were 1,002 experimental F16 animals produced by 78 double-mated breeding pairs producing 148 litters of 6.8 animals each. Males (N = 500) and females (N = 502) from each of these litters were evenly partitioned into cohorts fed a low-fat (247 males; 254 females) or a high-fat (253 males; 248 females) diet. After weaning at 21 days, animals were separated into sex-specific cages of four or five animals per cage, and placed on isocaloric high- (Harlan Teklad cat. no. TD88137) or low-(Research Diets cat. no. D12284; see Table 1) fat diets, dividing each sibship equally between treatments. Calories from fat are 15% in the low-fat diet and 43% in the high-fat diet.
Table 1.
High- and low-fat diet constituents
| Source | High fat | Low fat |
|---|---|---|
| Energy from fat | 42% | 15% |
| Casein (g/kg) | 195 | 197 |
| Sugars (g/kg) | 341 | 307 |
| Corn starch (g/kg) | 150 | 313 |
| Cellulose (g/kg) | 50 | 30 |
| Corn oil (g/kg) | — | 58 |
| Hydrogenated coconut oil (g/kg) | — | 7 |
| Anhydrous milkfat (g/kg) | 210 | — |
| Cholesterol (g/kg) | 1.5 | — |
| Total energy (kJ/g) | 18.95 | 16.99 |
Animals were weighed weekly from 1 to 20 weeks after which they were killed and necropsied. Carcass weight was recorded at necropsy and four fat depots were dissected and weighed: the reproductive, kidney, mesenteric, and inguinal fat depots. Carcass weight was measured at killing before any fat depots or organs were removed. We also analyzed the sum of the four fat depot weights (total depot weight).
Quantitative genetic analyses to obtain heritabilities, genetic, and phenotypic correlations were undertaken using a multivariate analysis of variance with carcass weight and the four fat depot weights as dependent variables and sex, diet, sex-by-diet interaction, and family as the independent variables. Genetic variances and covariances were calculated as twice the between-family variance (30), whereas phenotypic variances and covariances were obtained as the sum of the between- and within-family variances and covariances. Heritabilities and correlations were calculated from the variance/covariance matrices. In this design, heritabilities and correlations are pooled across the sex–diet cohorts. Cohort-specific values are given in Ehrich et al. (21).
We selected 1,536 polymorphic single-nucleotide polymorphisms (SNPs) from the Oxford/CTC database (http://www.well.ox.ac.uk/mouse/INBREDS/) for scoring with the Illumina GoldenGate Bead Array (Illumina, San Diego, CA). A total of 1,402 autosomal SNPs were reliably scored and used to create a map using R/QTL (32). The map and physical positions of the SNPs are listed in Supplementary Table S1 online. The map is measured in terms of recombination accumulated by the F16 generation. At short distances across the genome, the F16 map is eight times longer than the F2 map, so 1 F2 cM is equal to 8 F16 cM and 1 F16 cM is equivalent to 0.125 F2 cM.
Interval mapping was performed using the method of Haley and Knott (33). Additive (Xa) and dominance (Xd) scores were assigned to individuals at each marker; Xa = −1, 0, 0, 1 and Xd = 0, 1, 1, 0 for the SS, LS, SL and LL genotypes, respectively, where the first allele is inherited from the father and the second from the mother. We have added the genomic imprinting score (Xi) as another direct genetic effect (24). Genomic imprinting distinguishes the two reciprocal heterozygotes and is scored 0, +1, −1, 0 for the LL, LS, SL, and SS genotypes, respectively. In order to differentiate between LS and SL heterozygotes, it is necessary to determine the maternally and paternally derived alleles of F16 heterozygotes. This was accomplished using the Integer Linear Programming algorithm in PedPhase 2.0 (34) and the SNP genotypes of the F15 parents and their F16 offspring. Because the Integer Linear Programming algorithm is computationally intensive, it was necessary to divide the larger chromosomes into segments for this analysis.
The three genotype scores, Xa, Xd, and Xi were then used to impute scores in the intervals between markers using the equations provided by Haley and Knott (33) with the addition of newly derived equations for imputing imprinting genotype scores described in Supplementary Table S2 online. We imputed genotype scores every 1 F16 cM between the most proximal and distal markers on each chromosome. Genotype scores for the markers were intercalated at their map positions.
We used a complex mapping model including sex, diet, the sex-by-diet interaction, the direct effects of the genomic locations (Xa, Xd, and Xi), and their two- and three-way interactions with sex and diet as fixed effects, and family and its two- and three-way interactions with sex and diet as random effects using the Mixed Procedure (method = ML) in SAS (SAS, version 9.1; SAS Institute, Cary, NC);
The −2 ln(likelihood) of this model was compared to a base model including only sex, diet, and the sex-by-diet interaction terms using a χ2-test with 12 degrees of freedom. Probabilities were transformed into logarithm of the odds (LOD) scores using LOD = −log10(Prob). The regression coefficients estimated by this model are equivalent to the standard genotypic values from quantitative genetic theory (30). Each coefficient is identified by its genetic effect, additive (a), dominance (d) and genomic imprinting (i), combined, when appropriate, with the interacting factors of sex (as, ds, is), diet (ad, dd, id), and sex and diet (asd, dsd, isd). A genotypic value is defined as the average value of individuals carrying that genotype (). The additive genotypic value is half the difference between the two homozygotes () (ref. 30). The dominance genotypic value is the deviation of the average phenotype of the heterozygotes from the midpoint of the two homozygotes () (ref. 30). The imprinting genotypic value is half the difference between the reciprocal heterozygotes () (ref. 24). Inclusion of family and family-by-sex and/or -diet interaction random effects in the model adjusts its probability to account for autocorrelation due to family structure (24). The different sibships are treated as unrelated here, although there are small, diffuse relationships among them. The bias caused by this low degree of between-sibship autocorrelation is inconsequential given the maring program of the AI Line (24).
We corrected for multiple comparisons using the Bonferroni adjustment estimating the number of independent tests using the Li and Ji (35) procedure based on the eigenvalues of the correlation matrix of marker additive genotype scores. We corrected for multiple comparisons first at the genome-wide level (LOD >3.90) and then separately for each chromosome (for chromosome-wise thresholds see Supplementary Table S1 online), as suggested by Chen and Storey (36). With chromosome-wise significance thresholds, we expect a single false-positive result for each trait in a genome-wide scan. QTL support regions were determined using a one LOD drop from the peak LOD of the QTL.
We also examined our QTL results to determine whether total body mass (carcass weight) might account for our observed significant fat depot QTLs. For each QTL, for each trait, for each population cohort in which a significant result was obtained, we fit the original null model (sex, diet, sex-by-diet interaction) plus carcass weight. We then added the genotype scores (Xa, Xd, and Xi) to the model and used the difference in −2 ln (likelihood) as a χ2 value with three degrees of freedom to test whether the fit was significantly improved. If so, gene effects on obesity are independent of carcass weight to some extent. We also tested a further model that included the interactions between carcass weight and the genotype scores using the difference in −2 ln (likelihood) as a χ2 value with three degrees of freedom to test whether the fit was significantly improved by adding the interactions. A significant finding at this level indicates that the relationship between carcass weight and obesity differs depending on the genotype. This kind of effect defines a relationship QTL (rQTL; 37,38), a locus that affects the relationship between fat depot weight and carcass weight.
Results
Trait means for males and females fed a low- or high-fat diet are provided in Table 2. Overall, males carry an average of 45% more fat in their depots than do females when fed the low-fat diet. Females respond much more strongly to the high-fat diet (125% increase) than do males (64% increase) so that fat depot weights on a high-fat diet are only 10% heavier in males than in females. Heritabilities are moderate, ranging from 30% for the inguinal fat depot to 47% for carcass weight. Both phenotypic and genetic correlations among the depots and carcass weight are high (all r > 0.67; see Table 2). Genetic correlations are higher than their phenotypic counterparts, an expected result due to an upward bias in genetic correlation estimates (39). The high correlations predict substantial levels of pleiotropy at loci affecting the traits.
Table 2.
Mean fat depot weights (g) and s.e. for the four experimental cohorts, low-fat fed females, high-fat fed females, low-fat fed males, and high-fat fed males, and heritabilities, (on diagonal), genetic (above diagonal), and phenotypic correlations (below diagonal) between carcass weight and fat depots
| Trait | Low fat | s.e. | High fat | s.e. | High/low |
|---|---|---|---|---|---|
| Female | |||||
| Carcass weight | 30.00 | 0.51 | 41.29 | 0.51 | 1.38 |
| Reproductive | 1.55 | 0.09 | 3.87 | 0.09 | 2.50 |
| Renal | 0.71 | 0.05 | 1.76 | 0.05 | 2.47 |
| Mesenteric | 0.67 | 0.03 | 1.19 | 0.03 | 1.78 |
| Inguinal | 1.38 | 0.09 | 2.85 | 0.09 | 2.07 |
| All fat depots | 4.42 | 0.23 | 9.87 | 0.23 | 2.23 |
| Male | |||||
| Carcass weight | 40.78 | 0.51 | 50.52 | 0.51 | 1.24 |
| Reproductive | 1.94 | 0.09 | 3.22 | 0.09 | 1.66 |
| Renal | 1.03 | 0.05 | 1.80 | 0.05 | 1.75 |
| Mesenteric | 1.04 | 0.03 | 1.54 | 0.03 | 1.48 |
| Inguinal | 2.19 | 0.09 | 3.58 | 0.09 | 1.64 |
| All fat depots | 6.21 | 0.23 | 10.18 | 0.23 | 1.64 |
| Carcass weight | Reproductive | Renal | Mesenteric | Inguinal | |
| Correlations | |||||
| Carcass weight | 0.47 | 0.89 | 0.94 | 0.90 | 0.90 |
| Reproductive | 0.80 | 0.40 | 0.89 | 0.83 | 0.74 |
| Renal | 0.82 | 0.82 | 0.36 | 0.88 | 0.87 |
| Mesenteric | 0.80 | 0.72 | 0.71 | 0.32 | 0.88 |
| Inguinal | 0.73 | 0.67 | 0.72 | 0.73 | 0.30 |
All correlations and heritabilities are significant at the 0.001 level.
We found 40 obesity QTLs (see Table 3) distributed across the genome. Of these 40, 11 have effects on at least one trait that is significant at the genome-wide level, whereas the others are significant at the chromosome-wise level. There are 95 trait-specific effects grouped into these 40 locations (see Supplementary Table S3 online). The reproductive fat depot is the most commonly affected trait (26 QTLs), followed by total depot weight (20 QTLs), and carcass weight (17 QTLs). The mesenteric (13 QTLs), kidney (11 QTLs), and inguinal (8 QTLs) fat depots mapped to fewer locations, but still occur at a rate at least eight times higher than expected by chance given a chromosome-wise significance threshold. With the strict Bonferroni-corrected thresholds, 24 QTLs affect only a single fat depot, 10 affect two depots, 4 affect three or more depots, one affects only the total depot weight, and one affects only carcass weight with no effects on obesity.
Table 3.
D ietary obesity QTLs named Dobxy with x being the chromosome and y the QTL on the chromosome
| QTL | Traitsa | LODb | Posc | Prox | Dist | ad | d | i | ad | dd | id | as | ds | is | asd | dsd | isd | Old QTLe | Genesf | Obesity genesg |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dob1a | RITW(KM) | 3.878 | 77.9 | 76.56 | 78.16 | × | × | × | × | × | × | × | × | Bod1.1 | 170 | |||||
| Dob1b | RITW(KM) | 5.137 | 78.73 | 78.2 | 79.43 | × | × | × | × | × | × | × | Bod1.1 | 8 | ||||||
| Dob1c | RT(KI) | 4.699 | 172.98 | 170.99 | 173.09 | × | × | × | × | 29 | ||||||||||
| Dob2a | RMIT(K) | 3.724 | 6.67 | 3.38 | 8.24 | × | × | × | 36 | Camk1d | ||||||||||
| Dob2b | RT(KMI) | 3.928 | 71.25 | 70.34 | 71.85 | × | × | Adip9 | 17 | Bbs5 | ||||||||||
| Dob2c | M(I) | 4.223 | 85.75 | 81.63 | 88.19 | × | × | 170 | Fad2lp | |||||||||||
| Dob2d | RMW(KI) | 5.881 | 103.29 | 102.63 | 105.04 | × | × | × | × | × | 34 | |||||||||
| Dob3a | RKTW(MI) | 4.447 | 20.36 | 19.47 | 20.89 | × | × | × | × | Adip21 | 12 | |||||||||
| Dob3b | M(K) | 4.055 | 111.12 | 108.19 | 112.75 | × | × | 27 | ||||||||||||
| Dob4a | R(K) | 2.834 | 39.16 | 38.15 | 44.12 | × | × | × | × | Adip23 | 130 | |||||||||
| Dob4b | K(RI) | 2.802 | 129.78 | 128.8 | 130.34 | × | Adip11 | 43 | Fabp3 | |||||||||||
| Dob6a | M | 2.89 | 47.57 | 45.89 | 50.49 | × | × | 55 | Npy, Igf2bp3 | |||||||||||
| Dob6b | RW(KMI) | 3.842 | 91.7 | 91.05 | 92.57 | × | × | × | × | × | Adip2a | 17 | ||||||||
| Dob6c | RW(KMI) | 3.791 | 97.07 | 93.92 | 99.45 | × | × | × | × | × | 24 | |||||||||
| Dob6d | K(RM) | 3.721 | 111.26 | 110.45 | 112.54 | × | × | × | 7 | Grm7, Oxtr | ||||||||||
| Dob6e | K(R) | 2.966 | 116.88 | 114.79 | 117.56 | × | × | × | × | Adip2b | 39 | Hrh1, Pparg, Alox5 | ||||||||
| Dob7a | RKMITW | 3.414 | 65.96 | 63.8 | 66.81 | × | × | × | × | × | × | × | Adip3A | 10 | Oca2, Gabrb3, Snord116 | |||||
| Dob7b | RKT(I) | 4.337 | 80.34 | 79.93 | 82.44 | × | × | × | × | × | Adip25 | 10 | ||||||||
| Dob7c | RTW(KI) | 3.837 | 82.77 | 80.75 | 85.8 | × | × | × | Adip25 | 6 | ||||||||||
| Dob7d | RKTW(MI) | 3.877 | 87.85 | 87.24 | 90.15 | × | × | × | × | × | Adip25 | 58 | ||||||||
| Dob8a | RKMITW | 5.291 | 22.29 | 18.15 | 25.77 | × | × | × | × | Adip4 | 153 | |||||||||
| Dob8b | I(KM) | 2.684 | 64.98 | 63.93 | 68.4 | × | × | × | 21 | Cpe | ||||||||||
| Dob8c | M(K) | 2.806 | 85.97 | 82.91 | 87.44 | × | 70 | Ucp1 | ||||||||||||
| Dob9a | RM(K) | 3.264 | 26.41 | 24.17 | 26.83 | × | × | × | × | × | 15 | |||||||||
| Dob10a | W(I) | 2.884 | 96.56 | 93.22 | 103.73 | × | 47 | |||||||||||||
| Dob10b | RKMTW(I) | 2.94 | 112.99 | 111.75 | 114.03 | × | × | × | × | × | Adip15 | 4 | ||||||||
| Dob10c | IT(RKM) | 3.415 | 116.29 | 115.74 | 116.99 | × | × | × | × | × | 18 | |||||||||
| Dob11a | RT(KMI) | 3.917 | 47.44 | 47.08 | 48.06 | × | × | × | × | × | Adip16 | 2 | ||||||||
| Dob11b | RTW(KMI) | 4.211 | 99.71 | 98.91 | 100.97 | × | × | × | × | Wtn11.1b | 47 | Igfbp4, Hcrt, Stat5b, Stat3, Stard3, Rara, Becn1 | ||||||||
| Dob13a | R(M) | 2.705 | 43.92 | 43.01 | 45.03 | × | × | × | × | Adip18a | 12 | |||||||||
| Dob14a | RW(K) | 2.597 | 23.95 | 23.33 | 32.33 | × | × | × | Bod14.2 | 85 | Capn7 | |||||||||
| Dob15a | M(RK) | 2.687 | 67.71 | 66.42 | 69.8 | × | × | 14 | ||||||||||||
| Dob15b | K(RMI) | 2.899 | 97.67 | 97.09 | 100.95 | × | × | × | 100 | Vdr | ||||||||||
| Dob17a | RMTW(KI) | 3.389 | 30.79 | 29.32 | 31.56 | × | × | × | × | × | 30 | Glp1r, Ppard, Mapk14, Mapk13, Abcg1 | ||||||||
| Dob17b | M(KI) | 2.603 | 35.97 | 33.16 | 43.67 | × | × | 228 | Rxrb, Tnf | |||||||||||
| Dob17c | RTW(KMI) | 3.388 | 46.89 | 44.69 | 48.27 | × | × | × | × | Adip20b | 88 | Capn11, Pla2g7, Dpp9 | ||||||||
| Dob17d | RI | 3.932 | 72.87 | 70.75 | 73.69 | × | × | × | × | × | 23 | Capn13 | ||||||||
| Dob17e | RT(I) | 3.434 | 76.56 | 76.15 | 77.44 | × | × | × | × | × | × | 1 | Crim1 | |||||||
| Dob18a | T(RKM) | 2.788 | 71.8 | 71.42 | 72.65 | × | × | × | × | 1 | Dcc | |||||||||
| Dob19a | RKTW(MI) | 4.099 | 14.63 | 12.32 | 15.67 | × | × | × | × | × | Bod19.1a | 39 |
LOD, logarithm of odds; QTL, quantitative trait locus; RI, Recombinant Inbred.
Traits mapping to the QTL are listed as R (Reproductive fat depot weight), K (Kidney fat depot weight), M (Mesenteric fat depot weight), I (inguinal fat depot weight), T (Total fat depot weight), and N (Carcass weight at necropsy). Traits in parentheses are only significant at the point-wise (0.05) level without correction for multiple comparisons.
LOD is the highest LOD score at the location. LOD scores in boldface are significant at the genome-wide level, whereas the remainder are significant at the chromosome-wide level.
QTL Positions (POS) and Proximal (PROX) and Distal (Dist) confidence interval limits are given in Megabases.
Significant genotypic values for traits at the locus are indicated by a “×”. The first position in the headings refers to the genotypic value (a, additive; d, dominance; i, imprinting) while the second and third positions refer to interactions between the genotypic values and sex (s), diet (d), or sex-by-diet (sd) interactions.
Genes refers to the number of annotated genes in the interval.
Obesity genes lists the names of annotated obesity genes in the interval.
However, this is likely to be an underestimate of the true level of pleiotropy. Specific depots may not pass the Bonferroni-corrected threshold but may still be significant at a protected point-wise 5% threshold (LOD >1.3). If we accept as significant any fat depot significant at the point-wise threshold at locations where at least one depot is significant at the Bonferroni-adjusted level, a different picture emerges. Under these criteria, 47.5% of the 40 QTLs affect all four depots (see Table 3), 25% affect 3 of 4 depots, 22.5% affect two fat depots, and only 5% affect a single depot.
Traits at most QTLs show significant interactions with sex and/or diet (n = 37). Interactions with sex (n = 35) are slightly more common than interactions with diet (n = 31) but the majority of loci interact with both (n = 29). Context dependency is a nearly universal feature of these QTL. There are three direct genetic effects included in our analytical model, additive (a), dominance (d), and genomic imprinting (i) at each locus and, hence, 40 potential interactions with sex and/or diet for each genetic effect for each trait across the 40 QTLs. Of these 120 potential effects about one-third are null, not significant either as main effects or as any form of interaction with sex and/or diet (27.5% additive; 30% dominance; 42.5% imprinting). Approximately 11% of the effects are significant main effects that do not interact with sex and/or diet (12% additive; 10% dominance; 10% imprinting). The remaining 56% of the potential effects are interactions with sex, diet, or with both jointly (60% additive; 60% dominance; 47.5% imprinting). When genes have an effect, it is most often a context-dependent one.
The 40 QTLs have 131 trait-specific effects that are often restricted to sex or diet-based subpopulations rather being evident across the whole population. Only 13 QTL effects are manifest in the full population across all cohorts. Sixteen effects are restricted to animals of both sexes on a specific diet, 13 affecting only high-fat fed animals and 3 affecting only low-fat fed animals. Twenty-nine effects are restricted to a single sex but are evident on both diets, 20 in females and 9 in males. The remaining effects are restricted to a single cohort, with 58 effects restricted to high-fat fed females (HF), 7 effects exclusive to low-fat fed females (LF), 5 effects solely on highfat fed males (HM), and 3 effects restricted to the low-fat fed males (LM). Overall, more effects are found in females and in animals fed a high-fat diet than amongst males and animals fed a low-fat diet.
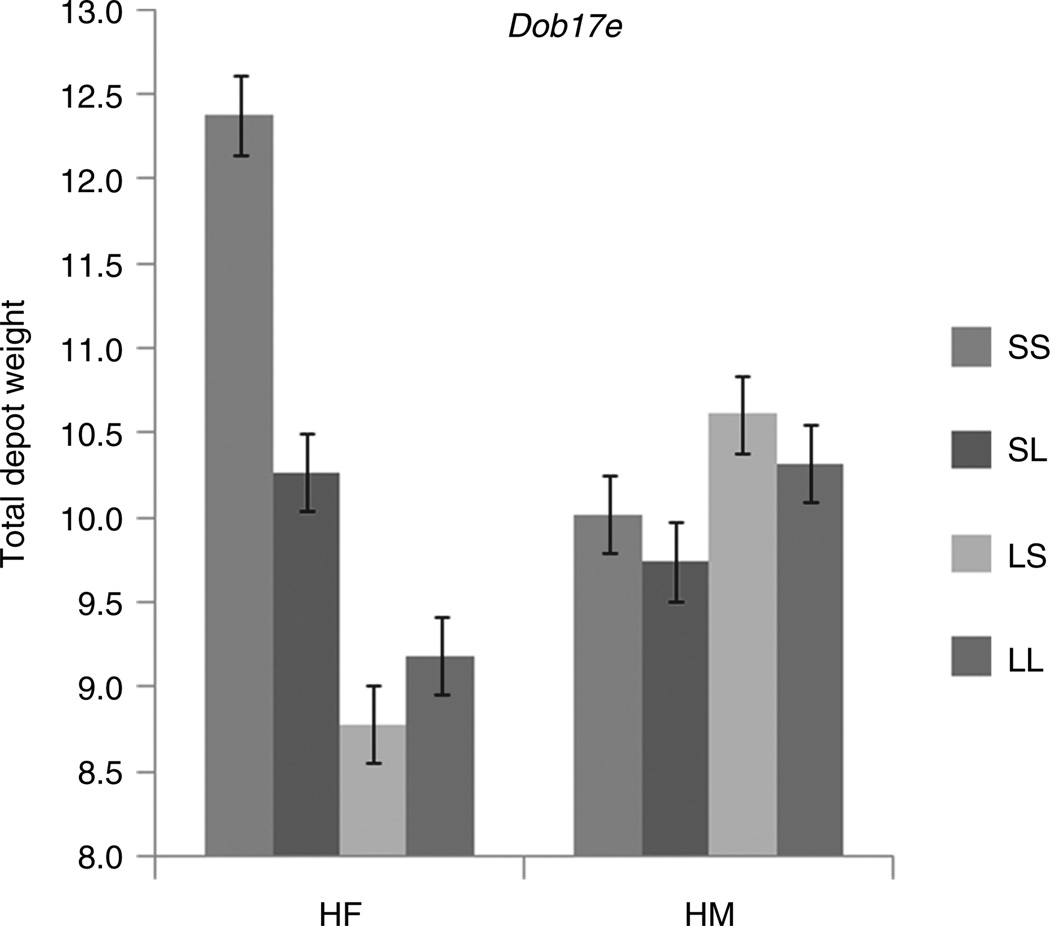
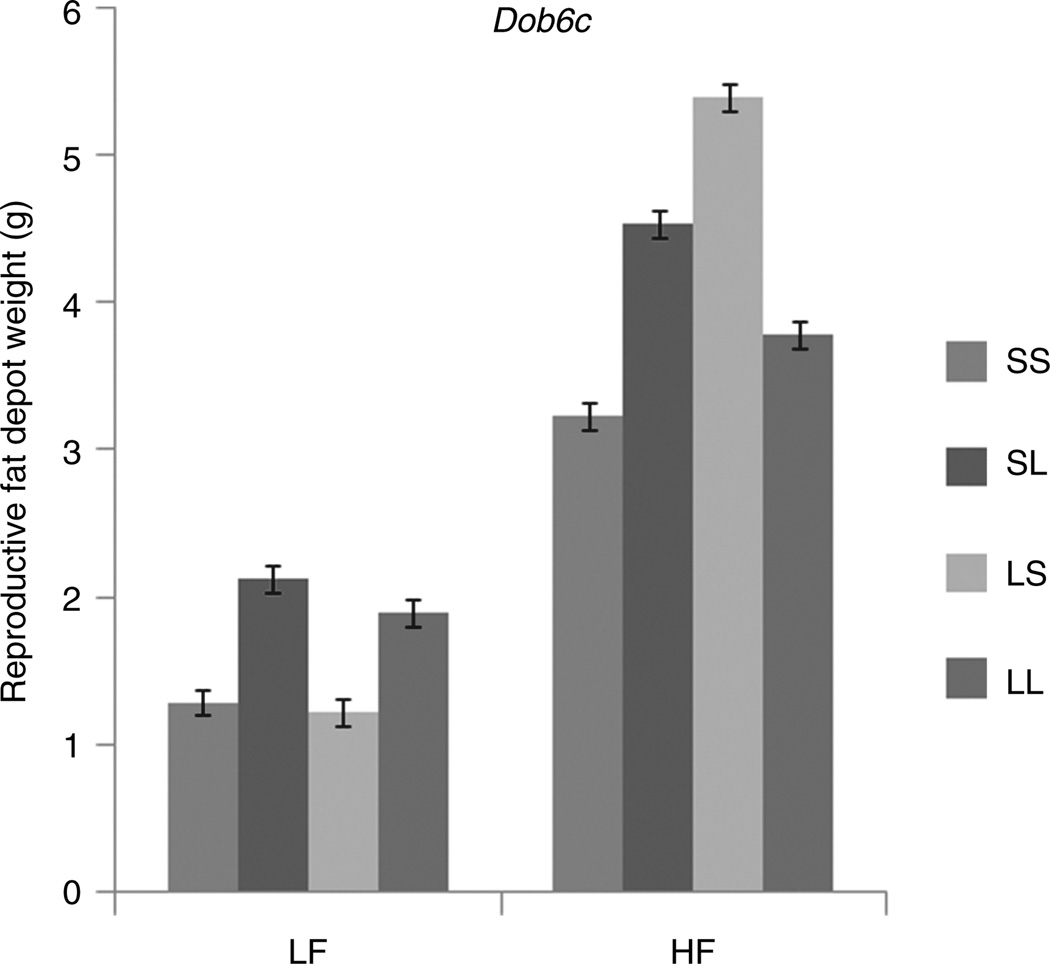
Eighteen loci have different effects in multiple sex–diet cohorts. For example, Dob17e (see Figure 1) has significant additive and dominance effects on total fat depot weight in HF and a significant imprinting effect in HM. There are also eight instances where the same genetic effect is significant in multiple cohorts but the effects are of opposite sign. For example Dob6c (see Figure 2) has a negative imprinting effect on reproductive fat depot weight in LF and a positive imprinting effect in HF. This indicates a substantially stronger effect of the high-fat diet on LS animals relative to SL animals.
Figure 1.
Genotypic values of total fat depot weight in high-fat fed males (HM) and females (HF) with contrasting imprinting genotypic values at locus Dob17e. In the females there are negative additive and negative imprinting genotypic values, whereas in males both values are positive. Error bars indicate ± 1.0 s.e.
Figure 2.
Genotypic values of reproductive fat depot weights in the high-fat (HF) and low-fat (LF) fed females at QTL Dob6c, where the genetic effects in the cohorts are distinct. Low-fat fed females have a significant positive additive effect and a significant negative imprinting effect, producing a pattern consistent with maternal expression, while high-fat fed females have no additive effect, a significant positive dominance effect and a significant positive imprinting effect. Error bars indicate ± 1.0 s.e.
The loci discovered here display a complex genetic architecture. Most of the 95 trait-specific QTLs have significant additive effects (80% of loci) followed in frequency by dominance (64%) and genomic imprinting (61%) effects. Significant additive effects average 0.28 s.d. units (range 0.1–0.7 s.d.). The LG/J allele results in heavier weights for 44 trait-specific QTLs, whereas the SM/J allele results in heavier weights at 31 trait-specific QTLs. In terms of grams of fat or body weight, homozygotes are distinguished, on average, by 1.2 g of reproductive fat, 0.76 g of inguinal fat, 0.27 g of mesenteric fat, 0.26 g of kidney fat, 2.6 g of fat summed across the depots, and 5.5 g of carcass weight.
Dominance values, like additive ones, tend to be relatively small with significant values averaging 0.30 s.d. units on an absolute scale. Positive and negative dominance effects are equally common. Most trait-specific QTLs do not have dominance effects (70 of 131), with 52 classified as codominant and 18 as null, involving neither additive nor dominance effects. At 16 trait-loci the LG/J allele is dominant to the SM/J allele, whereas the reverse is true for 7 loci. There are a large number of both overdominant (21; heterozygotes heavier with no additive effect) and underdominant (17; heterozygotes lighter with no additive effect) trait-specific effects.
Genomic imprinting is also quite common. Imprinting genotypic values average 0.23 s.d. units on an absolute scale. Unlike additive and dominance values, there is a significant tendency for imprinting values to be positive (38 positive and 22 negative effects) indicating that most often animals with the LS genotype, inheriting their LG/J allele from their father, are heavier than SL animals. We observe many different imprinting patterns, including 24 instances of parental imprinting with paternal and maternal expression equally common and 43 instances of dominance imprinting, including 25 examples of polar dominance (including seven cases that also displayed maternal expression) and 18 of bipolar dominance imprinting (no additive or dominance effects). The average differences between reciprocal heterozygotes in grams and the number of QTL effects included is: reproductive fat depot (0.90 g; 18), kidney fat depot (0.31 g, 6); mesenteric fat depot (0.24 g, 9), inguinal fat depot (0.64 g; 2), total fat depot weight (2.35 g, 9), and carcass weight (4.33 g, 13).
After correcting for carcass weight, 54% of the QTLs still had significant genetic effects on at least one of the fat depots in at least one affected population cohort (see Supplementary Table S3 online), although an independent effect was only maintained at about 30% of the QTL effects over all traits and cohorts. Of the 18 QTLs, 11 that were carcass weight-dependent had significant genotype by carcass weight interactions, indicating that the relationship between fat depot weight and carcass weight differed depending on the genotype and that genotypic effects on obesity changed depending on the carcass weight. The relationship of fat depot weights to carcass weight changes with genotype.
We can also examine these results in relation to the replication of the 24 reproductive fat depot QTLs (Adip1–25, Adip22 never assigned) mapped in the F2/3 and F9/10 populations (27,28,31). These are listed in Supplementary Table S4 online. Twelve of the 24 Adip loci map to dietary obesity loci significant at the chromosome-wise level. The remaining 12 Adip loci map to genomic locations that are significant at the point-wise level (LOD >1.30) in the AI Line. Point-wise thresholds can be used for replication of previously mapped QTL because they provide an a priori hypothesis generated with unrelated data. Therefore, significance thresholds at these QTL locations are protected from correction for multiple comparisons. All Adip loci are confirmed in this study.
The accumulation of recombination to the F16 generation allows us to map QTL positions with greater precision than before. Confidence regions for trait-specific QTLs range from 1 to 17 Mb, with a median confidence interval length of 3.1 Mb. About one-quarter of the intervals are <2.0 Mb. The numbers of annotated genes within these intervals ranges from 1 to 228 with a median number of 27 genes. A short list of obesity-related positional candidate genes is provided in Table 3.
Discussion
All groups of animals added to their fat depots when fed a high-fat diet with females being much more responsive than males. Earlier work on the parental strains, LG/J and SM/J, indicated that each strain consumed the same amount of the high- and the low-fat diets (19). This makes it unlikely that the effects noted here are due to differences in consumption levels or caloric intake but to differences in processing nutrients.
We have mapped an unprecedented number of obesity QTLs in the AI Line to highly restricted genomic regions. Surprisingly, all of the adiposity loci previously discovered for the reproductive fat depot in animals reared on a low-fat diet (27,28,31) replicated in this study. The dietary obesity loci mapped here also correspond to locations mapped in other studies (see Supplementary Table S5 online) utilizing strains both related and unrelated to LG/J and SM/J.
We also evaluated whether fat depot QTLs were independent of body mass (carcass weight). Carcass weight is strongly correlated with fat depot weights and mapped to 16 of the 39 fat depot QTLs, indicating substantial pleiotropy between body mass and fat depot weight. When carcass size was included as a covariate, about half of the 39 loci no longer had independent obesity effects (see Supplementary Table S3 online). This outcome could represent loci that have organism-wide effects common to all organs and that are not specific to fat deposition. Alternatively, the outcome could be due to the part–whole relationship between fat depot weight and carcass weight. On average, nearly 20% of an animal’s weight is dissectible fat in this population and in extreme cases this can reach 50%. Also, lipid deposits in other organs, such as liver, heart, and muscle may increase with increased fat depot weight rather than with other size factors. In a later generation of the AI Line (E.A. Norgard, personal communication), we measured long bone lengths, a measure of size not confounded with fat depot weight. There is only a 1% increase in long bone length among animals fed the high-fat diet and a correlation of only 0.2 (4% shared variation) between fat depot weights and limb lengths within cohorts. Hence, we believe that the part–whole relationship between fat depot weights and carcass weight is the likely source of their association and that the use of carcass weight as a covariate is not indicated. Furthermore, most of the carcass weight-dependent loci showed significant carcass weight by genotype interactions indicating that the genotypic effects on obesity are altered at different carcass weights. Such effects are referred to as relationship QTLs (37,38) as they affect the relationship (regression slope) of fat depot weight and body mass.
We have mapped QTLs with much greater precision than in earlier studies. In a mapping analysis of dietary obesity in the LGXSM Recombinant Inbred Lines, we located 20 trait-specific obesity QTLs in 8 locations to a resolution of 22 cM (ref. 22), 14 were diet-responsive and 6 were not. Here we mapped 95 trait-specific QTLs to 40 genomic locations at a resolution of 1–2 cM (~3 Mb). This is a dramatic improvement both in power to detect QTLs and in precision, localizing them to narrow regions of interest. The median number of genes in a QTL support interval in this study is 27 but many locations contain 10 or fewer genes. There are many genes in our regions that have annotations of relating to obesity or other aspects of the metabolic syndrome (see Table 3). These include several genes involved in PPAR signaling, including transcription factors Pparg (Dob6e), Ppard (Dob17a), and Rxrb (Dob17b), fatty acid transport genes Fabp3 (Dob4b) and Acsl3 (Dob1a), a fatty acid oxidation gene Acad8 (Dob9a), Angptl4 (Dob17b) involved in adipocyte differentiation, Ucp1 (Dob8c) involved in adaptive thermogenesis, and Aqp7 (Dob4a) involved in gluconeogenesis. Another pathway with multiple positional candidate genes is the p38 MAP kinase signaling pathway controlling cell cycle. This pathway includes positional candidate genes Mapk13 (Dob17a), Mapk14 (Dob17a), Dusp6 (Dob10a), Tnf (Dob17b), Daxx (Dob17b), Gadd45gip1 (Dob8c), and Hspa1l (Dob17b). Positional candidate genes in the leptin signaling pathway include Npy (Dob6a), Stat3 (Dob11b), Prkag1 (Dob15b), and G6pc (Dob11b). Tnf (Dob17b) signaling also involves Nfkbie (Dob17c) and links into the autophagy pathway, involving Atg7 (Dob6e) and Becn1 (Dob11b), through mTOR signaling. Clearly there are several different mechanisms potentially involved in the obesity differences between LG/J and SM/J. Those specified here may be most fruitful for follow- up with gene expression studies and genomic sequence.
Nearly 75% of the 40 obesity QTLs affected three or all four fat depots, when depots significant at a point-wise level are included at locations originally identified using Bonferroni-adjusted thresholds. Thus, most QTLs affect fat deposition generally rather than having depot-specific effects. This is consistent with the very strong genetic correlations among fat depot weights in this cross (see Table 2; refs. 20,21). Even so, the remaining 25% of the QTLs, affecting only one or two depots, may have distinct effects on different fat depots. Most of the QTL affect the reproductive fat depot. This may be due to the fact that the reproductive depot is the largest in mice and hence has a relatively smaller measurement error variance. This also supports the primary use of the reproductive fat depot in murine studies of obesity despite the lack of a comparable fat depot in humans in that most QTLs affecting the reproductive fat depot weight also affect other depots with homologues in humans.
Genetic (a and d) and genomic imprinting (i) effects were relatively small (0.1–0.8 s.d. units) and numerous. Most loci have additive effects and both dominance and genomic imprinting occur at nearly two-thirds of the loci. Effect distributions for all three genotypic values are negative exponential, indicating a high frequency of small effects and reduced frequency as the effect becomes larger. For additive effects, the LG/J allele leads to a heavier animal at 60% of the loci; this is a much lower percentage than found in our other studies of obesity on a low-fat diet (27,28,31). The relatively high percentage of loci where the SM/J allele leads to higher levels of fat is consistent with our studies of the parent strains showing that SM/J animals respond more strongly to a high-fat diet than LG/J animals (19). The most common dominance relationship is codominance with the heterozygote midway between the two homozygotes. Of the remaining loci showing significant dominance, 37% show full dominance with the LG/J allele being most often dominant to the SM/J allele. Surprisingly, 63% of the dominance effects are either overdominant or underdominant. Interaction between alleles at a single locus is very common for obesity in this population and often occurs in the absence of additive effects.
The results for genomic imprinting are the most intriguing and unexpected. Imprinting is common, occurring for traits at 61% of the loci. Although it has long been known that genomic imprinting can play a role in obesity, the genome-wide scope of such effects has not been realized. Here, the phenomenon is as prevalent as dominance. Relative to the total of 83 known imprinted loci in mice, the discovery of 24 imprinted obesity loci in this study indicates that the true number of imprinted genes is much higher than generally thought, a finding consistent with the bioinformatic study of Luedi et al. (40). Furthermore, our results show a preponderance of dominance imprinting over parental imprinting. Fully two-thirds of the effects are dominance imprinting, where the homozygotes are identical and either one (polar dominance) or both (bipolar dominance) heterozygotes deviate from the common homozygous value. The callipyge locus in sheep (41) is an especially dramatic example of dominance imprinting that has been examined at the molecular level. The dominance imprinting effects at callipyge are caused by the coinheritance of tightly linked maternally and paternally expressed loci (24,41). Mapping studies that only parameterize parental imprinting fail to discover these more complex imprinting effects (e.g., 42). Clearly, imprinting plays an important role in the genotype- phenotype relationship for obesity.
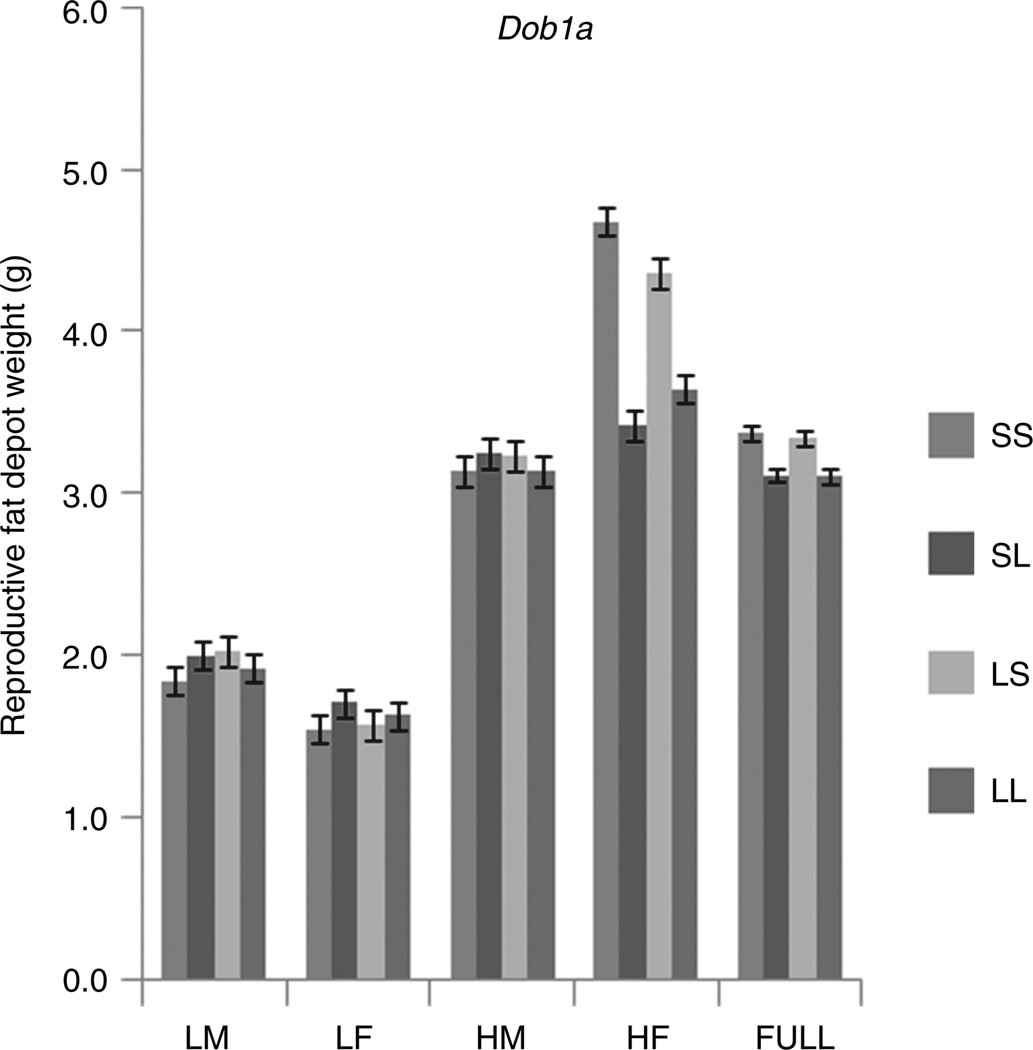
The most striking result of our study is that obesity loci show strong, nearly ubiquitous, context dependency. The effects of genes change depending on the sex they are expressed in and on the proportion of fat in the diet. Genetic variation among individuals in their response to the high-fat diet is remarkable. As with the obesity loci mapped in the Recombinant Inbred lines (22), the effect of nearly all obesity QTL mapped in the AI Line change depending on sex and diet. Over half the gene effects detected occur in only one of the four subpopulation cohorts (LF, HF, LM, HM) each making up only one-quarter of the total population. Most of these gene effects are of reduced magnitude and statistical significance in the general population when pooled over sexes and diets. For example, at Dob1a there are no effects in the LF, LM, or HM cohorts, the only significant effects being in the HF cohort (see Figure 3). If the population is analyzed by pooling these cohorts and ignoring interaction, the additive and imprinting effect sizes are reduced fourfold and are no longer very apparent or significant. Indeed, 76% of the trait-specific effects in this study would have been missed entirely if the analysis had relied on the genotypic values pooled over cohorts and ignored gene-by-sex and gene-by-diet interactions. Analyzing pooled data washes out cohort-specific effects.
Figure 3.
Genotypic values of reproductive fat depot weights in the four sex–diet-specific cohorts and in the full population, pooled across the cohorts (FULL), at QTL Dob1a. This locus has an effect only evident in the HF. There are no significant differences among the genotypes in the LM, HM, LF, or FULL population cohorts but there is a significant negative additive effect, with the S allele leading to a heavier weight, and a positive imprinting effect with maternal expression in the HF cohort. Error bars indicate ± 1.0 s.e. LF, low-fat fed females; LM, low-fat fed males; HF, high-fat fed females; HM, high-fat fed males; QTL, quantitative trait locus.
Some loci have more complex interaction patterns with multiple cohorts affected in different ways. For example, at Dob6c (see Figure 2), there are significant positive additive and negative imprinting genotypic values in the LF cohort but significant positive dominance and positive imprinting values in the HF cohort. All females are affected by this locus, however, the pattern of genotypic values is different depending on the diet. In the LF cohort the LG/J allele causes a heavier reproductive fat depot weight, there is no dominance, and there is maternal expression imprinting. In contrast, in the HF cohort we observe no additive effect, significant over-dominance, and a significant positive dominance imprinting effect. At QTL Dob17e (see Figure 1), both males and females are affected when fed a high-fat diet but in females the SM/J allele results in heavier total depot weights, the LG/J allele is dominant, and there is maternal imprinting () whereas in males there are no significant additive or dominance effects and the bipolar imprinting effect is the opposite of that found in females (). The genetics of obesity presents a rich array of possibilities and patterns, the clearest of which is that gene effects do not belong to the alleles themselves but are conditional on sex and dietary environment. It is not enough to consider these factors as covariates. Interactions have to be explicitly included in analytical models to determine their effects. Gene-by-environment interaction is not merely a nuisance factor but lies at the heart of the obesity epidemic determining who becomes obese and who remains relatively lean in an obesogenic environment.
Imprinting interactions are of special interest as they have rarely been mapped in genome scans. Genomic imprinting patterns are dynamic, changing with age (23,24), with sex (26), and, as described here, with dietary environment. The usual manner of thinking about imprinting effects is that they are set at or soon after fertilization by epigenetic marking that affects gene expression. The diversity of imprinting effect interactions detected in our study reveal imprinting patterns that are labile, changing with environmental exposures and perhaps being expressed differently in different tissues. Similarly, Jirtle et al. (43,44) have repeatedly found that levels of imprinting and its associated genomic signatures are affected by both maternal and postnatal environments. Waterland et al. (45) found that genomic imprinting at the Igf2 locus and the methylation status of the Igf2 differentially methylated region 2 (DMR2) in mice could be altered by a methyl-donor-deficient diet administered after weaning. Their results indicate that postweaning nutritional factors can have a permanent effect on the epigenetic regulation of imprinted genes. We have discovered similar widespread alterations of imprinting expression due to a high-fat postnatal diet. Whether this corresponds to alterations in epigenetic marks as well remains to be determined.
If genetic and genomic imprinting interactions like those detected in our current study also occur in human populations, it follows that it would be difficult to detect obesity QTLs in human populations. There is certainly a broad diversity of environmental and dietary exposure in human populations that often goes unmeasured. If most gene effects are conditional on dietary environment, as we describe here, their effects would be washed out in a human population survey because only those individuals experiencing the appropriate diet would show a genetic effect. These individuals would be analyzed along with the majority of the population who, experiencing a different dietary environment, do not display that gene effect. This may be one reason that human genome-wide association studies fail to find very many obesity genes and why they only explain a small part of the population genetic variance for obesity.
In conclusion, we have fine-mapped a large number of dietary obesity QTLs affecting fat depot weights. Some of these locations correspond to regions identified in our earlier mapping work and that of other researchers; however, many are novel locations. Many evocative positional candidate genes were identified for further research. We find that the effects of genes on obesity are nearly always dependent on the context, specifically, the sex of the individual and the amount of dietary fat ingested. We discovered that genomic imprinting effects on adult obesity are widespread, occurring at a majority of the loci and that they are labile in response to the hormonal environment engendered by sex and in response to the amount of dietary fat consumed. Further studies will refine the QTL intervals reported here and evaluate gene sequence and expression variation in positional candidate genes leading to experiments that will validate the identity of the quantitative trait genes responsible for these mapped QTLs.
Supplementary Material
Acknowledgments
This work was supported by funding from the National Institute of Diabetes and Digestive and Kidney Diseases (J.M.C., C.F.S., R01 DK055736; Washington University Nutrition Obesity Research Center (NORC), DK056341; Washington University Diabetes Research and Training Center (DRTC) DK020579), the National Institute of General Medical Sciences (J.M.C., GM065509), the National Heart, Lung, and Blood Institute (H.A.L., T32 HL091823), and the Biotechnology and Biological Sciences Research Council (J.B.W., BBSRC-BB/C/516936). We also thank Seth Crosby and the Washington University Genome Sequencing Center for their help in SNP genotyping.
Footnotes
Supplementary Material
Supplementary material is linked to the online version of the paper at http://www.nature.com/oby
Disclosure
The authors declared no conflict of interest.
REFERENCES
- 1.Heymsfield SB. How large is the energy gap that accounts for the obesity epidemic? Am J Clin Nutr. 2009;89:1717–1718. doi: 10.3945/ajcn.2009.27889. [DOI] [PubMed] [Google Scholar]
- 2.Swinburn BA, Sacks G, Lo SK, et al. Estimating the changes in energy flux that characterize the rise in obesity prevalence. Am J Clin Nutr. 2009;89:1723–1728. doi: 10.3945/ajcn.2008.27061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Musani SK, Erickson S, Allison DB. Obesity–still highly heritable after all these years. Am J Clin Nutr. 2008;87:275–276. doi: 10.1093/ajcn/87.2.275. [DOI] [PubMed] [Google Scholar]
- 4.Hindorff LA, Sethupathy P, Junkins HA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogardus C. Missing heritability and GWAS utility. Obesity (Silver Spring) 2009;17:209–210. doi: 10.1038/oby.2008.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Junyent M, Parnell LD, Lai CQ, et al. Novel variants at KCTD10, MVK, and MMAB genes interact with dietary carbohydrates to modulate HDL-cholesterol concentrations in the Genetics of Lipid Lowering Drugs and Diet Network Study. Am J Clin Nutr. 2009;90:686–694. doi: 10.3945/ajcn.2009.27738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warodomwichit D, Shen J, Arnett DK, et al. ADIPOQ polymorphisms, monounsaturated fatty acids, and obesity risk: the GOLDN study. Obesity (Silver Spring) 2009;17:510–517. doi: 10.1038/oby.2008.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allis CD, Jenuwein T, Reinberg D. Epigenetics. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2007. [Google Scholar]
- 9.Frontera M, Dickins B, Plagge A, Kelsey G. Imprinted genes, postnatal adaptations and enduring effects on energy homeostasis. Adv Exp Med Biol. 2008;626:41–61. doi: 10.1007/978-0-387-77576-0_4. [DOI] [PubMed] [Google Scholar]
- 10.Dong C, Li WD, Geller F, et al. Possible genomic imprinting of three human obesity-related genetic loci. Am J Hum Genet. 2005;76:427–437. doi: 10.1086/428438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorlova OY, Amos CI, Wang NW, et al. Genetic linkage and imprinting effects on body mass index in children and young adults. Eur J Hum Genet. 2003;11:425–432. doi: 10.1038/sj.ejhg.5200979. [DOI] [PubMed] [Google Scholar]
- 12.Lindsay RS, Kobes S, Knowler WC, Bennett PH, Hanson RL. Genome-wide linkage analysis assessing parent-of-origin effects in the inheritance of type 2 diabetes and BMI in Pima Indians. Diabetes. 2001;50:2850–2857. doi: 10.2337/diabetes.50.12.2850. [DOI] [PubMed] [Google Scholar]
- 13.Rance KA, Fustin JM, Dalgleish G, et al. A paternally imprinted QTL for mature body mass on mouse chromosome 8. Mamm Genome. 2005;16:567–577. doi: 10.1007/s00335-005-0012-4. [DOI] [PubMed] [Google Scholar]
- 14.Mantey C, Brockmann GA, Kalm E, Reinsch N. Mapping and exclusion mapping of genomic imprinting effects in mouse F2 families. J Hered. 2005;96:329–338. doi: 10.1093/jhered/esi044. [DOI] [PubMed] [Google Scholar]
- 15.Cheverud JM, Hager R, Roseman C, et al. Genomic imprinting effects on adult body composition in mice. Proc Natl Acad Sci USA. 2008;105:4253–4258. doi: 10.1073/pnas.0706562105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheverud JM, Routman EJ, Duarte FA, et al. Quantitative trait loci for murine growth. Genetics. 1996;142:1305–1319. doi: 10.1093/genetics/142.4.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kramer MG, Vaughn TT, Pletscher LS, et al. Genetic variation in body weight growth and composition in the intercross of Large (LG/J) and Small (SM/J) inbred strains of mice. Genet Mol Biol. 1998;21:211–218. [Google Scholar]
- 18.Cheverud JM, Pletscher LS, Vaughn TT, Marshall B. Differential response to dietary fat in large (LG/J) and small (SM/J) inbred mouse strains. Physiol Genomics. 1999;1:33–39. doi: 10.1152/physiolgenomics.1999.1.1.33. [DOI] [PubMed] [Google Scholar]
- 19.Ehrich TH, Kenney JP, Vaughn TT, Pletscher LS, Cheverud JM. Diet, obesity, and hyperglycemia in LG/J and SM/J mice. Obes Res. 2003;11:1400–1410. doi: 10.1038/oby.2003.189. [DOI] [PubMed] [Google Scholar]
- 20.Cheverud JM, Ehrich TH, Kenney JP, Pletscher LS, Semenkovich CF. Genetic evidence for discordance between obesity- and diabetesrelated traits in the LGXSM recombinant inbred mouse strains. Diabetes. 2004;53:2700–2708. doi: 10.2337/diabetes.53.10.2700. [DOI] [PubMed] [Google Scholar]
- 21.Ehrich TH, Kenney-Hunt JP, Pletscher LS, Cheverud JM. Genetic variation and correlation of dietary response in an advanced intercross mouse line produced from two divergent growth lines. Genet Res. 2005;85:211–222. doi: 10.1017/S0016672305007603. [DOI] [PubMed] [Google Scholar]
- 22.Cheverud JM, Ehrich TH, Hrbek T, et al. Quantitative trait loci for obesity- and diabetes-related traits and their dietary responses to high-fat feeding in LGXSM recombinant inbred mouse strains. Diabetes. 2004;53:3328–3336. doi: 10.2337/diabetes.53.12.3328. [DOI] [PubMed] [Google Scholar]
- 23.Hager R, Cheverud JM, Wolf JB. Relative contribution of additive, dominance, and imprinting effects to phenotypic variation in body size and growth between divergent selection lines of mice. Evolution. 2009;63:1118–1128. doi: 10.1111/j.1558-5646.2009.00638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolf JB, Cheverud JM, Roseman C, Hager R. Genome-wide analysis reveals a complex pattern of genomic imprinting in mice. PLoS Genet. 2008;4:e1000091. doi: 10.1371/journal.pgen.1000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolf JB, Cheverud JM. A framework for detecting and characterizing genetic background-dependent imprinting effects. Mamm Genome. 2009;20:681–698. doi: 10.1007/s00335-009-9209-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hager R, Cheverud JM, Leamy LJ, Wolf JB. Sex dependent imprinting effects on complex traits in mice. BMC Evol Biol. 2008;8:303. doi: 10.1186/1471-2148-8-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fawcett GL, Roseman CC, Jarvis JP, et al. Genetic architecture of adiposity and organ weight using combined generation QTL analysis. Obesity (Silver Spring) 2008;16:1861–1868. doi: 10.1038/oby.2008.300. [DOI] [PubMed] [Google Scholar]
- 28.Fawcett GL, Jarvis JP, Roseman CC, et al. Fine-mapping of obesity-related quantitative trait loci in an F(9/10) advanced intercross line. Obesity (Silver Spring) 2009 doi: 10.1038/oby.2009.411. e-pub ahead of print 12 November 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Darvasi A, Soller M. Advanced intercross lines, an experimental population for fine genetic mapping. Genetics. 1995;141:1199–1207. doi: 10.1093/genetics/141.3.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Falconer DS, Mackay T. Introduction to Quantitative Genetics. New York: Longman Press; 1997. [Google Scholar]
- 31.Cheverud JM, Vaughn TT, Pletscher LS, et al. Genetic architecture of adiposity in the cross of LG/J and SM/J inbred mice. Mamm Genome. 2001;12:3–12. doi: 10.1007/s003350010218. [DOI] [PubMed] [Google Scholar]
- 32.Browman KW, Sen Ś. A Guide to QTL Mapping with R/qtl. New York: Springer; 2009. [Google Scholar]
- 33.Haley CS, Knott SA. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity. 1992;69:315–324. doi: 10.1038/hdy.1992.131. [DOI] [PubMed] [Google Scholar]
- 34.Li J, Jiang T. Efficient inference of haplotypes from genotypes on a pedigree. J Bioinform Comput Biol. 2003;1:41–69. doi: 10.1142/s0219720003000204. [DOI] [PubMed] [Google Scholar]
- 35.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95:221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- 36.Chen L, Storey JD. Relaxed significance criteria for linkage analysis. Genetics. 2006;173:2371–2381. doi: 10.1534/genetics.105.052506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheverud JM, Ehrich TH, Vaughn TT, et al. Pleiotropic effects on mandibular morphology II: differential epistasis and genetic variation in morphological integration. J Exp Zool B Mol Dev Evol. 2004;302:424–435. doi: 10.1002/jez.b.21008. [DOI] [PubMed] [Google Scholar]
- 38.Pavlicev M, Kenney-Hunt JP, Norgard EA, et al. Genetic variation in pleiotropy: differential epistasis as a source of variation in the allometric relationship between long bone lengths and body weight. Evolution. 2008;62:199–213. doi: 10.1111/j.1558-5646.2007.00255.x. [DOI] [PubMed] [Google Scholar]
- 39.Cheverud JM. A comparison of genetic and phenotypic correlations. Evolution. 1988;42:958–968. doi: 10.1111/j.1558-5646.1988.tb02514.x. [DOI] [PubMed] [Google Scholar]
- 40.Luedi PP, Hartemink AJ, Jirtle RL. Genome-wide prediction of imprinted murine genes. Genome Res. 2005;15:875–884. doi: 10.1101/gr.3303505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Georges M, Charlier C, Cockett N. The callipyge locus: evidence for the trans interaction of reciprocally imprinted genes. Trends Genet. 2003;19:248–252. doi: 10.1016/S0168-9525(03)00082-9. [DOI] [PubMed] [Google Scholar]
- 42.Rowe SJ, Pong-Wong R, Haley CS, Knott SA, De Koning DJ. Detecting parent of origin and dominant QTL in a two-generation commercial poultry pedigree using variance component methodology. Genet Sel Evol. 2009;41:6. doi: 10.1186/1297-9686-41-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dolinoy DC, Das R, Weidman JR, Jirtle RL. Metastable epialleles, imprinting, and the fetal origins of adult diseases. Pediatr Res. 2007;61:30R–37R. doi: 10.1203/pdr.0b013e31804575f7. [DOI] [PubMed] [Google Scholar]
- 44.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waterland RA, Lin JR, Smith CA, Jirtle RL. Post-weaning diet affects genomic imprinting at the insulin-like growth factor 2 (Igf2) locus. Hum Mol Genet. 2006;15:705–716. doi: 10.1093/hmg/ddi484. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.