Abstract
Lung cancer in never-smokers ranks as the seventh most common cause of cancer death worldwide, and the incidence of lung cancer in non-smoking Korean women appears to be steadily increasing. To identify the effect of genetic polymorphisms on lung cancer risk in non-smoking Korean women, we conducted a genome-wide association study of Korean female non-smokers with lung cancer. We analyzed 440,794 genotype data of 285 cases and 1,455 controls, and nineteen SNPs were associated with lung cancer development (P < 0.001). For external validation, nineteen SNPs were replicated in another sample set composed of 293 cases and 495 controls, and only rs10187911 on 2p16.3 was significantly associated with lung cancer development (dominant model, OR of TG or GG, 1.58, P = 0.025). We confirmed this SNP again in another replication set composed of 546 cases and 744 controls (recessive model, OR of GG, 1.32, P = 0.027). OR and P value in combined set were 1.37 and < 0.001 in additive model, 1.51 and < 0.001 in dominant model, and 1.54 and < 0.001 in recessive model. The effect of this SNP was found to be consistent only in adenocarcinoma patients (1.36 and < 0.001 in additive model, 1.49 and < 0.001 in dominant model, and 1.54 and < 0.001 in recessive model). Furthermore, after imputation with HapMap data, we found regional significance near rs10187911, and five SNPs showed P value less than that of rs10187911 (rs12478012, rs4377361, rs13005521, rs12475464, and rs7564130). Therefore, we concluded that a region on chromosome 2 is significantly associated with lung cancer risk in Korean non-smoking women.
Keywords: Lung Neoplasms, Genome-Wide Association Study, Non-Smoking Women
INTRODUCTION
Lung cancer is the leading cause of cancer death worldwide with over 1 million deaths each year. Although cigarette smoking is the major cause of this malignancy, global statistics estimate that 15% of lung cancers in men and 53% in women are not directly attributable to smoking (1). The incidence of lung cancer in Korean women appears to be steadily increasing, as in Asian women generally, and the majority of these women are non-smokers. Moreover, lung cancer in Korean women appears to be predominantly adenocarcinoma. Gender, clinicopathological, and molecular differences in lung cancers arising in never-smokers and smokers indicate that lung cancer in non-smoking women might be a different disease (2, 3). Recent evidence for genetic influence on smoking behavior and nicotine dependence has prompted a search for susceptibility genes related with lung cancer development (4). However, no regions were replicated except for 5p15.33 as susceptibility loci for never-smoking lung cancer patients although three human genome regions at chromosome 5p15, 15q25, and 6p21 were found to be associated with susceptibility to lung cancer through several genome-wide association (GWA) studies (4-14). Furthermore, there was no information for Korean non-smoking women. For these reasons, we conducted a genome-wide association study (GWAS) of female non-smoking Koreans with lung cancer to identify low-penetrance alleles influencing lung cancer risk in Korean non-smoking women.
MATERIALS AND METHODS
Study populations
A total of 286 non-smoking women with newly diagnosed lung cancer at Kyungpook National University Hospital (30 cases), Korea University Anam Hospital (39 cases), Seoul National University Hospital (140 cases), and Seoul National University Bundang Hospital (77 cases) in Korea between 2001 and 2008 were recruited for GWAS. There were no age, histologic, or stage restrictions, and all lung cancer cases were histologically confirmed. The control subjects for GWAS were 1,462 non-cancer participants admitted to the Korean Association REsource (KARE) of Korea Centers for Disease Control and Prevention (KCDC). They were matched to the cases by sex and smoking status (case: control≒1:5). For the replication studies, we recruited new population, 293 cases and 495 controls for replication 1 (70 cases and 100 controls in Kyungpook National University Hospital; 64 cases in Korea University Anam Hospital; 115 cases in Seoul National University Hospital; and 44 cases and 395 controls in Seoul National University Bundang Hospital) and 546 cases and 744 controls for replication 2 (84 cases and 43 controls in Kyungpook National University Hospital; 44 cases in Korea University Anam Hospital; 90 cases in Seoul National University Hospital; 57 cases and 232 controls in Seoul National University Bundang Hospital; and 271 cases and 469 controls in Korea National Cancer Center). All participants were never-smoking women and information on age, sex, smoking history, and disease status for them was collected by trained interviewers working in each center.
GWA genotyping
DNA was extracted from whole blood sample using a QIAamp® DNA Blood Midi Kit (Qiagen, Valencia, CA, USA). SNP genotyping was performed using Affymetrix Genome-Wide Human SNP array 5.0 comprising 440,794 genome-wide SNPs (Affymetrix, Santa Clara, CA, USA) according to the manufacturer's protocol. Total genomic DNA (500 ng) were digested with Nsp I and Sty I restriction enzymes and ligated to specific adaptors which incorporate a universal PCR priming sequence. PCR amplification was performed with universal primers in a reaction optimized to amplify fragments between 200-1,100 base pairs. A fragmentation step then reduced the PCR product to segments of approximately 25-50 bp, which were then end-labeled using biotinylated nucleotides. The labeled product was hybridized to a chip, washed and detected. The images were analyzed using GCOS software (Affymetrix, Santa Clara, CA, USA). For quality control (QC), two algorithms were used: the QC call rates for each array exceeded 86% using the Dynamic Model algorithm, and genotype calls for each site were more than 98% using the birdseed v2 genotyping call algorithm.
GWA data mining
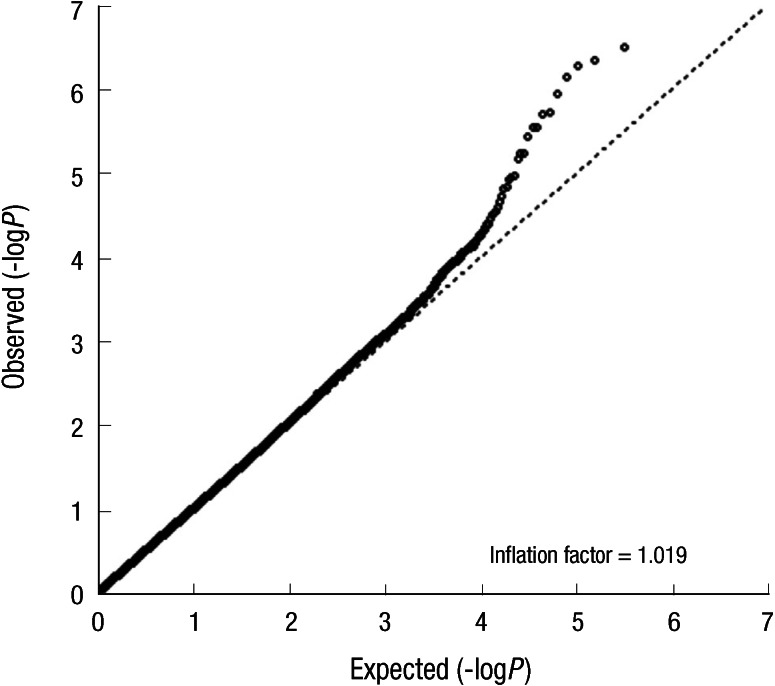
We genotyped 440,794 SNPs in 286 cases using Affymetrix GeneChip 5.0 and obtained chip data of 1,462 controls from KARE of KCDC. First, we checked relationships among subjects using graphical representation of relationship errors (GRR) software and excluded 8 subjects (7 controls from KARE and 1 case from Seoul National University Bundang Hospital) with mean of identical-by-state (IBS) allele sharing over a number of polymorphic loci for each pair of individuals more than 1.5 and finally analyzed 285 cases and 1,455 controls using PLINK v1.07 program. Cutoff P values of Hardy-Weinberg Equilibrium (HWE), call rate, and minor allele frequency were 0.001, 0.95, and 0.01, respectively, and 331,088 SNPs were used in the analysis. Because of our small sample size, we used a 10-fold cross-validation method (90% randomly repeated selection of total population for a validation, 10 trials) to protect against false positives in logistic regression analysis. In all analyses, we adjusted for age in the model. Finally we selected 19 SNPs under P value 0.0001 for replication in another sample sets after considering each genotype scatter plot. All nineteen SNPs were consistent in all ten times cross-validation under P value 0.001 and confirmed to be called accurately. A quantile-quantile plot using P values obtained from additive model in logistic regression analyses revealed no difference of the observed P values and those expected by chance (Fig. 1). A small inflation factor (λ) of 1.019 (or 1.000, based on the bottom 90% of quantile-quantile plot) indicated that the association we found is unlikely to have resulted from population stratification. Population structure was evaluated by PLINK v1.07, and the quantile-quantile plot was generated using R 2.13.0. We used the MACH 1.0.16 software to impute untyped SNPs using the linkage disequilibrium information from the HapMap phase II database (CHB+JPT was used as reference set, released 17 July 2006).
Fig. 1.
Quantile-quantile plot of P value.
Replication genotyping
From the GWA analysis, we selected 19 SNPs (rs10187911, rs13005521, rs1484322, rs755927, rs2191876, rs2378688, rs303451, rs41393946, rs7928853, rs7305016, rs8041151, rs17258206, rs17258247, rs9939608, rs7241996, rs10485620, rs2884026, rs925338, and rs8068946) associated with lung cancer development under cutoff P value 0.0001 and genotyped those SNPs in samples obtained from new populations (replication set 1 and replication set 2).
Replication 1. The 19 SNPs were evaluated in replication study. Of the 19 SNPs, 14 variants (rs10187911, rs13005521, rs2884026, rs1484322, rs755927, rs2191876, rs2378688, rs303451, rs41393946, rs7928853, rs7305016, rs8041151, rs17258206, rs17258247, rs925338, rs9939608, rs8068946, rs7241996, and rs10485620) were selected by MassARRAY AssayDesign software package (v3.1) and genotyped using SEQUENOM's MassArray iPLEX technology (SEQUENOM I') with negative controls following the manufacturer's instructions. Genotype calls were made using the default post-processing calling parameters in SEQUENOM's Typer 4.0 software. The genotypes of five SNPs (rs13005521, rs2191876, rs2884026, rs8068946, and rs925338) were performed using a polymerase chain reaction (PCR) and restriction fragment length polymorphism (RFLP) (Table 1) with negative controls. For quality control, the genotyping analysis was performed blind with regards to the subjects. In addition, approximately 5% of the samples were randomly selected to be genotyped again by SEQUENOM's MassArray iPLEX technology and PCR-RFLP, and the results showed 100% concordance.
Table 1.
Primers and PCR conditions for amplification and RFLP
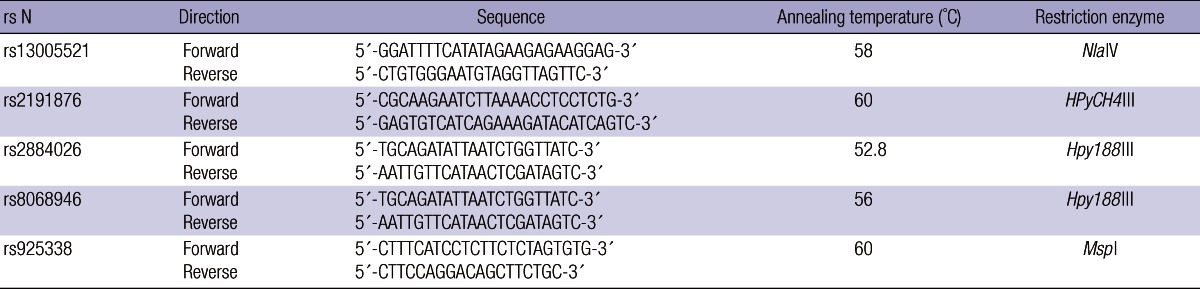
PCR, polymerase chain reaction; RFLP, restriction fragment length polymorphism.
Replication 2. Because only rs10187911 of replicated 19 SNPs showed statistical significance in replication set 1, only rs10187911 was genotyped using the TaqMan fluorogenic 5' nuclease assay (AB I') in replication set 2. The final volume of polymerase chain reaction (PCR) was 5 µL, containing 10 ng of genomic DNA and 2.5 µL TaqMan Universal PCR Master Mix, with 0.13 µL of 40X Assay Mix (Assay ID, C_30098973_10). Thermal cycle conditions were as follows: 50℃ for 2 min to activate the uracil N-glycosylase and to prevent carry-over contamination, 95℃ for 10 min to activate the DNA polymerase, followed by 45 cycles of 95℃ for 15 sec and 60℃ for 1 min. All PCR were performed using 384-well plates by a Dual 384-Well GeneAmp PCR System 9700 (AB I') and the endpoint fluorescent readings were performed on an ABI PRISM 7900 HT Sequence Detection System (AB I'). Duplicate samples and negative controls were included to ensure accuracy of genotyping.
Statistical analyses for replication study
Unconditional logistic regression was used to estimate odds ratios (OR) and 95% confidence interval (CI) after adjustment for age in lung cancer patients and control subjects. To find out whether each SNP site are on the HWE, the distributions of observed genotype frequency and expected genotype frequency calculated from observed allele frequency were compared using χ2 test. SAS, version 9.1 (SAS institute Inc., Cary, NC, USA), was used for statistical analysis.
Ethics statement
The study protocol was approved by the institutional review board at Seoul National University Hospital (IRB No. C-0903-037-274), and written informed consent was provided by all study participants.
RESULTS
We analyzed 285 cases and 1,455 controls using Affymetrix GeneChip 5.0. To protect against false positive, we used 0.001, 0.95, and 0.01 as cutoff P values of HWE, call rate, and minor allele frequency, respectively, as well as 10-fold cross-validation method, and then considered each genotype scatter plot. Finally we selected 19 SNPs under P value 0.0001 for replication in another sample sets. Table 2 shows summary of 19 SNPs significantly associated with lung cancer development in GWAS. For validation, the nineteen SNPs were replicated in another sample set, replication 1, which was composed of 293 cases and 495 controls, and only one SNP (rs10187911 on p16.3 of chromosome 2) was significantly associated with lung cancer development in dominant model (OR of TG or GG, 1.58, P = 0.025) (Table 3). We reconfirmed this SNP in another sample set, replication 2, composed of 546 cases and 744 controls (total 1,290) (recessive model, OR of GG, 1.32, P = 0.027) (Table 4). OR and P value in combined set (total set) were 1.37 and < 0.001 for additive model, 1.51 and < 0.001 for dominant model, and 1.54 and < 0.001 for recessive model (Table 4). We also assessed the effect of the SNP by center (Fig. 2). All five centers showed consistent results both in total population and in adenocarcinoma cases (additive model, ORtotal, 1.37 vs ORadenocarcinoma, 1.36).
Table 2.
Summary of nineteen SNPs significantly associated with lung cancer development in GWAS
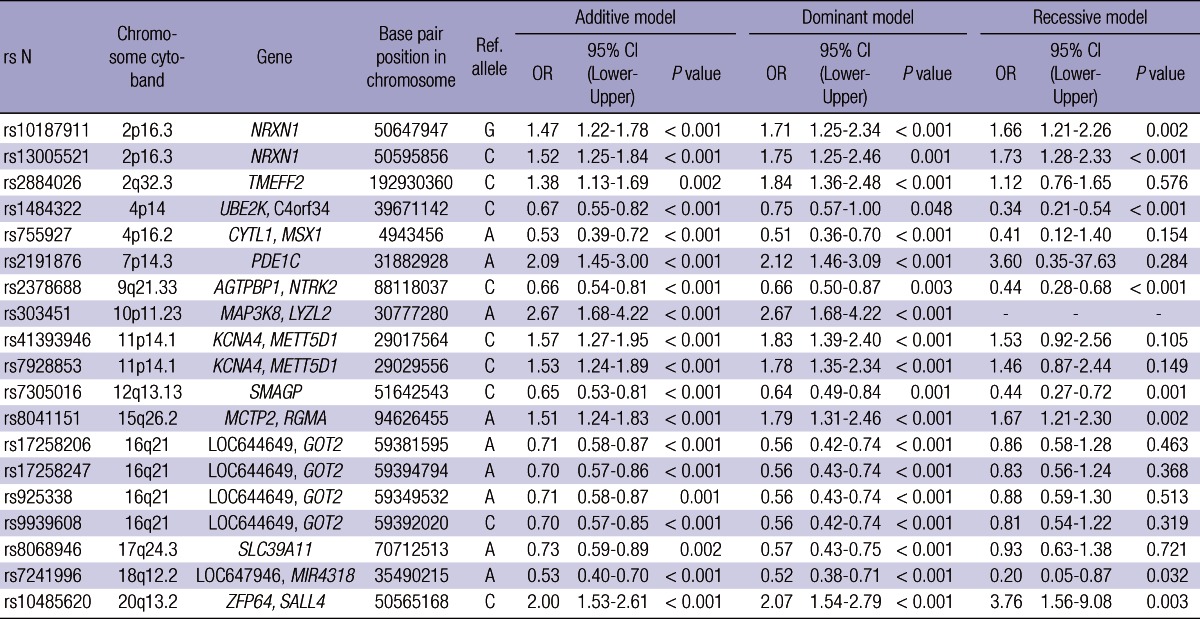
OR, odds ratio; CI, confidence interval; GWAS, genome-wide association study.
Table 3.
Summary of nineteen SNPs in another sample set, replication 1
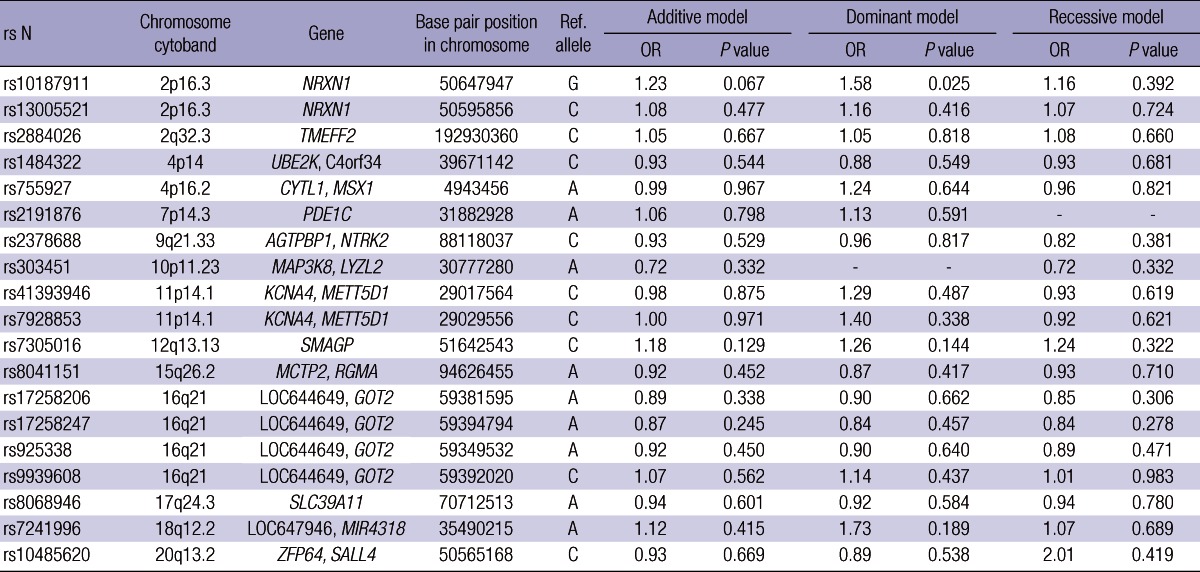
OR, odds ratio.
Table 4.
Summary of SNP (rs10187911) replicated in other validation sets, replication 1 and replication 2
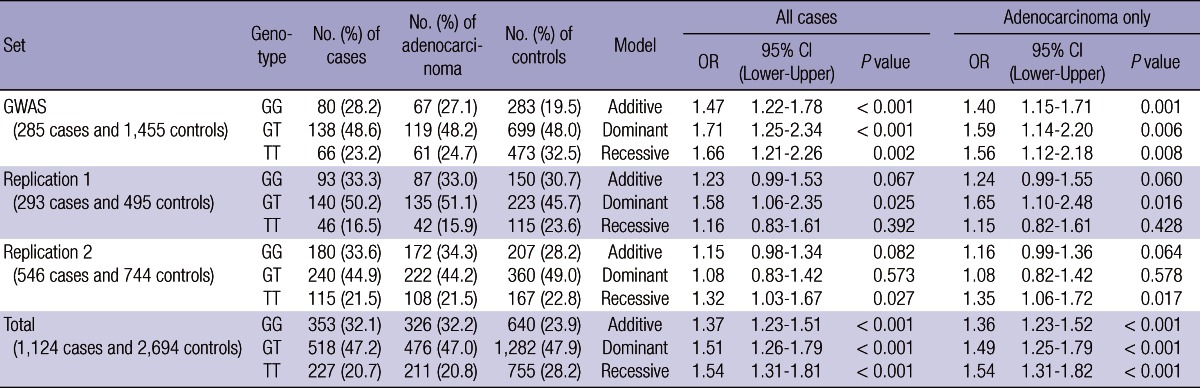
OR, odds ratio; CI, confidence interval; GWAS, genome-wide association study.
Fig. 2.
Summary of odds ratio (OR) and confidence interval (CI) by center. The number of cases, OR, and 95% CI were depicted in order in brackets.
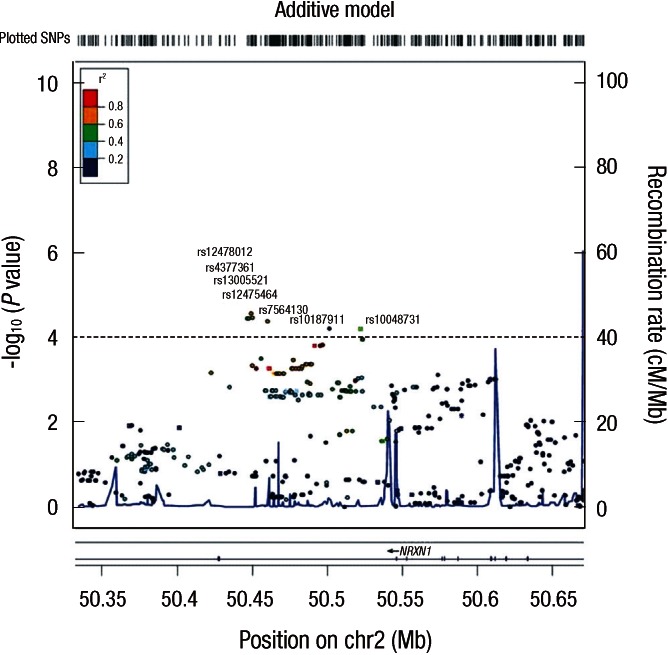
Because we confirmed the effect of rs10187911 on lung cancer development in all sets of GWAS, replication 1, replication 2, and total population, we tried to show regional plot near rs10187911 with genotypes estimated using HapMap data to increase the spectrum of variants tested for associations in this region (Fig. 3). After imputation, we found regional significance near rs10187911, and all rs10187911 and six SNPs existing in the same intron of NRXN1 gene with rs10187911 showed P value < 0.001. Among these six SNPs existing in the same intron of NRXN1 gene with rs10187911, five SNPs (rs12478012, rs4377361, rs13005521, rs12475464, and rs7564130) showed P value less than that of rs10187911 and one SNP (rs10048731) showed P value more than that of rs10187911.
Fig. 3.
Regional plot surrounding rs10187911.
DISCUSSION
We found a new SNP (rs10187911) at 2p16.3 as a susceptibility marker to lung cancer in never-smoking Korean females. Since most cases in our population were adenocarcinoma patients (92.4%), this SNP was significantly associated only with adenocarcinoma.
This SNP (rs1884396) is located on intron of NRXN1 gene, one of the neurexins that function as cell adhesion molecules during synaptogenesis and take part in intracellular signaling (15). NRXN1 is extraordinarily large, occupying 1,112,039 bp of DNA - nearly 0.04% of the entire human genome. Neurexin transcripts undergo alternative splicing at several sites (15). Each site of alternative splicing is defined as a position in the mRNA where sequence variations have been observed. Therefore, the SNP (rs1884396) on intron of NRXN1 gene may be responsible for regulation of splicing events, leading to protein diversity. In fact, NRXN1 is related with hemangiomas as well as autism spectrum disorders, mental retardation, language development delays, hypotonia, and the VACTERL association (16-19). Moreover, Nussbaum et al. suggested involvement of NRXN1 in nicotine dependence (20), associated with the 15q25 region in a previous lung cancer GWA study. Mutation of epidermal growth factor receptor (EGFR) is known to be important in chemotherapy of Asian lung cancer patients. Interestingly, NRXN1 encodes alpha type of isoform using upstream promoter and this isoform has an epidermal growth factor (EGF)-like sequence (16) which can bind to EGFR. Furthermore, lung injury by several chemicals may in part be inferred via curated interactions between NRXN1 and chemicals such as tetrachlorodibenzodioxin (TCDD), ozone (O3), and chlorine (Cl) (Comparative Toxicogenomics Database, http://ctd.mdibl.org/detail.go?type=gene&acc=9378; http://ctd.mdibl.org/detail.go?type=gene&acc=9378&view=ixn).
In this study, we imputed other SNPs not explored in our GWAS on lung cancer development using HapMap data and found significant associations with other SNPs surrounding rs10187911 (existing in the same intron) in NRXN1, indicating that the rs10187911 we found is a true indicator related with lung cancer development. However, we need to confirm which one among these significant SNPs is a real marker of lung cancer development in the future.
In this study, we did not confirm the previously reported SNPs from GWA studies in Asian population and found inconsistent results among several reports (21-25). A study of lung cancer in never-smoking Asian women (21) demonstrated that common genetic variants (rs2736100) in the TERT-CLPTM1L locus on chromosome 5p15.33 are associated with risk for lung adenocarcinoma. Li et al. (22) demonstrated novel genetic variants at 13q31.3 as susceptibility marker to never-smoker lung cancer in mostly American subjects. However, this study did not replicate significant association of 5p15.33 with lung cancers in never-smokers. Ahn et al. (23) suggested the 18p11.22 region as novel lung cancer susceptibility locus in never-smokers in Korean population. This study observed a P value of 0.008 for rs2736100 with no genome-wide significance level. Unfortunately, we could not evaluate the significance of rs2736100 because the locus was not included in 497,345 markers of our GWA study. We tried to estimate the effect of SNPs (rs2352028 at 13q31.3 and rs11080466 and rs11663246 at 18p11.22) reported from the Li et al.'s and Ahn et al.'s studies. However, rs2352028 and rs11663246 did not reach significance in our data (P = 0.838 for rs2352028 and P = 0.421 for rs11663246), and rs11080466 was not included in our markers. Another study of lung cancer in Han Chinese (24) suggested three new lung cancer susceptibility SNPs, rs753955 at 13q12.12 and rs17728461 and rs36600 at 22q12.2. However, rs753955 and rs17728461 did not reach significance in our data (P = 0.382 for rs753955 and P = 0.537 for rs17728461), and rs36600 was not included in our markers. A recent study of lung cancer in never-smoking women in Asia (25) indicated that rs7086803 of the VTI1A gene on chromosome 10q25.2 is associated with lung carcinogenesis. The inconsistent observations in several GWA studies might be attributed to different environmental exposures, ethnic differences (21, 22, 24), mixed histology (23, 24), sex (22-24), and smoking history (23, 24), or different SNP markers (21, 22, 25).
The incidence of lung cancer in non-smoking Asian women appears to be increasing and is predominantly adenocarcinoma. Based on clinico-pathological and molecular characteristics of lung cancer arising in never-smoking females, adenocarcinoma has been considered a different disease from other lung cancer types. In the present study, all of our subjects were Korean females and most of the subjects had adenocarcinoma. Therefore, rs10187911 located on intron of NRXN1 gene at 2p16.3 found in this study may be an important marker for adenocarcinoma development in never-smoking females.
There were some limitations to this study. First, the sample size in the present study was small. However, to protect against false positives resulting from small sample size, we used a 10-fold cross-validation method and confirmed that rs10187911 was replicated in all 10 trials with randomly repeated selection of total population. Furthermore, we replicated the same analyses in other sample sets, replication 1 and 2, and obtained consistent results for rs10187911. Second, we did not adjust for center and histology regardless of differences in histology by center. However, when we stratified the total population in the present study by center only for adenocarcinoma, we did not find any differences by center in the association between rs10187911 and lung cancer risk. Moreover, we found a small inflation factor indicating that the association we found is unlikely to result from difference by center.
Although the association of NRXN1 and lung cancer has not been explored in epidemiologic study, previously reported data (15-20) support a role for this gene in the tumorigenesis of lung cancer. Nonetheless, the exact import of NRXN1 in lung cancer development remains largely unknown, and further biological and epidemiologic studies are needed.
In conclusion, we have identified a novel genetic locus at 2p16.3 region associated with susceptibility of adenocarcinoma in Korean never-smoking females. Further replication studies in larger populations are necessary to clarify our hypothesis.
Footnotes
This study was supported by the National Research Foundation (NRF) of the Korea government (grant No. 2011-0016106), Sang Koo Hahn Research Fund of Seoul National University (grant No. 800-2010-0226), and the Korea Healthcare technology R&D Project (A092258) and the Korea Health 21 R&D Project (A010250) of Korea Ministry of Health and Welfare.
The authors have no conflicts of interest to disclose.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers: a different disease. Nat Rev Cancer. 2007;7:778–790. doi: 10.1038/nrc2190. [DOI] [PubMed] [Google Scholar]
- 3.Torok S, Hegedus B, Laszlo V, Hoda MA, Ghanim B, Berger W, Klepetko W, Dome B, Ostoros G. Lung cancer in never smokers. Future Oncol. 2011;7:1195–1211. doi: 10.2217/fon.11.100. [DOI] [PubMed] [Google Scholar]
- 4.Spitz MR, Amos CI, Dong Q, Lin J, Wu X. The CHRNA5-A3 region on chromosome 15q24-25.1 is a risk factor both for nicotine dependence and for lung cancer. J Natl Cancer Inst. 2008;100:1552–1556. doi: 10.1093/jnci/djn363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, Dong Q, Zhang Q, Gu X, Vijayakrishnan J, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 7.McKay JD, Hung RJ, Gaborieau V, Boffetta P, Chabrier A, Byrnes G, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J, et al. Lung cancer susceptibility locus at 5p15.33. Nat Genet. 2008;40:1404–1406. doi: 10.1038/ng.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rafnar T, Sulem P, Stacey SN, Geller F, Gudmundsson J, Sigurdsson A, Jakobsdottir M, Helgadottir H, Thorlacius S, Aben KK, et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat Genet. 2009;41:221–227. doi: 10.1038/ng.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Truong T, Hung RJ, Amos CI, Wu X, Bickeböller H, Rosenberger A, Sauter W, Illig T, Wichmann HE, Risch A, et al. Replication of lung cancer susceptibility loci at chromosomes 15q25, 5p15, and 6p21: a pooled analysis from the International Lung Cancer Consortium. J Natl Cancer Inst. 2010;102:959–971. doi: 10.1093/jnci/djq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landi MT, Chatterjee N, Yu K, Goldin LR, Goldstein AM, Rotunno M, Mirabello L, Jacobs K, Wheeler W, Yeager M, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet. 2009;85:679–691. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Broderick P, Webb E, Wu X, Vijayakrishnan J, Matakidou A, Qureshi M, Dong Q, Gu X, Chen WV, et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat Genet. 2008;40:1407–1409. doi: 10.1038/ng.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rafnar T, Sulem P, Besenbacher S, Gudbjartsson DF, Zanon C, Gudmundsson J, Stacey SN, Kostic JP, Thorgeirsson TE, Thorleifsson G, et al. Genome-wide significant association between a sequence variant at 15q15.2 and lung cancer risk. Cancer Res. 2011;71:1356–1361. doi: 10.1158/0008-5472.CAN-10-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kohno T, Kunitoh H, Shimada Y, Shiraishi K, Ishii Y, Goto K, Ohe Y, Nishiwaki Y, Kuchiba A, Yamamoto S, et al. Individuals susceptible to lung adenocarcinoma defined by combined HLA-DQA1 and TERT genotypes. Carcinogenesis. 2010;31:834–841. doi: 10.1093/carcin/bgq003. [DOI] [PubMed] [Google Scholar]
- 14.Pande M, Spitz MR, Wu X, Gorlov IP, Chen WV, Amos CI. Novel genetic variants in the chromosome 5p15.33 region associate with lung cancer risk. Carcinogenesis. 2011;32:1493–1499. doi: 10.1093/carcin/bgr136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rowen L, Young J, Birditt B, Kaur A, Madan A, Philipps DL, Qin S, Minx P, Wilson RK, Hood L, et al. Analysis of the human neurexin genes: alternative splicing and the generation of protein diversity. Genomics. 2002;79:587–597. doi: 10.1006/geno.2002.6734. [DOI] [PubMed] [Google Scholar]
- 16.Ching MS, Shen Y, Tan WH, Jeste SS, Morrow EM, Chen X, Mukaddes NM, Yoo SY, Hanson E, Hundley R, et al. Deletions of NRXN1 (neurexin-1) predispose to a wide spectrum of developmental disorders. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:937–947. doi: 10.1002/ajmg.b.31063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, Zhang H, Estes A, Brune CW, Bradfield JP, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bucan M, Abrahams BS, Wang K, Glessner JT, Herman EI, Sonnenblick LI, Alvarez Retuerto AI, Imielinski M, Hadley D, Bradfield JP, et al. Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS Genet. 2009;5:e1000536. doi: 10.1371/journal.pgen.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pedrosa E, Kaushik S, Lachman HM. ChIP-chip analysis of neurexins and other candidate genes for addiction and neuropsychiatric disorders. J Neurogenet. 2010;24:5–17. doi: 10.3109/01677060903305658. [DOI] [PubMed] [Google Scholar]
- 20.Nussbaum J, Xu Q, Payne TJ, Ma JZ, Huang W, Gelernter J, Li MD. Significant association of the neurexin-1 gene (NRXN1) with nicotine dependence in European- and African-American smokers. Hum Mol Genet. 2008;17:1569–1577. doi: 10.1093/hmg/ddn044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsiung CA, Lan Q, Hong YC, Chen CJ, Hosgood HD, Chang IS, Chatterjee N, Brennan P, Wu C, Zheng W, et al. The 5p15.33 locus is associated with risk of lung adenocarcinoma in never-smoking females in Asia. PLoS Genet. 2010;6:e1001051. doi: 10.1371/journal.pgen.1001051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, Sheu CC, Ye Y, de Andrade M, Wang L, Chang SC, Aubry MC, Aakre JA, Allen MS, Chen F, et al. Genetic variants and risk of lung cancer in never smokers: a genome-wide association study. Lancet Oncol. 2010;11:321–330. doi: 10.1016/S1470-2045(10)70042-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahn MJ, Won HH, Lee J, Lee ST, Sun JM, Park YH, Ahn JS, Kwon OJ, Kim H, Shim YM, et al. The 18p11.22 locus is associated with never smoker non-small cell lung cancer susceptibility in Korean populations. Hum Genet. 2012;131:365–372. doi: 10.1007/s00439-011-1080-z. [DOI] [PubMed] [Google Scholar]
- 24.Hu Z, Wu C, Shi Y, Guo H, Zhao X, Yin Z, Yang L, Dai J, Hu L, Tan W, et al. A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12.2 in Han Chinese. Nat Genet. 2011;43:792–796. doi: 10.1038/ng.875. [DOI] [PubMed] [Google Scholar]
- 25.Lan Q, Hsiung CA, Matsuo K, Hong YC, Seow A, Wang Z, Hosgood HD, 3rd, Chen K, Wang JC, Chatterjee N, et al. Genome-wide association analysis identifies new lung cancer susceptibility loci in never smoking women in Asia. Nat Genet. 2012;44:1330–1335. doi: 10.1038/ng.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]