Abstract
In fungi, ambient pH sensing involves the activation of the Pal/PacC signalling pathway. In the dermatophyte Trichophyton rubrum, pH-dependent secretion of keratinases, which are major virulence determinants, is affected by disruption of the pacC gene. Here, the transcription profiling of the genes coding for N- and O-linked mannosyltransferases, enzymes involved in protein glycosylation, was evaluated in T. rubrum in response to disruption of the pacC gene and growth in keratin, glucose, and glucose plus glycine. We show that transcription of these mannosyltransferase genes is affected by nutrients at acidic pH and by PacC.
Keywords: Glycosylation, Enzyme secretion, pH regulation, Transcription profiling
Highlights
▸ The PacC/Pal transduction pathway mediates the metabolic response to pH sensing. ▸ pH-dependent secretion of keratinases is modulated by the pacC gene in T. rubrum. ▸ The pacC gene modulates transcription of O- and N-mannosyltransferase genes at acidic pH. ▸ An O-mannosyltransferase gene is preferentially transcribed in keratin at pH 5.0. ▸ An N-mannosyltransferase gene is preferentially transcribed in non-buffered glucose at pH 5.0.
1. Introduction
Dermatophytes are the most common organisms infecting keratinized structures such as skin, hair, and nails, and their ability to degrade keratin is believed to be a major virulence factor [1,2]. A correlation between keratinolytic activity and pathogenesis has been proposed because dermatophytes secrete a battery of endo- and exo-proteases during infection that degrade keratinized structures into oligopeptides and free amino acids for use as nutrients [3,4]. It is likely that proteases with optimal activity at both acidic and alkaline pHs are important virulence factors in dermatophytes, and that their regulation during infection is crucial [5]. In the early stages, and in response to the acidic pH of human skin, the pathogen de-represses the synthesis of non-specific keratinases and proteases that have optimal activity at acidic pH. They act on skin proteins, producing peptides that are hydrolyzed into amino acids, which are then used by the fungus as sources of carbon, nitrogen, and sulfur. The metabolism of some amino acids promotes the alkalinization of the host microenvironment, making it suitable for the action of keratinases with optimal activity at alkaline pH. The dermatophyte Trichophyton rubrum rapidly responds to pH changes by modulating the expression of genes, allowing the use of skin proteins over a wide pH range, thereby enabling the development of infection and persistence of the dermatophyte in host tissues [6,7] Moreover, inactivation of the pacC gene, a component of the pH signalling pathway in T. rubrum, reduces the activity of secreted keratinases [3], indicating that the pacC gene is somehow involved in the regulation of keratinolytic activity, and consequently in the virulence and pathogenicity of this organism.
Protein secretion from a eukaryotic cell requires movement through the endoplasmic reticulum (ER) and the Golgi apparatus. In the course of trafficking, the secreted proteins undergo glycosylation, which is the major post-translational molecular event [8–14]. In secreted proteins, the glycosyl groups are usually attached to either an amide group (N-glycosylation) or a hydroxyl residue (O-glycosylation), which are mainly found on serine and threonine residues. During glycosylation, the oligosaccharide GlcNAc2Man9Glc3 is transferred to an Asn residue within the sequence Asn-XSer/Thr by an oligosaccharyltransferase), where X represents any amino acid except proline [15,16]. O-glycosylation occurs via several pathways. In higher eukaryotes, the main pathway utilizes sugar nucleotides and is located in the Golgi apparatus [17]. In yeasts, O-mannosylation begins in the ER lumen and, like N-glycosylation, it requires dolichol phosphate-activated sugar residues. The initial reaction is catalyzed by proteins from the evolutionarily conserved mannosyltransferase (Pmt) family [18,19]. Proteins secreted from yeast cells are usually heavily N- and/or O-glycosylated. In proteins that are glycosylated at both sites, it is not known whether N-glycosylation precedes O-mannosylation, or vice versa [20]. It is also unknown whether the O-mannosylation that takes place in the ER prevents N-glycosylation; however, there is some evidence for the opposite situation [21]. Altered glycosylation may affect the stability and half-life of proteins, thus changing their activities or affinities towards substrates [22,23].
Delineating the mechanisms underlying fungal adaptability to ambient variation is fundamental to an understanding of the mechanisms of pathogenicity and resistance to inhibitors in pathogenic organisms. This work was aimed at investigating the expression of genes encoding dolichyl-P-Man:Man(5)GlcNAc(2)-PP-dolichyl mannosyltransferase and an O-mannosyltransferase (referred to as the N-man and O-man genes, respectively) in the dermatophyte T. rubrum in response to nutrients, ambient pH, and disruption of the pacC gene. Our findings revealed a relationship between the expression of these two mannosyltransferase genes and the pacC gene in response to ambient pH and carbon source.
2. Materials and methods
2.1. Strains and growth conditions
T. rubrum clinical isolate H6 (ATCC MYA-3108) and a pacC-1 mutant that carries a disrupted pacC gene, which were used throughout this study, were selected as previously described [3,24]. The H6 and pacC-1 strains were cultivated on Sabouraud glucose agar for 15 days at 28 °C, and pacC-1 cultures were supplemented with 450 μg/ml hygromycin. Mycelia were collected with a sterile spatula, vortexed in saline solution [0.9% (w/v) NaCl] with 0.01% (v/v) Tween, filtered through fiberglass to remove mycelial debris, and then centrifuged to recover the conidia. Then, 106 conidia were transferred to 50 ml of Sabouraud broth, and germination was carried out at 28 °C for 72 h on an orbital shaker at 180 rpm (control). After incubation, the final pH of the culture medium was measured with a pH meter, and the resulting mycelia were harvested by filtration through sterilized Whatman paper (Whatman International, Maidstone, UK), washed with sterilized water, and transferred to minimal medium (MM) [25] at pH 5.0 or pH 8.0, which was, in some cases (as indicated), buffered with 50 mM sodium citrate or 50 mM Tris–HCl, respectively. MM was supplemented with glucose (50 mM) or glucose plus glycine (50 mM each) and sodium nitrate (70 mM). MM was also supplemented with keratin from bovine hooves (2.5 g/l) as a nutrient source. All cultures were incubated at 28 °C for 3, 6, and 24 h with agitation.
2.2. RNA extraction and cDNA synthesis
Mycelia obtained from each culture were harvested by filtration, and total RNA was extracted from approximately 100 mg of frozen mycelium using TRIzol™ reagent, and treated with RNase-free DNase I (Invitrogen, Carlsbad, CA), following the manufacturer's instructions. One microgram of DNase-treated RNA was reverse transcribed into cDNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. The cDNA from 3 independent biological experiments was quantified spectrophotometrically, and stored at −80 °C until PCR amplification.
2.3. Real-time PCR
For quantitative real-time PCR analyses, the genes encoding dolichyl-P-Man:Man(5)GlcNAc(2)-PP-dolichyl mannosyltransferase (N-man, TERG_06338; GenBank: XM_003233297) and O-mannosyltransferase (O-man, TERG_06465; GenBank: XM_003233430) were amplified from cDNA using the following primers (5′–3′): TAAACGACAGTGGTATGCCG (N-manFWD) and TGTAGCCTGTTGGGTTCTCT (N-manREV); CCATGGGACGTGTATACTC (O-manFWD) and CGTCATCATAGCAACATTCAG (O-manREV). Reactions were performed in three independent experiments using SYBR green PCR master mix (Applied Biosystems, Foster City, CA), in the StepOne Plus Real-Time PCR system. A 12.5-μl reaction was set up using 50 ng of cDNA and 300 nM of each primer, and the PCR cycle was as follows: 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Melting curve analyses were performed after each cycling to exclude primer dimers and nonspecific PCR products. Relative transcript quantities were calculated using the ΔΔCt method using the experimental condition that yielded the lowest Ct value (N-man, non-buffered H6 cultures in keratin, 6 h incubation) [26,27]. The T. rubrum α-actin (TERG_06637) and β-tubulin (TERG_07904) genes, which were used as endogenous reference genes, were amplified using the following oligonucleotides (sequence 5′–3′): AACGCCATCATGAAGTGT (actinFWD) and TCCTTCTGCATACGGTCAGA (actinREV); CCGTATGATGGCCACTTT (tubulinFWD) and CTGACCTGGGAAACGAAGAC (tubulinREV). Data normalization and analyses were performed using the GenEx 5 MultiD Analyses AB (www.multid.se). To confirm the identity of the genes analyzed in this work, as well as the endogenous reference genes, the PCR products were sequenced and analyzed by alignment with sequences retrieved from the Broad Institute Dermatophyte Comparative Database (http://www.broadinstitute.org/annotation/genome/dermatophyte_comparative/MultiHome.html).
3. Results
In vitro growth of the dermatophyte T. rubrum is dependent on the initial culture pH, with apparent optimal growth at pH 4.0–5.0, irrespective of carbon source (glucose, glycine, or protein). The initial pH of the T. rubrum cultures increased from 5.0 to a pH that ranged from 8.3 to 8.9 after 72–96 h of incubation in glycine or keratin. This effect was not observed when the fungus was cultivated with glucose as the carbon source, and the pH was maintained at approximately 5.0 [28,29]. Hydrolysis of keratin and other proteins releases amino acids, such as glycine, whose metabolism leads to the secretion of ammonia, thereby shifting the pH of the culture from acidic to alkaline [28]. Therefore, the metabolism of both glycine and keratin at pH 5.0 were alkalinizing events [28,29], even though the culture pH was still acidic after 24 h of incubation in glycine or keratin (Fig. 1). Moreover, disruption of the pacC gene in T. rubrum neither affects this pH shift (Fig. 1) [30] nor its growth on Sabouraud solid and in liquid media [3]. However, a marked decrease in the conidiation on Sabouraud solid medium and in the secretion of keratinolytic activity by the pacC-1 mutant, which correlated with its reduced capacity to infect human nails in vitro, were observed as compared to the control strain [3]. No ambient pH changes were observed when the fungus was cultured in buffered media (data not shown). T. rubrum grows poorly in culture medium with a starting pH of 8.0, even though the culture becomes acidified after incubation with glucose as a carbon source (Fig. 1). Growth in glycine or keratin dropped the pH of the culture to approximately 7 after the first 3–6 h of incubation, which increased to approximately pH 7.5 after 24 h of incubation (Fig. 1). An attractive hypothesis is that T. rubrum senses the alkaline environment and then acidifies the culture medium to an ambient pH at which its growth is stimulated. Metabolism of glycine or keratin leads to the secretion of ammonia, which shifts the culture pH to alkaline pH values. Therefore, to better understand the transcription of both the O-man and N-man genes during the first 24 h of incubation, when the pH of the medium was still acidic in non-buffered cultures (Fig. 1), we estimated the expression of both genes by qRT-PCR in different culture conditions.
Fig. 1.
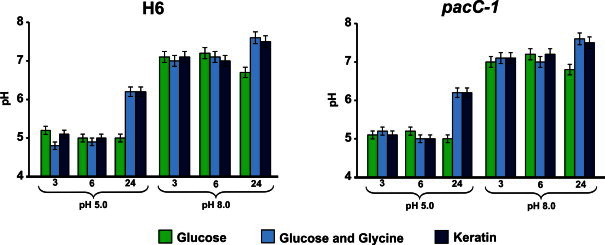
Evaluation of pH changes during the cultivation of Trichophyton rubrum with glucose, glucose plus glycine, or keratin, as the carbon source, for 3, 6 and 24 h. The initial pH of the culture medium was 5.0 or 8.0. The bars in the columns represent the standard deviation of data obtained from three independent experiments.
Transcription of the O-man and N-man genes in both the H6 and pacC-1 mutant strains was affected differently by the carbon source, the culture pH, and the time of incubation (Fig. 2). At pH 5.0, O-man was preferentially transcribed in both buffered and non-buffered keratin cultures, whereas N-man was apparently preferentially transcribed in non-buffered glucose cultures. The expression of N-man decreased as incubation time increased at pH 5.0, but the expression decreased more consistently in buffered glucose cultures (Fig. 2). Disruption of the pacC gene decreased transcription of O-man in both glucose and keratin at pH 5.0; however, O-man expression was enhanced in glucose plus glycine. Interestingly, the transcription of N-man in keratin cultures was enhanced in the pacC-1 mutant at pH 5.0. Therefore, at acidic pH, transcription of the O-man and N-man genes was positively and negatively affected, respectively, by the pacC gene in keratine cultures (Fig. 2). Moreover, transcription of the O-man and N-man genes in buffered cultures indicated that the O-man gene was preferentially transcribed at acidic pH in the presence of keratin compared to glucose or glucose plus glycine as the carbon sources, whereas transcription of the N-man gene was almost the same in both acidic and alkaline pH in the presence of keratin, and these properties were affected by disruption of the pacC gene (Fig. 2). It is also worth noting that while the disruption of the pacC gene resulted in changed transcription of the O-man gene in the different conditions at pH 5.0, it also resulted in the opposite changes for growth in glucose and glucose plus glycine at pH 8.0. Thus, our results suggest that the product of the pacC gene negatively affect both the transcription of the N-man gene in keratin at pH 5.0 and the O-man gene at pH 8.0 in the presence of different nutrients (Fig. 2) i.e., the pacC gene is functional irrespective of the culture conditions assayed.
Fig. 2.
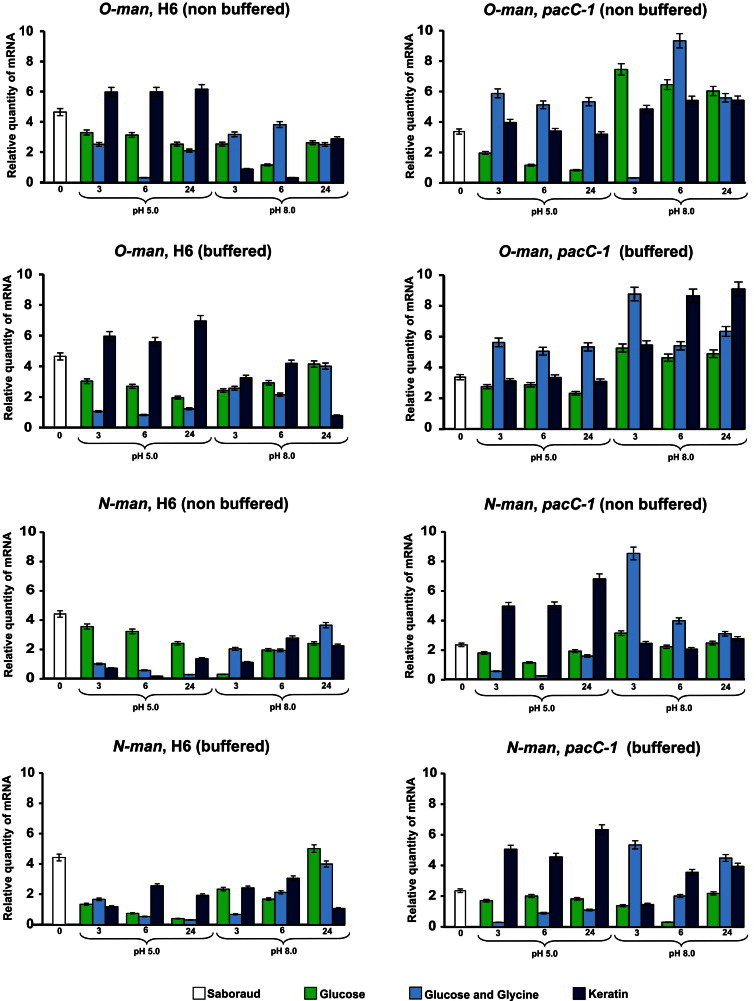
Expression of mannosyltransferase genes (O-man and N-man) in T. rubrum by quantitative real-time RT-PCR. Strains H6 and pacC-1 were cultured with glucose, glucose plus glycine, or keratin as the carbon source, for 3, 6, and 24 h. The initial pH of the culture medium was 5.0 or 8.0. The expression of the mannosyltransferase genes at 0 h is indicated (control). The bars in the columns represent the standard deviation of data obtained from three independent experiments.
4. Discussion
It has been well documented that the dermatophyte T. rubrum, as well as other filamentous fungi, acidifies the culture medium and represses the secretion of proteases during growth in glucose as the sole carbon source [31,32]. Moreover, during growth in glycine or keratin, the culture medium is alkalinized; however, this change is dependent on the initial pH of the culture, with an apparent optimum at pH 4.0–5.0. Interestingly, glycine utilization is apparently not repressed by glucose, because alkalinization of the culture medium occurs with glucose and glycine as carbon sources [29,33]. However, it is worth noting that during the first 24 h of cultivation, the pH of the medium is still acidic (Fig. 1), implying that the metabolism of glycine or keratin, an alkalinizing event, occurs during the first 24 h of cultivation, exclusively in an acidic environment.
In the model fungi Neurospora crassa and Aspergillus nidulans, one of the metabolic responses to the pH of the culture medium is the pH-dependent glycosylation of secreted enzymes [34–37]. For example, the level of glycosylation of the Pho-2 alkaline phosphatase synthesized by N. crassa at alkaline pH differs from that synthesized at acidic pH, which is approximately 13% and 21% for the Pho-2 enzyme purified from mycelium grown at pH 5.4 and 7.8, respectively [37,38]. The loss of enzymatic activity observed for the Pi-repressible alkaline phosphatase secreted at acidic pH is probably because the glycosylation of this enzyme is lower than that secreted at alkaline pH. We have also provided evidence that glycosylation of secreted enzymes, as documented for the Pi-repressible phosphatases in N. crassa and A. nidulans, is PacC-dependent [35].
In T. rubrum, disruption of the pacC gene, as in the pacC-1 mutant strain, resulted in decreased growth on human nails and decreased secretion of keratinolytic proteases in liquid medium when supplemented with keratin, which suggests that the keratinases secreted by T. rubrum are somehow regulated by the PacC protein [3]. PacC might be involved in the glycosylation of these keratinases through transcriptional modulation of O- and N-linked mannosyltransferases, a hypothesis supported by the results described here. Transcriptional profiling of both the O-man and N-man genes revealed a high level of complexity, because transcription of these genes was affected by nutrients, culture pH, and the functioning of the pacC gene. Disruption of the pacC gene increased the expression of N-man at pH 5.0 in keratin cultures. Moreover, if O-mannosylation precedes N-glycosylation in T. rubrum, as demonstrated in yeast [20], this physiological effect is dependent on the function of the pacC gene at acidic pH.
In conclusion, the genes encoding the O- and N-mannosyltransferases had different expression profiles, and the O-man gene was preferentially expressed at acidic pH when T. rubrum was grown on medium containing keratin. The balance between N-man and O-man expression levels in cultures at acidic pH may be under the control of the PacC transcription factor (in response to different carbon sources). Therefore, the product of the pacC gene of T. rubrum is functional at acidic pH. Moreover, transcription of the N-man and O-man genes might be required at different culture pHs for the glycosylation of transported proteins, according to the stage of infection, which suggests a possible role in cell adhesion and activation of signalling pathways regulating the production of enzymes that enable nutrient uptake for fungal development and maintenance in the host [39–41].
Acknowledgements
This work was supported by grants from the Brazilian funding agencies FAPESP, CNPq, CAPES and FAEPA. We thank M. Mazucato and S.H. Castrechini for skilled technical assistance.
References
- 1.Achterman R.R., White T.C. Dermatophyte virulence factors: identifying and analyzing genes that may contribute to chronic or acute skin infections. Int J Microbiol. 2012;2012:358305. doi: 10.1155/2012/358305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peres N.T.A. Transcriptional profiling reveals the expression of novel genes in response to various stimuli in the human dermatophyte Trichophyton rubrum. BMC Microbiol. 2010;10:39. doi: 10.1186/1471-2180-10-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferreira-Nozawa M.S., Silveira H.C.S., Ono C.J., Fachin A.L., Rossi A., Martinez-Rossi N.M. The pH signaling transcription factor PacC mediates the growth of Trichophyton rubrum on human nail in vitro. Med Mycol. 2006;44:641–645. doi: 10.1080/13693780600876553. [DOI] [PubMed] [Google Scholar]
- 4.Monod M. Secreted proteases from dermatophytes. Mycopathologia. 2008;166:285–294. doi: 10.1007/s11046-008-9105-4. [DOI] [PubMed] [Google Scholar]
- 5.Tsuboi R., Ko I.J., Takamori K., Ogawa H. Isolation of a keratinolytic proteinase from Trichophyton mentagrophytes with enzymatic activity at acidic pH. Infect Immun. 1989;57:3479–3483. doi: 10.1128/iai.57.11.3479-3483.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez-Rossi, N.M., Ferreira-Nozawa, M.S., Graminha, M.A.S., Nozawa, S.R., Fachin, A.L., Cervelatti, E.P., Prade, R.A., Rossi, A. (2004) in: Fungi in Human and Animal Health, vol. 9 (Kushwaha, R.K.S., Ed.), pp. 143–165, Scientific Publishers, Jodhpur, India.
- 7.Martinez-Rossi N.M., Persinoti G.F., Peres N.T., Rossi A. Role of pH in the pathogenesis of dermatophytoses. Mycoses. 2012;55:381–387. doi: 10.1111/j.1439-0507.2011.02162.x. [DOI] [PubMed] [Google Scholar]
- 8.Gentzsch M., Tanner W. The PMT gene family: protein O-glycosylation in Saccharomyces cerevisiae is vital. EMBO J. 1996;15:5752–5759. [PMC free article] [PubMed] [Google Scholar]
- 9.Gemmill T.R., Trimble R.B. Overview of N- and O-linked oligosaccharide structures found in various yeast species. Biochim Biophys Acta. 1999;1426:227–237. doi: 10.1016/s0304-4165(98)00126-3. [DOI] [PubMed] [Google Scholar]
- 10.Motteram J., Lovegrove A., Pirie E., Marsh J., Devonshire J., van de Meene A., Hammond-Kosack K., Rudd J.J. Aberrant protein N-glycosylation impacts upon infection-related growth transitions of the haploid plant-pathogenic fungus Mycosphaerella graminicola. Mol Microbiol. 2011;81:415–433. doi: 10.1111/j.1365-2958.2011.07701.x. [DOI] [PubMed] [Google Scholar]
- 11.Lommel M., Schott A., Jank T., Hofmann V., Strahl S. A conserved acidic motif is crucial for enzymatic activity of protein O-mannosyltransferases. J Biol Chem. 2011;286:39768–39775. doi: 10.1074/jbc.M111.281196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mouyna I. Members of protein O-mannosyltransferase family in Aspergillus fumigatus differentially affect growth, morphogenesis and viability. Mol Microbiol. 2010;76:1205–1221. doi: 10.1111/j.1365-2958.2010.07164.x. [DOI] [PubMed] [Google Scholar]
- 13.Shibata N., Kobayashi H., Suzuki S. Immunochemistry of pathogenic yeast, Candida species, focusing on mannan. Proc Jpn Acad Ser B Phys Biol Sci. 2012;88:250–265. doi: 10.2183/pjab.88.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lussier M., Sdicu A.M., Bussey H. The KTR and MNN1 mannosyltransferase families of Saccharomyces cerevisiae. Biochim Biophys Acta. 1999;1426:323–334. doi: 10.1016/s0304-4165(98)00133-0. [DOI] [PubMed] [Google Scholar]
- 15.Knauer R., Lehle L. The oligosaccharyltransferase complex from Saccharomyces cerevisiae. Isolation of the OST6 gene, its synthetic interaction with OST3, and analysis of the native complex. J Biol Chem. 1999;274:17249–17256. doi: 10.1074/jbc.274.24.17249. [DOI] [PubMed] [Google Scholar]
- 16.Helenius A., Aebi M. Intracellular functions of N-linked glycans. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 17.Brockhausen I. Substrate specificity and inhibition of UDP-GlcNAc:GlcNAc beta 1–2Man alpha 1–6R beta 1,6-N-acetylglucosaminyltransferase V using synthetic substrate analogues. Glycoconj J. 1995;12:371–379. doi: 10.1007/BF00731340. [DOI] [PubMed] [Google Scholar]
- 18.Strahl-Bolsinger S., Gentzsch M., Tanner W. Protein O-mannosylation. Biochim Biophys Acta. 1999;1426:297–307. doi: 10.1016/s0304-4165(98)00131-7. [DOI] [PubMed] [Google Scholar]
- 19.Lengeler K.B., Tielker D., Ernst J.F. Protein-O-mannosyltransferases in virulence and development. Cell Mol Life Sci. 2008;65:528–544. doi: 10.1007/s00018-007-7409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ecker M., Mrsa V., Hagen I., Deutzmann R., Strahl S., Tanner W. O-mannosylation precedes and potentially controls the N-glycosylation of a yeast cell wall glycoprotein. EMBO Rep. 2003;4:628–632. doi: 10.1038/sj.embor.embor864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harty C., Strahl S., Romisch K. O-mannosylation protects mutant alpha-factor precursor from endoplasmic reticulum-associated degradation. Mol Biol Cell. 2001;12:1093–1101. doi: 10.1091/mbc.12.4.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorka-Niec W., Kania A., Perlinska-Lenart U., Smolenska-Sym G., Palamarczyk G., Kruszewska J.S. Integration of additional copies of Trichoderma reesei gene encoding protein O-mannosyltransferase I results in a decrease of the enzyme activity and alteration of cell wall composition. Fungal Biol. 2011;115:124–132. doi: 10.1016/j.funbio.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 23.Maddi A., Free S.J. alpha-1,6-Mannosylation of N-linked oligosaccharide present on cell wall proteins is required for their incorporation into the cell wall in the filamentous fungus Neurospora crassa. Eukaryot Cell. 2010;9:1766–1775. doi: 10.1128/EC.00134-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fachin A.L., Maffei C.M.L., Martinez-Rossi N.M. In vitro susceptibility of Trichophyton rubrum isolates to griseofulvin and tioconazole. Induction and isolation of a resistant mutant to both antimycotic drugs. Mycopathologia. 1996;135:141–143. doi: 10.1007/BF00632334. [DOI] [PubMed] [Google Scholar]
- 25.Cove D.J. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim Biophys Acta. 1966;113:51–56. doi: 10.1016/s0926-6593(66)80120-0. [DOI] [PubMed] [Google Scholar]
- 26.Winer J., Jung C.K., Shackel I., Williams P.M. Development and validation of real-time quantitative reverse transcriptase–polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal Biochem. 1999;270:41–49. doi: 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
- 27.Jacob T.R., Peres N.T., Persinoti G.F., Silva L.G., Mazucato M., Rossi A., Martinez-Rossi N.M. rpb2 is a reliable reference gene for quantitative gene expression analysis in the dermatophyte Trichophyton rubrum. Med Mycol. 2012;50:368–377. doi: 10.3109/13693786.2011.616230. [DOI] [PubMed] [Google Scholar]
- 28.Ferreira-Nozawa M.S., Nozawa S.R., Martinez-Rossi N.M., Rossi A. The dermatophyte Trichophyton rubrum secretes an EDTA-sensitive alkaline phosphatase on high-phosphate medium. Braz J Microbiol. 2003;34:161–164. [Google Scholar]
- 29.Maranhão F.C.A., Paião F.G., Martinez-Rossi N.M. Isolation of transcripts over-expressed in human pathogen Trichophyton rubrum during growth in keratin. Microb Pathog. 2007;43:166–172. doi: 10.1016/j.micpath.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 30.Silveira H.C.S., Gras D.E., Cazzaniga R.A., Sanches P.R., Rossi A., Martinez-Rossi N.M. Transcriptional profiling reveals genes in the human pathogen Trichophyton rubrum that are expressed in response to pH signaling. Microb Pathog. 2010;48:91–96. doi: 10.1016/j.micpath.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 31.Meevootisom V., Niederpruem D.J. Control of exocellular proteases in dermatophytes and especially Trichophyton rubrum. Sabouraudia. 1979;17:91–106. doi: 10.1080/00362177985380141. [DOI] [PubMed] [Google Scholar]
- 32.Apodaca G., McKerrow J.H. Regulation of Trichophyton rubrum proteolytic activity. Infect Immun. 1989;57:3081–3090. doi: 10.1128/iai.57.10.3081-3090.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thedei Jr. G., Doubowetz T.H., Rossi A. Effect of carbon source and extracellular pH on the acidification of the culture medium and phosphatase excretion in Neurospora crassa. Braz J Med Biol Res. 1994;27:1129–1134. [PubMed] [Google Scholar]
- 34.Nozawa S.R., May G.S., Martinez-Rossi N.M., Ferreira-Nozawa M.S., Coutinho-Netto J., Maccheroni Jr. W., Rossi A. Mutation in a calpain-like protease affects the posttranslational mannosylation of phosphatases in Aspergillus nidulans. Fungal Genet Biol. 2003;38:220–227. doi: 10.1016/s1087-1845(02)00521-2. [DOI] [PubMed] [Google Scholar]
- 35.Nozawa S.R., Ferreira-Nozawa M.S., Martinez-Rossi N.M., Rossi A. The pH-induced glycosylation of secreted phosphatases is mediated in Aspergillus nidulans by the regulatory gene pacC-dependent pathway. Fungal Genet Biol. 2003;39:286–295. doi: 10.1016/s1087-1845(03)00051-3. [DOI] [PubMed] [Google Scholar]
- 36.Nahas E., Rossi A. Properties of a repressible alkaline-phosphatase secreted by the wild-type strain 74A of Neurospora crassa. Phytochemistry. 1984;23:507–510. [Google Scholar]
- 37.Palma M.S., Han S.W., Rossi A. Dissociation and catalytic activity of phosphate-repressible alkaline-phosphatase from Neurospora crassa. Phytochemistry. 1989;28:3281–3284. [Google Scholar]
- 38.Nozawa S.R., Thedei G., Crott L.S.P., Barbosa J.E., Rossi A. The synthesis of phosphate-repressible alkaline phosphatase does not appear to be regulated by ambient pH in the filamentous mould Neurospora crassa. Brazilian Journal of Microbiology. 2002;33:92–95. [Google Scholar]
- 39.Kotz A., Wagener J., Engel J., Routier F.H., Echtenacher B., Jacobsen I., Heesemann J., Ebel F. Approaching the secrets of N-glycosylation in Aspergillus fumigatus: characterization of the AfOch1 protein. PLoS One. 2010;5:e15729. doi: 10.1371/journal.pone.0015729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheth C.C., Hall R., Lewis L., Brown A.J., Odds F.C., Erwig L.P., Gow N.A. Glycosylation status of the C. albicans cell wall affects the efficiency of neutrophil phagocytosis and killing but not cytokine signaling. Med Mycol. 2010;49:513–524. doi: 10.3109/13693786.2010.551425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diaz-Jimenez D.F., Mora-Montes H.M., Hernandez-Cervantes A., Luna-Arias J.P., Gow N.A., Flores-Carreon A. Biochemical characterization of recombinant Candida albicans mannosyltransferases Mnt1, Mnt2 and Mnt5 reveals new functions in O- and N-mannan biosynthesis. Biochem Biophys Res Commun. 2012;419:77–82. doi: 10.1016/j.bbrc.2012.01.131. [DOI] [PMC free article] [PubMed] [Google Scholar]


