RNF111/Arkadia targets SUMOylated XPC for ubiquitylation, negatively regulating its association with damaged DNA
Abstract
Protein modifications by ubiquitin and small ubiquitin-like modifier (SUMO) play key roles in cellular signaling pathways. SUMO-targeted ubiquitin ligases (STUbLs) directly couple these modifications by selectively recognizing SUMOylated target proteins through SUMO-interacting motifs (SIMs), promoting their K48-linked ubiquitylation and degradation. Only a single mammalian STUbL, RNF4, has been identified. We show that human RNF111/Arkadia is a new STUbL, which used three adjacent SIMs for specific recognition of poly-SUMO2/3 chains, and used Ubc13–Mms2 as a cognate E2 enzyme to promote nonproteolytic, K63-linked ubiquitylation of SUMOylated target proteins. We demonstrate that RNF111 promoted ubiquitylation of SUMOylated XPC (xeroderma pigmentosum C) protein, a central DNA damage recognition factor in nucleotide excision repair (NER) extensively regulated by ultraviolet (UV)-induced SUMOylation and ubiquitylation. Moreover, we show that RNF111 facilitated NER by regulating the recruitment of XPC to UV-damaged DNA. Our findings establish RNF111 as a new STUbL that directly links nonproteolytic ubiquitylation and SUMOylation in the DNA damage response.
Introduction
Protein modification by ubiquitin and the small ubiquitin-like modifier (SUMO) play important, often interconnected, regulatory roles in numerous signaling pathways in eukaryotic cells (Kerscher et al., 2006; Gareau and Lima, 2010; Komander and Rape, 2012). Similar enzymatic cascades involving activating (E1), conjugating (E2), and ligase (E3) enzymes underlie protein modification by ubiquitin and SUMO (Kerscher et al., 2006). Although no consensus sequences surrounding ubiquitylation sites have been described, SUMOylation is frequently, but not always, targeted to K-X-E/D motifs or an inverted version of this sequence (Matic et al., 2010). Three different SUMO isoforms, SUMO1–3, are expressed in cells, and although SUMO2 and SUMO3 are 97% identical and thus often referred to as SUMO2/3, SUMO1 and SUMO2/3 only share ∼50% sequence identity (Gareau and Lima, 2010). Both ubiquitin and SUMO can be attached to target proteins as single moieties but additionally share the ability to form chains via internal lysine residues. Unlike ubiquitin, only a single lysine residue in SUMO that conforms to the SUMO consensus sequence is used for chain formation, and this ability is exclusive to SUMO2/3 (Tatham et al., 2001; Komander and Rape, 2012).
Different polyubiquitin chains have distinct cellular functions (Komander and Rape, 2012). Although most of the known ubiquitylation processes generate K48-linked chains, which target substrates for degradation by the 26S proteasome, protein ubiquitylation does not always promote destruction; in particular, K63-linked polyubiquitylation, catalyzed by the E2 enzyme Ubc13 in conjunction with its partner proteins Mms2 or Uev1, is a nondegradative modification used in a range of signaling pathways, including cellular stress responses such as DNA damage and inflammatory responses (Chen and Sun, 2009; Al-Hakim et al., 2010; Komander and Rape, 2012). The function of poly-SUMO chains is less well understood, but roles in processes such as chromosome segregation, DNA damage, and heat shock responses have been described (Schwartz et al., 2007; Golebiowski et al., 2009; Yin et al., 2012). Several cellular processes, including the DNA damage response, are intimately coregulated by ubiquitin- and SUMO-mediated signaling (Kerscher et al., 2006; Bergink and Jentsch, 2009; Bekker-Jensen and Mailand, 2011). The discovery of SUMO-targeted ubiquitin ligases (STUbLs) revealed a further, direct interplay between these modifications. By means of tandem SUMO-interacting motifs (SIMs; Hecker et al., 2006), STUbLs recognize poly-SUMOylated proteins and target them for K48-linked polyubiquitylation and degradation via their E3 ubiquitin ligase activities (Prudden et al., 2007; Sun et al., 2007). Accordingly, although SUMOylation is not a degradative modification per se, it can indirectly promote proteasomal destruction via STUbLs. Only a few STUBLs have been identified so far, including Slx5-Slx8 in Saccharomyces cerevisiae, Rfp1/Rfp2-Slx8 in Schizosaccharomyces pombe, and RNF4 in mammalian cells. All of these enzymes play important roles in maintenance of genome stability (Prudden et al., 2007; Sun et al., 2007; Galanty et al., 2012; Yin et al., 2012), consistent with the extensive involvement of both ubiquitin and SUMO in cellular responses to DNA damage.
In a search for new SUMO-binding proteins, we discovered that the human RNF111 ubiquitin ligase (also known as Arkadia) is a STUbL, which can promote nonproteolytic ubiquitylation of target proteins through cognate E2 enzymes such as Ubc13. We demonstrate that RNF111 has a physiological role in nucleotide excision repair (NER), catalyzing DNA damage–induced ubiquitylation of SUMOylated XPC (xeroderma pigmentosum C). Our findings reveal direct coupling between nonproteolytic ubiquitylation and SUMOylation in the DNA damage response.
Results and discussion
RNF111 recognizes poly-SUMO chains via tandem SIMs
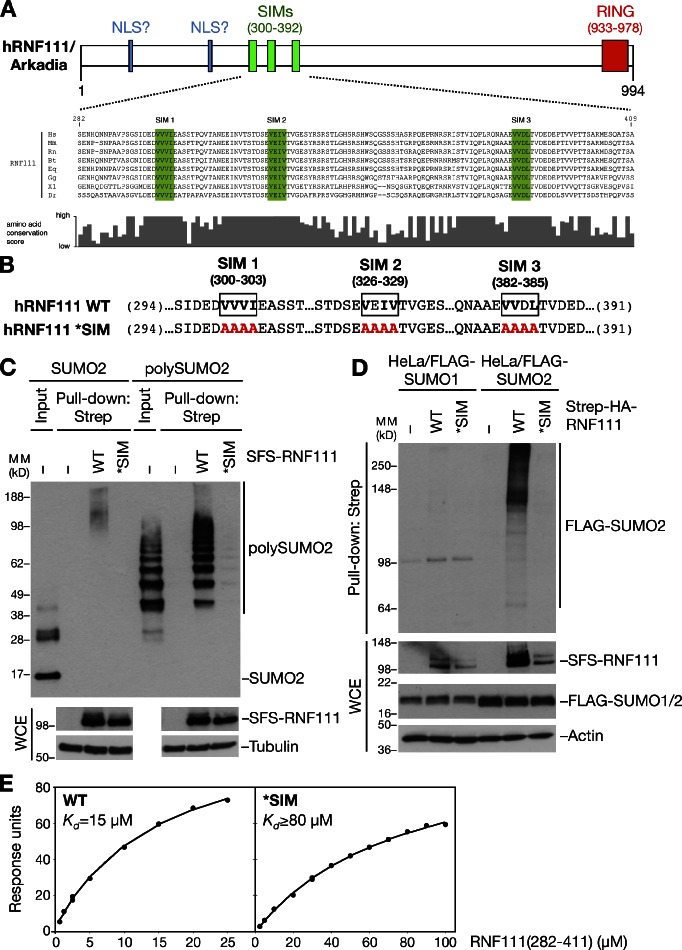
In a search for proteins containing SIMs, we noted that the human RNF111/Arkadia E3 ubiquitin ligase, which has been shown to function in amplification of TGF-β signaling pathways (Miyazono and Koinuma, 2011), contains three highly conserved, potential SIMs in its N-terminal region (Fig. 1, A and B). To test whether these putative SIMs are functional SUMO-binding modules, we generated an RNF111 mutant (*SIM) in which the core hydrophobic residues in each of the three SIMs were mutated to alanines, predicted to disrupt their SUMO-binding ability (Fig. 1 B; Hecker et al., 2006). We first assessed the SUMO-binding ability of ectopically expressed Strep-tagged forms of RNF111 wild type (WT) or *SIM purified on Strep-Tactin agarose. We found that RNF111 bound purified poly-SUMO2 chains with high affinity in vitro but was virtually unable to bind free SUMO2 (Fig. 1 C). This was fully dependent on the integrity of the SIM motifs, as the RNF111 *SIM mutant did not interact with poly-SUMO2 (Fig. 1 C). To test whether RNF111 binds to SUMOylated proteins in cells, we overexpressed RNF111 WT or *SIM in cells stably expressing FLAG-SUMO1 or 2 and analyzed their interactions in immunoprecipitation (IP) experiments. Consistent with in vitro binding experiments, RNF111 interacted with high–molecular weight SUMOylated species, but not free SUMO2, in a SIM-dependent manner (Fig. 1 D and not depicted). Moreover, RNF111 selectively interacted with proteins modified with SUMO2 but not SUMO1 (Fig. 1 D), in agreement with the notion that SUMO2, but not SUMO1, forms poly-SUMO chains in vivo (Tatham et al., 2001). Surface plasmon resonance analysis showed that the RNF111 SIM region bound directly to poly-SUMO2 with a Kd of ∼15 µM, whereas the *SIM mutations reduced binding to a Kd > 80 µM (Fig. 1 E). These data demonstrate that RNF111 interacts with poly-SUMOylated proteins via three N-terminal SIM motifs, in accordance with recent findings that showed an additive contribution of each SIM to poly-SUMO binding (Sun and Hunter, 2012).
Figure 1.
Human RNF111 binds to poly-SUMOylated proteins via an N-terminal SIM region. (A) Schematic of human RNF111/Arkadia. The RING domain, two putative NLSs (Episkopou et al., 2001), and three SUMO-interacting motifs (SIMs; top), conserved in higher vertebrates (bottom), are shown. Core hydrophobic SIM residues are highlighted in green. (B) Amino acid substitutions (highlighted in red) in the RNF111 SIM region to disrupt its SUMO-binding ability (*SIM). (C) S-FLAG-Strep–tagged RNF111 (SFS-RNF111) proteins expressed in U2OS cells were purified on Strep-Tactin Sepharose, incubated with purified SUMO2 or poly-SUMO2 (3–8), and washed extensively. Bound complexes were immunoblotted with the SUMO2 antibody. WCE, whole-cell extract. (D) HeLa cells stably expressing FLAG-SUMO isoforms were transfected with Strep-HA-RNF111 plasmids as indicated. Whole-cell extracts were subjected to Strep-Tactin pull-down and immunoblotting with the FLAG antibody. (E) Plasmon surface resonance analysis of poly-SUMO2 binding kinetics of RNF111 fragments spanning the SIMs. Data shown are from a single representative experiment out of three repeats. MM, molecular mass.
RNF111 promotes Ubc13–Mms2-dependent ubiquitylation
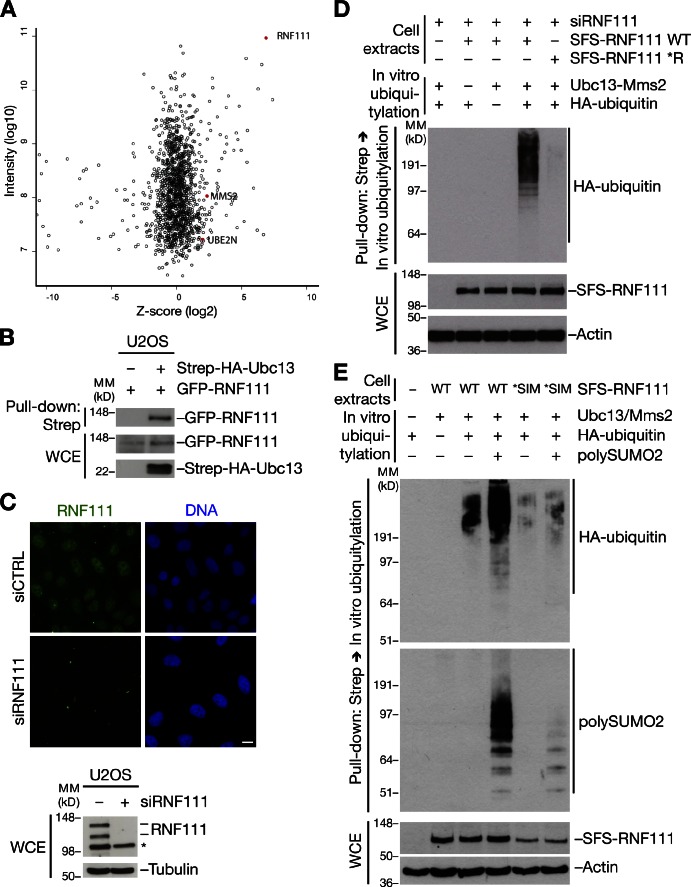
To gain insight into the functional significance of RNF111 SUMO binding, we performed quantitative mass spectrometry (MS)–based analysis of cellular RNF111-interacting proteins (Fig. 2 A and Fig. S1 A). Several potential RNF111-binding factors were identified by this approach, including components of the AP2 (clathrin adaptor 2) complex, consistent with the known role of RNF111 in regulating endocytosis via interaction with this complex (Fig. S1, A and B; Miyazono and Koinuma, 2011). Among the RNF111-associated proteins, we also found two E2 ubiquitin–conjugating enzymes: Ubc13–Mms2, which specifically catalyzes K63-linked ubiquitin chain formation, and UBE2O, a large E2 enzyme of unknown function (Fig. 2 A and Fig. S1 B). The presence of both Ubc13 and Mms2 lends strong support to the possibility that this complex is a physiological E2 partner for RNF111. We validated the interactions between RNF111 and Ubc13 or UBE2O by reciprocal co-IP analysis (Fig. 2 B and Fig. S2, A and B). In contrast, we did not observe binding of RNF4, the known mammalian STUbL, to Ubc13 under a range of conditions (Fig. S2, C and D).
Figure 2.
RNF111 has STUbL activity in the presence of Ubc13–Mms2. (A) MS-based analysis of RNF111-interacting proteins. U2OS and U2OS/GFP-RNF111 cells were grown in light and heavy SILAC medium, respectively. GFP-RNF111 and associated proteins enriched on GFP-Trap resin were analyzed by MS. Plot shows z scores (from SILAC heavy/light ratios) and total intensity of identified proteins. RNF111, Ubc13 (UBE2N), and Mms2 (MMS2) are highlighted. See also Fig. S1 (A and B). (B) U2OS cells were cotransfected with indicated combinations of GFP-RNF111 and Strep-HA-Ubc13 plasmids. Whole-cell extracts (WCE) were subjected to Strep-Tactin pull-down followed by immunoblotting with GFP and HA antibodies. (C) U2OS cells transfected with nontargeting (control [CTRL]) or RNF111 siRNAs were collected 72 h later and processed for immunostaining (top) or immunoblot (bottom) with RNF111 antibody. Asterisk indicates a nonspecific band. Bar, 10 µm. (D) Extracts of U2OS cells sequentially transfected with RNF111 siRNA and S-FLAG-Strep–tagged RNF111 (SFS-RNF111) plasmids were subjected to Strep-Tactin pull-down. Bound complexes were incubated with ubiquitylation reaction mixture containing E1, Ubc13–Mms2 complex, and HA-ubiquitin as indicated and washed extensively, and RNF111 E3 ligase activity was analyzed by immunoblotting with the HA antibody. (E) As in D, except that ubiquitylation reactions were performed in the presence or absence of poly-SUMO2 (3–8) chains followed by immunoblotting with HA and SUMO2 antibodies. MM, molecular mass.
Because RNF111 promotes degradation of factors in TGF-β signaling pathways, the interaction with Ubc13–Mms2 was unexpected, and we set out to investigate its physiological relevance. We noted that endogenous RNF111 is primarily localized in the nucleus (Fig. 2 C), suggesting that in addition to facilitating amplification of TGF-β signaling and endocytosis, RNF111 might have other important nuclear functions. To test whether RNF111 has E3 ligase activity in the presence of Ubc13–Mms2, we performed in vitro ubiquitylation assays using ectopic RNF111 immunopurified from cells. Because RNF111 appeared to form homodimers in cells (unpublished data), we depleted endogenous RNF111 to remove background E3 ligase activity of copurifying endogenous RNF111. We found that RNF111 was highly active as an E3 ligase in the presence of purified Ubc13–Mms2, as judged from its autoubiquitylation (Fig. 2 D). As expected, this required the integrity of the RNF111 RING domain (Fig. 2 D), whereas mutation of the SIMs did not impair intrinsic RNF111 E3 ligase activity (Fig. S2 E). In addition to Ubc13–Mms2, RNF111 was active with more generic E2 enzymes, such as UbcH5, as expected (Fig. S2 F). To test whether RNF111 has STUbL activity in the presence of Ubc13–Mms2, we analyzed the impact of SUMO2 on RNF111 E3 ligase activity. Strikingly, we found that poly-SUMO2 chains, but not free SUMO2, were efficiently targeted for Ubc13–Mms2-dependent ubiquitylation by RNF111 in a manner fully dependent on the integrity of the SIMs (Fig. 2 E and not depicted). We conclude from these experiments that RNF111 functions as a STUbL that employs Ubc13–Mms2 and likely other cognate E2 partners in ubiquitylation of SUMOylated substrates.
RNF111 promotes UV-induced ubiquitylation of XPC
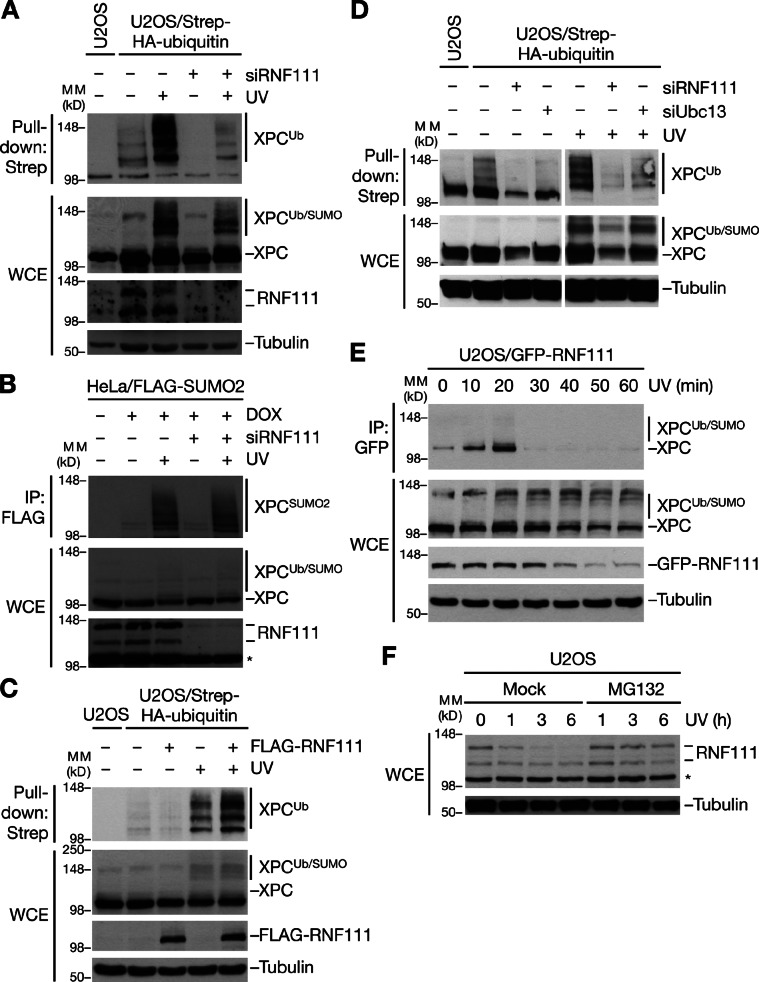
We next attempted to identify physiological substrates for the STUbL activity of RNF111. The NER factor XPC is known to undergo both SUMOylation and ubiquitylation in response to UV radiation, and the UV-induced ubiquitin chains on XPC do not appear to destine XPC for proteasomal destruction (Sugasawa et al., 2005; Wang et al., 2005). We reasoned that SUMOylated XPC might be a candidate target of the Ubc13–Mms2-dependent E3 ligase activity of RNF111. Indeed, knockdown of RNF111 by any of several independent siRNAs impaired UV-induced ubiquitylation but not SUMOylation of XPC (Fig. 3, A and B; and Fig. S3, A and B), suggesting that XPC is SUMOylated before ubiquitylation by RNF111. The slow-migrating, UV-inducible XPC species seen in immunoblots represent a mixture of ubiquitin- and SUMO-modified forms; hence, the dramatic decrease in XPC ubiquitylation but not SUMOylation in RNF111-depleted cells manifests less prominently in total XPC blots (Fig. 3 A). Consistent with a direct role of RNF111 in ubiquitylating XPC after UV, we found that elevated levels of RNF111 augmented the UV-induced increase in XPC-GFP ubiquitylation (Fig. 3 C). In contrast, depletion of RNF4, the known STUbL in mammalian cells, had no effect on UV-induced XPC ubiquitylation (Fig. S3 C). The ability of RNF111 to promote Ubc13–Mms2-dependent ubiquitylation prompted us to test whether UV-induced XPC ubiquitylation required Ubc13 function. Like RNF111 knockdown, depletion of Ubc13 decreased UV-induced XPC ubiquitylation substantially (Fig. 3 D and Fig. S3 D), suggesting that RNF111-dependent XPC ubiquitylation after UV exposure was, at least partially, mediated by Ubc13-dependent, nonproteolytic ubiquitylation.
Figure 3.
RNF111 promotes UV-induced ubiquitylation of XPC. (A) U2OS or U2OS/Strep-HA-ubiquitin cells transfected with control (−) or RNF111 siRNAs were exposed or not exposed to UV and collected 1 h later, and XPC ubiquitylation was analyzed by immunoblotting Strep-Tactin pull-downs of whole-cell extracts (WCE) with the XPC antibody. (B) HeLa/FLAG-SUMO2 cells transfected with control (−) or RNF111 siRNAs and left untreated or induced to express FLAG-SUMO2 by addition of doxycycline (DOX) were exposed or not exposed to UV and collected 1 h later. Cells were lysed under denaturing conditions, and XPC SUMOylation was analyzed by immunoblotting of FLAG IPs with XPC antibody. (C) U2OS/Strep-HA-ubiquitin cells transfected with empty vector (−) or FLAG-RNF111 plasmid were exposed or not exposed to UV and collected 1 h later. XPC ubiquitylation was analyzed as in A. (D) XPC ubiquitylation in U2OS/Strep-HA-ubiquitin cells depleted of RNF111 or Ubc13 was analyzed as in A. Ubc13 knockdown efficiency is shown in Fig. S3 D. (E) Extracts of U2OS/GFP-RNF111 cells collected at the indicated times after UV radiation were subjected to GFP IP followed by immunoblotting with XPC antibody. (F) Extracts of U2OS cells incubated with or without MG132, exposed to UV 30 min later, and collected at the indicated times after UV were analyzed by immunoblotting with the RNF111 antibody. Asterisks denote a nonspecific band. MM, molecular mass.
To further probe the basis of RNF111-dependent XPC ubiquitylation in response to UV, we asked whether RNF111 and XPC interact in cells. Indeed, UV induced prominent, but transient, interaction between RNF111 and XPC at early time points after UV (Fig. 3 E). Interestingly, like several known NER factors, both endogenous and ectopic RNF111 underwent partial degradation after UV in a proteasome-dependent manner, which, however, did not require the intrinsic E3 ligase activity of RNF111 (Fig. 3, E and F; and Fig. S3 E). In general, the kinetics of UV-induced RNF111 interaction with XPC and degradation correlated with that of XPC ubiquitylation after UV exposure (Fig. 3, E and F; and Fig. S3 F).
RNF111 selectively ubiquitylates SUMOylated XPC
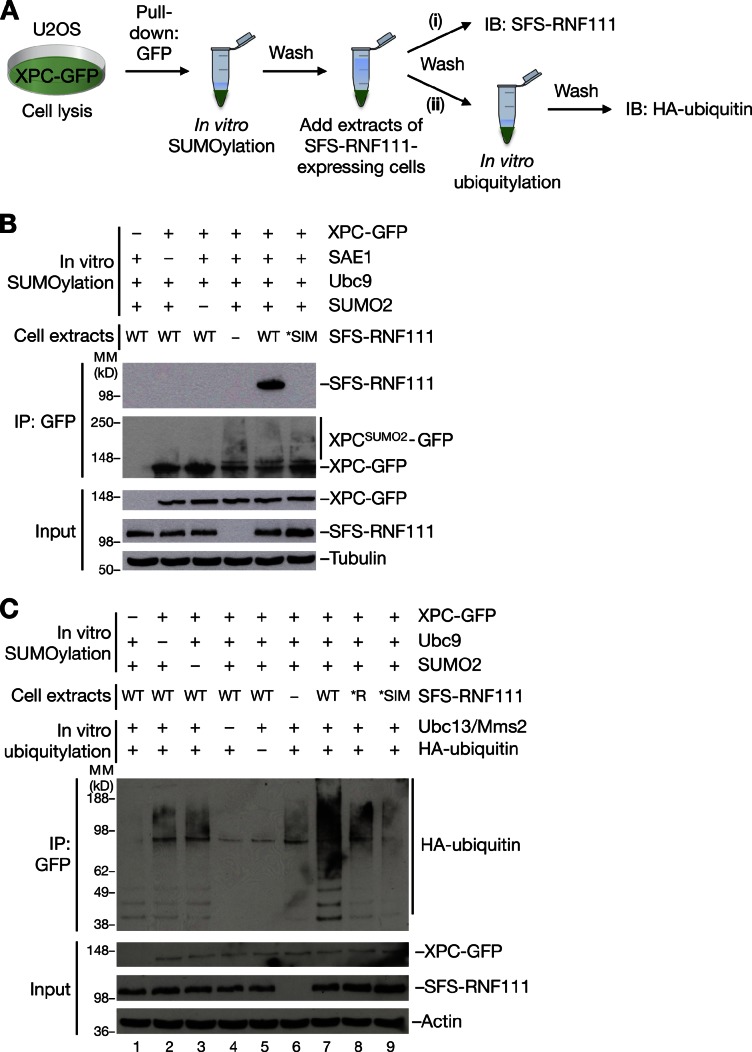
The aforementioned findings suggested that RNF111 targets SUMOylated XPC for ubiquitylation in response to UV. Hence, we tested whether RNF111 specifically interacts with SUMO-modified XPC via its SIMs, using a strategy wherein GFP-tagged XPC immunopurified from cells was SUMOylated in vitro and then incubated with extracts of cells transfected with WT or mutant forms of ectopic RNF111 (Fig. 4 A). Under these conditions, RNF111 efficiently interacted with XPC, but only if XPC had been pre-SUMOylated, and this required the integrity of the RNF111 SIMs (Fig. 4 B), in agreement with the notion that RNF111 specifically recognizes SUMOylated XPC. We next tested whether RNF111 functions as a STUbL for XPC. To do this, we extended the setup to monitor SUMO-dependent RNF111-XPC binding, by subjecting the bound complexes to an in vitro ubiquitylation assay in the presence of Ubc13–Mms2 as an E2 (Fig. 4 A). Although a background level of Ubc13–Mms2-dependent ubiquitylation of XPC-GFP could be seen in the absence of ectopically expressed RNF111 (Fig. 4 C, compare lanes 1–6), the addition of RNF111 WT markedly enhanced XPC ubiquitylation (Fig. 4 C, compare lanes 6 and 7) but only if XPC had been pre-SUMOylated (Fig. 4 C, compare lanes 2, 3, and 7). Importantly, this increase in RNF111-dependent XPC ubiquitylation required the functional integrity of both the RNF111 RING and SIM domains (Fig. 4 C, compare lanes 7–9). These data suggest that RNF111 acts as a STUbL for XPC, catalyzing its nonproteolytic ubiquitylation in response to UV damage.
Figure 4.
RNF111 ubiquitylates XPC in a SUMOylation-dependent manner. (A) Outline of in vitro SUMO-binding and STUbL assays. XPC-GFP expressed in U2OS cells was immunopurified on GFP-Trap resin and subjected to in vitro SUMOylation. After washing, the XPC-GFP–containing beads were incubated with extracts of cells transfected or not transfected with S-FLAG-Strep-RNF111 (SFS-RNF111) constructs, washed again, and processed for immunoblotting (IB) of bound SFS-RNF111 with FLAG antibody (i) or subjected to in vitro ubiquitylation followed by washing and immunoblotting with the HA antibody to analyze ubiquitin ligase activity (ii). (B) SUMOylation-dependent binding of RNF111 to XPC, analyzed as described in A. (C) Analysis of SUMOylation-dependent XPC ubiquitylation by RNF111 was performed as described in A. MM, molecular mass.
RNF111 promotes NER by regulating the interaction of XPC with damaged DNA
Because RNF111 promotes ubiquitylation of XPC after UV, we asked whether RNF111 regulates NER. Although UV-induced ubiquitylation of XPC has been suggested to increase its DNA-binding affinity (Sugasawa et al., 2005), the exact role of this modification in NER is unclear. Previous work suggested that XPC is ubiquitylated by CRL4DDB2, an E3 ligase complex functioning as a proximal sensor of UV lesions in DNA (Sugasawa et al., 2005). It is possible that XPC is ubiquitylated by both CRL4DDB2 and RNF111 in response to UV. Indeed, using MS, we found that XPC ubiquitylation involves a variety of ubiquitin chains and ≥15 individual ubiquitylation sites (unpublished data; Povlsen et al., 2012); hence, the nature and regulation of XPC ubiquitylation appears to be highly complex, likely involving several E3 ligases. To determine whether RNF111 loss affects NER, we measured UV-induced DNA repair synthesis (UDS) in RNF111−/− mouse embryonic fibroblasts (MEFs; Mavrakis et al., 2007). Strikingly, these MEFs showed a marked reduction in UDS, as was also observed in XPC−/− MEFs (Fig. 5 A). Moreover, using two independent siRNAs, we found that RNF111 knockdown resulted in increased accumulation of XPC-GFP to locally UV-irradiated chromatin, whereas knockdown of DDB2 had the opposite effect, as previously observed (Fig. 5, B and C; Nishi et al., 2009). Hence, although DDB2 and RNF111 have opposing effects on XPC accumulation at UV lesions, interfering with the proper kinetics of XPC interaction with damaged chromatin by inactivation of either E3 reduces the efficiency of NER. These data suggest that RNF111 has a physiological role in promoting NER by regulating ubiquitylation of XPC and its association with damaged DNA.
Figure 5.
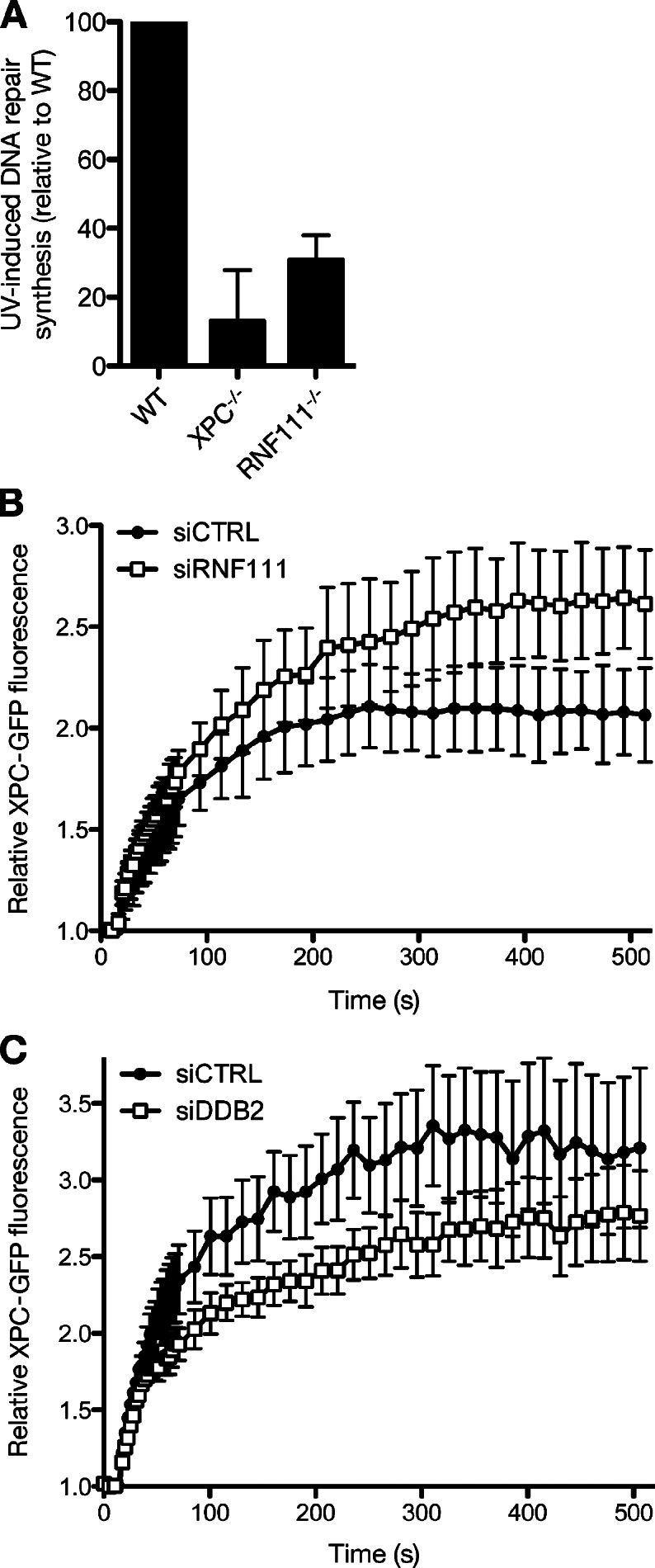
RNF111 promotes NER by regulating XPC recruitment to UV-damaged DNA. (A) UDS of the indicated MEF cell lines, determined by EdU incorporation for 3 h after exposure to 16 J/m2 UV-C. Error bars indicate SDs of three independent experiments. (B) Cells stably expressing XPC-GFP were transfected with indicated siRNAs and locally exposed to laser-induced UV-C damage. XPC-GFP fluorescence intensity at the damaged area relative to predamage intensity was recorded in time using live-cell confocal imaging (mean of three independent experiments, n = 8 cells per experiment, ±SD). (C) As in B, except that cells were transfected with control (CTRL) or DDB2 siRNA. Results of a representative experiment (n = 8 cells per sample, ±SEM) are shown.
Our findings show that RNF111 is a STUbL that promotes nonproteolytic ubiquitylation of at least a subset of its substrates, including XPC, implying that STUbL activity is not confined to RNF4 in higher vertebrates and that STUbLs do not always target substrates for proteasomal degradation. Although Ubc13–Mms2 appears to be a major cognate E2 enzyme for RNF111 in cells, RNF111 also interacts with other E2 enzymes and is known to promote ubiquitin-dependent degradation of TGF-β signaling factors (Koinuma et al., 2003; Levy et al., 2007; Nagano et al., 2007). Hence, depending on the context, RNF111 may work with different E2s to promote degradative or nonproteolytic ubiquitylation of SUMOylated substrate proteins. Despite the fact that both RNF4 and RNF111 interact with poly-SUMOylated proteins through tandem SIMs, they appear to have largely nonoverlapping roles in the cell. For instance, RNF4, but not RNF111, was dispensable for UV-induced ubiquitylation of XPC, whereas RNF111 was not recruited to laser microirradiation-induced DNA double-strand breaks, unlike RNF4 (unpublished data; Galanty et al., 2012; Yin et al., 2012). This distribution of labor between RNF4 and RNF111 in targeting distinct subsets of SUMOylated factors may reflect differences in the SUMO-binding properties of their tandem SIMs, which have a distinct configuration, as well as differential target-binding specificity contributed by other domains in these proteins.
Although our comprehensive analysis of RNF111-binding factors in unperturbed cells uncovered several E2 partner proteins, we did not detect any known components of TGF-β signaling pathways, nor XPC. Given the involvement of RNF111 in regulating these proteins, we speculate that processes mediated by the RNF111 STUbL activity may, in many cases, be induced by stimuli such as TGF-β or UV treatment, which may promote SUMOylation of specific factors and thus trigger their RNF111-mediated ubiquitylation. This is consistent with previous findings that elevated levels of RNF111 only cause degradation of SnoN in TGF-β–stimulated cells (Levy et al., 2007). Based on the large and heterogeneous group of proteins identified by MS as putative RNF111-interacting proteins, we propose that RNF111, like RNF4, is a multifunctional STUbL regulating a diverse range of cellular signaling processes, determined to a large extent by the SUMOylation state of target proteins. This scenario reconciles the involvement of RNF111 in radically different cellular processes, such as TGF-β signaling and endocytosis (Miyazono and Koinuma, 2011), and NER. Whether the ability of RNF111 to ubiquitylate proteins in the former processes involves its STUbL activity remains to be addressed. Our findings shed further light on how STUbLs directly couple ubiquitylation and SUMOylation in important cellular signaling pathways.
Materials and methods
Plasmids and siRNA
Full-length human RNF111 cDNA was amplified by PCR and inserted into pEGFP-C1 (Takara Bio Inc.) and pcDNA4/TO (Invitrogen) containing N-terminal Strep-HA or S-FLAG-Strep tags to generate mammalian expression constructs for GFP-, Strep-HA–, and S-FLAG-Strep–tagged RNF111, respectively. The RNF111 *RING (W963A) point mutation was introduced using the site-directed mutagenesis kit (QuikChange; Agilent Technologies). The RNF111 *SIM mutations (VVVI(300–303)AAAA, VEIV(326–329)AAAA, and VVDL(382–385)AAAA) were introduced by replacing part of the coding sequence of human RNF111 (nucleotides 665–1,677 of the RNF111 ORF) with a synthetic gene spanning this region and containing the mutated *SIM sequence using the unique KpnI and EcoNI sites in RNF111. All constructs were verified by sequencing. Constructs expressing Strep-HA–tagged Ubc13 and GFP-XPC were described previously (Bekker-Jensen et al., 2010). Plasmid transfections were performed using GeneJuice (EMD Millipore) according to the manufacturer’s instructions. siRNA transfections were performed with Lipofectamine RNAiMAX (Invitrogen) as described. siRNA target sequences used in this study were control, 5′-GGGAUACCUAGACGUUCUA-3′; RNF111 (#1), 5′-GGAUAUUAAUGCAGAGGAA-3′; RNF111 (#4), 5′-GGAUAUGAAGAGUGAGAUU-3′; Ubc13, 5′-GAGCAUGGACUAGGCUAUA-3′; XPC, 5′-GCAAAUGGCUUCUAUCGAAUU-3′; DDB2, 5′-CCCAGAUCCUAAUUUCAAA-3′; RNF4 (#1), 5′-GCUAAUACUUGCCCAACUU-3′; and RNF4 (#2), 5′-GACAGAGACGUAUAUCUGA-3′.
Cell culture
Human U2OS and HeLa cells were cultured in DMEM containing 10% fetal bovine serum. SV40-immortalized XP4PA cells stably expressing XPC-GFP (Hoogstraten et al., 2008) were cultured in DMEM containing 5% fetal bovine serum and 2 mM l-glutamine. RNF111−/− primary mouse fibroblasts of mixed 129Sv/MF1 genetic backgrounds (provided by V. Episkopou, Imperial College London, London, England, UK; Mavrakis et al., 2007), and XPC−/− MEFs in which exons 4–7 of the XPC gene were deleted (Sands et al., 1995) were cultured in a 1:1 ratio of Ham’s F10 and DMEM supplemented with 10% fetal calf serum and 1% nonessential amino acids. To generate cell lines stably expressing GFP-tagged WT and mutant RNF111 alleles, U2OS cells were cotransfected with GFP-RNF111 constructs and pBabe-puromycin plasmid, and positive clones were selected with 1 µg/ml puromycin. A stable U2OS/Strep-HA-ubiquitin cell line (Danielsen et al., 2011) was generated by selecting cells transfected with Strep-HA-ubiquitin expression plasmid in medium containing 400 µg/ml G418 until resistant clones grew out. Stable HeLa cell lines expressing FLAG-SUMO1/2 in a doxycycline-inducible manner (Danielsen et al., 2012) were generated by cotransfection of HeLa/FRT/TRex cells (Invitrogen) with pcDNA5/FRT/TO-3×FLAG-SUMO1/2 and pOG44 followed by selection with 200 µg/ml Hygromycin B. Unless stated otherwise, cells were exposed to 30 J/m2 UV and collected 1 h later.
MS-based analysis of RNF111-interacting proteins
For stable isotope labeling by amino acids in cell culture (SILAC) labeling, U2OS or U2OS/GFP-RNF111 cells were cultured for 14 d in Eagle’s minimum essential medium (Sigma-Aldrich) supplemented with l-arginine and l-lysine or l-arginine-U-13C6-15N4 and l-lysine-U-13C6-15N2 (Cambridge Isotope Laboratories), respectively (Ong et al., 2002). Cells were lysed in EBC buffer supplemented with protease and phosphatase inhibitor cocktails (Roche), and GFP-RNF111 and its interacting proteins were enriched using GFP-Trap resin. Proteins were resolved by SDS-PAGE and in-gel digested with trypsin. Peptide fractions were analyzed on a quadrupole mass spectrometer (Q Exactive; Orbitrap; Thermo Fisher Scientific) equipped with a nanoflow HPLC system (Thermo Fisher Scientific; Michalski et al., 2011). Raw data files were analyzed using MaxQuant software (version 1.2.2.9; Cox and Mann, 2008). Parent ion and MS2 spectra were searched against protein sequences obtained from the UniProt knowledge base using the Andromeda search engine (Cox et al., 2011). Spectra were searched with a mass tolerance of 6 ppm in MS mode and 20 ppm in higher-energy C-trap dissociation MS2 mode, strict trypsin specificity, and allowing up to two missed cleavage sites. Cysteine carbamidomethylation was included as a fixed modification, and N-terminal protein acetylation was included as variable modification. The dataset was filtered based on posterior error probability to arrive at a false discovery rate <1% for peptide spectrum matches and protein groups. For calculation of z scores, the protein group ratios were logarithmized, and the standard deviation was estimated separately for ratios below and above 0 based on the 0.159 and 0.841 quantile (Cox and Mann, 2008).
Immunochemical methods and antibodies
Immunoblotting, IP, and Strep-Tactin pull-downs were performed as previously described (Poulsen et al., 2012). In brief, cells were lysed in EBC buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, and 0.5% NP-40) or denaturing buffer (20 mM Tris, pH 7.5, 50 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.5% NP-40, 0.5% sodium deoxycholate, and 0.5% SDS) supplemented with protease and phosphatase inhibitors and incubated on ice for 10 min, and lysates were cleared by centrifugation for 10 min at 20,000 rpm. Lysates were incubated with FLAG agarose (Sigma-Aldrich), GFP-Trap agarose (ChromoTek), or Strep-Tactin Sepharose (IBA BioTAGnology) for 1.5 h on an end-over-end rotator at 4°C, washed five times with EBC buffer or denaturing buffer, and resuspended in 2× Laemmli sample buffer.
Antibodies used in this study included mouse monoclonals to RNF111 (M05; Abnova), GFP (sc-9996) and β-actin (sc-130301; Santa Cruz Biotechnology, Inc.), and FLAG (F1804; Sigma-Aldrich), rat monoclonal to HA (Roche), and rabbit polyclonals to XPC (Bethyl Laboratories, Inc.), SUMO1 (ab32058), SUMO2/3 (ab3742), β-tubulin (ab6046; Abcam), and Ubc13 (4919; Cell Signaling Technology). Rabbit polyclonal RNF4 antibody was a gift of J. Palvimo (University of Eastern Finland, Kuopio, Finland).
Immunofluorescence staining, microscopy, and laser microirradiation
Cells were fixed in 4% formaldehyde, permeabilized with PBS containing 0.2% Triton X-100 for 5 min, and incubated with primary antibodies diluted in DMEM for 1 h at room temperature. After staining with secondary antibodies (Alexa Fluor 488 and 568; Life Technologies) for 30 min, coverslips were mounted in Vectashield mounting medium (Vector Laboratories) containing nuclear stain DAPI. Confocal images were acquired on a microscope (LSM 510; Carl Zeiss) mounted on a confocal laser-scanning microscope (Axiovert 100M; Carl Zeiss) equipped with Plan-Neofluar 40×/1.3 NA oil immersion objective. Dual-color confocal images were acquired with standard settings using laser lines 488 and 543 nm for excitation of Alexa Fluor 488 and Alexa Fluor 568 dyes (Molecular Probes/Invitrogen), respectively. Band pass filters 505–530 and 560–615 nm were used to collect the emitted fluorescence signals. Image acquisition and analysis was performed with LSM ZEN software (Carl Zeiss). Raw images were exported as TIF files, and if adjustments in image contrast and brightness were applied, identical settings were used on all images of a given experiment.
In vitro ubiquitylation, SUMOylation, and binding experiments
To analyze in vitro binding of RNF111 to SUMO, S-FLAG-Strep-RNF111 constructs were overexpressed in U2OS cells, purified on Strep-Tactin Sepharose, and incubated with purified free SUMO1, SUMO2, or poly-SUMO chains (3–8; all obtained from Boston Biochem) for 2 h at 4°C. Bound complexes were washed extensively in EBC buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, and 0.5% NP-40), and immobilized material was resolved by SDS-PAGE and analyzed by immunoblotting.
For in vitro RNF111 ubiquitylation assays, S-FLAG-Strep-RNF111 purified from cells as described in the previous section and incubated in 20 µl ubiquitylation assay buffer (50 mM Tris, pH 7.5, 5 mM MgCl2, 2 mM NaF, 2 mM ATP, and 0.6 mM DTT) supplemented with 60 ng E1, 300 ng E2 (Ubc13–Mms2 complex or UbcH5c), and 5 µg HA-ubiquitin (all obtained from Boston Biochem) for 1 h at 37°C. Reactions were stopped by addition of Laemmli sample buffer, resolved by SDS-PAGE, and immunoblotted with the HA antibody.
For in vitro SUMOylation and STUbL assays, XPC-GFP ectopically expressed in U2OS cells was captured on GFP-Trap resin and incubated with 100 ng SAE1/2, 200 ng Ubc9, and 3 µg SUMO2 (all obtained from Boston Biochem) in ubiquitylation assay buffer for 1 h at 37°C. The beads were washed extensively in EBC buffer and incubated with extracts of U2OS cells transfected with WT or mutant versions of S-FLAG-Strep-RNF111 for 2 h at 4°C. The immobilized material was then washed in EBC and processed for immunoblotting or subjected to in vitro ubiquitylation assay as described in the previous section.
For surface plasmon resonance analysis, recombinant His6-tagged fragments (WT and *SIM) of human RNF111 (encompassing amino acids 282–411) were expressed in Escherichia coli and purified on an ÄKTAxpress system (GE Healthcare). The His6 tag was removed with tobacco etch virus protease, and the RNF111 fragments were further purified using reverse-phase chromatography on an UltiMate 3000 system (Dionex), using C18 columns (Phenomenex). Eluted proteins were lyophilized, and their masses were verified by SDS-PAGE and MS. Poly-SUMO2 chains (3–8) were immobilized on a CM5 sensor chip using standard amine-coupling chemistry. Before titration experiments, the RNF111(282–411) fragments were dialyzed in running buffer (10 mM Hepes, pH 7.4, 150 mM NaCl, and 0.005% P20). After each titration point, the surface was regenerated using 10 mM glycine, pH 2.5. All data were collected on an instrument (T200; Biacore) at 25°C and analyzed using the T200 evaluation software (Biacore), in which the data were fitted to a steady-state model.
UDS and XPC-GFP accumulation kinetics assays
UDS was performed as described previously (Limsirichaikul et al., 2009; Schwertman et al., 2012). In brief, MEFs were seeded on coverslips 3 d before the UDS assay and cultured in medium without serum to reduce the number of S-phase cells. Cells were exposed to 16 J/m2 UV-C and labeled with 5-ethynyl,2′-deoxyuridine (EdU) for 3 h. Subsequently, cells were fixed with 3.7% formaldehyde, and EdU incorporation was visualized using Alexa Fluor 594 nm (Click-iT) according to manufacturer’s protocol (Invitrogen). UDS was quantified in ≥75 cells by measuring the overall nuclear fluorescence using ImageJ software (National Institutes of Health). Images were obtained using a microscope (LSM-700; Carl Zeiss).
Kinetic study of XPC-GFP accumulation was performed in SV40-transformed XP4PA cells stably expressing XPC-GFP as described previously (Dinant et al., 2007). In brief, cells were cultured on 25-mm quartz coverslips (SPI Supplies) and imaged on a laser-scanning confocal microscope (SP5; Leica) using an Ultrafluar quartz 100×, 1.35 NA glycerol immersion lens (Carl Zeiss) at 37°C and 5% CO2. Imaging medium was the same as culture medium. For UV laser irradiation, a 2-mW pulsed (7.8 kHz) diode pumped solid-state laser emitting at 266 nm (DPSL; Rapp OptoElectronic) was connected to the microscope (SP5) with all-quartz optics. Treated nuclei were imaged using the same scanning speed, zoom factor, and laser power. Images were acquired using the LAS AF software (Leica). Data analysis was performed using the ImageJ software package. Measured fluorescence levels were determined in the specific region of the damage in the nucleus over time and corrected for background values. Resulting curves show the relative amount of protein in the damaged area over time and were normalized to 1 for the data points before damage.
Online supplemental material
Fig. S1 shows MS-based analysis of RNF111-interacting proteins. Fig. S2 shows analysis of the interplay between RNF111 and E2 ubiquitin–conjugating enzymes. Fig. S3 shows analysis of RNF111-dependent ubiquitylation of XPC in response to UV. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201212075/DC1.
Supplementary Material
Acknowledgments
We thank Drs. V. Episkopou and J. Palvimo for providing reagents.
This work was supported by the Novo Nordisk Foundation, Danish Medical Research Council, Danish Cancer Society, Netherlands Organisation for Health Research and Development TOP grant (912.08.031), and the Lundbeck Foundation.
Footnotes
Abbreviations used in this paper:
- EdU
- 5-ethynyl,2′-deoxyuridine
- IP
- immunoprecipitation
- MEF
- mouse embryonic fibroblast
- MS
- mass spectrometry
- NER
- nucleotide excision repair
- SILAC
- stable isotope labeling by amino acids in cell culture
- SIM
- SUMO-interacting motif
- STUbL
- SUMO-targeted ubiquitin ligase
- SUMO
- small ubiquitin-like modifier
- UDS
- UV-induced DNA repair synthesis
- WT
- wild type
References
- Al-Hakim A., Escribano-Diaz C., Landry M.C., O’Donnell L., Panier S., Szilard R.K., Durocher D. 2010. The ubiquitous role of ubiquitin in the DNA damage response. DNA Repair (Amst.). 9:1229–1240 10.1016/j.dnarep.2010.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker-Jensen S., Mailand N. 2011. The ubiquitin- and SUMO-dependent signaling response to DNA double-strand breaks. FEBS Lett. 585:2914–2919 10.1016/j.febslet.2011.05.056 [DOI] [PubMed] [Google Scholar]
- Bekker-Jensen S., Rendtlew Danielsen J., Fugger K., Gromova I., Nerstedt A., Lukas C., Bartek J., Lukas J., Mailand N. 2010. HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat. Cell Biol. 12:80–86 10.1038/ncb2008 [DOI] [PubMed] [Google Scholar]
- Bergink S., Jentsch S. 2009. Principles of ubiquitin and SUMO modifications in DNA repair. Nature. 458:461–467 10.1038/nature07963 [DOI] [PubMed] [Google Scholar]
- Chen Z.J., Sun L.J. 2009. Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell. 33:275–286 10.1016/j.molcel.2009.01.014 [DOI] [PubMed] [Google Scholar]
- Cox J., Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26:1367–1372 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., Mann M. 2011. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10:1794–1805 10.1021/pr101065j [DOI] [PubMed] [Google Scholar]
- Danielsen J.M., Sylvestersen K.B., Bekker-Jensen S., Szklarczyk D., Poulsen J.W., Horn H., Jensen L.J., Mailand N., Nielsen M.L. 2011. Mass spectrometric analysis of lysine ubiquitylation reveals promiscuity at site level. Mol. Cell. Proteomics. 10:M110.003590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielsen J.R., Povlsen L.K., Villumsen B.H., Streicher W., Nilsson J., Wikström M., Bekker-Jensen S., Mailand N. 2012. DNA damage–inducible SUMOylation of HERC2 promotes RNF8 binding via a novel SUMO-binding Zinc finger. J. Cell Biol. 197:179–187 10.1083/jcb.201106152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinant C., de Jager M., Essers J., van Cappellen W.A., Kanaar R., Houtsmuller A.B., Vermeulen W. 2007. Activation of multiple DNA repair pathways by sub-nuclear damage induction methods. J. Cell Sci. 120:2731–2740 10.1242/jcs.004523 [DOI] [PubMed] [Google Scholar]
- Episkopou V., Arkell R., Timmons P.M., Walsh J.J., Andrew R.L., Swan D. 2001. Induction of the mammalian node requires Arkadia function in the extraembryonic lineages. Nature. 410:825–830 10.1038/35071095 [DOI] [PubMed] [Google Scholar]
- Galanty Y., Belotserkovskaya R., Coates J., Jackson S.P. 2012. RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev. 26:1179–1195 10.1101/gad.188284.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gareau J.R., Lima C.D. 2010. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 11:861–871 10.1038/nrm3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golebiowski F., Matic I., Tatham M.H., Cole C., Yin Y., Nakamura A., Cox J., Barton G.J., Mann M., Hay R.T. 2009. System-wide changes to SUMO modifications in response to heat shock. Sci. Signal. 2:ra24 10.1126/scisignal.2000282 [DOI] [PubMed] [Google Scholar]
- Hecker C.M., Rabiller M., Haglund K., Bayer P., Dikic I. 2006. Specification of SUMO1- and SUMO2-interacting motifs. J. Biol. Chem. 281:16117–16127 10.1074/jbc.M512757200 [DOI] [PubMed] [Google Scholar]
- Hoogstraten D., Bergink S., Ng J.M., Verbiest V.H., Luijsterburg M.S., Geverts B., Raams A., Dinant C., Hoeijmakers J.H., Vermeulen W., Houtsmuller A.B. 2008. Versatile DNA damage detection by the global genome nucleotide excision repair protein XPC. J. Cell Sci. 121:2850–2859 10.1242/jcs.031708 [DOI] [PubMed] [Google Scholar]
- Kerscher O., Felberbaum R., Hochstrasser M. 2006. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22:159–180 10.1146/annurev.cellbio.22.010605.093503 [DOI] [PubMed] [Google Scholar]
- Koinuma D., Shinozaki M., Komuro A., Goto K., Saitoh M., Hanyu A., Ebina M., Nukiwa T., Miyazawa K., Imamura T., Miyazono K. 2003. Arkadia amplifies TGF-beta superfamily signalling through degradation of Smad7. EMBO J. 22:6458–6470 10.1093/emboj/cdg632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D., Rape M. 2012. The ubiquitin code. Annu. Rev. Biochem. 81:203–229 10.1146/annurev-biochem-060310-170328 [DOI] [PubMed] [Google Scholar]
- Levy L., Howell M., Das D., Harkin S., Episkopou V., Hill C.S. 2007. Arkadia activates Smad3/Smad4-dependent transcription by triggering signal-induced SnoN degradation. Mol. Cell. Biol. 27:6068–6083 10.1128/MCB.00664-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limsirichaikul S., Niimi A., Fawcett H., Lehmann A., Yamashita S., Ogi T. 2009. A rapid non-radioactive technique for measurement of repair synthesis in primary human fibroblasts by incorporation of ethynyl deoxyuridine (EdU). Nucleic Acids Res. 37:e31 10.1093/nar/gkp023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matic I., Schimmel J., Hendriks I.A., van Santen M.A., van de Rijke F., van Dam H., Gnad F., Mann M., Vertegaal A.C. 2010. Site-specific identification of SUMO-2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Mol. Cell. 39:641–652 10.1016/j.molcel.2010.07.026 [DOI] [PubMed] [Google Scholar]
- Mavrakis K.J., Andrew R.L., Lee K.L., Petropoulou C., Dixon J.E., Navaratnam N., Norris D.P., Episkopou V. 2007. Arkadia enhances Nodal/TGF-beta signaling by coupling phospho-Smad2/3 activity and turnover. PLoS Biol. 5:e67 10.1371/journal.pbio.0050067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalski A., Damoc E., Hauschild J.P., Lange O., Wieghaus A., Makarov A., Nagaraj N., Cox J., Mann M., Horning S. 2011. Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol. Cell. Proteomics. 10:M111. 011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazono K., Koinuma D. 2011. Arkadia—beyond the TGF-β pathway. J. Biochem. 149:1–3 10.1093/jb/mvq133 [DOI] [PubMed] [Google Scholar]
- Nagano Y., Mavrakis K.J., Lee K.L., Fujii T., Koinuma D., Sase H., Yuki K., Isogaya K., Saitoh M., Imamura T., et al. 2007. Arkadia induces degradation of SnoN and c-Ski to enhance transforming growth factor-beta signaling. J. Biol. Chem. 282:20492–20501 10.1074/jbc.M701294200 [DOI] [PubMed] [Google Scholar]
- Nishi R., Alekseev S., Dinant C., Hoogstraten D., Houtsmuller A.B., Hoeijmakers J.H., Vermeulen W., Hanaoka F., Sugasawa K. 2009. UV-DDB-dependent regulation of nucleotide excision repair kinetics in living cells. DNA Repair (Amst.). 8:767–776 10.1016/j.dnarep.2009.02.004 [DOI] [PubMed] [Google Scholar]
- Ong S.E., Blagoev B., Kratchmarova I., Kristensen D.B., Steen H., Pandey A., Mann M. 2002. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics. 1:376–386 10.1074/mcp.M200025-MCP200 [DOI] [PubMed] [Google Scholar]
- Poulsen M., Lukas C., Lukas J., Bekker-Jensen S., Mailand N. 2012. Human RNF169 is a negative regulator of the ubiquitin-dependent response to DNA double-strand breaks. J. Cell Biol. 197:189–199 10.1083/jcb.201109100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povlsen L.K., Beli P., Wagner S.A., Poulsen S.L., Sylvestersen K.B., Poulsen J.W., Nielsen M.L., Bekker-Jensen S., Mailand N., Choudhary C. 2012. Systems-wide analysis of ubiquitylation dynamics reveals a key role for PAF15 ubiquitylation in DNA-damage bypass. Nat. Cell Biol. 14:1089–1098 10.1038/ncb2579 [DOI] [PubMed] [Google Scholar]
- Prudden J., Pebernard S., Raffa G., Slavin D.A., Perry J.J., Tainer J.A., McGowan C.H., Boddy M.N. 2007. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 26:4089–4101 10.1038/sj.emboj.7601838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sands A.T., Abuin A., Sanchez A., Conti C.J., Bradley A. 1995. High susceptibility to ultraviolet-induced carcinogenesis in mice lacking XPC. Nature. 377:162–165 10.1038/377162a0 [DOI] [PubMed] [Google Scholar]
- Schwartz D.C., Felberbaum R., Hochstrasser M. 2007. The Ulp2 SUMO protease is required for cell division following termination of the DNA damage checkpoint. Mol. Cell. Biol. 27:6948–6961 10.1128/MCB.00774-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwertman P., Lagarou A., Dekkers D.H., Raams A., van der Hoek A.C., Laffeber C., Hoeijmakers J.H., Demmers J.A., Fousteri M., Vermeulen W., Marteijn J.A. 2012. UV-sensitive syndrome protein UVSSA recruits USP7 to regulate transcription-coupled repair. Nat. Genet. 44:598–602 10.1038/ng.2230 [DOI] [PubMed] [Google Scholar]
- Sugasawa K., Okuda Y., Saijo M., Nishi R., Matsuda N., Chu G., Mori T., Iwai S., Tanaka K., Tanaka K., Hanaoka F. 2005. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell. 121:387–400 10.1016/j.cell.2005.02.035 [DOI] [PubMed] [Google Scholar]
- Sun H., Hunter T. 2012. Poly-small ubiquitin-like modifier (PolySUMO)-binding proteins identified through a string search. J. Biol. Chem. 287:42071–42083 10.1074/jbc.M112.410985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Leverson J.D., Hunter T. 2007. Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 26:4102–4112 10.1038/sj.emboj.7601839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatham M.H., Jaffray E., Vaughan O.A., Desterro J.M., Botting C.H., Naismith J.H., Hay R.T. 2001. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276:35368–35374 10.1074/jbc.M104214200 [DOI] [PubMed] [Google Scholar]
- Wang Q.E., Zhu Q., Wani G., El-Mahdy M.A., Li J., Wani A.A. 2005. DNA repair factor XPC is modified by SUMO-1 and ubiquitin following UV irradiation. Nucleic Acids Res. 33:4023–4034 10.1093/nar/gki684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Seifert A., Chua J.S., Maure J.F., Golebiowski F., Hay R.T. 2012. SUMO-targeted ubiquitin E3 ligase RNF4 is required for the response of human cells to DNA damage. Genes Dev. 26:1196–1208 10.1101/gad.189274.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.