Abstract
Phospholipid-esterified oxylipins are newly described families of bioactive lipids generated by lipoxygenases in immune cells. Until now, assays for their quantitation were not well developed or widely available. Here, we describe a mass spectrometric protocol that enables accurate measurement of several, in particular hydro(pero)xyeicosatetraenoic acids (H(p)ETEs), hydroxyoctadecadienoic acids (HODEs), hydroxydocosahexaenoic acids (HDOHEs) and keto-eicosatetraenoic acids (KETEs), attached to either phosphatidylethanolamine (PE) or phosphatidylcholine (PC). Lipids are isolated from cells or tissue using a liquid phase organic extraction, then analyzed by HPLC-tandem mass spectrometry (LC/MS/MS) in multiple reaction monitoring (MRM) mode. The protocol can simultaneously monitor up to 23 different species. Generation of standards takes 2 days approximately. Following this, extraction of 30 samples takes approximately 3 hrs, with LC/MS/MS run time of 50 min per sample.
Keywords: Lipid biochemistry, eicosanoid, phospholipid, mass spectrometry, macrophage, neutrophil, platelet
INTRODUCTION
Eicosanoids and related oxylipins have long been known as important free acid signalling molecules. They are generated by three enzyme families, cyclooxygenase (COX), lipoxygenase (LOX) and cytorochrome P450 (CYP), and signal in health and disease in all organs1. More recently, they have been shown to form more complex molecules, where the free acid is esterified to a larger group via the carboxyl terminus2-5. For example, we identified families of phospholipid (PL)-esterified HETEs generated by circulating immune cells and resident macrophages2-5. Each cell type generates 4 - 6 distinct molecular species including both PE and PC derivatives (Figure 1). Positional isomers formed exclusively depend on the lipoxygenase (LOX) isoform expressed, with platelets, neutrophils and monocytes generating 12-, 5- and 15-HETE-containing lipids respectively2,3,5(and unpublished data). Related oxylipins that contain either hydroperoxyeicosatetraenoic acid (HpETE-PLs), oxidized docosahexanoic acid (hydroxydocosahexaenoic acid, HDOHE-PLs), oxidized linoleate (hydroxyoctadecadienoic acid, HODE-PLs) or keto derivatives (keto-eicosatetraenoic acid, KETE-PLs) have also been detected in platelets and monocytes(unpublished data). The lipids tend to stay membrane associated and preliminary studies indicate that their signalling actions occur at or close to the plasma membrane. For example, 12-HETE-PC enhances activity of coagulation proteins, while HETE-PEs suppress activation of the LPS receptor, Toll-like receptor 4 (TLR4) in human monocytes3,5. Generally, these lipids are generated in response to agonist stimulation, and so are absent in healthy primary immune cells. One exception is the family of 12/15-LOX-derived 12-HETE-PEs detected in murine peritoneal macrophages. These are present at high concentrations (~1 ng/lavage) basally, but disappear during bacterial infection, returning again during resolution3. In contrast, they are actively generated in a murine lung allergy model, coinciding with the timepoint of IL-4/13 generation and eosinophil influx3. Esterified 5-HETE has also been detected in vivo during human and murine bacterial peritonitis, paralleling neutrophil influx and activation(unpublished data).
Figure 1. Structures of some of the molecular species of oxidized phospholipids acutely generated by human and murine immune cells.
Thus, esterified oxylipins are a new class of signalling lipid generated in vivo, and as such standardized methods to study them are not yet available. Sensitive and specific assays for their identification, characterization and quantitation are required to facilitate research into their generation and action in mammalian cells. In particular, in studies on inflammation and immune disorders, where they might impact on the nature and progression of the response. This paper describes protocols for direct quantitation of these lipids in extracts from cells or tissue. These can be used for in vitro or in vivo analysis of their generation during human or animal disease progression, including studies on their mechanism of action. Furthermore, protocols for synthesis of standards provide methods for generation and purification of sufficient amounts for undertaking biological studies.
Free eicosanoids are also analyzed using LC/MS/MS approaches, however due to the difference in structure of these new molecules, methods for their analysis differ considerably in terms of extraction procedure, LC solvent/gradient composition, and MS/MS transitions monitored-9. Other methods for lipid analysis including antibody-based methods (e.g. ELISA) or HPLC-UV are not suitable for esterified oxylipins. ELISAs require specific antibodies and due to the structural similarity of these lipids with other phospholipids including unoxidized species, it is unlikely that antibodies that would discriminate sufficiently could be generated. In the case of LC-UV, the conjugated dienes and keto groups absorb in the low UV range (235-280 nm), however sensitivity/selectivity is insufficient for their analysis in complex extracts from cells or tissue.
The methods outlined require primary standards that are generated as part of the procedure. Two strategies are employed for generation of hydroxy- or hydroperoxylipid-containing standards. (i) Single positional isomers are synthesised for some species (e.g. 15-H(p)ETE-PL, 13-H(p)ODE-PL) using soybean lipoxygenase (modified from10). The enzymatic approach is not of use for generating other positional isomers since commercially available plant LOXs, or recombinant mammalian isoforms that oxidise at other sites are not suitable (low activity, or generate more than one positional isomer). (ii) For other positional isomers, we use an air oxidation method that yields defined purified mixtures (modified from11). In this, air oxidation (room air) in the presence of the vitamin E analog, pentamethylchromanol (PMC), gives a good yield of primary hydroperoxide products. The inclusion of PMC promotes reduction of lipid peroxyl radicals to hydroperoxides, and minimizes hydroperoxide decomposition, which can lead to lower yield and contamination with decomposition products. With both methods, the initial product is the lipid hydroperoxide, which can be reduced to the hydroxide using tin chloride (SnCl2), if desired. This is then purified from unoxidized parent material and decomposition products using HPLC. For KETE-PLs, we oxidize HETE-PLs using Dess-Martin periodinane (DMP). All standards are purified using HPLC, to remove secondary oxidation products and unoxidized substrate phospholipid.
Extraction of esterified eicosanoids from cells or tissue requires care to ensure that artifactual oxidation does not occur during tissue processing. Precautions include low temperature and addition of antioxidants and metal chelators. These recommended steps are detailed herein.
PRINCIPLES
The assay is based on the use of LC/MS/MS in MRM mode. Lipids are extracted from samples of tissue culture supernatant or other biological fluids, cells, or tissue homogenates, using a hexane:isopropanol:acetic acid liquid extraction procedure. Just prior to extraction, internal standards are added to all samples. These are phospholipids of the same class, that are not normally present. Typically dimyristoyl-phosphatidylethanolamine (DMPE) and dimyristoyl-phosphatidylcholine (DMPC) are used for PE and PC, respectively, with both added simultaneously as a mixture to each sample. For studies on immune cells in vitro, at least triplicate samples are analyzed, although with human or murine tissue, more may be required for sufficient power. Lipid extracts are then analyzed using reverse phase LC/MS/MS, monitoring for parent → daughter transitions that are specific for the molecule of interest. We use a Luna (Phenomenex) column at 200 μl/min with a methanol/acetonitrile/ammonium acetate gradient. Long separation times (50 min) are needed as these complex mixtures contain many lipids with the same molecular weight (isobaric), and retention time is important for accurate identification. Molecular species are identified based on both retention time and MRM transition. For some molecules, daughter ions specific for positional isomers of eicosanoids can be utilized (e.g. m/z 115 for the 5-HETE-PE positional isomer). Quantitation is then achieved using standard curves generated using synthetic primary standards. Methods for generation of these are provided in this protocol. Alternatively, they are available for research purposes on a collaborative basis from our group, including all positional isomers of HETEs, HODEs, HDOHEs or 15-KETE attached to either PC or PE at sn2 (carbon 2 of the glycerol moiety). Contact the corresponding author (o-donnellvb@cardiff.ac.uk) for further information. Some of these are generated as purified single standards and others as defined mixtures of positional isomers.
In our laboratory, we use an Applied Biosystems 4000 Q-Trap in triple quadrupole mode. In practice the methods can be applied to any tandem mass spectrometer, although older instruments may be less sensitive. This should not generally be a problem however, since platelets, neutrophils and monocytes generate relatively large amounts of these lipids. Careful tuning and optimization of instruments using purified standards is always recommended to ensure maximum sensitivity for detection and should be conducted according to instrument manufacturers’ procedures. This includes determination of appropriate collision energies for fragmentation. The protocols would not be suited to a single quad instrument since both selectivity and sensitivity would be significantly compromised, due to the presence of co-eluting isobaric lipids. A good technical level of expertise in LC/MS/MS and HPLC separation methodologies would be essential for the successful set-up and use of these protocols.
MATERIALS
Reagents
di-14:0-PE (Avanti Polar Lipids, Alabaster, AL, cat. no. 850745)
di-14:0-PC (Avanti Polar Lipids, Alabaster, AL, cat. no. 850345)
16:0/20:4-PC (Avanti Polar Lipids, Alabaster, AL, cat. no. 850459)
18:0/20:4-PE (Avanti Polar Lipids, Alabaster, AL, cat. no. 850804)
18:0/22:6-PE (Avanti Polar Lipids, Alabaster, AL, cat. no. 850806)
16:0/18:2-PE (Avanti Polar Lipids, Alabaster, AL, cat. no. 850756)
Soybean lipoxygenase, type V (Sigma-Aldrich, cat. no. L6632)
Hexane, 2-propanol, acetonitrile, methanol, chloroform, water, all HPLC grade (Fisher Scientific). <CAUTION> organic solvents need to be handled in a fume hood and stored in appropriately vented cabinets.
Discovery C18 column (25 cm c 4.6 mm, 5 μm) (Fisher Scientific, cat. no. 504971)
Spherisorb ODS2 column (15 cm × 4.6 mm, 5 μm) (Waters, cat. no. PSS839853)
Luna C18 (2) column (15 cm × 2 mm, 3 μm) (Phenomenex, cat. no. 00F-4251-B0)
Free acid eicosanoid standards all from Cayman Chemical: 15-HETE (cat. no. 34720), 12-HETE (34570), 5-HETE (34230), 11-HETE (34500), 8-HETE (34340), 20-HDOHE (33750), 17-HDOHE (33650), 14-HDOHE (33550), 11-HDOHE (33450), 8-HDOHE (33350), 16-HDOHE (33600), 13-HDOHE (33500), 10-HDOHE (33400), 7-HDOHE (33300), 4-HDOHE (33200), 13-HODE (38600), 9-HODE (38400).
Dess-Martin Periodinane, 97%. <CRITICAL> Allow bottle to fully warm to room temperature before opening (Sigma Aldrich, cat. no. 274623)
Anhydrous dichloromethane (Sigma Aldrich, cat. no. 270997)
2,2,5,7,8-Pentamethyl-6-chromanol (Sigma Aldrich, cat. no. 430676) <CRITICAL> Make up fresh solutions in water on the day of use.
Stannous chloride (Sigma-Aldrich, cat. no. 204722) <CRITICAL> Make up fresh solutions in water on the day of use.
Butylated hydroxytoluene (Sigma-Aldrich, W218405) <CRITICAL> Make up fresh solutions in water on the day of use.
Equipment Setup
Protective equipment
<CAUTION> Use gloves and lab coat when working with biological samples, and ensure that work using solvents is carried out in a fume hood.
LC/MS/MS system
Mass spectrometry was carried out using an Applied Biosystems 4000 Q-Trap mass spectrometer operating in MRM mode. Ionisation was by electrospray, utilising the Turbo V probe. HPLC separation utilised a Shimadzu SIL-HT autosampler, online degasser, and LC-10ADVPμ binary pump system. A column oven operating at 22° was used, although this is not essential. Data was acquired and analyzed using Analyst 1.2. Before analysis of any new compound, it is essential that the instrument is tuned according to the manufacturers’ protocols. On our instrument, collision energies of −40 to −50 V are typically required for MS/MS of oxidized phospholipids.
Since phospholipids are especially “sticky” molecules, it is important to ensure that extensive washing steps are used between sample injections. On our instrument, we employ a 1 min rinse at 35 μl/sec both before and after aspiration, and our needle is teflon-coated for low carryover. It is also essential to run methanol blanks at defined periods as needed (e.g. between every 10-20 samples) to confirm that carryover is not taking place. Methanol blanks are also run at the start and end of all batches. A further check can be made by subjecting cell-free buffers to extraction, alongside cell samples and analyzing these for the presence of the lipids.
When analyzing concentrated samples, e.g. standards, it is important to make serial dilutions of standards and start with the lowest concentration, otherwise contamination of the lines/source can occur. We find that the best way to remove contamination with phospholipid standards is with a 20 min wash with chrlorform:methanol (50:50) at 1 ml/min. However, due to the corrosive nature of chloroform, it is important to purge all lines immediately after carrying out this cleaning procedure.
HPLC system
HPLC with UV detection was carried out using a Gilson HPLC system comprising 811D dynamic mixer, 306 pumps, 805 manometric module, and an Agilent 1100 series UV detector. No column oven was used. Data was acquired and analyzed using Uvikon.
Glassware
When working with organic solvents, clean glassware is essential. Keep beakers and cylinders aside from general laboratory use, and clean by rinsing with HPLC grade solvents (water, methanol, chloroform, in that order) and leave to dry. Do not use glassware that has been cleaned using detergents. Extraction tubes (from Chromacol, catalogue number: 10-SV T928) and glass pasteur pipettes are single use only, but tube lids can be recycled by rinsing well with methanol and chloroform. Defined volumes of solvents are aliquoted into tubes using bottle top dispensers (Fisher Chemical Company) that provide an efficient and safe method for working with organic solvents.
Drying Techniques
When evaporating small amounts of solvent, two methods can be used. A dryblock (Techne DB.3) set a maximum 30 °C with either nitrogen or argon gas will evaporate multiple samples. Alternatively we generally use the RapidVap system (Labconco), a shaking vacuum dryer and tends to evaporate faster than blowing with inert gas.
PROCEDURES
Synthesis of primary standards for quantitation (approximately 2 days per procedure)
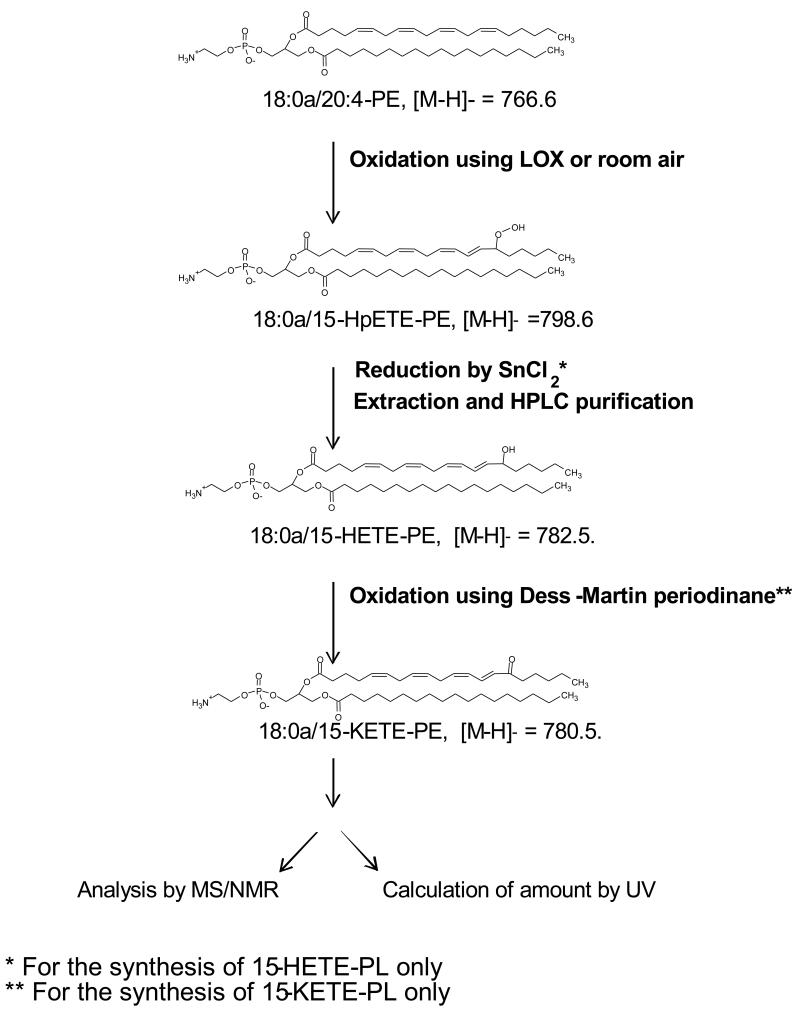
1| Prepare the required standards as described in Options A, B or C. Option A describes generation of purified esterified H(p)ETEs or H(p)ODEs using soybean LOX oxidation of substrates, resulting in single positional isomers, specifically 15-H(p)ETE-PL or 13-H(p)ODE-PL. Option B is for the generation and purification of positional isomer mixtures of H(p)ETE-, H(p)ODE- or H(p)DOHE-PE or -PC standards. Positional isomers do not separate on reverse phase HPLC sufficiently for integration, thus, they are used as a mixture of known composition. The total amount of these standards is determined using absorbance with ε = 28 mM−1,cm−1, with the proportion of each individual positional isomer determined after saponification and quantitation of relative amounts of free eicosanoid using authentic standards. This then allows calculation of the ng amount of each positional isomer in the mixture. In Option C, KETE-PLs are generated by oxidation of the corresponding HETE-PLs (either single or mixed positional isomers). Fresh Dess-Martin periodinane (DMP, Sigma Aldrich, Poole, UK) and anhydrous dichloromethane are essential for successful reaction12. The synthesis scheme is summarized in Figure 2, and NMR and MS/MS data is provided in Figures and Tables in the Supplementary Data section.
Figure 2. Summarized scheme for synthesis of standards.
A. Generation and purification of 15S-H(p)ETE-PL or 13S-H(p)ODE-PL
i. Dry purified phospholipid (e.g. 18:0a/20:4-PE, 16:0a/20:4-PC, or containing 18:2 at sn2, from Avanti Polar lipids) under inert gas (blow with nitrogen or argon using a glass pasteur pipette in a 50 ml Falcon tube), then resuspend in 26 ml buffer (10 mM deoxycholate, 0.2 M borate buffer, pH 9.0) to give a final concentration of 1 mM lipid. Transfer to a large glass (e.g. 250 ml) conical/Erlenmeyer flask to give a large surface area.
ii. Add soybean 15-LOX type V or IV (5.2 KU/ml) and incubate for 30 min at room temperature with stirring and bubbling with 95% O2. If desired, reduce the hydroperoxides to corresponding alcohols by addition of 1 mM SnCl2 for 10 min at room temperature (261 μl of 19 mg/ml in water) after oxidation by LOX.
<CRITICAL STEP> Bubbling with O2 is essential, since its depletion will cause hydroperoxidase activity of LOX, with degradation of the hydroperoxide product.
iii. Extract the lipids by adding 65 ml lipid extraction solvent (1 M acetic acid, 2-propanol, hexane (2:20:30, v/v/v)) to the sample, then vortex for 1 min. Then add 65 ml hexane and vortex again. Work in a fume hood.
iv. Decant as much of the upper hexane layer as possible into a conical flask, then pour the rest (aqueous phase) into clean glass tubes and centrifuge at low speed for 5 min. Using a glass pasteur pipette, recover the hexane (upper phase) and add to what is already decanted. Then pour the aqueous phase back into the conical flask for a second extraction.
v. Add 65 ml hexane to the aqueous phase, and vortex. Repeat the extraction and removal of hexane upper layer as described above. Combine all hexane fractions into the conical flask and dry under a stream of inert gas (nitrogen or argon supplied via a glass pasteur pipette). Keeping the flask warm by placing in a dryblock at max 30 °C aids evaporation. Periodically swirl to recover lipids that have dried onto the sides of the vessel and once the sample is less than 4 ml, transfer to smaller glass vial to finish drying. Alternatively, a separatory funnel may be useful for extraction with these large volumes, if available.
vi. Resuspend the lipid in 0.5 ml methanol and store at −80 °C (under inert gas) until purification.
<PAUSE POINT> The crude mixture can be stored overnight before purification, at −80° under inert gas.
vii. Purify using reverse phase HPLC using a Discovery C18 column (25 cm × 4.6 mm, 5 um) at 1 ml/min, and gradient of 50 % to 100 % mobile phase B (A: water, 5 mM ammonium acetate, B: methanol, 5 mM ammonium acetate) over 15 min, then held at 100 % B for 20 min. Elution is monitored at 205 nm (unoxidized lipid) and 235 nm (HETE-PL).
viii. Oxidized phospholipids typically elute at 22-24 min, and the 15-HETE- or 13-HODE-PL will elute as a single peak (Figure 3). For both, the S:R ratio will typically be around 9:1, characteristic of the enantiomeric specificity of soybean LOX (not shown). Collect into a glass vial, dry under inert gas or using a RapidVap and resuspend in a small volume of methanol (e.g. 100 μl).
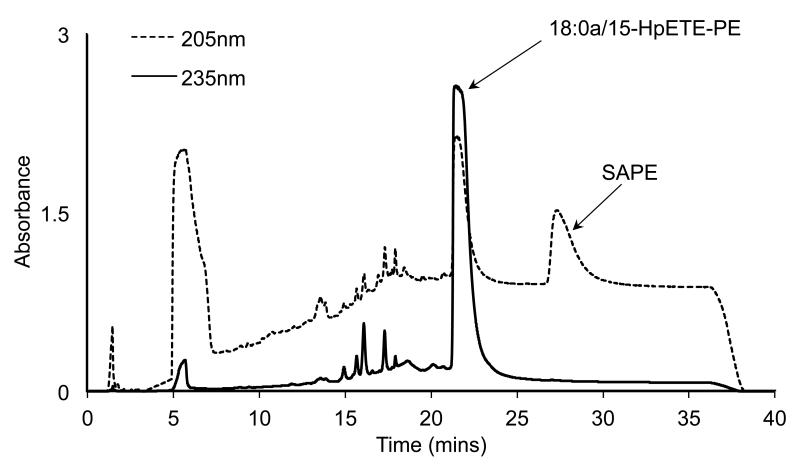
Figure 3. Purification of 18:0a/15-HpETE-PE using reverse phase HPLC-UV.
SAPE was oxidized using soybean LOX as described, then lipids extracted and 18:0a/15-HpETE-PE purified using reverse phase HPLC, with monitoring at 235 nm (product) and 205 nm (substrate). The esterified eicosanoid elutes around 21-22 min.
ix. Quantify the amount of esterified eicosanoid by absorbance at 235 nm of the purified lipid mixture using the extinction coefficient, ε = 28 mM−1,cm−1, and store at −80 °C under N2 in methanol13. For this, dilute a small amount into methanol and scan in a quartz cuvette from 200-400 nm using a scanning spectrophotometer.
<PAUSE POINT> Reduced hydroxy-PL standards are stable for several months, although we find that concentrations gradually decrease due to adsorption to the glass vial. In contrast, hydroperoxides are not stable and should be used within 1-2 weeks of generation. Stability may be enhanced by addition of 100 μM BHT to the storage vial.
B Generation and purification of positional isomer mixtures of H(p)ETE-, H(p)ODE- or H(p)DOHE-PE or -PC standards
i. Add 64 μl 10 mM pentamethylchromanol (PMC) in chloroform to 5 mg phospholipid (e.g. 16:0a/20:4-PC, 18:0a/20:4-PE, or with 18:2 or 22:6 at sn2) in 1.5 ml chloroform, then dry under N2 in a clean glass tube.
ii. Incubate the sample at 37 °C for 24 hrs then resuspend in 200 μl methanol containing 20 mg/ml butylated hydroxytoluene (BHT).
iii. Reduce hydroperoxides using SnCl2 if desired as described in step 1A(ii).
iv. Extract lipids by addition of 800 μl methanol, 2 ml chloroform, 0.5 ml water, vortex, then centrifuge to separate the layers. Remove the bottom layer using a glass pasteur pipette, then dry and resuspend in 200 μl methanol.
<PAUSE POINT> The crude mixture can be stored overnight before purification, at −80° under inert gas.
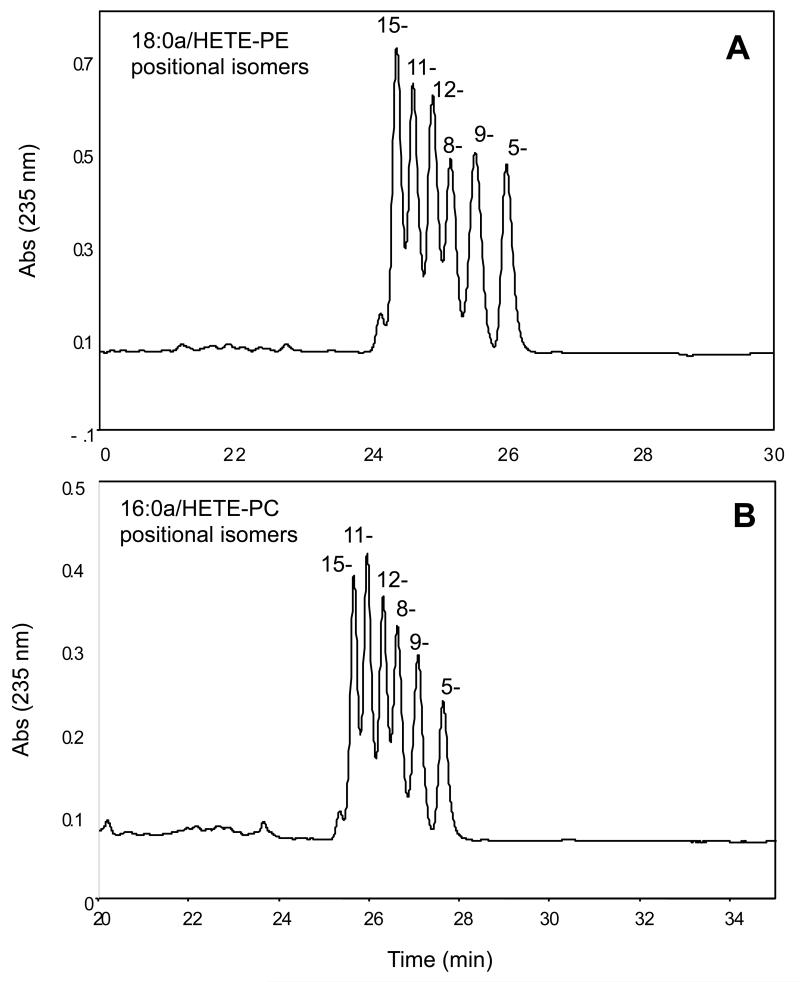
v. Purify the esterified eicosanoids using reverse phase HPLC as described in 1A(vii), monitoring absorbance at 205 nm (parent) and 235 nm (conjugated diene of esterified eicosanoid). At this stage, H(p)ETE-PLs will appear at 235 nm as 6 unresolved peaks each representing a single positional isomer (Figure 4). HODE-PLs and HDOHE-PLs contain 2 and 10 separate positional isomers, respectively, however these are not easily resolved using LC-UV.
Figure 4. Purification of 18:0a/15-HETE-PE using reverse phase HPLC-UV.
SAPE was oxidized using air oxidation, then reduced using SnCl2. Panel A. Chromatogram showing elution of the six 18:0a/15-HETE-PE positional isomers during purification. Panel B. Chromatogram showing elution of the six 16:0a/15-HETE-PC positional isomers during purification.
vi. Total HETE-PLs in each standard preparation is quantified by absorbance at 235 nm, using ε1mm,1cm = 28 au. For this, dilute a small amount into methanol and scan in a quartz cuvette from 200-400 nm using a scanning spectrophotometer. The relative amounts of each isomer are then determined the analysis of the free acids following saponification.
vii. Saponification Remove a small amount of standard (10 μl) and suspend in 1.5 ml of propan-2-ol, and add 1.5 ml 0.2 M NaOH and incubate at 60°C for 30 min under N2.
viii. Acidify to pH 3.0 with 0.5 M HCl, and add 3 ml hexane, vortex, and centrifuge at 260 g for 5 min.
ix. Remove upper layer and re-extract using 3 ml hexane and combine hexane extracts.
x. Dry hexane extracts, resuspend in 100 μl MeOH and store under N2 at −80°C until analysis by LC/MS/MS for free eicosanoids (below).
<PAUSE POINT> Samples can be stored for several weeks if have been reduced to hydroxylipids, but hydroperoxy-standards should be analyzed within a few days.
xi. Analysis of positional isomers as free acids Separate free eicosanoid positional isomers, using a Spherisorb ODS2, 5 mm, 150 × 4.6 mm column (Waters Ltd, Herts, UK) with a gradient of 50 – 90 % B over 15 min (A, water:acetonitrile:acetic acid, 75:25:0.1; B, methanol:acetonitrile:acetic acid, 60:40:0.1) and a flow rate of 1 ml/min. Elution is monitored using MS/MS transitions listed in Table 1.
Table 1. Precursor and product ions of HETEs, HODEs and HDOHE positional isomers analyzed using LC/MS/MS.
| Eicosanoid | Precursor m/z | Product m/z |
|---|---|---|
| 15-HETE | 319.2 | 219.1 |
| 12-HETE | 319.2 | 179.1 |
| 5-HETE | 319.2 | 115.1 |
| 11-HETE | 319.2 | 167.1 |
| 8-HETE | 319.2 | 155.1 |
| 20-HDOHE | 343.2 | 187.2 |
| 17-HDOHE | 343.2 | 273.2 |
| 14-HDOHE | 343.2 | 205.2 |
| 11-HDOHE | 343.2 | 149.2 |
| 8-HDOHE | 343.2 | 109.2 |
| 16-HDOHE | 343.2 | 233.2 |
| 13-HDOHE | 343.2 | 193.2 |
| 10-HDOHE | 343.2 | 153.2 |
| 7-HDOHE | 343.2 | 141.2 |
| 4-HDOHE | 343.2 | 101.2 |
| 13-HODE | 295.2 | 171.1 |
| 9-HODE | 295.2 | 195.1 |
xii. Quantitate using standard curves generated with positional isomer standards for free eicosanoids that are commercially available from Cayman Chemical, and listed under Reagents.
xiii. Once relative amounts of each free acid positional isomer have been determined, the absolute amount of each can be calculated based on the total yield of HETE-PLs as determined using absorbance in step 1B(vi), above.
<PAUSE POINT> Stability information is as for Option A.
C. Generation and purification of KETE-PL standards
i. Dry down 100 μg purified HETE-PL (either mixed or single isomers) in a glass vial to remove the methanol, and reconstitute in 100 μl anhydrous dichloromethane. Add 700 μg of Dess-Martin periodinane (DMP) and incubate at room temperature for 30 minutes in the dark, with regular agitation.
<CRITICAL STEP> It is essential that DMP is allowed to fully warm up to room temperature before opening the stock bottle. If this is not done, then additional oxidations of the HETE take place resulting in artifactual product formation.
ii. Dry the sample under N2 and add 1 ml HPLC grade water along with 2.5 ml lipid extraction solvent (1 M acetic acid, 2-propanol, hexane (2:20:30, v/v/v). Vortex vigorously, and then add 2.5 ml hexane. Vortex and centrifuge (260 g, 5 min), then recover the lipids using a glass pasteur pipette by decanting the upper hexane layer.
iii. Re-extract the lower aqueous layer with 2.5 ml hexane, vortexing, centrifuging as above and decanting the upper hexane layer. Combine this with the first hexane extraction.
iv. Dry under stream of N2 or in vacuum dryer (Labconco RapidVap) and reconstitute into 100 μl methanol.
<PAUSE POINT> The crude mixture can be stored overnight before purification, at −80° under inert gas.
v. Purify KETE-PL as described for esterified eicosanoids (1A(vii)), using reverse phase HPLC, monitoring elution at 280 nm (typically around 27 min). Quantify total KETE-PL by measuring absorbance of the purified lipid mixture using ε = 22.96 mM−1.cm−1 (for the conjugated carbonyl, determined using 15-KETE free acid) and store at −80 °C under N2 in methanol. For this, dilute a small amount into methanol and scan in a quartz cuvette from 200-400 nm using a scanning spectrophotometer.
vi. When isomeric mixtures of HETE-PLs are used to generate KETE-PLs, the relative amount of each KETE-PL will mirror that of the HETE-PL substrate, and thus can then be used to calculate the absolute amount of each positional isomer in the mixture.
<PAUSE POINT> Store as for other standards above. Stability information is not yet available for these lipids, however we anticipate that they may be less stable than hydroxy-phospholipids due to the presence of the electrophillic carbonyl group. We recommend at least at monthly intervals that the concentration and purity be re-checked by LC/MS/MS analysis (as detailed in Steps 12-14 below).
Tissue preparation (up to 1 day depending on number of samples)
Homogenization of solid tissue samples (e.g. lung, liver)
2| After tissue harvest, samples need to be snap frozen in cryovials as quickly as possible, then stored at −80 °C until analysis.
<CRITICAL STEP> Care needs to be taken with fresh tissue samples to ensure artifactual oxidation does not take place. This includes freezing samples as quickly as possible after harvest.
3| Weigh the frozen tissue (up to 0.5 g), then place in an ice-cold mortar (pre-cooled in freezer) on ice.
4| Add up to 1 ml phosphate-buffered saline, pH 7.4, containing 100 μM each diethylene triamine pentaacetic acid (DTPA) and butylated hydroxytoluene (BHT), followed by some liquid N2 (e.g. 10 ml).
<CRITICAL STEP> Addition of antioxidants/chelators during homogenisation is essential to prevent artifactual oxidation. Also, liquid N2 is added after the buffer so that the solution freezes and making homogenisation easier.
5| Homogenize using the pestle until a smooth homogenate is achieved. Extract as described below.
6| Cells or supernatant samples are extracted without the above steps.
<PAUSE POINT> The homogenate can be stored overnight before extraction, at −80° under inert gas, although to minimize freeze-thawing, this is not recommended if possible.
Lipid extraction from samples
7| Add internal standards, at 10 ng/sample: di-14:0-phosphatidylethanolamine (DMPE) and/or di-14:0-phosphatidylcholine (DMPC). For quantitation of PEs or PCs, we use DMPE, or DMPC, respectively.
8| If preferred, the more unstable hydroperoxides can be reduced to the alcohols, by addition of 10 μl SnCl2 (100 mM in water) per ml sample, for 10 min at room temp before extraction.
9| Add the sample to a glass extraction tube containing 2.5 ml lipid extraction solvent (1 M acetic acid, 2-propanol, hexane (2:20:30, v/v/v)) per 1 ml sample. Cap and vortex vigorously, and then add 2.5 ml hexane. Vortex and centrifuge (260 g, 5 min), then recover the lipids using a glass pasteur pipette by harvesting the upper hexane layer.
<CRITICAL STEP> Extraction is greatly enhanced by vigorous vortexing, of at least 30 sec per sample, as well as 30 sec manual shaking of the sample before centrifugation.
10| Re-extract the lower aqueous layer with additional 2.5 ml hexane. Vortex, centrifuge as above and harvest the upper hexane layer. Combine this with the first hexane extraction.
11| Dry the combined hexane layers using either N2, or a vacuum dryer (e.g. Labconco RapidVap), with warming not greater than 30 °C, then resuspend in 100 μl mobile phase (50:50 solvents A, B as listed below in step 12) and store under N2 at −80°C until analysis by LC/MS/MS, preferably within a week of generating the samples.
<CRITICAL STEP> Resuspend carefully, taking care to recover all lipids from the side of the vial. This is important for recovery. If samples appear cloudy or contain precipitate, this must be removed before LC/MS/MS. Centrifugation or spinning through 0.2 μ filters (e.g. Spin-X filter, Corning, USA) will be required. If this is not done, samples may clog the LC column and cause pressure buildup.
<PAUSE POINT> Samples can be stored before analysis, but this time should be kept as short as possible to minimise decay of oxidized lipids. In particular, when analyzing hydroperoxy-lipids in complex cell extracts. we recommend the samples are analyzed immediately after extraction.
LC/MS/MS analysis
Reverse phase LC/MS/MS of esterified eicosanoids (50 min per sample)
12| Analyze samples using reverse phase chromatography. We use a Luna 3 μm C18 (2) 150 × 2 mm column (Phenomenex, Ltd, Torrance, CA) with a gradient of 50-100 % B over 10 min followed by 30 min at 100 % B (Solvent A: methanol:acetonitrile:water, 1 mM ammonium acetate, 60:20:20. Solvent B: methanol, 1 mM ammonium acetate) with a flow rate of 200 μl/min.
13| On our instrument (Applied Biosystems 4000 Q-Trap), the typical optimum system settings to determine esterified HETEs are declustering potential (DP) −140V, collision energy (CE) −45V. However, this will differ on other models and other manufacturers’ instruments and needs to be determined by the investigator according to the standard protocols for their instrument. It is important that the user tunes the instrument separately for each analyte and standard (including collision energy) to be measured, to obtain the optimum settings for analysis.
14| MRM transitions to monitor and retention times these lipids on our system are shown in Table 2. Figures 5 - 8 show the LC/MS/MS elution of positional isomer standards for HETE, HDOHE, HODE and KETE-PE lipids analysed using this method.
Table 2. Precursor and product ions, and retention time of some esterified HETEs, HODEs, HDOHEs and KETE standards.
Retention times refer to their elution using the phospholipid LC/MS/MS method (Luna column).
| Lipid name | Precursor m/z |
Product m/z | Retention time (min) |
|---|---|---|---|
| 18:0a/HETE-PE | 782.6 | 319.2 | 24.5-26.5 |
| 18:0a/15-HETE-PE | 782.6 | 219.1 | 24.8 |
| 18:0a/12-HETE-PE | 782.6 | 179.1 | 25.4 |
| 18:0a/5-HETE-PE | 782.6 | 115.1 | 26.4 |
| 18:0a/11-HETE-PE | 782.6 | 167.1 | 25.1 |
| 18:0a/8-HETE-PE | 782.6 | 155.1 | 25.6 |
| 16:0a/HETE-PC | 782.6 | 319.2 | 22-24 |
| 16:0a/15-HETE-PC | 782.6 | 219.1 | 22.2 |
| 16:0a/12-HETE-PC | 782.6 | 179.1 | 22.9 |
| 16:0a/5-HETE-PC | 782.6 | 115.1 | 23.9 |
| 16:0a/11-HETE-PC | 782.6 | 167.1 | 22.6 |
| 16:0a/8-HETE-PC | 782.6 | 155.1 | 23.2 |
| 18:0a/HDOHE-PE | 806.6 | 343.2 | 22-25 |
| 18:0a/20-HDOHE-PE | 806.6 | 187.2 | 22.8 |
| 18:0a/17-HDOHE-PE | 806.6 | 273.2 | 23.0 |
| 18:0a/14-HDOHE-PE | 806.6 | 205.2 | 23.3 |
| 18:0a/11-HDOHE-PE | 806.6 | 149.2 | 23.6 |
| 18:0a/8-HDOHE-PE | 806.6 | 109.2 | 24.1 |
| 18:0a/16-HDOHE-PE | 806.6 | 233.2 | 22.9 |
| 18:0a/13-HDOHE-PE | 806.6 | 193.2 | 23.1 |
| 18:0a/10-HDOHE-PE | 806.6 | 153.2 | 23.4 |
| 18:0a/7-HDOHE-PE | 806.6 | 141.2 | 23.8 |
| 18:0a/4-HDOHE-PE | 806.6 | 101.2 | 24.9 |
| 16:0a/HODE-PE | 730.6 | 295.2 | 20.6 |
| 16:0a/13-HODE-PE | 730.6 | 171.1 | 20.6 |
| 16:0a/9-HODE-PE | 730.6 | 195.1 | 20.6 |
| 18:0a/15-KETE-PE | 780.6 | 317.1 | 23.8 |
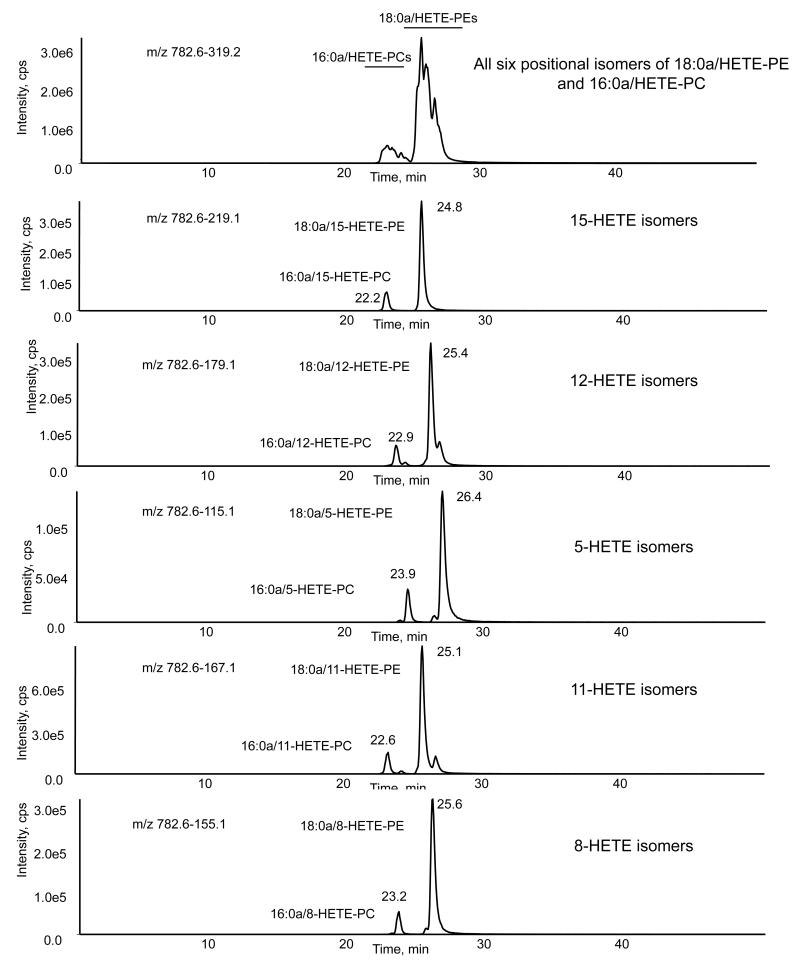
Figure 5. LC/MS/MS of a mixture of standards comprising 6 positional isomers of 16:0a/HETE-PC and 18:0a/HETE-PE, all with m/z 782.6.
HETE-PE and -PC lipids were generated by air oxidation, as described in Materials and Methods, and purified by reverse phase HPLC. A sample containing both HETE-PEs and -PCs was separated using LC/MS/MS as described in Materials and Methods. Elution was monitored either by 782 → 319 (all positional isomers) or by product ions specific for each positional isomer.
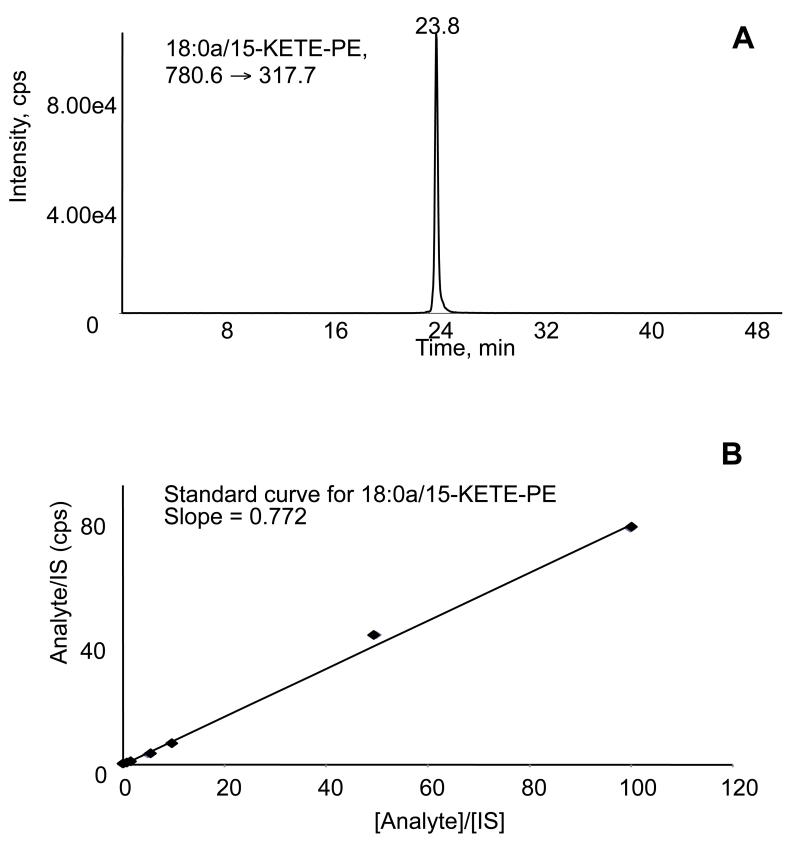
Figure 8. LC/MS/MS and standard curve for 18:0a/15-KETE-PE.
Panel A. LC/MS/MS elution profile of KETE-PE standard, showing single peak at 23.8 min. Panel B. A standard curve was created by analyzing a fixed amount of DMPE with varying amounts of 15-KETE-PE, using LC/MS/MS.
<CRITICAL POINT> It is essential to include a number of blanks (typically methanol), especially at the start and end of each batch, to monitor for carryover. If carryover occurs, extensive wash steps between samples may need to be introduced. See Equipment Setup for further information on our washing procedure.
LC/MS/MS analysis
Standard curves and quantitation (1 hr to set up standard curve vials, approximately 9-10 hrs to analyse using LC/MS/MS, post acquisition analysis 1-2 days depending on number of samples)
15| Generate a standard curve by serial dilution of primary esterified eicosanoid analytes (A) (e.g. HETE-PE, KETE-PE, HETE-PC, etc) in methanol that contains an equal amount of internal standard (IS) per vial (e.g. DMPE, DMPC). We typically work with a range that gives the following amounts: 10 ng, 5 ng, 1 ng, 500 pg, 100 pg, 50 pg, 10 pg, 5 pg, 1 pg primary standard per 10 μl injection, with 100 pg internal standard.
16| When using mixtures of positional isomers, amounts of each will be slightly different and need to be accounted for during quantitation.
17| Analyse 10 μl of each standard using reverse phase LC/MS/MS as described above for esterified eicosanoids.
18| Integrate peaks for all lipids. Calculate ratio of A:IS for ng amounts of lipid in each standard vial and for A:IS (cps) for each standard run. Plot A:IS (cps) vs A:IS (ng) as shown by examples in Figures 6,7. Calculate slope for each analyte.
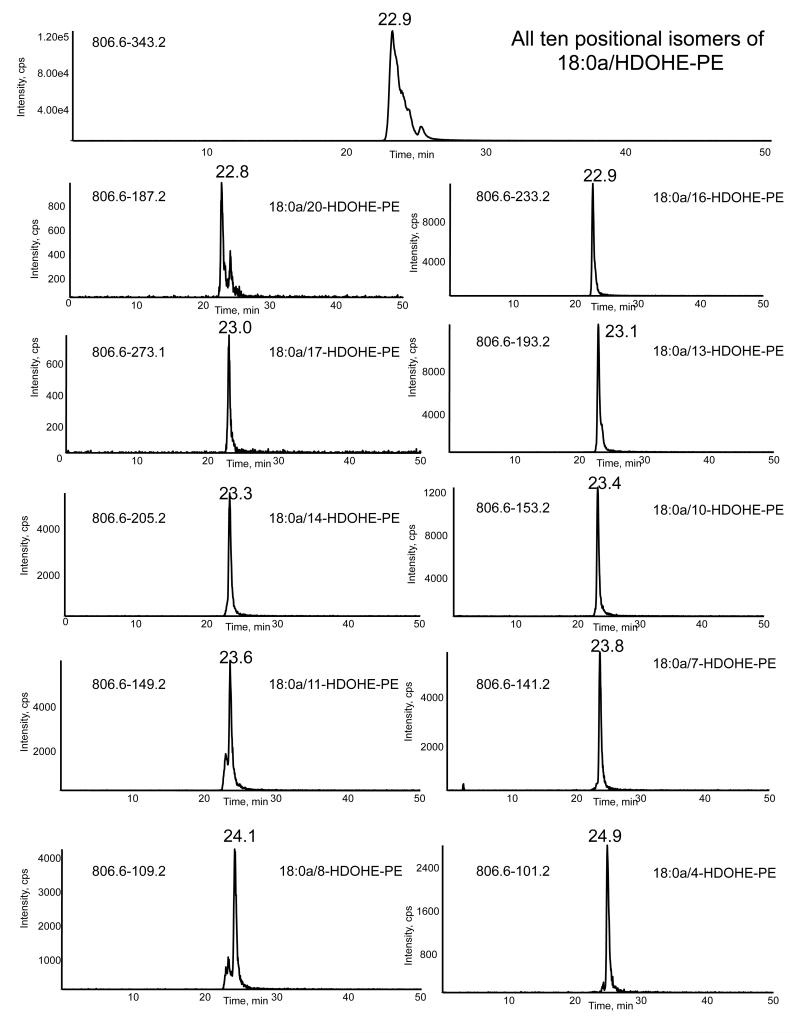
Figure 6. LC/MS/MS of a mixture of standards comprising 10 positional isomers of 18:0a/HDOHE-PE, all with m/z 806.6.
HDOHE-PE lipids were generated by air oxidation, as described in Materials and Methods, and purified by reverse phase HPLC. A sample was separated using LC/MS/MS as described in Materials and Methods. Elution was monitored either by m/z 806 → 343 (all positional isomers) or by product ions specific for each positional isomer.
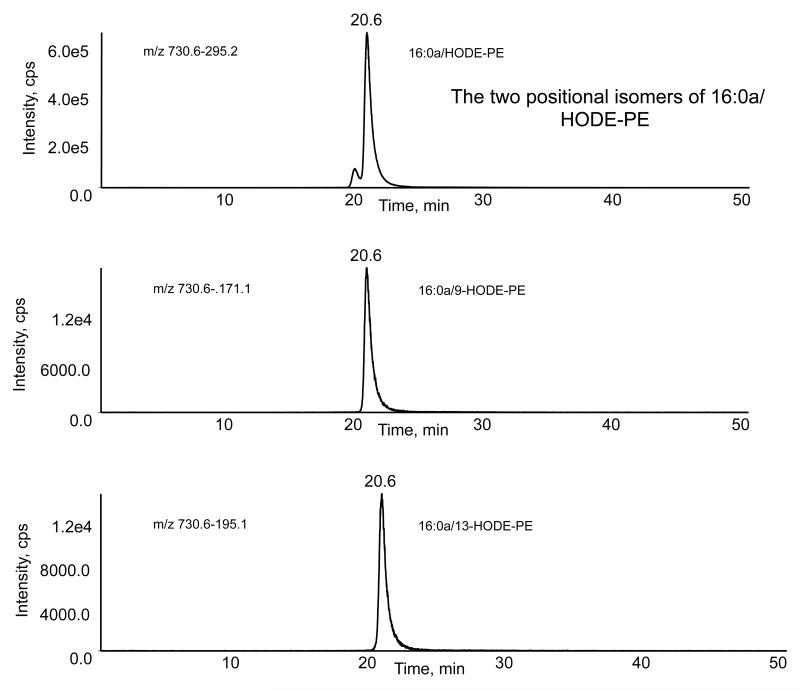
Figure 7. LC/MS/MS of a mixture of standards comprising 2 positional isomers of 16:0a/HODE-PE, with m/z 730.6.
HODE-PE lipids were generated by air oxidation, as described in Materials and Methods, and purified by reverse phase HPLC. A sample was separated using LC/MS/MS as described in Materials and Methods. Elution was monitored either by m/z 730 → 295 (all positional isomers) or by product ions specific for each positional isomer.
19| For all samples, analyze 10 - 20 μl (depending on sensitivity), then integrate area, and use the following equation to determine amounts of each analyte in all samples:
20| Data is given as ng/sample extracted and should then be normalized per cell number or tissue wet weight.
TROUBLESHOOTING
As mentioned, clean glassware is essential and organic solvents should not be used with regular laboratory plasticware since they will leach compounds from the plastic which can interfere, in particular in the low UV range employed for conjugated dienes. This could especially be a problem when purifying newly synthesized standards using LC-UV.
The data shown here utilized an Applied Biosystems 4000 Q-Trap instrument, considered to be very sensitive when detecting in MRM mode (1 – 10 pg on column of oxidized phospholipids). When setting up these procedures using a different platform, careful examination of the standard curve will determine the best range for analysis of the lipids, and in some cases, a lower sensitivity may be apparent. If this is the case, adjustment of standard amounts in the curves may be warranted, to cover a different range of analyte concentrations than we present here.
As internal standards, we typically use di-14:0-PE and PC since we have determined that they are absent in samples. If different cells or tissues are to be analyzed, for example, plants, fish or other organisms, then a lipid extract should first be checked using MS to ensure that the internal standard is not present as a constituent lipid. If it is, then a different/absent one should be chosen instead. It is important not to use one that differs markedly, e.g. with very two short fatty acid chains, since this may extract with different efficiency to the analyte. If using different internal standards to those listed here, their extraction efficiency should be determined and compared to the analytes of interest to ensure they are comparable.
ANTICIPATED RESULTS
These methods require tandem mass spectrometry and the ability to work with organic solvents (fume hood, dispensers, protective equipment). A special knowledge of working with lipids may help but is not essential. In this method, we use primary standards and a structurally-related internal standard to quantitate families of lipid mediators in cells/tissue samples. The primary standards are generated as either a single isomer (in only a few cases) or by air oxidation of lipid, as a defined mixture of positional isomers. In the case of HETEs, we typically get a fairly even distribution although with the 11-, 12- and 15-isomers predominating to some extent (Figure 4 A,B). NMR and MS/MS data for some of the lipids is provided in the supplementary data section.
Typically, immune cells generate 4-6 different individual lipids (e.g. 18:0a/15-HETE-, 18:0p/15-HETE-, 18:1p/15-HETE- and 16:0p/15-HETE-PE for human monocytes)2-5. For each family of the same phospholipid class (e.g. PE or PC), a primary standard that corresponds in terms of the correct eicosanoid positional isomer, and headgroup is used. Thus, for human monocytes, 18:0a/15-HETE-PE standard is used to quantitate all 4 monocyte lipids, monitoring MRM for m/z parent → 219.1, a daughter ion fragment that arises from internal fragmentation of 15-HETE. Similarly for human platelets, we use 18:0a/12-HETE-PE for the four 12-HETE-PEs and 16:0a/12-HETE-PC for the two 12-HETE-PCs (monitoring m/z parent → 179.1) (Table 3). It would be preferable to utilise identical primary standards for all cell-derived lipids, however, purified substrate lipid is not currently available for all, especially in the case of plasmalogens.
Table 3. Choice of primary and internal standards to use with particular immune cells or LOX isoforms.
| Cell type/LOX isoform expressed |
Primary Std (HETE-PEs) |
Internal Std (HETE-PEs) |
Primary Std (HETE-PCs) |
Internal Std (HETE-PCs) |
|---|---|---|---|---|
| Human monocytes induced with IL-4 or 15-LOX1 |
18:0a/15- HETE-PE |
DMPE | 16:0a/15- HETE-PC |
DMPC |
| Human neutrophils or 5-LOX |
18:0a/HETE- PE mixed isomer preparation |
DMPE | 16:0a/HETE- PC mixed isomer preparation |
DMPC |
| Human platelets or murine peritoneal macrophages, or 12-, or 12/15- LOXs |
18:0a/HETE- PE mixed isomer preparation |
DMPE | 16:0a/HETE- PC mixed isomer preparation |
DMPC |
This protocol covers the quantitation of known lipids using defined standards in immune cells. Thus the transitions for those lipids are as given in Table 1. In contrast, with samples from other tissues that have not been analyzed before, it is very useful to first characterize what esterified oxylipins are present, in case the headgroup or sn1 lipids differ from those described herein. To do this, we used precursor LC/MS/MS, scanning for parent ions that generate the carboxylate anions of the oxylipins2-5. For structural confirmation, we typically generate mixtures of esterified HETE or HDOHE by either LOX or air oxidation of brain or egg phospholipid mixtures and analyse cell extracts by a number of methods including MS/MS, MS3 and normal phase and chiral chromatography. Inclusion of methods for these approaches is beyond the scope of this protocol, but they are summarized in our related papers2-5. We are also very willing to provide technical support to others wishing to characterize new related lipids in other types of tissue not mentioned in this protocol.
Phospholipid ionization efficiency can decrease with increasing mass, particularly over large mass ranges. However, these lipids are close enough in molecular weight that this should not impact significantly on accuracy of quantitation. In support, we previously analyzed total PE-esterified 15-HETE in monocytes by (i) purifying PE using normal phase LC, then (ii) saponifying and quantifying 15-HETE using HPLC, and found very similar results to this method for total 15-HETE-PE2. It is important to use the correct eicosanoid positional isomer, as quantification uses internal HETE fragmentation ions rather than the HETE carboxylate anion. Although positional isomers are partially separated in our LC system, use of internal daughter ions increases selectivity for detection. For example, for 15-HETE-PE lipids, parent → m/z 219 is used rather than m/z 319 which would detect all 6 positional isomers. This approach works well for HETE, HODE and HDOHE positional isomers for which specific internal fragment ions have been identified (Table 2). However in the case of KETE-PL, it was not possible to identify diagnostic daughter ions with confidence. In this case, we utilise the carboxylate anion, 317.2 and detect all positional isomers simultaneously.
As these are primary standards, an internal standard that is absent from the cell lipids is added before extraction, depending on the PL class being quantified (e.g. DMPE or DMPC are used for PE or PC lipids, respectively). Table 3 lists which primary and internal standards to use for each class of lipid in various immune cells.
The separation of oxidized phospholipids requires long run times of 50 min per sample. This is essential since extracts from cells/tissue contain thousands of lipids, that may include one or more isobaric species with the same MRM transition. In samples from immune cells, large peaks are sometimes observed eluting later, around the same time as unoxidized phospholipids (not shown). Based on their retention times, these are structurally distinct from the lipids of interest, and need to be separated so they are not included in the analysis. Thus, direct infusion approaches are not suitable for analysis and quantitation of these lipids. In the future, new generation ultra high pressure LC systems may be able to significantly reduce run times significantly. Depending on the peak width (typically up to 1 min maximum for analytes on our system), the method can be used for up to 30 MRM transitions within the same run, but more than this would compromise sensitivity.
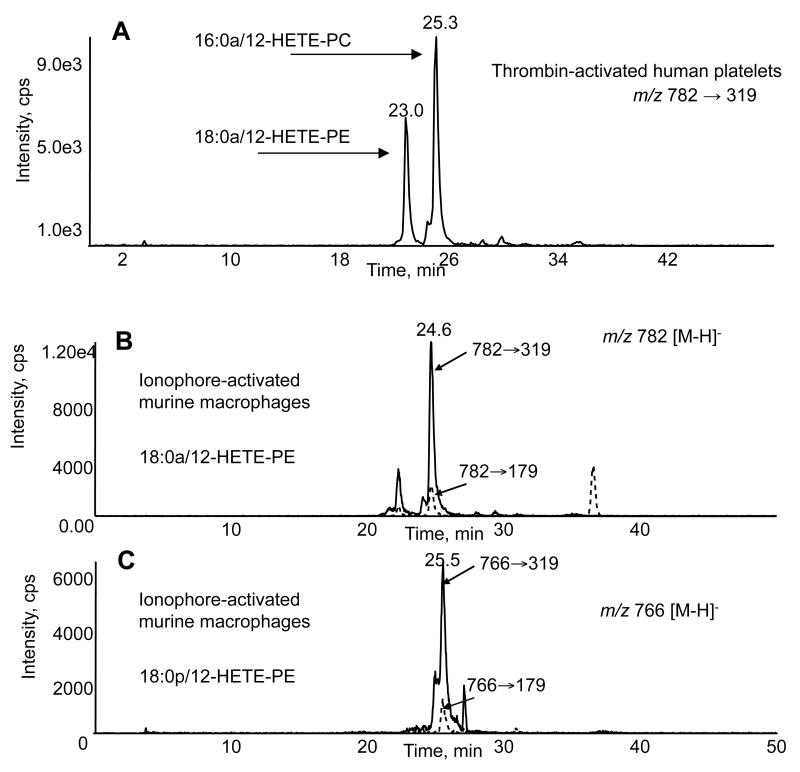
Using the extraction method described herein, recoveries of standards and analytes (DMPE, 18:0a/15-HETE-PE and 16:0a/15-HETE-PC) are generally better than the classical Bligh and Dyer method, which was around 60 - 62 % (Supplementary Table 2, and data not shown). PE lipids extracted more efficiently than PCs, further illustrating the need to use the appropriate internal standard (e.g. headgroup) for quantitation. We also evaluated a method using methyl-tert-butyl-ether, but recoveries were lower for oxidized phospholipids14. For standard curves, the ratio of the signals is plotted as a function of ratio of amounts. Responses are linear over 10,000-fold range (1 pg - 10 ng), with r2 values all above 0.99, and a detection limit of 1-10 pg on column (Figures 8, 9, Supplementary Table 2). Co-efficients of variation for analytes were 2 - 4 % (inter-sample) and 2 - 7 % (intra-sample) (Supplementary Table 2). Using this approach, we estimated amounts of HETE-PE in human platelets, monocytes and murine peritoneal macrophages (Table 4). Figure 10 shows typical detection of 12-HETE-PE generation by human platelets activated by thrombin. Levels are in the same order of magnitude to free HETEs generated, indicating this to be a significant metabolic pathway for LOXs in mammalian cells5.
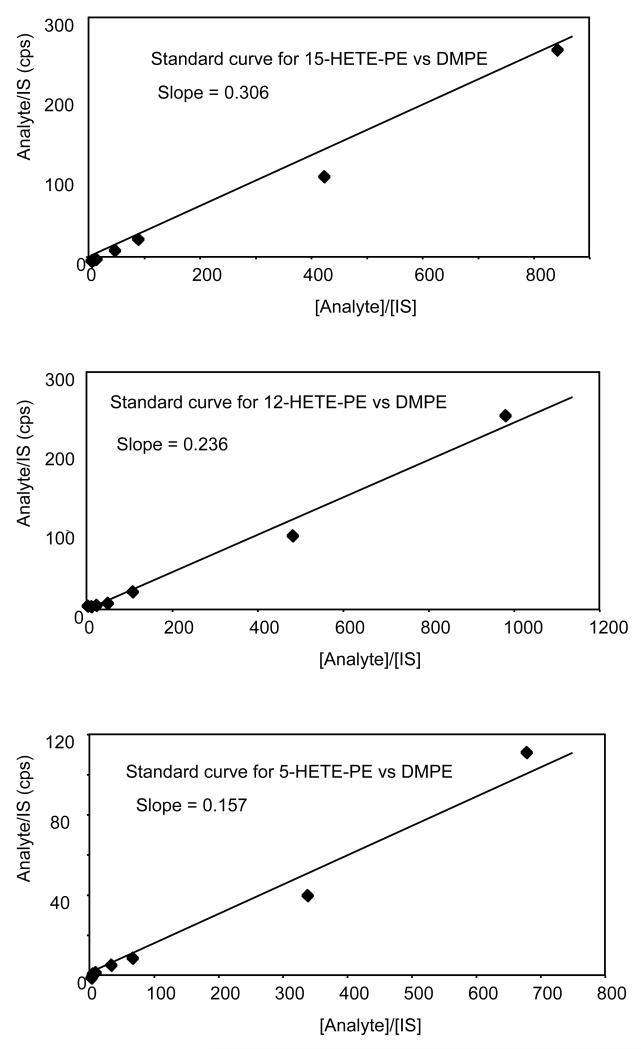
Figure 9. Generation of standard curves for 5-, 12- and 15-HETE-PE isomers.
Standard curves were created by adding a fixed amount of DMPE to varying amounts of the standard HETE-PE positional isomer mixture, generated by air oxidation, and analyzed using LC/MS/MS as described in Materials and Methods.
Table 4. Amounts of HETE-PEs and free HETEs detected in human and murine immune cells.
| Cell type | Free HETE | HETE-PC (total) | HETE-PE (total) |
|---|---|---|---|
| Human platelets activated using thrombin (0.2 U/ml, 15 min) ng/4×107 cells, n = 5, mean ±SEM |
65.5 ±17.6 | 18.35 ± 4.61 | 5.85 ± 1.42 |
| Human IL-4-treated monocytes activated with ionophore (10 μM A23187, 15 min), ng/4 × 106 cells |
257 | Not detected | 150.8 |
| Murine peritoneal lavage from healthy C57BL/6 mice, ng/lavage |
18.5 ± 1.03 | Not detected | 5.5 ± 0.2 |
Figure 10. Detection of HETE-PEs and PCs in activated immune cells using LC/MS/MS.
Panel A. Lipid extracts from thrombin-activated human platelets were separated as described, with detection of m/z 782.6 → 319.2. Panel B. Lipid extracts from ionophore-activated murine peritoneal macrophages were analyzed for 18:0a/12-HETE-PE with detection using m/z 782.6 → 319.2 or 179.2, as shown. Panel C. Lipid extracts from ionophore-activated murine peritoneal macrophages were analyzed for 18:0p/12-HETE-PE with detection using m/z 766.6 → 319.2 or 179.2, as shown.
Supplementary Material
Abbreviations
- HETE
hydroxyeicosatetraenoic acid
- HODE
hydroxyoctadecadienoic acid
- HDOHE
hydroxydocosahexaenoic acid
- PE
phosphatidylethanolamine
- PC
phosphatidylcholine
- PL
phospholipid
- LOX
lipoxygenase
- DHA
docosahexaenoic acid
- LA
linoleic acid
- ACD
acid-citrate-dextrose
- DMPE
dimyristoyl-PE
- DMPC
dimyristoyl-PC
- Q
quadrupole
- DP
declustering potential
- CE
collision energy
- DMP
Dess-Martin periodinane
- BHT
butylated hydroxytoluene
- LC/MS/MS
liquid chromatography-tandem mass spectrometry
- COX
cyclooxygenase
- SnCl2
stannous chloride
- KETE
ketoeicosatetraenoic acid
- DTPA
diethylene triamine pentaacetic acid
Footnotes
Competing financial statements. The authors declare no competing financial interests.
References
- 1.Haeggstrom JZ, Rinaldo-Matthis A, Wheelock CE, et al. Biochem Biophys Res Commun. 2010;396(1):135. doi: 10.1016/j.bbrc.2010.03.140. [DOI] [PubMed] [Google Scholar]
- 2.Maskrey BH, Bermudez-Fajardo A, Morgan AH, et al. J Biol Chem. 2007;282(28):20151. doi: 10.1074/jbc.M611776200. [DOI] [PubMed] [Google Scholar]
- 3.Morgan AH, Dioszeghy V, Maskrey BH, et al. J Biol Chem. 2009;284(32):21185. doi: 10.1074/jbc.M109.021634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morgan LT, Thomas CP, Kuhn H, O’Donnell VB. Biochemical Journal. 2010 Jul; doi: 10.1042/BJ20100415. in press. [DOI] [PubMed] [Google Scholar]
- 5.Thomas CP, Morgan LT, Maskrey BH, et al. J Biol Chem. 2010;285(10):6891. doi: 10.1074/jbc.M109.078428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masoodi M, Nicolaou A. Rapid Commun Mass Spectrom. 2006;20(20):3023. doi: 10.1002/rcm.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kingsley PJ, Rouzer CA, Saleh S, et al. Anal Biochem. 2005;343(2):203. doi: 10.1016/j.ab.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 8.Maskrey BH, O’Donnell VB. Biochem Soc Trans. 2008;36(Pt 5):1055. doi: 10.1042/BST0361055. [DOI] [PubMed] [Google Scholar]
- 9.O’Donnell VB, Maskrey B, Taylor GW. Methods Mol Biol. 2009;462:5. [PubMed] [Google Scholar]
- 10.Brash AR, Ingram CD, Harris TM. Biochemistry. 1987;26(17):5465. doi: 10.1021/bi00391a038. [DOI] [PubMed] [Google Scholar]
- 11.Milne GL, Porter NA. Lipids. 2001;36(11):1265. doi: 10.1007/s11745-001-0841-2. [DOI] [PubMed] [Google Scholar]
- 12.Dess DB, Martin JC. Journal of Organic Chemistry. 1983;48:4155. [Google Scholar]
- 13.Niki E, kawakami A, Yamamoto Y, Kamiya Y. Bulletin of the Chemical Society of Japan. 1985;58(7):5. [Google Scholar]
- 14.Matyash V, Liebisch G, Kurzchalia TV, et al. J Lipid Res. 2008;49(5):1137. doi: 10.1194/jlr.D700041-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Related manuscripts
- 1.Morgan AH, Dioszeghy V, Maskrey BH, et al. J Biol Chem. 2009;284(32):21185. doi: 10.1074/jbc.M109.021634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morgan LT, Thomas CP, Kuhn H, O’Donnell VB. Biochemical Journal. 2010 Jul; doi: 10.1042/BJ20100415. in press. [DOI] [PubMed] [Google Scholar]
- 3.Thomas CP, Morgan LT, Maskrey BH, et al. J Biol Chem. 2010;285(10):6891. doi: 10.1074/jbc.M109.078428. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.