SUMMARY
Interleukin-22 (IL-22) is central to host protection against bacterial infections at barrier sites. Both innate lymphoid cells (ILCs) and T cells produce IL-22. However, the specific contributions of CD4+ T cells and their developmental origins are unclear. We found that the enteric pathogen Citrobacter rodentium induced sequential waves of IL-22 producing ILCs and CD4+ T cells that were each critical to host defense during a primary infection. Whereas IL-22 production by ILCs was strictly IL-23–dependent, development of IL-22 producing CD4+ T cells occurred via an IL-6–dependent mechanism that was augmented by, but not dependent on, IL-23, and was dependent on both transcription factors T-bet and AhR. Transfer of CD4+ T cells differentiated with IL-6 in the absence of TGF-β (“Th22” cells) conferred protection of infected IL-22-deficient mice whereas transferred Th17 cells did not. These findings establish Th22 cells as an important component of mucosal anti-microbial host defense.
INTRODUCTION
Effective host protection is characterized by integrated responses of innate and adaptive arms of immunity. At barrier sites (eg. skin, respiratory and intestinal tracts), production of IL-22 by both innate and adaptive lymphoid cells is important in host defense through its actions on epithelial cells (Ouyang et al., 2011; Wolk et al., 2010). Because expression of IL-22 and its receptor segregate between hematopoietic and non-hematopoietic cells, respectively, the IL-22–IL-22R axis represents a critical link in the integration of immunity with barrier defense (Aujla et al., 2008; Wolk et al., 2004; Zheng et al.,2008).
Cellular sources of IL-22 are diverse. IL-22–expressing innate lymphoid cells (ILCs) have been identified in mice and humans, particularly natural killer (NK)-like cells and lymphoid tissue-inducer (LTi) cells (Colonna, 2009; Sonnenberg et al., 2011a). IL-22 producing ILCs express the transcription factors RORγt and AhR, and have a shared requirement for IL-23 to produce IL-22 (Sawa et al., 2010; Takatori et al., 2009). Mice deficient for IL-23 fail to resist infection by intestinal or pulmonary bacterial pathogens (Aujla et al., 2008; Mangan et al., 2006; Zheng et al., 2008). In the well-studied Citrobacter rodentium model (Higgins et al., 1999; Mundy et al., 2005), deficiency of IL-23 is fatal early in infection due to impaired induction of intestinal IL-22 (Cella et al., 2009; Sonnenberg et al., 2011b; Zheng et al., 2008). While specific contributions of different IL-22 producing ILCs remain to be defined, their role in early barrier protection appears indispensable.
CD4+ T cells also produce IL-22. Originally described as a product of Th1 cells (Gurney, 2004), Th17 cells are also an important source of IL-22 (Liang et al., 2006; Zheng et al., 2007). In common with ILCs, IL-22 expression by Th17 cells is positively regulated in by the transcription factors RORγt and AhR (Schraml et al., 2009; Veldhoen et al., 2008). A subset referred to as Th22 cells has been described in humans (Duhen et al., 2009; Trifari et al., 2009). Human Th22 cells are characterized by production of IL-22 with little or no IL-17, and appear to have a counterpart in mice based on ex vivo studies that have shown that naive CD4+ T cells differentiated with IL-6 under conditions of no addition of TGFβ express high IL-22 and negligible IL-17 compared to conventional Th17 cells differentiated in the presence of IL-6 plus TGFβ (Zheng et al., 2007). A similar reciprocal pattern of IL-22 and IL-17 expression was linked to a “pathogenic” subset of murine Th17 cells induced under similar conditions (Ghoreschi et al., 2010). However, the normal function of this cell population is not well understood.
While ILCs are a critical source of IL-22 during mucosal infection, less is known regarding specific contributions of IL-22 produced by CD4+ T cells. Here, we have examined the relative contribution of ILCs and CD4+ T cells in host protection against the enteric bacterial pathogen, Citrobacter rodentium. We find that while early contribution of ILC-derived IL-22 induced by IL-23 is critical to curb bacterial loads pending development of pathogen-induced CD4+ T cells, the latter are critical for late host protection via an IL-6 dependent mechanism that is augmented by, but not strictly dependent on, IL-23. Further, CD4+ T cells appear to be the major producers of IL-22 in the infected intestinal tissues, with cells differentiated in the absence of TGFβ (referred to herein as Th22 cells) providing more potent protection on a per cell basis than classical Th17 cells. The transcriptome of Th22 cells differed significantly from Th17 cells and both AhR and T-bet were required for optimal IL-22 production by Th22 cells and their host-protective function. These findings identify Th22 cells as important contributors to mucosal host defense, and suggest overlap between the functional programming of these cells with that of the recently described subset of Th17 cells proposed to be key contributors to autoimmune disease.
RESULTS
IL-23-Independent, IL-22-Dependent Protection against C. rodentium Infection
We previously identified an essential role for IL-23 in the protective host response against C. rodentium infection (Mangan et al., 2006). Challenge of IL-23-deficient mice (Il23a−/−) with high doses of C. rodentium (≥0.5 × 109 cfu) is uniformly fatal, whereas wild-type (WT) mice develop a transient colitis before clearing infection. In the course of titering inocula of C. rodentium in Il23a−/− mice (Fig. 1A), we found that at lower doses (0.5−1.0 × 107 cfu) >90% of mice survived. To monitor bacterial loads, a strain of C. rodentium expressing luciferase was used to enable real-time whole body imaging (Wiles et al., 2006). In contrast to Il23a−/− mice infected with high-dose C. rodentium, nearly all Il23a−/− mice infected with low dose cleared infection with similar kinetics to WT mice. This was despite greater than a log-fold higher bacterial load at the peak of infection (Fig. 1B and 1C ). Thus, IL-23 is essential for protection at high doses of C. rodentium, but is dispensable at low doses.
Figure 1.
IL-23-independent, IL-22-dependent pathway to protection from C. rodentium Infection
(A) Survival rates of Il23a−/− mice infected with high-dose (2 × 109 cfu) or low-dose (0.5 × 106 cfu) C. rodentium (n= 4–5 mice per group). Data are from one of three similar experiments.
(B) Serial whole-body imaging of Il23a−/− mice inoculated with high- or low-dose of the luminescent strain of C. rodentium (strain ICC180) by gastric gavage and imaged at the indicated days PI.
(C) Colonization kinetics of WT or IL23a−/− mice inoculated with indicated doses of C. rodentium. Light unit counts/sec directly correlate with the bacterial load (data not shown). Data are means +/− SEM. *p<0.01, **p<0.001.
(D) Survival rates of IL23a−/− mice infected with low-dose C. rodentium and dosed IP with 150 µg of neutralizing anti-IL-22 mAb or isotype control every other day starting at d0 or d5. Data are representative of two or more experiments with a minimum of 3–4 mice per group.
Blockade of IL-22 in Il23a−/− mice challenged with low-dose infection was uniformly fatal, whether antibody was administered coincident with infection (day 0), or after the peak of the innate immune response (d5; Fig. 1D). In extension of a previous study (Zheng et al., 2008), we found that mice deficient for global IL-17 signaling (Il17ra−/−) cleared infection to high-dose challenge with similar kinetics to WT mice, albeit with significantly higher bacterial loads over the mid-coarse of infection (Fig. S1). Thus, while IL-17 signaling appears to be important in restraining bacterial growth during infection, IL-17 cytokines, including IL-17A, IL-17F and IL-17C, are not essential for clearance. We also found that Ifng−/− mice cleared infection with comparable kinetics and bacterial loads to WT mice (Fig. S1). These results implicate an IL-23-independent, IL-22-dependent pathway that contributes to protection against C. rodentium.
IL-6 is Indispensable for Development of IL-22–Producing CD4+ T cells
IL-23 is required for production of IL-22 by ILCs (Cella et al., 2009; Sonnenberg et al., 2011b; Zheng et al., 2008). Whereas published data are in agreement that IL-6 induces the development of IL-22 producing CD4+ T cells from naive precursors in the absence of TGFβ (Duhen et al., 2009; Rutz et al., 2011; Zheng et al., 2007), disparate results concerning the role for IL-23 to induce development of IL-22 producers in mouse and human have been reported (Duhen et al., 2009; Zheng et al., 2007). In view of our findings that implicated an IL-23–independent pathway to IL-22–mediated clearance (Fig. 1), we reexamined this issue.
In agreement with previous reports (Rutz et al., 2011; Zheng et al., 2007), IL-6 alone induced the development of CD4+ T cells that expressed high amounts of IL-22 mRNA and protein, but not IL-17A, and optimal development of IL-22 producers was suppressed by TGFβ in favor of Th17 development under APC-free conditions (Fig. S2A). Addition of IL-23 to T cells differentiated by IL-6 alone significantly enhanced their production of IL-22, in accord with heightened expression of the IL-23 receptor (IL-23R) induced by IL-6 or IL-6+IL-23 in the absence of TGFβ Fig S2A–C). However, IL-23 alone did not induce the development of IL-22 producing T cells from naïve precursors (Fig. S2D–F). Indeed, the reported ability of IL-23 to induce development of IL-22 producing CD4+ T cells appears to be due to production of IL-6 by splenic APCs (Zheng et al., 2007), as IL-22 was induced by addition of IL-23 alone only under conditions of IFNγ and IL-4 neutralization (Figs. 2A–C), and was ablated either by use of Il6−/− APCs or addition of neutralizing anti-IL-6 (Fig. 2A–C, and S2D–F). Thus, IL-6 is essential for the differentiation of IL-22-producing CD4+ T cells from naïve precursors, whereas IL-23 is not. Rather, actions of IL-23 to enhance IL-22 appear primarily limited to T cells primed by IL-6 under conditions of low or absent TGFβ co-signaling — not through its direct actions on naïve T cells. In view of similar differentiation requirements and functional properties of these cells with those of human Th22 cells (Duhen et al., 2009), we refer to them as ―“Th22” cells, to distinguish them from conventional Th17 cells differentiated with TGFβ.
Figure 2.
IL-6 is indispensable for development of IL-22-producing CD4+ T cells
(A) Naïve OTII-Tg CD4 T cells were sorted and activated by Ova peptide for 3d using splenic APCs from Il23a−/− or Il6−/− mice, with or without addition of the indicated neutralizing mAbs. Recovered cells were stained for surface CD4 and intracellular IL-22 and analyzed by flow cytometry. Numbers indicate the frequencies of IL-22+ CD4+ T cells.
(B, C) Pooled frequency data from three or more independent experiments performed as in (A). Data are means +/− SEM (*p<0.05, **p<0.01).
IL-23–Dependent Induction of IL-22 and Proinflammatory Cytokines Predominates Early in C. rodentium Infection
Based on the foregoing, we postulated that the innate and adaptive immune responses contributed sequential production of IL-22 from ILCs and CD4+ T cells during primary infection, induced principally by IL-23 and IL-6, respectively. To characterize the relative contributions of IL-23 and IL-6 to the innate versus adaptive responses to C. rodentium infection, WT, Il6−/−, Il23a−/− and Il22−/− mice were challenged with high-dose luminescent C. rodentium and monitored for survival and colonization kinetics until immune-deficient mice began dying (Fig. 3 A–C). By d3 post-infection (PI), Il23a−/− mice had the greatest bacterial loads until beginning to succumb on d8. Colonization kinetics and peak bacterial loads were similar for Il6−/− and Il22−/− mice: both groups had greater bacterial loads than WT controls, but less than Il23a−/− mice. All Il23a−/− mice succumbed by d12 PI (Fig. 3C). Infected Il6−/− and Il22−/− mice had significantly improved survival compared to Il23a−/− mice, but the majority still succumbed by d14 PI. All WT mice survived and cleared the infection by 21 d PI (Fig. 1C, and data not shown).
Figure 3.
IL-23 governs early protective immunity to C. rodentium
(A) Whole-body imaging of WT, Il23a−/−, Il22−/− and Il6−/− mice inoculated with 2 × 109 cfu of luminescent strain of C. rodentium and imaged at the indicated days PI.
(B) Colonization kinetics of WT, Il23a−/−, Il22−/− and Il6−/− mice inoculated with 2 × 109 cfu of luminescent strain of C. rodentium. Data are means +/− SEM. *p<0.05, **p<0.01.
(C) Survival rates of WT, Il23a−/−, Il22−/− and Il6−/− mice inoculated with 2 × 109 cfu of C. rodentium.
(D) Histopathology of distal colonic tissues from WT, Il23a−/−, Il22−/− and Il6−/− mice collected eight days PI. H&E-stained sections (scale bars: 100 mm, left panel; 25 mm, right panels). White arrows denote individual clusters of C. rodentium attached to the apical surface of the colonic epithelium (WT) or dense colonies that extend into colonic crypts and penetrate the epithelium (Il22−/− and Il6−/−) or deeply invade the necrotic mucosa (Il23a−/−). Black arrows denote dying colonic epithelial cells.
(E) Histopathological scoring of colons from infected WT, Il23a−/−, Il22−/− and Il6−/− mice was performed at d8 PI as per Supplemental Experimental Procedures. Data are means +/− SEM from two or more experiments (n=3–4 mice per group). *p<0.05, **p<0.01.
(F) Multiplexed cytokine ELISA of the indicated cytokines and chemokines from supernatants of colonic tissue homogenates from WT, Il23a−/−, Il6−/−, Il22−/− mice collected d3 PI (24h cultures). Data are means +/− SEM from two or more experiments (n=3–4 mice per group). *p<0.05, **p<0.01.
C. rodentium infection involves the descending colon and cecal patch in WT mice (Fig. S3), sites at which the bacterium attaches to the apical surface of the epithelium. At d3 PI, increased bacteria colonized the epithelium of Il23a−/− mice, although no other differences were evident between groups (data not shown). However, by d8 PI there were severe multifocal ulcerations of the descending colon in Il23a−/− mice, with translocation of dense colonies of C. rodentium as well as multifocal transmural inflammation — none of which were evident in WT mice (Fig. 3D, E) (Mangan et al., 2006). Despite severe, multifocal injury in Il23a−/− mice, the degree of mucosal inflammation in non-ulcerated areas was significantly reduced (Fig. 3D, E, and data not shown), resulting in reduced recoveries of T cells and other mononuclear cells from colons of Il23a−/− mice (>40% reduction vs WT).
Compared to WT mice, colons of infected Il6−/− and Il22−/− mice had significantly increased epithelial injury and ulceration, which was comparable in both groups, although significantly reduced and more focal than in Il23a−/− mice (Fig. 3D, E). This was associated with significantly greater numbers of invasive bacterial colonies. Mucosal inflammation was increased in both Il6−/− and Il23a−/− mice compared to Il23a−/− mice, as was crypt hyperplasia (Fig. 3E). T cell recoveries were similar in WT, Il6−/− and Il22−/− mice, consistent with comparable inflammation scores. Thus, at the peak of infection (d8), when the adaptive immune response had become predominant, Il6−/− and Il22−/− mice had similarly impaired host protection against C. rodentium, which, although substantial, was less severe than that of Il23−/− mice. The greater severity of disease and mortality in Il23−/− mice imply that there are host-protective effects of IL-23 during the early course of infection that are not mediated by IL-6 or IL-22 alone.
To define differences in the inflammatory response of Il23−/− versus Il6−/− and Il22−/− mice that might contribute to the distinct survival and bacterial clearance, production of key cytokines and chemokines at the peak of the innate response was compared (Fig. 3F). Notably, IL-6 was highly attenuated in colon explants of Il23a−/− mice compared to both WT and Il22−/− mice, akin to the defect in IL-22 production in the absence of IL-23. Significant reductions in IL-1β and TNFα, as well IFNγ, were associated with the loss of IL-6 production in Il23−/− mice, suggesting that multiple proinflammatory signals downstream of IL-23 might contribute to reductions in mucosal IL-6. Notably, marked reductions of IL-1β and TNFα found in colons of infected Il23a−/− mice might also contribute to reductions in IL-22, as both cytokines can enhance IL-22 induction by ILCs. Conversely, normal production of IL-1β and TNFα was found in the involved colonic segments of Il6−/− and Il22−/− mice.
Decreases in chemokines (eg CCL2 and CXCL1) and proinflammatory cytokines in Il23a−/− mice likely contributed to the more limited inflammation in the colons of these mice, despite profound loss of barrier function (Fig. 3F, and data not shown). Of the additional cytokines examined, GM-CSF and G-CSF were the only ones that were significantly abrogated in both Il23a−/− and Il6−/− mice compared to WT, with significantly greater reductions in Il23a−/− mice. Thus, IL-23, but not IL-6, is essential for induction of key proinflammatory cytokines and IL-22 during the early course of infection, consistent with its indispensable role in coordination of the mucosal innate immune response (Buonocore et al., 2010; Cella et al., 2009; Sonnenberg et al., 2011b; Zheng et al., 2008).
IL-6–Dependent Production of IL-22 by CD4+ T cells Predominates Late in C. rodentium Infection
To better define the contribution of IL-6 to IL-22 expression, kinetics of IL-22 production by mice deficient for IL-23 or IL-6 were compared to WT controls. As early as d2 PI, reduced IL-22 was detected in Il23a−/− mice. This became significant on d3 and was reduced through d8 when the infected mice began to succumb (Fig. 4A). Despite a modest, progressive increase in IL-22 in Il23a−/− mice, this was insufficient to prevent bacterial dissemination and death (Fig. 3A–E). Reduction of IL-22 was also observed in Il6−/− mice (Fig. 4A), albeit delayed relative to that of Il23a−/− mice; it was not significant until d5 PI, when decreased IL-22+ CD4+ T cells were observed (Fig. 4B, C). Of note, significant production of IL-22 was induced by anti-CD3 as early as d3 PI (Fig. S4), indicating presence of IL-22+ T cells when IL-22 production by ILCs is dominant (Sonnenberg et al., 2011b; Zheng et al., 2008).
Figure 4.
IL-6–induced CD4+ T cells are major source of protective IL-22 late in C. rodentium infection
(A) WT, Il23a−/− and Il6−/− mice were inoculated with 2 × 109 cfu C. rodentium and colonic tissues collected at the indicated days PI for assessment of production of IL-22 in homogenate supernatants cultured for 24h. Data are means +/− SEM (*p<0.05, **p<0.01).
(B) Colonic LP cells of WT, Il23a−/− and Il6−/− mice infected with C. rodentium were isolated on the indicated days PI and analyzed by flow cytometry for expression of intracellular IL-22 and IL-17A by CD3+CD4+ gated cells. Numbers indicate the frequencies of cells in each quadrant.
(C) Flow cytometric frequency data for LPL isolates of WT, Il23a−/− and Il6−/− mice generated as in (B) and pooled from three independent experiments (n=3–4 mice per group). Data are means +/− SEM (*p<0.05, **p<0.01).
(D) Survival rates of C. rodentium infected WT mice that were untreated or treated with neutralizing IL-22 mAb on days 0 and 3 PI, or on days 6, 7 and 8 PI.
(E) Survival rates of Il6−/− mice infected with C. rodentium and treated with rIL-22-Fc protein (50 µg IP) or vehicle alone at d5 PI.
(F) Survival rates of C. rodentium infected Il22−/− mice treated with rIL-22Fc protein (50 µg IP) or vehicle alone at days 0 and 3, or days 5 and 7 PI.
(G) Survival rates of C. rodentium infected Il22−/− mice treated with rIL-22Fc protein (50 µg IP) or vehicle alone at days 0 and 3, or days 5 and 7 PI.
All data are representative of three or more experiments with a minimum of 3–4 mice per group.
By d8 PI, the deficit in IL-22 was significantly greater in Il6−/− than in Il23−/− mice (Fig. 4A). This correlated with a more pronounced decrease in frequencies of IL-22+ T cells in the lamina propria (LP) of Il6−/− mice (Fig. 4B, C) and was consistent with a predominance of IL-22+CD4+ T cells relative to ILCs (CD3−CD4− and CD3−CD4+ cells) in WT mice (Fig. S4A, B). Indeed, of the total IL-22+ cells over the first 8 days of infection, CD4+ T cells were significantly more abundant. Further, IL-22+IL-17− (single-positive; SP) CD4+ T cells were more frequent than IL-22+IL-17+ (DP) cells at each time point in infected WT mice (Fig. S4C), and, under conditions of ex vivo co-stimulation with IL-23, SP cells expressed the highest IL-22 per cell indicating they were likely the dominant source of IL-22 (Fig. S4A). Accordingly, IL-22 production in WT mice at d8 PI was almost a log-fold higher than at d3 (605pg/ml vs. 74pg/ml) (Fig. 4A), indicating that IL-22 contributed by CD4+ T cells surpassed that contributed by ILCs earlier in the infection. This was consistent with significantly more IL-22 elicited by anti-CD3 than IL-23 stimulation of colonic lymphoid cells (Fig. 4D), and the observation that IL-22 production in Il23a−/− mice was IL-6-dependent and therefore of T-cell origin (Fig. S4D). The critical role for IL-6 in the absence of IL-23 was highlighted by uniform fatality of low-dose C. rodentium infection in Il23a−/− mice treated with neutralizing antibody to IL-6 (Fig. S4E). Note that although the relative deficits of IL-22 produced by Il23a−/− and Il6−/− mice at d8 (Fig. 4A) appear more modest than might be expected from the far greater decrease in IL-22+ CD4+ T cells in the latter (Fig. 4B), when normalized for the considerably reduced numbers of T cells recruited to the colons of Il23a−/− mice (Fig. 3E, and data not shown), the greater deficit of IL-22 in Il6−/− mice more closely mirrored the reduction in IL-22+ T cells in these mice.
Because IL-21 has been identified as an autocrine factor downstream of IL-6 that promotes Th17 development and IL-22 expression by Th17 cells (Nurieva et al., 2007), we sought to determine whether IL-21 is important in host protection and the induction of CD4+ T cell-derived IL-22. Despite delayed clearance and elevated bacterial loads between weeks 2–5 PI (Fig. S4F, G), and modest reductions in mucosal IL-22+CD4+ T cells in Il21−/− mice, there was no increased mortality compared to WT controls and treatment of these mice with anti-IL-6 resulted in significant reductions in IL-22+CD4+ T cells and increased mortality (Fig. S4H, I, and data not shown). Thus, although IL-21 is required for efficient clearance of C. rodentium, in its absence the IL-6–dependent T cell response is sufficient to control bacterial loads and prevent mortality.
The importance of T cell-derived IL-22 to host protection was reflected in the marked decline of surviving WT mice treated with anti-IL-22 after d5 of infection (Fig. 4E). Whether administered during the onset of infection (days 0 and 3) or on days 6, 7 and 8, after the peak of the innate response, IL-22 blockade in WT animals resulted in comparable survival kinetics to that of Il22−/− and Il6−/− mice (Fig. 3C). Conversely, administration of either IL-22-Fc after the peak of the innate response rescued both Il22−/− and Il6−/− mice (Fig. 4F, G). Because Il6−/− mice retain IL-22–producing ILCs but are deficient in IL-22–producing T cells, IL-6–dependent induction of IL-22+CD4+ T cells appears critical for host protection after the early wave of IL-22 production by ILCs has peaked.
Reciprocal Expression of T-bet, AhR and RORγt in Th22 vs Th17 Cells
To identify factors that might distinguish Th22 and Th17 developmental programs, comparative transcriptome analysis was performed (Fig. 5A, B). Among 611 genes with at least a 2-fold difference in expression between the two subsets (Fig. 5A), notable reciprocal differences were found between transcripts encoding RORγt, RORα and the aryl hydrocarbon receptor (AhR), and T-bet by Th17 and Th22 cells, respectively (Fig. 5B). This was in contrast to IRF4 and BATF (Huber et al., 2008; Schraml et al., 2009), which were not significantly different between the two populations (data not shown). Expression differences were validated by quantitative RT-PCR (Fig. 5C, and data not shown). Thus, transcripts encoding RORγt (Rorc) and AhR (Ahr) were increased in polarized Th17 cells, with increasing expression differences correlating with the dose of TGFβ, whereas T-bet (Tbx21) was increased in polarized Th22 cells.
Figure 5.
Th22 and Th17 cells are distinguished by reciprocal expression of T-bet, AhR and RORγt
(A, B) FACS-sorted naïve OTII-Tg CD4+ T cells were activated by Ova peptide in presence of CD4+ T-cell depleted splenic APCs from Il23a−/− mice in the presence of IL-6 alone (Th22 polarization) or IL-6+TGFβ Thi7 polarization. CD4+ T cells were purified by magnetic sorting after 60h of culture and RNA was isolated for microarray-based gene expression analysis. Shown is a heat map of genes with more than 4-fold expression differences between the two populations (A) and transcriptome profiling of Th22- or Th17-polarized cells by scatterplot analysis (B).
(C) Naïve CD4+ T cells isolated from C57BL/6 mice were activated by anti-CD3 and anti-CD28 in presence of IL-6 and IL-23 (Th22 polarization), IL-6 and the indicated concentrations of TGFβ Thi7 polarization), or absence of added cytokines (Th0). On day 3 the cells were further stimulated with PMA/ionomycin for 6 hours and analyzed by RT-PCR for Rorc ,Tbx21 and Ahr transcripts. Data are means +/− SEM (*p<0.05, **p<0.01).
(D) Naïve OTII CD4+ T cells were simulated with Ova peptide in presence of IL-6, IL-6+IL-23 or IL-6+TGFβ and analyzed by flow cytometry for frequencies of IL-22+ CD4+ T cells on day 3 following stimulation for 4 hours with PMA/I in the presence of brefeldin A. The expression of T-bet and RORγt within the IL-22+ CD4+ gates was determined by intracellular staining (histograms).
All data are representative of three or more experiments with a minimum of 3 mice per group.
Differential expression of T-bet and RORγt in Th22 and Th17 cells was verified by flow cytometry, gating on the IL-22+ fraction of each (Fig. 5D). Consistent with gene expression data IL-22+ Th22 cells expressed T-bet, which was not detected in IL-22+ cells Th17 cells; T-bet expression was modestly, but reproducibly increased by IL-23. Conversely, RORγt expression by IL-22+ cells was markedly increased in Th17 polarized cells, and was reduced in Th22 polarized cells, irrespective of IL-23 addition. Consonant with these results, IFNγ did not inhibit, but modestly enhanced, Th22 cell development (data not shown), in contrast to the potent inhibitory effects of IFNγ on the development of Th17 cells (Harrington et al., 2005). In sum, these observations establish that distinct transcription factor networks underpin Th22 versus Th17 development and suggest a T-bet-dependent mechanism for enhanced IL-22 expression by developing Th22 cells. These data further suggest that induction of IL-22 expression might be differentially regulated by T-bet and AhR in the context of the Th22 and Th17 pathways, respectively.
Th22 Cells are Protective against C. rodentium Infection via a T-bet- and AhR-dependent Mechanism
In view of the foregoing findings, we postulated that Th22 cells were protective against C. rodentium infection and required T-bet for their optimal development and function. Further, we posited that, in contrast to Th17 cells, AhR might be dispensable for IL-22 expression by Th22 cells. We therefore examined the expression of Il22 transcripts from Th17 and Th22 cells generated in vitro from WT, Tbx21−/−, or Ahr−/− mice (Fig. 6A). Notably, deficiency of T-bet significantly compromised expression of Il22 transcripts in Th22 cells, but had no significant impact on Il22 expression by Th17 cells (Fig. 6A). However, differentiation of Th17 cells in the presence of IL-23, which induced ~4-fold higher amounts of T-bet (Fig. S5A), augmented their IL-22 production in a T-bet-dependent manner (Fig. S5B, C).
Figure 6.
IL-22 production by Th22 cells is T-bet- and AhR-dependent
(A-C) FACS-sorted naïve CD4 T cells (CD4+CD25-CD62Lhigh CD44low) from WT, Tbx21−/− or Ahr−/− mice were activated with anti-CD3/CD28 in presence of IL-6 and IL-23 (Th22 polarization), IL-6 and different concentrations of TGFβ Thi7 polarizations, or absence of cytokines (Th0). On day 3 CD4+ T cells were re-stimulated with PMA/ionomycin for 6 hours and analyzed by RT-PCR for Il22 (A), Ahr and Tbx21 (B), or maf (C) transcripts. Expression values are normalized to Th0 controls. Data are means +/− SEM (*p<0.05). (D) Naïve CD4 T cells from WT or Tbx21−/− mice were differentiated under Th0, Th22 or Th17 conditions. Where indicated, the AhR-antagonist, CH-223191 (3µM), was added at the initiation of cultures. IL-22 protein was quantified by ELISA from culture supernatants of T cells treated with PMA/ionomycin for 48 hours. Data are means +/− SEM (*p<0.01). (E, F) WT and Tbx21−/− mice were inoculated with C. rodentium and sacrificed on d8 PI for analysis of IL-22 production from supernatants of colon homogenates cultured ex vivo for 24h (E; *p<0.05), or for analysis of expression of intracellular IL-22 and IFN-γ by CD3+CD4+ cells (F). Numbers in each quadrant indicate frequencies (CD3 and CD4 gated).
(G) Colonization kinetics of WT and Tbx21−/− mice infected with 2 × 109 luminescent C. rodentium bacterial strain and assessed by whole body imaging on the indicated days PI. Data are means +/− SEM (*p<0.05, **p<0.01).
All data are representative of two or more experiments.
In agreement with previous reports (Veldhoen et al., 2008), AhR was essential for Il22 induction in Th17 cells. Thus, although Il22 transcripts were substantially lower in Th17 cells than Th22 cells, they were reduced to background in AhR-deficient Th17 cells (Fig. 6A). Unexpectedly, despite lower expression of Ahr by Th22 cells (Fig. 5C), AhR deficiency also significantly reduced their expression of Il22. Reciprocal expression of T-bet and AhR in Th22 and Th17 cells suggested that actions of T-bet to enhance Il22 expression were unlikely due to increased expression of Ahr downstream of T-bet. Indeed, expression levels of Ahr were increased in T-bet-deficient Th22 cells (Fig. 6B). Conversely, Tbx21 was increased in AhR-deficient Th22 cells. Finally, c-Maf, is a repressor of Il22 transcription (Rutz et al., 2011), raising the possibility that T-bet might suppress c-Maf as a mechanism to enhance Il22 transcription. However, while Tbx21 and maf were reciprocally expressed by Th22 and Th17 cells (Fig. 5B), maf transcripts were not affected by T-bet deficiency in either Th22 or Th17 cells (Fig. 6C), indicating that IL-22 enhancing actions of T-bet were not due to suppression of maf.
Collectively, these results indicated that optimal Il22 induction by Th22 cells is both T-bet- and AhR-dependent, suggesting cooperative actions of these factors in enhancing Il22 transcription. To directly test this, WT and T-bet–deficient Th22 cells were treated with the AhR antagonist, CH-223191, to determine whether the residual expression of IL-22 found in T-bet–deficient T cells was AhR-dependent (Fig. 6D). Pharmacologic inhibition of AhR in T-bet-deficient Th22 cells further reduced IL-22 to near background levels. IL-22 produced by Th17 cells from WT and T-bet–deficient mice were also ablated by AhR antagonism, indicating that at the high levels of expression AhR found in Th17 cells, Il22 expression is induced independently of T-bet.
To determine whether IL-22 production by T cells was T-bet and AhR dependent in vivo, WT, Tbx21−/− and Ahr−/− mice were inoculated with C. rodentium. In contrast to WT and Tbx21−/− mice, nearly all Ahr−/− mice rapidly succumbed to infection, consistent with this factor’s role in IL-22 production by ILCs (Kiss et al., 2011). However, in the few Ahr−/− mice that survived long enough to assess the T cell response, the frequency of IL-22+CD4+ T cells was significantly reduced, supporting the in vitro findings (Fig. 6A, S6, and data not shown). In contrast, the majority of Tbx21−/− mice survived infection, and when examined 8d PI were found to produce more than 2-fold less IL-22 than WT controls (Fig. 6E), and frequencies of IL-22+CD4+ T cells in Tbx21−/− mice were reduced ~3-fold (Fig. 6F). Notably, although T-bet deficiency compromised both total IL-22+ T cells and IL-22/IL-17 DP T cells, frequencies of IL-17 SP cells were not affected, consistent with in vitro findings (Fig. 6A, D) and T-bet–independent IL-22 induction in conventional Th17 cells, but not Th22 cells.
In accord with decreased IL-22–producing CD4+ T cells in Tbx21−/− mice, bacterial loads were significantly greater during the adaptive phase of the response (Fig. 6G). Nevertheless, few of the Tbx21−/− mice innoculated with high dose C. rodentium succumbed to infection due to unimpaired ILC responses, and bacterial loads were gradually controlled due to the development of an unimpaired, protective antibody response in these mice (data not shown).
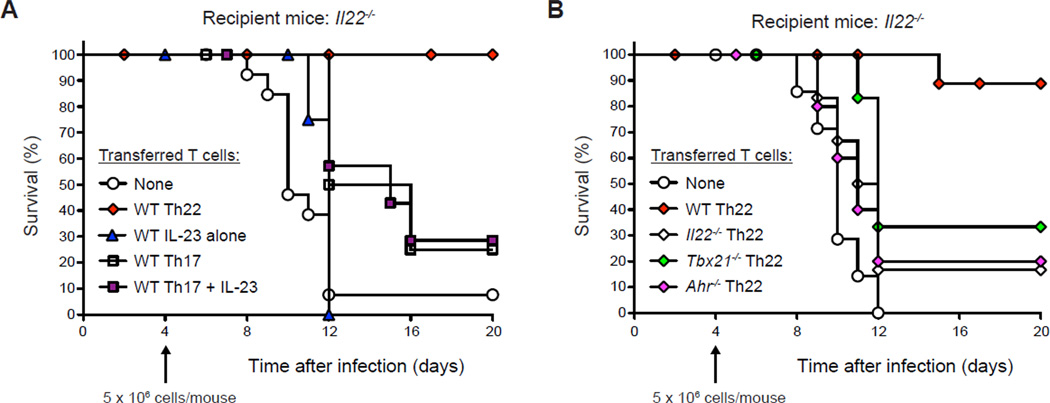
To bypass the contribution of ILC-derived IL-22 and more directly examine the protective function of Th22 and Th17 cells, transfers of different populations of polarized CD4+ T cells into IL-22-deficient mice were performed (Fig. 7A, B). Naïve WT CD4+ T cells were differentiated with IL-6 alone (Th22 polarizing conditions), IL-23 alone, or with IL-6+TGFβwithorwithoutadditionoflL̃23 (Th17 polarizing conditions), and then transferred into C. rodentium-infected Il22−/− mice after the peak of the innate response (Fig. 7A). Whereas Il22−/− recipients of Th22 cells were nearly completely protected compared to mice that received no T cells (10% vs. 85% mortality; p<0.01), recipients of IL-23-induced CD4+ T cells were not protected (100% mortality), in accord with their lack of IL-22 production (Figs. 2A and S2A, D). In contrast to recipients of Th22 cells, recipients of Th17 cells showed only modest, albeit significant, improvement in mortality and survival times whether treated with IL-23 or not.
Figure 7.
Th22 cells provide superior host protection to Th17 cells
(A, B) Naïve CD4+ T cells from WT mice were stimulated in vitro for 72h with anti-CD3+anti-CD28 with addition of IL-6 (Th22), IL-23 (IL-23 alone), or IL-6+TGFβ, without or with IL-23 addition (Th17 and Th17+IL-23, respectively), prior to isolation and transfer (A). Naïve CD4+ T cells from WT, Il22−/−, Tbx21−/− and Ahr−/− mice were similarly stimulated in vitro with addition of IL-6 (WT Th22, Il22−/− Th22, Tbx21−/− Th22 and Ahr−/− Th22, respectively), prior to isolation and transfer (B). N ≥ 5 mice per group; representative of two or more experiments.
Consistent with a requirement for T cell-derived IL-22 for protection, transfers of Il22−/− T cells polarized under Th22 conditions failed to enhance survival (Fig. 7B). Notably, recipients of Th22 cells derived from either Tbx21−/− or Ahr−/− mice had similarly impaired protection, consistent with important roles for both T-bet and AhR in expression of IL-22 by Th22 cells. Thus, Th22 cells provide greater host protection against lethal C. rodentium infection than Th17 cells, via an IL-22–dependent mechanism that requires T-bet and AhR for optimal anti-pathogen defense.
DISCUSSION
In this study we define an important role for Th22 cells in mucosal defense against the enteric bacterial pathogen, Citrobacter rodentium. Whereas Th17 cells were thought previously to be central to anti-bacterial protection at barrier sites, including the intestines (Aujla et al., 2008; Mangan et al., 2006; Ouyang et al., 2008; Sonnenberg et al., 2011a; Zheng et al., 2008), we find that Th22 cells differentiated in an IL-6–dependent, TGF-β–independent manner are more effective in host protection than Th17 cells. Programming of Th22 cells for high expression of IL-22 by IL-6 was independent of IL-23, but IL-23 enhanced production by Th22 cells primed by IL-6. Thus, although IL-23−induced expression of IL-22 by ILCs is indispensable for protection at high inoculating doses of this pathogen, we find that IL-23−deficient mice are protected against infection with lower inocula via an IL-6–dependent mechanism that is linked to development of Th22 and Th17 cells and is independent of production of IL-22 by ILCs. Importantly, whereas Th17 cells were strictly dependent on AhR for expression of IL-22, Th22 cells required co-expression of AhR and T-bet for optimal expression of IL-22, and both transcription factors were necessary for host protection by Th22 cells.
Anti-pathogenic responses are characterized by integration of the innate and adaptive arms of immunity. Recent studies have emphasized the critical role of different populations of ILCs in protection against C. rodentium challenge, including NK-like cells (NK-22) and CD4+ lymphoid tissue inducer (LTi) cells (Colonna, 2009; Sonnenberg et al., 2011a). While ILCs provide a rapid source of IL-22 that is essential for early host protection, we find that CD4+ T cells become the dominant source of IL-22 in the intestinal LP where the anti-microbial response is concentrated at the height of infection. While the current study does not define the relative contribution of IL-22–producing CD4+ T cells derived from Th22 cell precursors, Th17 cell precursors, or both, it does establish a critical role for IL-22–producing CD4+ T cells in epithelial barrier protection as the innate response declines. Based on greater production of IL-22 by Th22 cells derived ex vivo, the higher frequency of IL-22+ CD4+ T cells that produce IL-22 independently of IL-17A or IFNγ co-expression, and the superior protective function of Th22 cells transferred into infected IL-22–deficient recipients, we favor the view that Th22 cells are the principal contributors of IL-22 derived from the adaptive response, although this will require further study. Irrespective of developmental origin, however, our findings highlight a sequential model of IL-22–dependent bacterial clearance wherein early production of IL-22 by ILCs gives way to production of IL-22 by CD4+ T cells that become the dominant source of IL-22 and are required for complete clearance of infection.
Because ILCs are strictly dependent on IL-23 for their production of IL-22, the discovery herein that IL-23−deficient mice survive infection by low doses of C. rodentium via an IL-22–dependent mechanism provoked inquiry into ILC–independent mechanisms of pathogen clearance, and led to a re-examination of mechanisms by which IL-6 deficiency confers increased mortality (Dann et al., 2008; Zheng et al., 2008). Given that IL-6 is required for Th22 and Th17 differentiation, the finding that CD4+ T cells become the dominant source of IL-22 concordant with the decline in bacterial load provides a basis for the impaired survival of IL-6–deficient mice despite a lack of significantly diminished production of IL-22 by ILCs. It was reported previously that IL-6 produced by the intestinal epithelium and macrophages conferred protection against C. rodentium infection largely through its anti-apoptotic effects on the epithelium (Dann et al., 2008). While direct actions of IL-6 on the intestinal epithelium no doubt contribute to host protection, our finding that delivery of IL-22-Fc conferred protection of infected IL-6–deficient mice supports a more critical role for the effects of IL-6 on induction on IL-22 producing T cells. This is consistent with the remarkably similar survival, colonization kinetics and colonic histopathology of IL-6– and IL-22–deficient mice inoculated with C. rodentium, and reinforces the importance of IL-22 derived from CD4+ T cells as a critical component of the host’s response to maintain barrier integrity.
It is notable that resistance to C. rodentium infection is more impaired in IL-23−deficient mice than either IL-22– or IL-6–deficient mice. This implies that IL-23 exerts host-protective effects that are independent of its actions to induce IL-22 and IL-6, production of which in the intestinal mucosa are severely compromised in the absence of IL-23. Thus, in addition to directly impairing IL-22 production by ILCs and mature Th22 and Th17 cells, and perhaps indirectly impairing development of Th22 and Th17 cells through diminished IL-6 induction, deficiency of IL-23 compromises anti-bacterial resistance through additional mechanisms. As noted above, IL-23–dependent induction of IL-6 that directly promotes epithelium sparing as well as the induction of IL-1β and TNF-α that promotes epithelial production of anti-microbial peptides via an IL-17C–dependent autocrine loop are likely contributory (Ramirez-Carrozzi et al., 2011; Song et al., 2011). Further, effects of reduced local chemokine production, as exemplified by CXCL1 (KC), among others (data not shown), impair recruitment of innate and adaptive immune cells that contribute to barrier defense and eradication of the pathogen (Aujla et al., 2008; Kolls et al., 2008; Ouyang et al., 2008). Suffice to say that in view of the range of proinflammatory cytokines and chemokines that are uniquely diminished in the intestines of infected IL-23–deficient mice, the protective effects mediated by IL-23 are likely multifactorial and will require further study.
Whereas IL-6 was essential for the host-protective development of IL-22 producing CD4+ T cells, IL-21 proved dispensable. Despite a decrement in the frequency of IL-22+CD4+ T cells in IL-21–deficient mice challenged with C. rodentium, there was no increased mortality despite delayed pathogen clearance. We speculate that the delayed clearance is due to an impaired C. rodentium-specific antibody response, as systemic and mucosal antibody production is important for host protection in this model and IL-21-deficiency results in impaired IgG antibody responses (Bry and Brenner, 2004; Ozaki et al., 2002; Yoshida et al., 2006). In viral infections, IL-21 has been shown to be essential for preventing chronic infection and supporting optimal anti-viral antibody responses (Elsaesser et al., 2009; Eto et al., 2011; Yi et al., 2009), and in infection with Listeria monocytogenes, IL-21 deficiency was associated with increased IL-17 production and unimpaired pathogen clearance (Ertelt et al., 2010). Findings herein define a marginal role for IL-21 the development of IL-22+CD4+ T cells and suggest that its principal role in host-protective mucosal immunity might be more related to its role in providing B cell help.
Comparative transcriptome analysis of Th22 and Th17 cells revealed differential expression of several hundred genes, among which were several key transcription factors. This included reciprocal expression of transcripts encoding T-bet and RORγt, consistent with TGFβ’s established actions in suppressing Tbx21 and enhancing Rorc expression, and led to identification of T-bet as an important factor in driving high-level expression of IL-22 and host protection by Th22 cells. In contrast to T-bet, we found no significant difference in expression of Eomes, a related T-box family member expressed in T cells, suggesting that Eomes is unlikely to contribute to Th22 development. Remarkably, despite the low levels of AhR in Th22 cells, AhR also contributed significantly to Il22 expression in Th22 cells such that combined loss of T-bet and AhR actions resulted in nearly complete abrogation of Il22 expression. Regardless of higher levels of AhR in Th17 cells, optimal IL-22 induction is severely compromised, possibly due to TGFβ, which might inhibit IL-22 induction by repressing T-bet and enhancing other transcription factors, including c-Maf (Rutz et al., 2011).
Because neither T-bet nor AhR positively regulated the expression of the other, their actions on Il22 expression appear to be cooperative. This is in contrast to Th17 cells, which are strictly dependent on AhR for Il22 expression (Veldhoen et al., 2008). Thus, while our findings confirm that Th22 cells can express Il22 independently of AhR (Rutz et al., 2011), AhR clearly contributes to the transcriptional regulation of Il22 in both Th17 and Th22 cells and is a dominant transcription factor regulating Il22 transcription. It should be noted that Il22 is closely linked to the Ifng gene on chromosome 10 in mice (and to IFNG on chromosome 12 in humans), suggesting that T-bet, originally described as a critical regulator of Ifng gene expression, might have more extended regional actions on this gene cluster. This could offer an explanation for the original description of IL-22 as a product of Th1 cells (Gurney, 2004), which also co-express AhR, and has implications for Il22 expression in Th17 cells as well, in which enhanced expression of Il22 downstream of IL-23 is contingent upon STAT1- and/or STAT4-dependent expression of T-bet (unpublished observations). In this regard, it has been shown that ectopic expression of T-bet in Th17 cells enhanced Il22 expression while suppressing Il17a and Il17f (Lazarevic et al., 2011). Given the role of IL-23 induced T-bet in the late divergence of Th17 cells towards expression of Ifng (Lee et al., 2009), it would appear that this mechanism might facilitate the development of Th22 type cells from Th17 precursors, a subject of current investigation.
In agreement with a recent report (Rutz et al., 2011), we found higher expression of the AP-1 transcription factor family member Maf (c-Maf) by Th17 cells. In view of our finding that low expression of maf by Th22 cells was not reversed by T-bet deficiency, the actions of T-bet to promote Il22 expression by Th22 cells are not due to derepression of c-Maf. In addition to their decreased expression of Maf, Th22 cells also expressed significantly lower levels of Rorc compared to Th17 cells, consistent with T-bet’s actions to repress Rorc expression by direct binding to the Rorc locus and complexing with Rorc positive regulator, Runx1 (Lazarevic et al., 2011; Mukasa et al., 2010), expression of which was also significantly decreased in Th22 cells (data not shown). Whether RORγt is required for Il22 expression by Th22 cells despite its relatively low expression, akin to AhR, remains to be determined. However, these data establish both AhR and T-bet as critical positive regulators of Il22 expression in Th22 cells and suggest that the enhanced expression of Il22 downstream of IL-23 in both Th22 and Th17 cells are linked to its induction of Tbx21 expression by activation of STAT1, STAT4, or both.
Notably, the transcriptome of Th22 cells described herein is remarkably similar to that of the ‘pathogenic’ Th17 cells whose development from naïve CD4+ T cells was induced by actions of IL-6, IL-23 and IL-1β in the absence of TGFβ (Ghoreschi et al., 2010). These cells, referred to as Th17(23) cells, were distinguished from conventional Th17 cells differentiated with TGFβ, termed Th17(β) cells, by their distinct gene expression profile and greater pathogenicity in a transfer model of experimental allergic encephalomyelitis (EAE). Given their similarities to Th17(23) cells, this begs the question of whether Th22 cells might under certain circumstances also contribute to pathogenicity in the intestine and other tissue sites. IL-22–producing CD4+ T cells were recently reported to be pathogenic in a transfer model of colitis that used CD45RBlo cells depleted of Treg cells (Kamanaka et al., 2011). Given that Th22 cells were originally described in humans (Duhen et al., 2009; Trifari et al., 2009) and that elevations of IL-22 are found in the intestines of patients with inflammatory bowel disease (IBD) as well as patients with psoriasis and rheumatoid arthritis (Andoh et al., 2005; Ikeuchi et al., 2005; Wolk et al., 2004; Wolk et al., 2007; Wolk et al., 2006), it will be important to determine whether Th22 cells are contributors to pathogenicity in at least some variants of human autoimmune disease and IBD previously thought to be linked to conventional Th17 cells.
EXPERIMENTAL PROCEDURES
Mice
Mice were purchased from the Jackson Laboratories and/or bred at our facility: C57BL/6J (B6); B6.Cg-Tg(TcraTcrb)425Cbn/J (OTII TCR transgenic); B6.129S6-Tbx21tm1Glm/J (Tbx21−/−); B6.129S2-Il6tm1Kopf/J (Il6−/−); B6.129S7-Ifngtm1Ts/J (Ifng−/−); B6. Il17ra−/−; B6.Il23a−/− (kindly provided by Schering-Plough Biopharma); B6.129S5-Il22tm1Lex mice (Il22−/−; kindly provided by Genentech); B6.129S5-Il21tm1Lex (Il21−/−) and B6.Ahr−/−. All animals were bred and maintained in accordance with institutional animal care and use committee regulations.
Citrobacter rodentium Infection and Treatments
Citrobacter rodentium strain, DBS100 (ATCC), was used for all inoculations with the exception of experiments in which whole body imaging was performed. For imaging experiments, the bioluminescent C. rodentium strain ICC180 (derived from DBS100) was used (generously provided by Drs. Gadi Frankel and Siouxsie Wiles, Imperial College London) (Wiles et al., 2006). Mice were inoculated with 1–2 × 109 cfu (high dose) or 0.5 ×106 cfu (low dose) in a total volume of 200 µl of PBS via gastric gavage. In vivo neutralization studies used 150 µg per dose of anti-IL-22 mAb (Genentech, clone 8E11), anti-IL-6 mAb (clone MP5–20F3) or isotype control mAb, each injected IP. For in vivo delivery of IL-22, hIL-22.Fc protein (Pfizer) was given IP at 50 µg per mouse.
Bioluminescence Imaging
Mice were anesthetized with isoflurane and placed in a supine position in a custom built chamber for imaging with a IVIS-100 system and Living Image Software (Xenogen, Inc.). Baseline images were collected prior to gavage with 2 × 109 cfu C. rodentium strain ICC180 and whole body images were taken at a binning of 4 over 1 to 3 min at the indicated times during infection. Luminescence emitted from the same gate in individual mice was quantified as counts per second and pseudocolor images representing light intensity generated.
Gene Expression Analysis
Naïve OTII CD4+ T cells were sorted and activated by OVAp and T-cell depleted splenocytes from Il23a−/− mice in the presence of IL-6 alone (Th22) or under IL-6 + TGFβThi7. T cells were isolated after 60 hours of culture and DNase-free RNA was isolated for microarray analysis using the Affymetrix Mouse Gene ST 1.0 Array as previously described (Lee et al., 2009) and analyzed per Supplemental Procedures.
Adoptive transfer experiments
Naïve CD4+ T cells from pooled spleens and lymph nodes of WT, Il22−/− or Tbx21−/− mice were purified by FACS sorting and differentiated for 3 days under Th22 or Th17 conditions (see Supplemental Procedures for details). Recovered T cells were transferred by IP injection (5 × 106) cells per mouse) into recipients inoculated with C. rodentium 4 days prior to cell transfer and were analyzed for survival.
Statistical Analysis
Statistical analyses were performed using Student’s t-test and p<0.05 was considered significant.
Supplementary Material
Highlights.
IL-23–deficient mice are protected from low-dose Citrobacter rodentium infection.
CD4+ T cell derived IL-22 is required for C. rodentium infection protection.
Th22 cells provide more effective anti-bacterial defense than Th17 cells.
T-bet acts cooperatively with AhR in Il22 induction in Th22 cells.
Acknowledgments
The authors are grateful to Dr. David Randolph and members of the Weaver laboratory for their helpful comments and suggestions. We thank Drs. Josh Mezrich and Chris Bradfield (Univ. Wisconsin) for provision of AhR-deficient mice, and Dr. David Crossman for assistance with analysis of gene expression data. We also thank the UAB Digestive Diseases Research Developmental Center (DDRDC), the UAB Small Animal Imaging Facility, UAB Genomics Core, UAB Center for AIDS Research Flow Cytometry Core, and UAB Epitope Recognition and Immunoreagent Core Facility for genotyping of gene-targeted mice, imaging studies, gene expression studies, flow cytometric sorting, and antibody preparations, respectively. This work was supported by grants from the NIH (C.T.W. and R.D.H.) and the Crohn’s and Colitis Foundation of America (R.B. and D.B.O.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Andoh A, Zhang Z, Inatomi O, Fujino S, Deguchi Y, Araki Y, Tsujikawa T, Kitoh K, Kim-Mitsuyama S, Takayanagi A, et al. Interleukin-22, a member of the IL-10 subfamily, induces inflammatory responses in colonic subepithelial myofibroblasts. Gastroenterology. 2005;129:969–984. doi: 10.1053/j.gastro.2005.06.071. [DOI] [PubMed] [Google Scholar]
- Aujla SJ, Chan YR, Zheng M, Fei M, Askew DJ, Pociask DA, Reinhart TA, Mcallister F, Edeal J, Gaus K, et al. IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nat. Med. 2008;14:275–281. doi: 10.1038/nm1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bry L, Brenner MB. Critical role of T cell-dependent serum antibody, but not the gut-associated lymphoid tissue, for surviving acute mucosal infection with Citrobacter rodentium, an attaching and effacing pathogen. J. Immunol. 2004;172:433–441. doi: 10.4049/jimmunol.172.1.433. [DOI] [PubMed] [Google Scholar]
- Buonocore S, Ahern PP, Uhlig HH, Ivanov II, Littman DR, Maloy KJ, Powrie F. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature. 2010;464:1371–1375. doi: 10.1038/nature08949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella M, Fuchs A, Vermi W, Facchetti F, Otero K, Lennerz JKM, Doherty JM, Mills JC, Colonna M. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 2009;457:722–725. doi: 10.1038/nature07537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonna M. Interleukin-22-producing natural killer cells and lymphoid tissue inducer-like cells in mucosal immunity. Immunity. 2009;31:15–23. doi: 10.1016/j.immuni.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Dann SM, Spehlmann ME, Hammond DC, Iimura M, Hase K, Choi LJ, Hanson E, Eckmann L. IL-6-dependent mucosal protection prevents establishment of a microbial niche for attaching/effacing lesion-forming enteric bacterial pathogens. J. Immunol. 2008;180:6816–6826. doi: 10.4049/jimmunol.180.10.6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhen T, Geiger R, Jarrossay D, Lanzavecchia A, Sallusto F. Production of interleukin 22 but not interleukin 17 by a subset of human skin-homing memory T cells. Nat. Immunol. 2009;10:857–863. doi: 10.1038/ni.1767. [DOI] [PubMed] [Google Scholar]
- Elsaesser H, Sauer K, Brooks DG. IL-21 is required to control chronic viral infection. Science. 2009;324:1569–1572. doi: 10.1126/science.1174182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertelt JM, Johanns TM, Rowe JH, Way SS. Interleukin (IL)-21-independent pathogen-specific CD8+ T-cell expansion, and IL-21-dependent suppression of CD4+ T-cell IL-17 production. Immunology. 2010;131:183–191. doi: 10.1111/j.1365-2567.2010.03287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eto D, Lao C, Ditoro D, Barnett B, Escobar TC, Kageyama R, Yusuf I, Crotty S. IL-21 and IL-6 are critical for different aspects of B cell immunity and redundantly induce optimal follicular helper CD4 T cell (Tfh) differentiation. PLoS ONE. 2011;6:e17739. doi: 10.1371/journal.pone.0017739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoreschi K, Laurence A, Yang X-P, Tato CM, Mcgeachy MJ, Konkel JE, Ramos HL, Wei L, Davidson TS, Bouladoux N, et al. Generation of pathogenic TH17 cells in the absence of TGF-β signalling. Nature. 2010;467:967–971. doi: 10.1038/nature09447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurney AL. IL-22, a Th1 cytokine that targets the pancreas and select other peripheral tissues. Internat. Immunopharm. 2004;4:669–677. doi: 10.1016/j.intimp.2004.01.016. [DOI] [PubMed] [Google Scholar]
- Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, Weaver CT. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat. Immunol. 2005;6:1123–1132. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- Higgins LM, Frankel G, Douce G, Dougan G, MacDonald TT. Citrobacter rodentium infection in mice elicits a mucosal Th1 cytokine response and lesions similar to those in murine inflammatory bowel disease. Infect. Immun. 1999;67:3031–3039. doi: 10.1128/iai.67.6.3031-3039.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber M, Brüstle A, Reinhard K, Guralnik A, Walter G, Mahiny A, von Löw E, Lohoff M. IRF4 is essential for IL-21-mediated induction, amplification, and stabilization of the Th17 phenotype. Proc. Natl. Acad. Sci. USA. 2008;105:20846–20851. doi: 10.1073/pnas.0809077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi H, Kuroiwa T, Hiramatsu N, Kaneko Y, Hiromura K, Ueki K, Nojima Y. Expression of interleukin-22 in rheumatoid arthritis: potential role as a proinflammatory cytokine. Arthrit. Rheum. 2005;52:1037–1046. doi: 10.1002/art.20965. [DOI] [PubMed] [Google Scholar]
- Kamanaka M, Huber S, Zenewicz LA, Gagliani N, Rathinam C, O’Connor W, Wan YY, Nakae S, Iwakura Y, Hao L, Flavell RA. Memory/effector (CD45RBlo) CD4 T cells are controlled directly by IL-10 and cause IL-22-dependent intestinal pathology. J. Exp. Med. 2011;208:1027–1040. doi: 10.1084/jem.20102149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss EA, Vonarbourg C, Kopfmann S, Hobeika E, Finke D, Esser C, Diefenbach A. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science. 2011;334:1561–1563. doi: 10.1126/science.1214914. [DOI] [PubMed] [Google Scholar]
- Kolls JK, McCray PB, Chan YR. Cytokine-mediated regulation of antimicrobial proteins. Nat. Rev. Immunol. 2008;8:829–835. doi: 10.1038/nri2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarevic V, Chen X, Shim J-H, Hwang ES, Jang E, Bolm AN, Oukka M, Kuchroo VK, Glimcher LH. T-bet represses TH17 differentiation by preventing Runx1-mediated activation of the gene encoding RORγt. Nat. Immunol. 2011;12:96–104. doi: 10.1038/ni.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YK, Turner H, Maynard CL, Oliver JR, Chen D, Elson CO, Weaver CT. Late developmental plasticity in the T helper 17 lineage. Immunity. 2009;30:92–107. doi: 10.1016/j.immuni.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang SC, Tan X-Y, Luxenberg DP, Karim R, Dunussi-Joannopoulos K, Collins M, Fouser LA. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J. Exp. Med. 2006;203:2271–2279. doi: 10.1084/jem.20061308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan PR, Harrington LE, O’Quinn DB, Helms WS, Bullard DC, Elson CO, Hatton RD, Wahl SM, Schoeb TR, Weaver CT. Transforming growth factor-p induces development of the TH17 lineage. Nature. 2006;441:231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- Mukasa R, Balasubramani A, Lee YK, Whitley SK, Weaver BT, Shibata Y, Crawford GE, Hatton RD, Weaver CT. Epigenetic instability of cytokine and transcription factor gene loci underlies plasticity of the T helper 17 cell lineage. Immunity. 2010;32:616–627. doi: 10.1016/j.immuni.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy R, MacDonald TT, Dougan G, Frankel G, Wiles S. Citrobacter rodentium of mice and man. Cell. Microbiol. 2005;7:1697–1706. doi: 10.1111/j.1462-5822.2005.00625.x. [DOI] [PubMed] [Google Scholar]
- Nurieva R, Yang XO, Martinez G, Zhang Y, Panopoulos AD, Ma L, Schluns K, Tian Q, Watowich SS, Jetten AM, Dong C. Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature. 2007;448:480–483. doi: 10.1038/nature05969. [DOI] [PubMed] [Google Scholar]
- Ouyang W, Kolls JK, Zheng Y. The biological functions of T helper 17 cell effector cytokines in inflammation. Immunity. 2008;28:454–467. doi: 10.1016/j.immuni.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang W, Rutz S, Crellin NK, Valdez PA, Hymowitz SG. Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Ann. Rev. Immunol. 2011;29:71–109. doi: 10.1146/annurev-immunol-031210-101312. [DOI] [PubMed] [Google Scholar]
- Ozaki K, Spolski R, Feng CG, Qi C-F, Cheng J, Sher A, Morse HC, Liu C, Schwartzberg PL, Leonard WJ. A critical role for IL-21 in regulating immunoglobulin production. Science. 2002;298:1630–1634. doi: 10.1126/science.1077002. [DOI] [PubMed] [Google Scholar]
- Ramirez-Carrozzi V, Sambandam A, Luis E, Lin Z, Jeet S, Lesch J, Hackney J, Kim J, Zhou M, Lai J, et al. IL-17C regulates the innate immune function of epithelial cells in an autocrine manner. Nat. Immunol. 2011;12:1159–1166. doi: 10.1038/ni.2156. [DOI] [PubMed] [Google Scholar]
- Rutz S, Noubade R, Eidenschenk C, Ota N, Zeng W, Zheng Y, Hackney J, Ding J, Singh H, Ouyang W. Transcription factor c-Maf mediates the TGF-β-dependent suppression of IL-22 production in TH17 cells. Nat. Immunol. 2011;12:1238–1245. doi: 10.1038/ni.2134. [DOI] [PubMed] [Google Scholar]
- Sawa S, Cherrier M, Lochner M, Satoh-Takayama N, Fehling HJ, Langa F, Di Santo JP, Eberl G. Lineage relationship analysis of RORγt+ innate lymphoid cells. Science. 2010;330:665–669. doi: 10.1126/science.1194597. [DOI] [PubMed] [Google Scholar]
- Schraml B, Hildner K, Ise W, Lee W, Smith W, Solomon B, Sahota G, Sim J, Mukasa R, Cemerski S, et al. The AP-1 transcription factor Batf controls TH17 differentiation. Nature. 2009 doi: 10.1038/nature08114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Zhu S, Shi P, Liu Y, Shi Y, Levin SD, Qian Y. IL-17RE is the functional receptor for IL-17C and mediates mucosal immunity to infection with intestinal pathogens. Nat. Immunol. 2011;12:1151–1158. doi: 10.1038/ni.2155. [DOI] [PubMed] [Google Scholar]
- Sonnenberg GF, Fouser LA, Artis D. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat. Immunol. 2011a;12:383–390. doi: 10.1038/ni.2025. [DOI] [PubMed] [Google Scholar]
- Sonnenberg GF, Monticelli LA, Elloso MM, Fouser LA, Artis D. CD4+ lymphoid tissue-inducer cells promote innate immunity in the gut. Immunity. 2011b;34:122–134. doi: 10.1016/j.immuni.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatori H, Kanno Y, Watford WT, Tato CM, Weiss G, Ivanov II, Littman DR, O’shea JJ. Lymphoid tissue inducer-like cells are an innate source of IL-17 and IL-22. J. Exp. Med. 2009;206:35–41. doi: 10.1084/jem.20072713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifari S, Kaplan CD, Tran EH, Crellin NK, Spits H. Identification of a human helper T cell population that has abundant production of interleukin 22 and is distinct from TH-17, TH1 and TH2 cells. Nature Immunology. 2009;10:864–871. doi: 10.1038/ni.1770. [DOI] [PubMed] [Google Scholar]
- Veldhoen M, Hirota K, Westendorf AM, Buer J, Dumoutier L, Renauld J-C, Stockinger B. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453:106–109. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- Wiles S, Pickard KM, Peng K, MacDonald TT, Frankel G. In vivo bioluminescence imaging of the murine pathogen Citrobacter rodentium . Infect. Immun. 2006;74:5391–5396. doi: 10.1128/IAI.00848-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolk K, Kunz S, Witte E, Friedrich M, Asadullah K, Sabat R. IL-22 increases the innate immunity of tissues. Immunity. 2004;21:241–254. doi: 10.1016/j.immuni.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Wolk K, Witte E, Hoffmann U, Doecke W-D, Endesfelder S, Asadullah K, Sterry W, Volk H-D, Wittig BM, Sabat R. IL-22 induces lipopolysaccharide-binding protein in hepatocytes: a potential systemic role of IL-22 in Crohn’s disease. J. Immunol. 2007;178:5973–5981. doi: 10.4049/jimmunol.178.9.5973. [DOI] [PubMed] [Google Scholar]
- Wolk K, Witte E, Wallace E, Döcke W-D, Kunz S, Asadullah K, Volk H-D, Sterry W, Sabat R. IL-22 regulates the expression of genes responsible for antimicrobial defense, cellular differentiation, and mobility in keratinocytes: a potential role in psoriasis. Eur. J. Immunol. 2006;36:1309–1323. doi: 10.1002/eji.200535503. [DOI] [PubMed] [Google Scholar]
- Wolk K, Witte E, Witte K, Warszawska K, Sabat R. Biology of interleukin-22. Sem. Immunopathol. 2010;32:17–31. doi: 10.1007/s00281-009-0188-x. [DOI] [PubMed] [Google Scholar]
- Yi JS, Du M, Zajac AJ. A vital role for interleukin-21 in the control of a chronic viral infection. Science. 2009;324:1572–1576. doi: 10.1126/science.1175194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M, Masuda A, Kuo TT, Kobayashi K, Claypool SM, Takagawa T, Kutsumi H, Azuma T, Lencer WI, Blumberg RS. IgG transport across mucosal barriers by neonatal Fc receptor for IgG and mucosal immunity. Sem. Immunopathol. 2006;28:397–403. doi: 10.1007/s00281-006-0054-z. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Danilenko DM, Valdez P, Kasman I, Eastham-Anderson J, Wu J, Ouyang W. Interleukin-22, a TH17 cytokine, mediates IL-23-induced dermal inflammation and acanthosis. Nature. 2007;445:648–651. doi: 10.1038/nature05505. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ, Ouyang W. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.