Abstract
MicroRNAs (miRNAs) are critical post-transcriptional regulators of gene expression that control osteoblast mediated bone formation and osteoclast-related bone remodelling. Deregulation of miRNA mediated mechanisms is emerging as an important pathological factor in bone degeneration (e.g., osteoporosis) and other bone-related diseases. MiRNAs are intriguing regulatory molecules that are networked with cell signaling pathways and intricate transcriptional programs through ingenuous circuits with remarkably simple logic. This overview examines key principles by which miRNAs control differentiation of osteoblasts as they evolve from mesenchymal stromal cells during osteogenesis, or of osteoclasts as they originate from monocytic precursors in the hematopoietic lineage during osteoclastogenesis. Of particular note are miRNAs that are temporally up-regulated during osteoblastogenesis (e.g., miR-218) or osteoclastogenesis (e.g., miR-148a). Each miRNA stimulates differentiation by suppressing inhibitory signalling pathways (‘double-negative’ regulation). The excitement surrounding miRNAs in bone biology stems from the prominent effects that individual miRNAs can have on biological transitions during differentiation of skeletal cells and correlations of miRNA dysfunction with bone diseases. MiRNAs have significant clinical potential which is reflected by their versatility as disease-specific biomarkers and their promise as therapeutic agents to ameliorate or reverse bone tissue degeneration.
Keywords: Osteoblast, osteoclast, osteoporosis, skeletal development, bone mineral density, osteogenesis, mesenchymal stem cell, miRNA
Introduction
MicroRNAs (miRNAs) have important regulatory roles in bone cell growth, differentiation and function. These short non-coding RNAs are produced from larger precursors upon cleavage by the RNA processing enzyme Dicer and have emerged as critical post-transcriptional regulators of gene expression. They function by binding to specific microRNA (miRNA) recognition sequences (‘seed sequences’) located in the 3′ untranslated regions (3′ UTRs) of target mRNAs. By this mechanism, miRNAs can suppress expression of target proteins by enhancing mRNA degradation and/or by directly interfering with protein translation. Evidence from an ever-increasing number of studies indicates that the biological regulatory potential of miRNAs is functionally comparable to that of transcription factors binding to 5′ regulatory regions of genomic loci.
A large number of papers (>20,000) has been published on miRNAs since their initial discovery about twenty years ago by the research groups of Ambros and Ruvkun [reviewed in 1]. A respectable subset of these (>600) has focused on the roles of miRNAs in bone and cartilage tissues. Several excellent reviews have covered different aspects of miRNA functions in skeletal development and disease [2–7], including their regulatory roles in the growth, differentiation and function of both osteoblasts and osteoclasts, as well as chondrocytes and other mesenchymal cell types (e.g., adipocytes and myoblasts). This review will highlight key approaches that have been informative for our understanding of the biological, cellular and molecular roles of miRNAs in relation to normal bone homeostasis or osteoporosis. In particular, we discuss recent papers suggesting that deregulation of miRNA networks (e.g., due to polymorphisms that alter miRNA target sites or mutations that delete miRNA coding sequences) can predispose to bone diseases.
MicroRNAs control both osteoblastogenesis and osteoclastogenesis
Bone metabolism reflects a delicate balance in anabolic activities of osteoblasts and the catabolic actions of osteoclasts. MiRNAs contribute to the formation and activities of both of these cell types as evidenced by loss-of-function analyses of Dicer, an enzyme essential for miRNA maturation. A general null mutation of Dicer results in embryonic lethality and specifically prevents formation of a normal vertebrate body plan at gastrulation [8]. Subsequently, conditional Dicer null alleles were developed that permit Dicer gene inactivation by removal of an essential segment of the Dicer locus that inactivates the enzyme (i.e., Type III ribonuclease) [e.g., 9–10]. In these conditional null alleles, cell type specific expression of the cyclization recombinase (Cre; originally isolated from Enterobacteria phage P1) results in the removal of targeted gene segments via flanking LoxP recombination sequences. Different conditional Dicer null alleles have been used to prevent miRNA maturation in different bone cell compartments [11–14].
General functions of microRNAs in osteoblasts
The importance of mature miRNA expression in osteoblasts was established by selective ablation of the conditional Dicer allele in osteoblasts using a transgene that expresses Cre under control of two distinct osteoblast-related promoters [11]. Cre was expressed from the promoter of either the rat collagen type 1 (COL1A1) gene, which is expressed during early stages of osteoblast phenotype commitment, or the human bone gamma-carboxyglutamic acid-containing protein/osteocalcin (BGLAP/OC) gene, which is uniquely expressed in post-proliferative osteoblasts. Dicer gene deletion in mesenchymal cells using the COL1A1 promoter (i.e., a specific 2.3 kbp fragment from the 5′ region of the COL1A1 gene that is active in fetal osteogenic mesenchymal cells), caused lethality during late gestation (after E14.5), thereby precluding a definitive assessment of the direct role of Dicer during osteoblastogenesis during later developmental stages. More informative results were obtained with mice in which Dicer was excised in mature osteoblasts expressing the BGLAP promoter. These mice are viable and show delayed bone mineralization at birth. However, this delay is no longer detectable in 1-month old mice, and a prominent increase in cortical bone is evident in 2-month old mice. The observed prominent increase in cortical bone volume is not due to reduced bone resorption. Rather, it is appears to be intrinsic to mature osteoblasts and linked to increased synthesis and/or deposition of collagens in the extracellular matrix [11]. Hence, production of mature miRNAs by Dicer is necessary for normal pre-natal skeletal development and control of post-natal bone growth.
Studies by Raaijmakers and others examined the role of miRNAs in mice in which the conditional Dicer allele was eliminated by Cre excision under control of the promoter of the gene for the osteoblast-related transcription factor Osterix(OSX)/SP7 [12]. In these studies, Dicer was deleted in immature progenitor cells that are pre-committed to the osteoblastic lineage (i.e., from a developmental perspective, SP7/Cre is active after COL1A1/Cre but before BGLAP/Cre). As expected, loss of Dicer at this intermediate developmental stage compromises osteogenic lineage progression and osteoblast maturation, as well as generates alterations in the structure of the mineralized bone matrix. However, these studies also revealed that Dicer deficiency in mice affects the formation of the hematopoietic stem cell niche in bone, thus confounding further characterization of the skeletal phenotype [12].
General functions of microRNAs in osteoclasts
The essential role of miRNAs in osteoclasts was established by genetic ablation of Dicer in the macrophage/osteoclast lineage [13–14]. One study expressed Cre under control of the CD11b promoter, which eliminates Dicer in mononucleated progenitor cells early during osteoclast differentiation [13]. These mice exhibit mild osteopetrosis that is due to a lower number of osteoclasts and reduced bone resorption. It is not clear whether the mildness of the phenotype is technical in nature (e.g., CD11b may not be optimally expressed in osteoclast precursors), but loss of Dicer in this mouse model nonetheless reduces bone turn-over. Mechanistically, osteoclast differentiation is mediated by receptor activator for NFkappaB ligand (RANKL) and macrophage colony stimulating factor (M-CSF). The reduction in osteoclast number upon Dicer deletion is linked to decreased expression of the receptor for M-CSF (M-CSFR; gene symbol: CSF1R). In the studies by Noda and colleagues, Dicer was deleted in mature osteoclasts by expressing Cre under control of the cathepsin K (CTSK) promoter [14]. Similar to the studies by the Hruska group, loss of mature miRNAs in osteoclasts increases bone mass due to a reduction in the number of mature multi-nucleated osteoclasts and diminished bone resorption. The results from both studies clearly indicate that Dicer-dependent maturation of miRNAs is necessary for normal control of bone resorption.
Biological roles of microRNAs in controlling lineage-commitment and progression
To understand the biological roles of specific miRNAs in osteogenesis and skeletal remodelling, investigators have pursued two main approaches. The majority of published miRNA projects use a classical molecular ‘bottom-up’ approach, which examines temporal expression of known miRNAs – a list that continues to expand each year as more high-throughput RNA sequencing data become available. Temporal modulations (i.e., up- or down-regulation) in miRNA expression are then correlated with different mesenchymal cell fates or stages of cellular differentiation in each of the skeletal lineages. These studies have been performed in many different biological contexts relevant to bone, including during early lineage-commitment in mesenchymal stromal cells (also referred to as ‘mesenchymal stem cells’ or MSCs) from different sources (e.g., bone marrow, adipose-tissue) and during differentiation of committed osteoblasts or osteoclasts. Comprehensive reviews of earlier studies have been presented elsewhere [2–7] and here we present recent advances that exemplify general principles. It is of note that most of these expression profiling studies were carried out in just two main species used for skeletal research (i.e., mouse and human). There is still a remarkable paucity of data on miRNA expression in other animal models (e.g., rat, goat, pig) that would be useful for pre-translational studies in skeletal tissue repair or mechanotransduction. One main conclusion that can be drawn from the collective body of miRNA expression profiling studies using skeletal cells is that long-term changes in the biological states of mesenchymal cells invariably correspond with major alterations in miRNA expression programs.
MicroRNA regulation of mesenchymal cell fate determination
Initial expression analyses of a panel of representative microRNAs identified three microRNAs (miR-96, miR-124 and miR-199a) that are differentially expressed during mesenchymal lineage-commitment in mouse marrow stromal cells (MSCs)[15,16]. Selective up-regulation of each of these miRNAs was evident during osteogenesis (miR-96 and miR-199b), chondrogenesis (miR-199b) and/or adipogenesis (miR-96 and miR-124). MicroRNAs that are modulated can be examined for putative mRNA targets using well-established algorithms that predict the presence of miRNA binding sites in 3′UTRs based on a number of biochemical parameters, including sequence complementarity with miRNA seed sequences, conservation of seed sequences among vertebrate species and kinetic parameters related to the strength of RNA/RNA hybridization. Suomi and colleagues predicted [15] and subsequently validated [16] the concept that lineage-specific miRNAs enhance directed differentiation of multipotent cells into one phenotype by blocking alternative mesenchymal cell fates (e.g., enhancing osteogenic while blocking adipogenic and myogenic differentiation). A related study using human MSCs established that miR-138 is down-regulated during osteogenesis [17]. These studies showed that miR-138 blocks osteogenesis by repressing focal adhesion kinase, thus affecting signalling via integrins and the ERK pathway [17]. Thus, down-regulation of miR-138 serves to enable integrin/FAK/ERK signalling and this link may predict a role in mechanotransduction. Taken together, these studies illustrate the general principle that the biological activity of miRNAs in cell fate determination is based in part on their ability to regulate the expression of cell signalling components, lineage-related transcription factors, and phenotype-specific proteins during the initial stages of phenotype commitment in mesenchymal stem cells.
MicroRNA regulation of osteoblastogenesis
A series of related expression profiling studies established the importance of specific miRNAs in controlling differentiation of bone-anabolic osteoblasts. For example, Li and colleagues established microRNA expression programs during osteoblastic differentiation in two different cell culture models [18, 19]. Subsequent studies by Hassan and co-workers refined emerging mechanistic concepts for miRNA control of osteoblast differentiation [20–21]. One key finding is that the osteogenic morphogen BMP2 controls the switch between bone and muscle differentiation by controlling miRNA expression [18]. BMP2 achieves this primarily by suppressing miRNAs that would otherwise inhibit the osteogenic program, while to a lesser degree blocking the myogenic program. For example, BMP2 induced osteogenic differentiation of mouse C2C12 mesenchymal cells decreases expression of a set of anti-osteogenic miRNAs during the initial stages of trans-differentiation (i.e., miR-133 and miR-135). These two miRNAs collectively suppress the transcriptional activity of RUNX2 and its BMP2 responsive co-factor SMAD5. This BMP2 mediated ‘double-negative’ regulation (i.e., inhibition of osteogenic inhibitors) permits the initiation and sustained propagation of a BMP2/RUNX2/SMAD regulatory axis that induces osteoblast differentiation [18].
MicroRNA profiling was also performed with pre-committed mouse MC3T3 osteoblasts at successive stages of osteoblast differentiation [19]. These experiments identified a set of microRNAs (miR-29, miR-let-7, and miR-26) that regulate the synthesis of extracellular matrix (ECM) proteins in osteoblasts. For example, miR-29b suppresses production of collagens type I, IV and V (respectively, COL1A1, COL4A2 and COL5A3) to attenuate extracellular matrix biosynthesis and mineralization. Perhaps more interestingly, this study also suggested that miR-29b may support osteoblast maturation potentially by controlling the TGFβ pathway (e.g., relieving the inhibitory effects of TGFβ3 and ACVR2A), Wnt signaling (e.g., suppression the inhibitory β–catenin interacting protein CTNNBIP1), MAPK signaling (e.g., suppressing the inhibitory action of the dual specificity protease DUSP2) and/or epigenetic mechanisms (e.g., down-regulation of the transcriptionally repressive histone/lysine deacetylase HDAC4)[19]. These studies also revealed that osteoblast differentiation is promoted via transcriptional suppression of the miR cluster 23a~27a~24-2 by Runt-related transcription factor 2 (RUNX2) [20]. Each of these miRs inhibits SATB2, which supports bone formation together with RUNX2 and other bone-related transcription factors. Another discovery emerging from these miRNA profiling studies with osteoblasts is that microRNAs (miR-218) can promote osteoblast differentiation by enhancing Wnt signaling [21]. The osteo-inductive properties of miR-218 are based on its ability to decrease the production of three different Wnt inhibitors (i.e., Sclerostin (SOST), Dickkopf2 (DKK2), and secreted frizzled-related protein2 (SFRP2)]. In each case, miR-218 acts through its cognate binding sites in the respective 3′UTRs, and the loss of these inhibitors primes cells for the osteoblast-promoting effects of Wnt signaling [21].
Taken together, the general theme that emerged from these studies is that microRNAs do not just attenuate pathways, but can actively promote differentiation by ‘double-negative’ regulation (Fig. 1A). Down-regulation of miRNAs in the osteogenic lineage may directly relieve inhibition of osteogenic regulatory pathways. In contrast, the biological purpose of up-regulating osteogenic miRNAs is to eliminate anti-osteogenic pathways. Collectively, the reciprocal modulation of miRNAs with distinct molecular functions generates the requisite specificity for osteogenic lineage progression from immature osteoprogenitors towards mature osteoblasts.
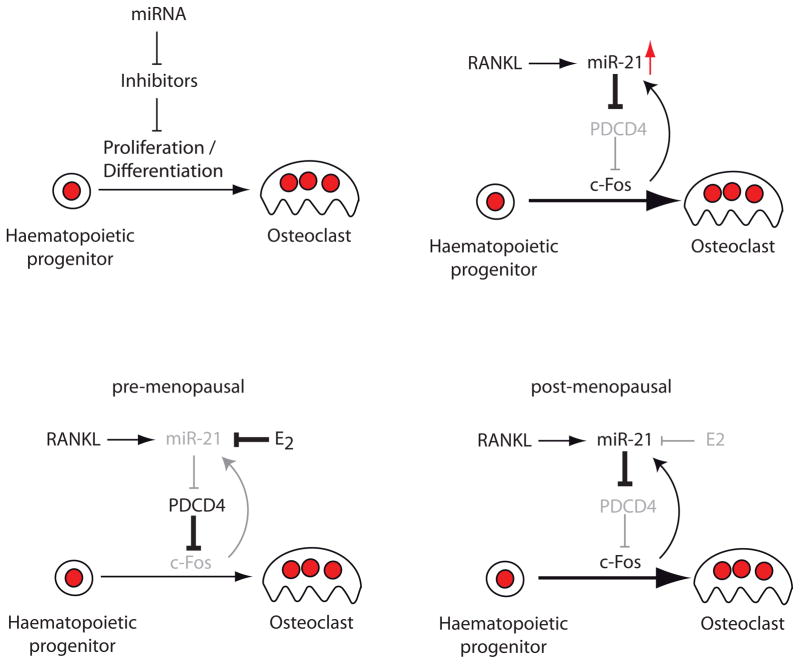
Figure 1. MicroRNAs stimulate osteoblast and osteoclast differentiation through ‘double negative’ circuits.
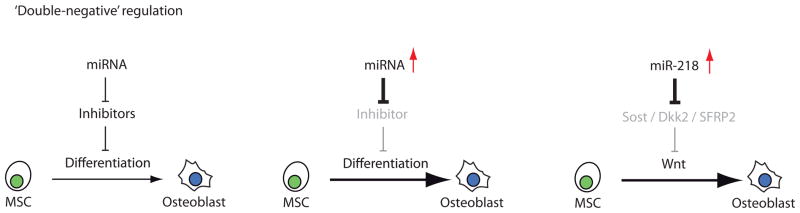
MicroRNAs control osteoblastogenesis (A) and osteoclastogenesis (B) through regulatory circuits in which microRNA interactions with the 3′UTRs of target mRNAs suppress (‘first negative’) the translation of proteins that inhibit (‘second negative’) the differentiation of osteoblasts or osteoclasts. Panels A and B illustrate two proposed circuits for either bone forming or bone promoting cells. Multiple other molecular circuits are known (or otherwise likely to exist) that may cross-regulate the factors shown in the diagram.
MicroRNA regulation of osteoclastogenesis
The best studied miRNAs that are involved in differentiation of osteoclasts are miR-21, miR-155 and miR-223 [12,13, 22–24]. Elevated expression of miR-223 in mouse RAW264.7 osteoclast precursor cells blocks maturation into tartrate-resistant acid phosphatase (TRAP)-positive multinucleated osteoclast [13]. Notwithstanding this inhibitory activity, miR-223 can stimulate normal osteoclastogenesis through a transcriptional relay mechanism that promotes expression of the CSFR1/M-CSFR receptor [22]. Essentially, the Ets-related transcription factor PU.1 (purine-rich binding protein 1; gene symbol: SPI1) stimulates miRNA-223 expression, while miRNA-223 suppresses the levels of the transcriptional repressor Nuclear Factor IA (NFI-A). The loss of this repressor permits transcriptional activation of the M-CSFR gene [22]. Consistent with the proposed pro-osteoclastogenic role of miR-223, this miRNA is upregulated during induction of differentiation in RAW264.7 cells [25]. The paradoxical results of miR-223 being both supportive and inhibitory for osteoclastogenesis is reminiscent of the opposing results for miR-218 which enhances osteoblastogenesis while attenuating the bone-specific activity of RUNX2 (see below). In both cases, negative feedback circuits may be triggered to prevent precocious differentiation or to avoid hyper-catalytic differentiation of bone cells, which in each case would perturb normal bone deposition and remodeling.
Subsequent miRNA expression studies indicated that osteoclastogenesis is enhanced by a positive feedback loop involving transcription factor AP1 (through effects on the cellular homolog of the FBJ murine osteosarcoma viral oncogene, c-Fos), miR-21 and programmed cell death 4 (PDCD4) protein [23]. In this ‘double negative’ circuit, RANKL induces miR-21 which down-regulates PDCD4 protein levels (Fig. 1B). Diminished PDCD4 expression in turn removes repression from c-Fos, thus allowing for execution of the osteoclastogenic program (Fig. 1B). The perpetuation of the initial RANKL signal is subsequently ensured by c-Fos dependent activation of miR-21 which then is predicted to minimize PDCD4 levels [23]. MicroRNA expression profiling studies show indeed that miR-21 is highly expressed during osteoclast differentiation [25], suggesting it is a pro-osteoclastogenic miRNA.
The induction of miR-21 during osteoclastogenesis is antagonized by estrogen [24], which is an osteo-protective steroid hormone that attenuates bone-resorption. Estrogen normally reduces miR-21 expression in osteoclasts and this reduction derepresses the expression of the miR-21 target Fas-ligand (FasL; gene symbol: FASLG). This autocrine production of FasL provokes osteoclastic apoptosis through its cognate Fas/CD95 receptor [24] thus reducing osteoclast activity. This finding suggests that miR-21 may play a role in reducing bone resorption in pre-menopausal women with robust estrogen levels.
Additional studies by Noda and co-workers focused on miR-155 [14], which is responsive to the osteoclastogenic factors RANKL and TNFalpha [13,25]. Like miR-21 (see above), miR-155 is induced by c-Fos in response to the anti-osteoclastogenic factor IFN-β [26]. The induction of miR-155 by IFN-β suppresses the signalling inhibitor SOCS1 and transcription factor MITF, which are both positive regulators of osteoclast differentiation [26]. Hence, miR-155 is a component of a negative feedback circuit that includes c-Fos and MITF. Additional feed-back regulatory mechanisms involving c-Fos are linked to the decline of miR-29b during human osteoclast differentiation [27]. Forced elevation of miR-29b impairs formation of TRAP positive cells and alters osteoclast specific transcriptional programs linked to both c-Fos and NFATC1 [27].
Apart from changes in miRNA expression of miR-21, miR-155 and miR-223, osteoclasts express miR-29b, miR-34c and miR-378, as well as miR-146a, and miR-210. It is of note that at least two of these miRNAs expressed in osteoclasts (i.e., miR-29b, miR34a/miR34c), attenuate osteoblast differentiation and/or regulate RUNX2 protein expression in osteoblasts (see below). Because the osteoblast specific factor RUNX2 is not expressed in immature or differentiated osteoclasts [28], this finding may simply reflect a fortuitous similarity. Nonetheless, several reports indicate that MSC-derived extracellular vesicles contain miRNAs and are involved in inter-cellular communication [29, 30]. Therefore, miRNAs in osteoclasts could be transferred to osteoblast through microvesicles and function as coupling factors to attenuate osteoblast differentiation and activity.
Other miRNAs expressed in osteoclasts have been characterized in different biological contexts. For comparison, miR-146a is expressed in chondrocytes [31], while miR-210 is a major regulator of the hypoxia-response [32]. The specific roles for either miR-146a or miR-210 in osteoclasts remain to be explored. The utilization of similar miRNAs in bone producing and bone resorbing cells reflects their versatility in regulating molecular pathways in different biological contexts.
The stimulatory role of miR-148a in osteoclastogenesis was recently revealed in studies using circulating CD14+ peripheral blood mononuclear cells (PBMCs) [33]. Expression of miR148a is normally enhanced upon induction of osteoclastogenesis with M-CSF and RANKL. However, precocious expression of miR-148a in osteoclast precursor cells stimulates differentiation. One molecular pathway by which miR148a controls the phenotypic conversion of mononuclear cells into osteoclasts is through direct effects on the 3′UTR for a key repressive transcription factor, V-maf musculoaponeurotic fibrosarcoma oncogene homolog B (MAFB). MAFB normally prevents induction of osteoclast differentiation in response to RANKL-induced osteoclastogenesis [33]. Hence, analogous to the pro-osteogenic miR-218 in osteoblasts [21], the pro-osteoclastic miR-148a positively controls osteoclast lineage progression through a ‘double-negative’ mechanism.
MicroRNA control of skeletal master regulators
Apart from expression profiling studies (‘bottom-up’ approach), recent studies have effectively used bioinformatic methods to focus on miRNAs that control the activities of master transcription factors (‘top-down’ approach). Master transcription factors of bone tissue (e.g., RUNX2, SP7/Osterix) are defined as such because genetic mutations cause major skeletal phenotypes and these factors have cell autonomous effects on differentiation in vitro. Several bone-related transcription factors are translated from mRNAs with remarkably long 3′UTRs. For example, the protein coding sequence for the osteogenic transcription factor RUNX2 is only ~1500 nucleotides, but its 3′UTR is twice as long (~3000 nucleotides).
Studies by Zhang and colleagues established that the 3′UTR of RUNX2 contains seed sequences for at least 11 different miRNAs (i.e., miR-23a, miR-30c, miR-34c, miR-133a, miR-135a, miR-137, miR-204, miR-205, miR-217, miR-218, and miR-338) [31]. All of these miRNAs control osteoblast differentiation [34] and are expressed to different degrees in both osteogenic and non-osteogenic cells. Interestingly, several of these microRNAs (i.e., miR-23a, miR-133a, miR-135a and miR-218) were independently identified in parallel studies within the same research group using different experimental approaches [18–21], reflecting the strength of concurrently identifying critical microRNAs using both ‘bottom-up’ and ‘top-down’ approaches.
The functional role of miR-218 is somewhat paradoxical, because this microRNA suppresses the expression of RUNX2 [31], yet stimulates osteoblastogenesis [21]. Under short-term basal conditions, elevation of miR-218 can block the 3′ UTR of RUNX2 (as is evident in luciferase-3′UTR promoter assays)[31]. Upon sustained induction of differentiation for more than a week, miR-218 is a potent stimulator of osteoblastogenesis [21,34]. One mechanistic model that clarifies these findings poses that miR-218 primarily promotes differentiation by enhancing Wnt signalling as part of a positive feed-forward loop, while attenuating RUNX2 activity as part of a negative feed-back loop to control osteoblast differentiation. The findings with miR-218 exemplify the principle that miRNAs can form intersecting loops to provide both positive and negative cross-talk between different regulatory pathways.
Seven of the eleven miRNAs (i.e., miR-23a, miR-30c, miR-34c, miR-133a, miR-135a, miR-205, and miR-217) that are known to target the osteogenic factor RUNX2, also regulate the chondrogenic GATA transcription factor tricho-rhino-phalangeal syndrome I (TRPS1) [35]. Of this group of miRNAs, miR23a and miR34a not only target RUNX2 and TRPS1 [35], but also a third transcription factor referred to as scaffold-associated AT-binding protein SATB2 [20]. Interestingly, TRPS1 and SATB2 both are upstream regulators of RUNX2 [discussed in 20,35] and the three transcription factors together may be interlocked in a microRNA-transcription factor circuit that controls a putative osteochondro-progenitor stage.
Each of the seven RUNX2/TRPS1 targeting miRNAs effectively blocks lineage progression in both pre-committed osteoblasts and pre-committed chondrocytes. More interestingly, when administered to multi-potent mesenchymal progenitor cells, each of these miRNAs diverted mesenchymal progenitor cells into an adipogenic cell fate. Furthermore, the quantitative effects of each miRNA on the expression of distinct mesenchymal markers were different and complementary to some degree [35]. These studies together provide a robust experimental demonstration of the concept that groups of related miRNAs may function together in regulating shared biological programs in skeletal cells.
The finding that RUNX2 is controlled by multiple miRNAs is corroborated by several other studies. For example, Chen and colleagues have shown that miR-204 regulates RUNX2 protein levels in mesenchymal progenitor cells [37]. In addition, complementary studies show that the BMP2/SMAD/RUNX2 axis is not only controlled by miR133 and miR135 [18, 34], but also linked to regulatory circuits involving miR-3960/miR-2861 [38] and miR20a [39] during osteoblast differentiation. Furthermore, other recent studies indicate that Osterix/SP7 expression is controlled by miR-93 [40] and miR-637 [41], while the protein levels of another osteogenic factor, ATF4, are attenuated by the anti-osteoblastic activity of miR-214 [42]. Taken together, there is a multiplicity of miRNAs that suppress the expression of distinct osteogenic master transcription factors.
MiRNAs have dramatic effects on the cell survival and/or differentiation, yet typically have only minor effects on their target proteins (e.g., master transcription factors) in laboratory experiments. MiRNAs have quantitatively modest effects (less than 2 fold) in molecular assays that are used to determine their function as post-transcriptional regulators (e.g., Luciferase-3′ UTR reporter assays). Changes in mRNA levels as analyzed by real-time reverse-transcriptase quantitative PCR require that the miRNA destabilizes the mRNA, which is not always the case. Determination of protein levels in response to miRNA administration is robust but protein immuno-blotting is not quantitative. Furthermore, the results of these molecular assays may not necessarily match up with each other due to a number of valid technical differences. Without improvements in the sensitivity and robustness of experimental read-outs in molecular assays that monitor miRNA effects, future studies may need to rely more on biological outcomes. The latter integrates the combined effects of miRNAs on multiple pathways and can have dramatic ‘fold-change effects’ in the biological phenotype of cells, rather than the modest ‘percentage effects’ observed for single proteins or signaling events.
MiRNA Dysfunction in Osteoporosis
Because miRNAs play important biological roles in the normal development and function of osteoblasts and osteoclasts, many studies have explored the pathological role of miRNAs in bone degeneration by examining their inhibitory effects on osteoblastogenesis or excessive stimulation of osteoclastogenesis [2]. As of yet, there are few completed studies that link bone diseases to defects in miRNA related mechanisms. The studies detailed below indicate that dysfunction of miRNA dependent mechanism can occur by various means and contribute to development of osteoporosis. From a therapeutic perspective, in vivo approaches that promote the activities of pro-osteoblastic miRNAs (e.g., miR-218 targeting Wnt inhibitors) [21] or inhibit pro-osteoclastic miRNAs (e.g., preventing miR-148a from blocking MAFB) [33] are attractive and experimentally realistic in, respectively, stimulating bone formation or mitigating excessive bone resorption.
MiRNA mutations causing osteoporosis
Deregulation of miRNA expression can affect both bone formation and bone resorption. One clinically relevant study revealed that a homozygous mutation in the genomic locus coding for miR-2861 causes a rare form of familial osteoporosis in two adolescent patients [43]. In cell culture models, expression of miR-2861 is induced by BMP2 in mouse osteoblasts. This enhancement of miR-2861 inhibits the epigenetic regulator histone deacetylase 5 (HDAC5). This enzyme, which removes acetyl groups from lysine residues in histones and other regulatory proteins (e.g., RUNX2), is inhibitory for osteoblast differentiation. Loss of miR-2816 expression that was apparent in bone biopsies correlates with elevated HDAC5 levels and decreased RUNX2 levels. Hence, dysfunction of miRNAs can disrupt skeletal remodelling during post-natal growth and contribute to osteoporosis by diminishing the bone anabolic functions of osteoblasts.
MiRNA binding site polymorphisms associated with osteoporosis
Genetic polymorphisms naturally occur in predicted miRNA targeting sites (poly-miRTs or poly-miRTSs) within the 3′ UTRs of target mRNAs. Variation in femoral neck bone mineral density in a large group of subjects (~3000) is statistically associated with natural genetic variation in three poly-miRTs (rs6854081, rs1048201, and rs7683093) [44]. These poly-miRTs contain seed sequences for miR-146a and miR-146b in the 3′UTR of the fibroblast growth factor 2 (FGF2) mRNA. Polymorphism-related changes in the expression of FGF2, which is a major mitogen for osteogenic cells and promotes osteoblast differentiation through MAPK signalling [45,46], correlate with differences in bone mineral density [44]. Thus, miRNA related gene polymorphism in bone-related genes may predispose to osteoporosis susceptibility by affecting the activity of osteoblasts and perhaps also osteoclasts.
Deregulation of miRNA expression and osteoporosis
Bone remodelling requires active communication between osteoblasts and osteoclasts. Corroborating previous studies that characterized the miR-34 family [18, 34–36], Bae and colleagues showed that BMP2 is a robust inducer of miR-34c [47]. Initial studies showed that miR-34c blocks osteoblast differentiation when exogenously expressed [34]. Transgenic mice in which miR-34c was selectively expressed in osteoblasts had defects in osteoblast proliferation and mineralization that reflect osteoporotic changes observed in aging mice [47]. Effects of transgenically expressed miR-34c on osteoblast proliferation are not surprising given the important cell growth suppressive role of the miR-34 target RUNX2 [34, 48, 49], the general effect of miR-34c that is linked to the tumor suppressor p53 [50] and prior mouse studies showing a role for miR-34 in skeletogenesis [36]. The study by Bae et al. [47], also examined the intriguing connection with Notch signalling in osteoblasts. MiR-34c directly regulates Notch1, Notch2 and Jag1 in osteoblasts to control differentiation of osteoclasts, presumably through cell/cell contact [47]. Thus, this study provides experimental evidence for miRNA-related deregulation of osteoblast/osteoclast communication that is linked to osteoporosis in a transgenic rodent model.
MiRNAs as circulatory biomarkers for osteoporosis
MiRNAs can be secreted in extracellular vesicles (microvesicles or exosomes) and can be monitored as biomarkers for a multitude of disease states [51,52]. Many studies (>200) have explored the presence of circulatory miRNAs in patient serum for different clinical conditions. Recent studies examined osteoclast-related miRNA markers in human peripheral blood mononuclear cells [33, 53], which represent a circulatory reservoir of osteoclast precursors. One of these studies showed that expression of miR-148a in CD14-positive circulating monocytes is elevated in lupus patients and correlates with low bone mineral density, consistent with results from molecular studies on the pro-osteoclastic effects of miR-148a [33]. Examination of a panel of miRNAs in circulating monocytes from a small patient cohort suggests that miR-133a is associated with lower bone mineral density [53]. Further investigations on miRNAs present in human monocytes may yield additional miRNAa that correlate with differences in bone mineral density that could be used as molecular markers for osteoporosis.
DISCUSSION & CONCLUSIONS
MiRNAs have emerged as key regulators of bone formation, remodeling and degeneration. Despite their fundamental importance and clinical promise, there is a paucity of mouse models to establish the in vivo roles played by miRNAs in skeletal development in animal models. In addition, there are no published studies examining miRNA functions in mature osteocytes or in response to mechanotransduction, which is critical for normal bone adaptation. Furthermore, studies on intercellular communication via extracellular vesicles and miRNA transfer are essential to understand the complex and versatile regulation of osteoblasts and osteoclast differentiation by miRNA. Despite these shortcomings, a great deal of progress has been made and there is clearly reason for optimism that this knowledge can be applied to clinical problems.
MiRNAs are integral components - and indeed key regulatory nodes - of molecular networks that respond to a multitude of developmental signaling pathways (e.g., BMP/TGFβ, WNT, Notch, FGF2). Many miRNAs control the activities of principal transcription factors that control bone development and remodeling, and many examples of transcription factors controlling miRNAs (either as independent transcription units or embedded in host genes) will continue to emerge. While there are undoubtedly exceptions to this simple rule, the default logic of miRNAs is to suppress the expression of target genes. Hence, miRNA-mediated stimulation of most biological processes proceeds by suppressing the activity of inhibitory factors. A number of such examples of ‘double negative’ regulation have been identified and a subset has been discussed in this overview. Bioinformatics and systems biology approaches will be necessary to establish the full complement of regulatory possibilities related to miRNA/mRNA interactions. The intercalation of in silico predictions with rapidly emerging miRNA, mRNA and protein expression data during skeletal development and in clinically samples (based on high-throughput RNA sequencing and proteomics approaches) will ultimately guide translational studies that transform fundamental knowledge into clinical practice.
The current experimental body of work demonstrates that miRNAs are critical post-transcriptional regulators of gene expression that control osteoblast-dependent bone formation and osteoclast-related bone remodeling. The studies discussed here indicate that deregulation of miRNA mediated mechanisms is pathologically linked to bone degeneration (e.g., osteoporosis) and other bone-related diseases. Biological, cellular and molecular principles have been established to account for how miRNAs control differentiation of osteoblasts from mesenchymal stem cells and differentiation of osteoclasts from monocytic/hematopoietic precursors. The clinical relevance of miRNAs is evident from the dramatic effects that individual miRNAs can have in driving differentiation of osteoblasts or osteoclasts. Clinical applications of this knowledge include their potential use as biomarkers that are linked to bone mineral density and as RNA-based drugs that can be leveraged to prevent bone tissue degeneration.
Acknowledgments
We thank the members of our laboratories for critical comments and sharing of ideas. This work was supported by National Institutes of Health Grant R01AR049069 (to AvW).
Footnotes
Disclosure
AJ van Wijnen declares no conflicts of interest.
J van de Peppel:
JP van Leeuwen:
JB Lian:
GS Stein:
JJ Westendorf:
MJ Oursler:
HJI Sampen:
H Taipaleenmaki:
E Hesse:
S Riester:
S Kakar:
Contributor Information
Andre J. van Wijnen, Email: vanwijnen.andre@mayo.edu.
Jeroen van de Peppel, Email: h.vandepeppel@erasmusmc.nl.
Johannes P. van Leeuwen, Email: j.vanleeuwen@erasmusmc.nl.
Jane B. Lian, Email: jane.lian@uvm.edu.
Gary S. Stein, Email: gary.stein@uvm.edu.
Jennifer J. Westendorf, Email: westendorf.jennifer@mayo.edu.
Merry-Jo Oursler, Email: oursler.merryjo@mayo.edu.
Hee-Jeong Im Sampen, Email: Hee-Jeong_Sampen@rush.edu.
Hanna Taipaleenmaki, Email: h.taipaleenmaeki@uke.de.
Eric Hesse, Email: e.hesse@uke.de.
Scott Riester, Email: riester.scott@mayo.edu.
Sanjeev Kakar, Email: kakar.sanjeev@mayo.edu.
References
Papers of particular interest, published recently, have been highlighted as:
•• Of major importance
• Of importance
- 1.Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001 Dec 28;107(7):823–6. doi: 10.1016/s0092-8674(01)00616-x. Review. [DOI] [PubMed] [Google Scholar]
- 2••.Lian JB, Stein GS, van Wijnen AJ, Stein JL, Hassan MQ, Gaur T, Zhang Y. MicroRNA control of bone formation and homeostasis. Nat Rev Endocrinol. 2012 Jan 31;8(4):212–27. doi: 10.1038/nrendo.2011.234. Review. Comprehensive review of the microRNA literature on skeletal development and disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miyaki S, Asahara H. Macro view of microRNA function in osteoarthritis. Nat Rev Rheumatol. 2012 Sep;8(9):543–52. doi: 10.1038/nrrheum.2012.128. Epub 2012 Aug 14. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xia Z, Chen C, Chen P, Xie H, Luo X. MicroRNAs and their roles in osteoclast differentiation. Front Med. 2011 Dec;5(4):414–9. doi: 10.1007/s11684-011-0168-0. Epub 2011 Dec 27. Review. [DOI] [PubMed] [Google Scholar]
- 5.Goldring MB, Marcu KB. Epigenomic and microRNA-mediated regulation in cartilage development, homeostasis, and osteoarthritis. Trends Mol Med. 2012 Feb;18(2):109–18. doi: 10.1016/j.molmed.2011.11.005. Epub 2011 Dec 17. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taipaleenmäki H, Bjerre Hokland L, Chen L, Kauppinen S, Kassem M. Mechanisms in endocrinology: micro-RNAs: targets for enhancing osteoblast differentiation and bone formation. Eur J Endocrinol. 2012 Mar;166(3):359–71. doi: 10.1530/EJE-11-0646. Epub 2011 Nov 14. Review. [DOI] [PubMed] [Google Scholar]
- 7•.Laine SK, Hentunen T, Laitala-Leinonen T. Do microRNAs regulate bone marrow stem cell niche physiology? Gene. 2012 Apr 10;497(1):1–9. doi: 10.1016/j.gene.2012.01.045. Epub 2012 Jan 28. Review. Important review that addresses miRNA dependent cell/cell communication in the bone marrow micro-environment. [DOI] [PubMed] [Google Scholar]
- 8.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003 Nov;35(3):215–7. doi: 10.1038/ng1253. Epub 2003 Oct 5. Erratum in: Nat Genet. 2003 Nov;35(3):287. [DOI] [PubMed] [Google Scholar]
- 9.Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci U S A. 2005 Aug 2;102(31):10898–903. doi: 10.1073/pnas.0504834102. Epub 2005 Jul 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mudhasani R, Zhu Z, Hutvagner G, Eischen CM, Lyle S, Hall LL, Lawrence JB, Imbalzano AN, Jones SN. Loss of miRNA biogenesis induces p19Arf-p53 signaling and senescence in primary cells. J Cell Biol. 2008 Jun 30;181(7):1055–63. doi: 10.1083/jcb.200802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11••.Gaur T, Hussain S, Mudhasani R, Parulkar I, Colby JL, Frederick D, Kream BE, van Wijnen AJ, Stein JL, Stein GS, Jones SN, Lian JB. Dicer inactivation in osteoprogenitor cells compromises fetal survival and bone formation, while excision in differentiated osteoblasts increases bone mass in the adult mouse. Dev Biol. 2010 Apr 1;340(1):10–21. doi: 10.1016/j.ydbio.2010.01.008. Epub 2010 Jan 15. Key study showing a post-natal bone phenotype in microRNA deficient mice. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raaijmakers MH, Mukherjee S, Guo S, Zhang S, Kobayashi T, Schoonmaker JA, Ebert BL, Al-Shahrour F, Hasserjian RP, Scadden EO, Aung Z, Matza M, Merkenschlager M, Lin C, Rommens JM, Scadden DT. Bone progenitor dysfunction induces myelodysplasia and secondary leukaemia. Nature. 2010 Apr 8;464(7290):852–7. doi: 10.1038/nature08851. Epub 2010 Mar 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13••.Sugatani T, Hruska KA. Impaired micro-RNA pathways diminish osteoclast differentiation and function. J Biol Chem. 2009 Feb 13;284(7):4667–78. doi: 10.1074/jbc.M805777200. Epub 2008 Dec 5. First in vivo demonstration of a requirement for Dicer in osteoclast-related bone remodelling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14•.Mizoguchi F, Izu Y, Hayata T, Hemmi H, Nakashima K, Nakamura T, Kato S, Miyasaka N, Ezura Y, Noda M. Osteoclast-specific Dicer gene deficiency suppresses osteoclastic bone resorption. J Cell Biochem. 2010 Apr 1;109(5):866–75. doi: 10.1002/jcb.22228. Important study revealing the biological contribution of Dicer to mature osteoclasts. [DOI] [PubMed] [Google Scholar]
- 15.Suomi S, Taipaleenmäki H, Seppänen A, Ripatti T, Väänänen K, Hentunen T, Säämänen AM, Laitala-Leinonen T. MicroRNAs regulate osteogenesis and chondrogenesis of mouse bone marrow stromal cells. Gene Regul Syst Bio. 2008 Apr 22;2:177–91. doi: 10.4137/grsb.s662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laine SK, Alm JJ, Virtanen SP, Aro HT, Laitala-Leinonen TK. MicroRNAs miR-96, miR-124, and miR-199a regulate gene expression in human bone marrow-derived mesenchymal stem cells. J Cell Biochem. 2012 Aug;113(8):2687–95. doi: 10.1002/jcb.24144. [DOI] [PubMed] [Google Scholar]
- 17••.Eskildsen T, Taipaleenmäki H, Stenvang J, Abdallah BM, Ditzel N, Nossent AY, Bak M, Kauppinen S, Kassem M. MicroRNA-138 regulates osteogenic differentiation of human stromal (mesenchymal) stem cells in vivo. Proc Natl Acad Sci U S A. 2011 Apr 12;108(15):6139–44. doi: 10.1073/pnas.1016758108. Epub 2011 Mar 2. Major study providing proof for miRNA effects on mesenchymal lineage-differentiation in mice using a subcutaneous growth model. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Z, Hassan MQ, Volinia S, van Wijnen AJ, Stein JL, Croce CM, Lian JB, Stein GS. A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc Natl Acad Sci U S A. 2008 Sep 16;105(37):13906–11. doi: 10.1073/pnas.0804438105. Epub 2008 Sep 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z, Hassan MQ, Jafferji M, Aqeilan RI, Garzon R, Croce CM, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J Biol Chem. 2009 Jun 5;284(23):15676–84. doi: 10.1074/jbc.M809787200. Epub 2009 Apr 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hassan MQ, Gordon JA, Beloti MM, Croce CM, van Wijnen AJ, Stein JL, Stein GS, Lian JB. A network connecting Runx2, SATB2, and the miR-23a~27a~24-2 cluster regulates the osteoblast differentiation program. Proc Natl Acad Sci U S A. 2010 Nov 16;107(46):19879–84. doi: 10.1073/pnas.1007698107. Epub 2010 Oct 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21••.Hassan MQ, Maeda Y, Taipaleenmaki H, Zhang W, Jafferji M, Gordon JA, Li Z, Croce CM, van Wijnen AJ, Stein JL, Stein GS, Lian JB. miR-218 directs a Wnt signaling circuit to promote differentiation of osteoblasts and osteomimicry of metastatic cancer cells. J Biol Chem. 2012 Dec 7;287(50):42084–92. doi: 10.1074/jbc.M112.377515. Epub 2012 Oct 11. First identification of a miRNA with osteogenic or osteomimetic properties in, respectively, osseous and non-osseous cels. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sugatani T, Hruska KA. MicroRNA-223 is a key factor in osteoclast differentiation. J Cell Biochem. 2007 Jul 1;101(4):996–9. doi: 10.1002/jcb.21335. [DOI] [PubMed] [Google Scholar]
- 23.Sugatani T, Vacher J, Hruska KA. A microRNA expression signature of osteoclastogenesis. Blood. 2011 Mar 31;117(13):3648–57. doi: 10.1182/blood-2010-10-311415. Epub 2011 Jan 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24••.Sugatani T, Hruska KA. Down-Regulation of miR-21 biogenesis by estrogen action contributes to osteoclastic apoptosis. J Cell Biochem. 2012 Dec 13; doi: 10.1002/jcb.24471. [Epub ahead of print].Exciting study that clarifies the interplay between miRNAs and estrogen levels. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kagiya T, Nakamura S. Expression profiling of microRNAs in RAW264.7 cells treated with a combination of tumor necrosis factor alpha and RANKL during osteoclast differentiation. J Periodontal Res. 2012 Oct 18; doi: 10.1111/jre.12017. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 26.Zhang J, Zhao H, Chen J, Xia B, Jin Y, Wei W, Shen J, Huang Y. Interferon-β-induced miR-155 inhibits osteoclast differentiation by targeting SOCS1 and MITF. FEBS Lett. 2012 Sep 21;586(19):3255–62. doi: 10.1016/j.febslet.2012.06.047. Epub 2012 Jul 4. [DOI] [PubMed] [Google Scholar]
- 27.Rossi M, Pitari MR, Amodio N, Di Martino MT, Conforti F, Leone E, Botta C, Paolino FM, Del Giudice T, Iuliano E, Caraglia M, Ferrarini M, Giordano A, Tagliaferri P, Tassone P. miR-29b negatively regulates human osteoclastic cell differentiation and function: Implications for the treatment of multiple myeloma-related bone disease. J Cell Physiol. 2012 Dec 18; doi: 10.1002/jcp.24306. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 28.Saltman LH, Javed A, Ribadeneyra J, Hussain S, Young DW, Osdoby P, Amcheslavsky A, van Wijnen AJ, Stein JL, Stein GS, Lian JB, Bar-Shavit Z. Organization of transcriptional regulatory machinery in osteoclast nuclei: compartmentalization of Runx1. J Cell Physiol. 2005 Sep;204(3):871–80. doi: 10.1002/jcp.20329. [DOI] [PubMed] [Google Scholar]
- 29•.Collino F, Deregibus MC, Bruno S, Sterpone L, Aghemo G, Viltono L, Tetta C, Camussi G. Microvesicles derived from adult human bone marrow and tissue specific mesenchymal stem cells shuttle selected pattern of miRNAs. PLoS One. 2010 Jul 27;5(7):e11803. doi: 10.1371/journal.pone.0011803. Early study that provides evidence for cell/cell contact through miRNA containing microvesicles. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30•.Chen TS, Lai RC, Lee MM, Choo AB, Lee CN, Lim SK. Mesenchymal stem cell secretes microparticles enriched in pre-microRNAs. Nucleic Acids Res. 2010 Jan;38(1):215–24. doi: 10.1093/nar/gkp857. Epub 2009 Oct 22. Concurrent early study also showing microvesicle-related secretuib of miRNAs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X, Gibson G, Kim JS, Kroin J, Xu S, van Wijnen AJ, Im HJ. MicroRNA-146a is linked to pain-related pathophysiology of osteoarthritis. Gene. 2011 Jul 1;480(1–2):34–41. doi: 10.1016/j.gene.2011.03.003. Epub 2011 Mar 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang X, Le QT, Giaccia AJ. MiR-210--micromanager of the hypoxia pathway. Trends Mol Med. 2010 May;16(5):230–7. doi: 10.1016/j.molmed.2010.03.004. Epub 2010 Apr 29. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33••.Cheng P, Chen C, He HB, Hu R, Zhou HD, Xie H, Zhu W, Dai RC, Wu XP, Liao EY, Luo XH. MiR-148a regulates osteoclastogenesis via targeting MAFB. J Bone Miner Res. 2012 Dec 7; doi: 10.1002/jbmr.1845. [Epub ahead of print]. Compelling paper providing evidence for a ‘double negative’ circuit that promotes osteoclast differentiation. [DOI] [PubMed] [Google Scholar]
- 34•.Zhang Y, Xie RL, Croce CM, Stein JL, Lian JB, van Wijnen AJ, Stein GS. A program of microRNAs controls osteogenic lineage progression by targeting transcription factor Runx2. Proc Natl Acad Sci U S A. 2011 Jun 14;108(24):9863–8. doi: 10.1073/pnas.1018493108. Epub 2011 May 31. Comprehensive study showing that osteogenic activity of transcription factor Runx2 is controlled by a multiplicity of miRNAs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35••.Zhang Y, Xie RL, Gordon J, LeBlanc K, Stein JL, Lian JB, van Wijnen AJ, Stein GS. Control of mesenchymal lineage progression by microRNAs targeting skeletal gene regulators Trps1 and Runx2. J Biol Chem. 2012 Jun 22;287(26):21926–35. doi: 10.1074/jbc.M112.340398. Epub 2012 Apr 27. Key paper revealing a miRNA-transcription factor network that regulates mesenchymal lineage allocation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36••.Wei J, Shi Y, Zheng L, Zhou B, Inose H, Wang J, Guo XE, Grosschedl R, Karsenty G. miR-34s inhibit osteoblast proliferation and differentiation in the mouse by targeting SATB2. J Cell Biol. 2012 May 14;197(4):509–21. doi: 10.1083/jcb.201201057. Epub 2012 May 7. First paper to demonstrate a skeletal phenotype linked to a specific microRNA expressed in osteogenic cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang J, Zhao L, Xing L, Chen D. MicroRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells. 2010 Feb;28(2):357–64. doi: 10.1002/stem.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu R, Liu W, Li H, Yang L, Chen C, Xia ZY, Guo LJ, Xie H, Zhou HD, Wu XP, Luo XH. A Runx2/miR-3960/miR-2861 regulatory feedback loop during mouse osteoblast differentiation. J Biol Chem. 2011 Apr 8;286(14):12328–39. doi: 10.1074/jbc.M110.176099. Epub 2011 Feb 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang JF, Fu WM, He ML, Xie WD, Lv Q, Wan G, Li G, Wang H, Lu G, Hu X, Jiang S, Li JN, Lin MC, Zhang YO, Kung HF. MiRNA-20a promotes osteogenic differentiation of human mesenchymal stem cells by co-regulating BMP signaling. RNA Biol. 2011 Sep-Oct;8(5):829–38. doi: 10.4161/rna.8.5.16043. Epub 2011 Jul 28. [DOI] [PubMed] [Google Scholar]
- 40.Yang L, Cheng P, Chen C, He HB, Xie GQ, Zhou HD, Xie H, Wu XP, Luo XH. miR-93/Sp7 function loop mediates osteoblast mineralization. J Bone Miner Res. 2012 Jul;27(7):1598–606. doi: 10.1002/jbmr.1621. [DOI] [PubMed] [Google Scholar]
- 41.Zhang JF, Fu WM, He ML, Wang H, Wang WM, Yu SC, Bian XW, Zhou J, Lin MC, Lu G, Poon WS, Kung HF. MiR-637 maintains the balance between adipocytes and osteoblasts by directly targeting Osterix. Mol Biol Cell. 2011 Nov;22(21):3955–61. doi: 10.1091/mbc.E11-04-035. Epub 2011 Aug 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang X, Guo B, Li Q, Peng J, Yang Z, Wang A, Li D, Hou Z, Lv K, Kan G, Cao H, Wu H, Song J, Pan X, Sun Q, Ling S, Li Y, Zhu M, Zhang P, Peng S, Xie X, Tang T, Hong A, Bian Z, Bai Y, Lu A, Li Y, He F, Zhang G, Li Y. miR-214 targets ATF4 to inhibit bone formation. Nat Med. 2013 Jan;19(1):93–100. doi: 10.1038/nm.3026. [DOI] [PubMed] [Google Scholar]
- 43.Li H, Xie H, Liu W, Hu R, Huang B, Tan YF, Xu K, Sheng ZF, Zhou HD, Wu XP, Luo XH. A novel microRNA targeting HDAC5 regulates osteoblast differentiation in mice and contributes to primary osteoporosis in humans. J Clin Invest. 2009 Dec;119(12):3666–77. doi: 10.1172/JCI39832. doi: 0.1172/ JCI39832. Epub 2009 Nov 16. Erratum in: J Clin Invest. 2010 Jan;120(1):395. Liao, Er-Yuan [removed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44•.Lei SF, Papasian CJ, Deng HW. Polymorphisms in predicted miRNA binding sites and osteoporosis. J Bone Miner Res. 2011 Jan;26(1):72–8. doi: 10.1002/jbmr.186. Solid study that characterizes a miRNA dependent polymorphisms linked to bone mineral density in human patients. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiao G, Jiang D, Gopalakrishnan R, Franceschi RT. Fibroblast growth factor 2 induction of the osteocalcin gene requires MAPK activity and phosphorylation of the osteoblast transcription factor, Cbfa1/Runx2. J Biol Chem. 2002 Sep 27;277(39):36181–7. doi: 10.1074/jbc.M206057200. Epub 2002 Aug 28. [DOI] [PubMed] [Google Scholar]
- 46.Teplyuk NM, Haupt LM, Ling L, Dombrowski C, Mun FK, Nathan SS, Lian JB, Stein JL, Stein GS, Cool SM, van Wijnen AJ. The osteogenic transcription factor Runx2 regulates components of the fibroblast growth factor/proteoglycan signaling axis in osteoblasts. J Cell Biochem. 2009 May 1;107(1):144–54. doi: 10.1002/jcb.22108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47•.Bae Y, Yang T, Zeng HC, Campeau PM, Chen Y, Bertin T, Dawson BC, Munivez E, Tao J, Lee BH. miRNA-34c regulates Notch signaling during bone development. Hum Mol Genet. 2012 Jul 1;21(13):2991–3000. doi: 10.1093/hmg/dds129. Epub 2012 Apr 12. Compelling paper that examines the miRNA dependent interplay between osteoclasts and osteoblasts. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pratap J, Galindo M, Zaidi SK, Vradii D, Bhat BM, Robinson JA, Choi JY, Komori T, Stein JL, Lian JB, Stein GS, van Wijnen AJ. Cell growth regulatory role of Runx2 during proliferative expansion of preosteoblasts. Cancer Res. 2003 Sep 1;63(17):5357–62. [PubMed] [Google Scholar]
- 49.Teplyuk NM, Galindo M, Teplyuk VI, Pratap J, Young DW, Lapointe D, Javed A, Stein JL, Lian JB, Stein GS, van Wijnen AJ. Runx2 regulates G protein-coupled signaling pathways to control growth of osteoblast progenitors. J Biol Chem. 2008 Oct 10;283(41):27585–97. doi: 10.1074/jbc.M802453200. Epub 2008 Jul 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He L, He X, Lowe SW, Hannon GJ. microRNAs join the p53 network--another piece in the tumour-suppression puzzle. Nat Rev Cancer. 2007 Nov;7(11):819–22. doi: 10.1038/nrc2232. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, Benjamin H, Kushnir M, Cholakh H, Melamed N, Bentwich Z, Hod M, Goren Y, Chajut A. Serum microRNAs are promising novel biomarkers. PLoS One. 2008 Sep 5;3(9):e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008 Jul 29;105(30):10513–8. doi: 10.1073/pnas.0804549105. Epub 2008 Jul 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53••.Wang Y, Li L, Moore BT, Peng XH, Fang X, Lappe JM, Recker RR, Xiao P. MiR-133a in human circulating monocytes: a potential biomarker associated with postmenopausal osteoporosis. PLoS One. 2012;7(4):e34641. doi: 10.1371/journal.pone.0034641. Epub 2012 Apr 10. First clinically relevant paper indicating the potential utility of circulating microRNAs in predicting differences in bone mineral density in human patients. [DOI] [PMC free article] [PubMed] [Google Scholar]