Abstract
Current advances in plant genotyping lead to major progress in the knowledge of genetic architecture of traits of interest. It is increasingly important to develop decision support tools to help breeders and geneticists to conduct marker-assisted selection methods to assemble favorable alleles that are discovered. Algorithms have been implemented, within an interactive graphical interface, to 1) trace parental alleles throughout generations, 2) propose strategies to select the best plants based on estimated molecular scores, and 3) efficiently intermate them depending on the expected value of their progenies. With the possibility to consider a multi-allelic context, OptiMAS opens new prospects to assemble favorable alleles issued from diverse parents and further accelerate genetic gain.
Key words: marker-assisted selection, plant breeding, QTL, multiparental designs, gene pyramiding
Molecular markers have lead since the 1980s to a rapidly growing body of information regarding quantitative trait loci (QTL) or genes controlling the variation of traits of biological/economical importance. For simple traits involving very few genes or QTL, associated markers can be applied for diagnostic purposes in early screening process (e.g., selection against disease susceptibility) or targeted replacement of chromosomal segments by means of marker-assisted backcrossing. For complex traits, Lande and Thompson (1990) advocated the use of markers significantly associated with QTL for predicting the genetic value of candidate plants (molecular score) in marker-assisted selection (MAS) programs in order to assemble most favorable alleles. Many studies investigated strategies of MAS theoretically and highlighted in particular the benefits of this strategy for accelerating genetic gain. It has been applied experimentally with success, in particular in private companies (Xu and Crouch 2008). This was conducted first mainly in a context of biparental populations derived from the cross between two inbred lines. By addressing a broader diversity, multiparental designs 1) increase the power and the accuracy of QTL detection; 2) enable to estimate simultaneously the different parental allele effects and to identify the most favorable ones for selection (Rebaï and Goffinet 2000; Blanc et al. 2006, 2008). Recently, two main types of multiparental designs have received specific interest in the plant breeding community to increase the resolution of QTL mapping by the joint use of dense genotyping of parental lines and linkage analysis in the progenies: the Nested Association Mapping design (NAM; Yu et al. 2008) and the Multiparent Advanced Inter-Cross design (MAGIC; Cavanagh et al. 2008). Such designs successfully led to the fine mapping of QTL in numerous species (Buckler et al. 2009; Poland et al. 2011; Cook et al. 2012 for maize; Kover et al. 2009 for arabidopsis; Huang et al. 2012 for wheat) and revealed multiallelic variation for a majority of QTL. Thanks to the development of dense marker genotyping, it becomes now possible for numerous species to search for marker–trait associations directly in diversity panels of inbred lines. Genome-wide associations (GWA) mapping will certainly lead to fine-mapped QTL that will be of interest in breeding programs. Meanwhile, genomic selection (GS; Meuwissen et al. 2001) has been proposed as a way to predict genetic value based on markers located all over the genome, without aiming at identifying causal polymorphisms. This approach received considerable attention in the plant breeding community (Jannink et al. 2010) and is often presented as an alternative for MAS based on QTL results. GS is certainly a good way to handle in selection QTL of small effect that are hardly detectable. However, GS does not aim explicitly at monitoring the assembly of favorable alleles. New QTL mapping approaches, including GWA, clearly contribute to the identification of alleles of interest for QTL with most important effects on the variation, even for complex traits (Hamblin et al. 2011). Such information is generally available for several traits and environmental conditions, leading to a possibly high total number of loci. The objective of OptiMAS is to valorize such results by helping breeders to create a given ideal genotype (ideotype) assembling favorables alleles from diverse parental origin.
OptiMAS provides help to geneticists and breeders for making rapid and efficient selection decisions through a user-friendly interface, considering the general framework of multiparental designs and complex pedigree structure generally observed in applied crop breeding programs. OptiMAS computes the probabilities of parental alleles transmission throughout the pedigree (potentially many generations of crossing or selfing) taking into account genotypic information at different generations when available. Using these probabilities, OptiMAS proposes easy ways for identifying the best candidates for selection and the best mating designs taking into account complementarities among selected plants to optimize the chance of obtaining superior genotypes in the next generations. To our knowledge, such a tool is not yet available for public research. OptiMAS, therefore, appears promising to accelerate genetic gain in plant breeding programs and facilitate biological investigations.
Features and Functionalities
OptiMAS main algorithms (described in documentation available as Supplementary Material) have been deployed to trace parental alleles along generations, using information given by markers located in the vicinity of the estimated QTL/gene positions. Probabilities of allele transmission are computed in different MAS schemes and mating designs (intercrossing, selfing, backcrossing, double haploids, recombinant inbred lines) with the possibility of considering generations without genotypic information. Then, strategies are proposed to select the best plants considering estimated molecular scores and to efficiently intermate them based on the expected value of their progenies. These functionalities have been defined in connection with a panel of users working on different species and tested on two reference datasets provided with the tool.
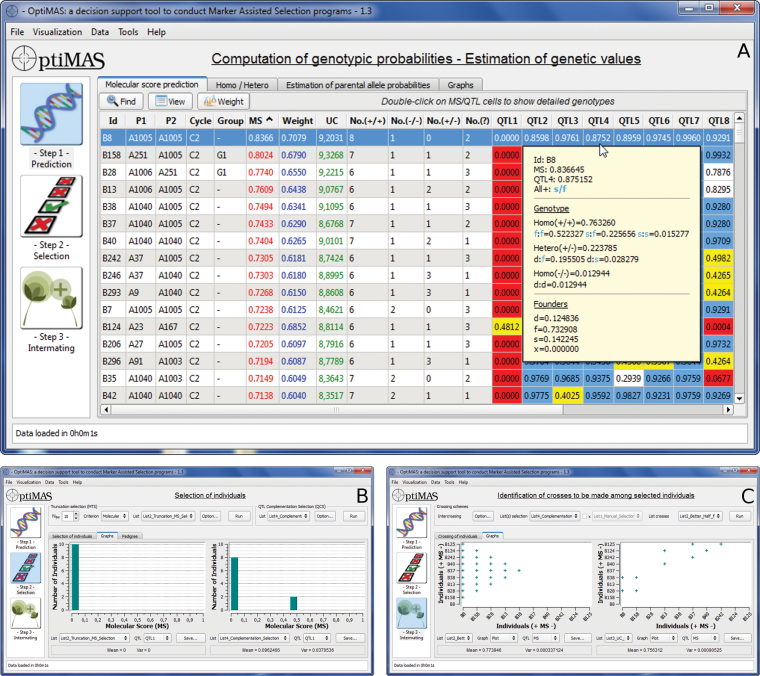
Two input files are needed to run the program: 1) the map file specifying information of markers, QTL, and identification of favorable alleles, issued from a QTL mapping analysis (possibly considering several traits, environments, etc.); and 2) the genotypes/pedigree file including individuals, pedigree information, and genotypic data. To visualize and analyze the results, OptiMAS includes in a graphical user interface (GUI) three different modules, corresponding to the different steps of a selection program (see Figure 1):
Figure 1.
OptiMAS graphical interface showing the three different steps of the selection process. (A) Prediction of global scores (MS, Weight, UC) and genotype probabilities at QTL (with detailed view for one cell). (B) Selection (comparison between two lists of selected individuals). Graphs display the distribution of the molecular scores (here for QTL1). (C) Intermating (comparison of two mating schemes). Individuals are ranked on the two axes based on their genetic value (MS from highest to lowest). Left side graph displays the outcome of the better-half procedure illustrating that crosses between individuals having lowest MS have been avoided (i.e., B37 to B125). Right side graph illustrates the outcome of selection of the “best” crosses based on the UC considering constraints (here each candidate can contribute only twice).
Step 1: Computation of Genotypic Probabilities—Estimation of Genetic Values
Taking into account all information available (pedigree, distance between loci, genotypic data), OptiMAS computes for each QTL the probability of all possible phased genotypes (also called diplotypes) corresponding to the union of parental gametes. The tool provides for each individual the probabilities of being homozygous or heterozygous for parental alleles at each QTL. Based on the classification of parental alleles into favorable and unfavorable categories, a molecular score (expected probability of favorable allele) is computed for each QTL. Individual molecular scores are then combined into a global genetic value by assigning identical or different weights to QTL (MS/Weight columns in Figure 1A). A colored view of the molecular score table is displayed to identify more easily QTL for which a given individual is considered as fixed or not for the targeted allele(s). In this table, the number of QTL homozygous for (un)favorable allele(s) or heterozygous or with uncertain genotype are also given (see No.(+/+), No.(−/ −), No.(+/ −) and No.(?) columns in Figure 1A). Graphs are generated to show the distribution of several indicators (QTL molecular scores at individual QTL, global genetic values, etc.) and their evolution over the different cycles of selection.
Step 2: Selection of Individuals
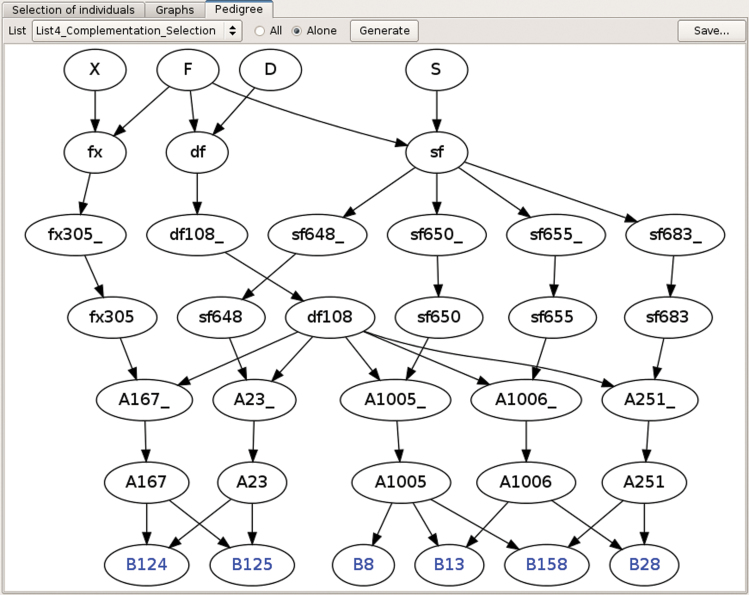
Different options are available to select candidates for producing the next generation. Truncation selection can be performed based on 1) the above described genetic value, or 2) a utility criterion (UC), which considers the probabilities of obtaining superior progenies following gametic segregation (UC column in Figure 1A). For a same MS, the UC favors individuals with no unfavorable alleles fixed. QTL complementation selection (QCS) can be conducted to take into account complementarities between candidate individual(s) regarding the favorable alleles they carry (Hospital et al. 2000). The QCS aims at preventing the loss of rare favorable allele(s) especially important when a high number of QTL is considered. Different lists of selected plants can be compared in two parallel tables and via graphs showing the distribution of above-mentioned indicators (see Figure 1B). All lists can be adjusted manually. A visualization tool of the pedigree of the selected plants is also provided (see Figure 2).
Figure 2.
Display of pedigree for a list of six selected individuals (B124, B125, B8, B13, B158, and B28).
The pedigree representation is useful to follow the contribution of selected individuals over generations of selection and to prevent possible bottlenecks (individuals coming from a reduced number of parents at a given generation), in order to limit risk of drift (which may lead, for instance, to the fixation of an undesired phenotypic type for traits not considered in the MAS process). It also can be used to maintain diversity for selection on traits complementary to those considered for the MAS process.
Step 3: Identification of Crosses among Selected Individuals
Considering that list(s) of selected individuals has been previously established, it is necessary to identify the crosses to be made to develop the next generation. We addressed crosses between individuals of a single list (diallel design) or two complementary lists (factorial design). The diallel situation can be managed with three options: 1) the automatic definition of the whole list of possible crosses according to a half-diallel; 2) the “better-half” strategy (Bernardo et al. 2006), which consists of avoiding crosses between selected individuals with the lowest scores; and 3) application of constraints on the contribution of parents and/or on the maximum number of crosses to be done. In this last case, best crosses are determined according to either the (weighted) molecular score or the UC. In each case, OptiMAS computes the expected molecular scores of the progeny. Then, lists of crosses to be done created via the different methods can be analyzed and compared via graphs (see Figure 1C).
All analysis outputs and the different lists of selected individuals and crosses can be exported in plain-text, tab-delimited format in order to save results to possibly reload the analysis later, or use output results in other tools (e.g., field nursery manager). Graphs and pedigree can be exported as png, eps, or svg files.
Implementation
Two versions of the tool have been developed. The first one, managing computationally intensive processes for step 1, is written in C-ANSI and runs in command line, which provides a convenient integration with custom analysis pipelines and databases. The second version integrates the C program and additional functionalities within a GUI coded in C++ using Qt, Qwt, and Graphviz libraries. Installable versions are distributed to run under most modern GNU/Linux, Windows (XP/7), and Mac OS X (10.5 or later with Intel processor) systems.
Although completely standalone, with data imported via simple plain-text formats, OptiMAS is part of the Integrated Breeding Platform (IBP), a web-based workflow system providing analytical tools and services to help breeders to improve crops for greater food security in the developing world (https://www.integratedbreeding.net).
Future Work
Future developments will handle 1) the QTL position uncertainty in score computation; 2) the addition of quantitative score(s) for global breeding value and/or background effects (e.g., genomic selection; Jannink et al. 2010); 3) allelic effects at QTL in order to compute expected gain for different traits with the possibility to weight them to compute indexes; 4) the development of a simulation procedure to produce a “virtual” next generation; and 5) a wizard to help users who want to run automatically the basic options of the tool.
Availability
OptiMAS is free software distributed under the GNU General Public License. The source code, example datasets, documentation, instructions for use, and executables are available from http://moulon.inra.fr/optimas.
Supplementary material
Supplementary material can be found at http://www.jhered.oxfordjournals.org/.
Funding
This program is part of the IBP within Generation Challenge Programme (GCP) and is funded by the Bill and Melinda Gates Foundation.
Supplementary Material
Acknowledgments
We are grateful to colleagues within INRA and the GCP for enlightening user-oriented input to this project. This development has benefited from the advices and beta-testing of Delphine Fleury and Mark Sawkins.
References
- Bernardo R, Moreau L, Charcosset A. 2006. Number and fitness of selected individuals in marker-assisted and phenotypic recurrent selection. Crop Sci. 46:1972–1980. [Google Scholar]
- Blanc G, Charcosset A, Mangin B, Gallais A, Moreau L. 2006. Connected populations for detecting quantitative trait loci and testing for epistasis: an application in maize. Theor Appl Genet. 113:206–224. [DOI] [PubMed] [Google Scholar]
- Blanc G, Charcosset A, Veyrieras JB, Gallais A, Moreau L. 2008. Marker-assisted selection efficiency in multiple connected populations: a simulation study based on the results of a QTL detection experiment in maize. Euphytica. 161:71–84. [Google Scholar]
- Buckler ES, Holland JB, Bradbury PJ, Acharya CB, Brown PJ, Browne C, Ersoz E, Flint-Garcia S, Garcia A, Glaubitz JC, et al. 2009. The genetic architecture of maize flowering time. Science. 325:714–718. [DOI] [PubMed] [Google Scholar]
- Cavanagh C, Morell M, Mackay I, Powell W. 2008. From mutations to MAGIC: resources for gene discovery, validation and delivery in crop plants. Curr Opin Plant Biol. 11:215–221. [DOI] [PubMed] [Google Scholar]
- Cook JP, McMullen MD, Holland JB, Tian F, Bradbury P, Ross-Ibarra J, Buckler ES, Flint-Garcia SA. 2012. Genetic architecture of maize kernel composition in the nested association mapping and inbred association panels. Plant Physiol. 158:824–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamblin MT, Buckler ES, Jannink JL. 2011. Population genetics of genomics-based crop improvement methods. Trends Genet. 27:98–106. [DOI] [PubMed] [Google Scholar]
- Hospital F, Goldringer I, Openshaw S. 2000. Efficient marker-based recurrent selection for multiple quantitative trait loci. Genet Res. 75:357–368. [DOI] [PubMed] [Google Scholar]
- Huang BE, George AW, Forrest KL, Kilian A, Hayden MJ, Morell MK, Cavanagh CR. 2012. A multiparent advanced generation inter-cross population for genetic analysis in wheat. Plant Biotechnol J. 10:826–839. [DOI] [PubMed] [Google Scholar]
- Jannink JL, Lorenz AJ, Iwata H. 2010. Genomic selection in plant breeding: from theory to practice. Brief Funct Genomics. 9:166–177. [DOI] [PubMed] [Google Scholar]
- Kover PX, Valdar W, Trakalo J, Scarcelli N, Ehrenreich IM, Purugganan MD, Durrant C, Mott R. 2009. A Multiparent Advanced Generation Inter-Cross to fine-map quantitative traits in Arabidopsis thaliana . PLoS Genet. 5:e1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R, Thompson R. 1990. Efficiency of marker-assisted selection in the improvement of quantitative traits. Genetics. 124:743–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen TH, Hayes BJ, Goddard ME. 2001. Prediction of total genetic value using genome-wide dense marker maps. Genetics. 157:1819–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland JA, Bradbury PJ, Buckler ES, Nelson RJ. 2011. Genome-wide nested association mapping of quantitative resistance to northern leaf blight in maize. Proc Natl Acad Sci U S A. 108:6893–6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebaï A, Goffinet B. 2000. More about quantitative trait locus mapping with diallel designs. Genet Res. 75:243–247. [DOI] [PubMed] [Google Scholar]
- Xu Y, Crouch JH. 2008. Marker-assisted selection in plant breeding:. from publications to practice. Crop Sci. 48:391–407. [Google Scholar]
- Yu J, Holland JB, McMullen MD, Buckler ES. 2008. Genetic design and statistical power of nested association mapping in maize. Genetics. 178:539–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.