Abstract
Development of methods for rapid screening and stratification of subjects after exposure is an integral part of countermeasures against radiation. The potential demographic and exposure history-related heterogeneity of exposed populations warrants robust biomarkers that withstand and reflect such differences. In this study, the effect of aging and repeated exposure on the metabolic response to sub-lethal irradiation was examined in mice using UPLC-ESI-TOF mass spectrometry. Aging attenuated post-exposure elevation in excretions of DNA damage biomarkers as well as N1-acetylspermidine. Although N1-acetylspermidine and 2’-deoxyuridine elevation was highly correlated in all age groups, xanthine and N1-acetylspermidine elevation was poorly correlated in older mice. These results may reflect the established decline in DNA damage-repair efficiency associated with aging and indicate a novel role for polyamine metabolism in the process. Although repeated irradiation at long intervals did not affect the elevation of N1-acetylspermidine, 2’-deoxyuridine, and xanthine, it did significantly attenuate the elevation of 2’-deoxycytidine and thymidine compared to a single exposure. However, these biomarkers were found to identify exposed subjects with accuracy ranging from 82% (xanthosine) to 98% (2’-deoxyuridine), irrespective of their age and exposure history. This indicates that metabolic biomarkers can act as robust noninvasive signatures of sub-lethal radiation exposure.
Keywords: Ionizing radiation, age, exposure history, biomarker, metabolomics, UPLC-ESIQTOF-MS, DNA damage-repair, polyamine metabolism
Introduction
Mass exposure to ionizing radiation due to sabotage, accident or natural disaster, such as the 2011 earthquake and tsunami in Japan, poses a grave threat to public health. The expanding landscape of nuclear facilities around the world, which are often located in earthquake-prone zones or near seashores, and the evolution of extreme weather patterns due to climate change may increase the risk of radiation leaks in the future. In addition, the establishment of nuclear facilities in densely populated parts of the world, such as in India, amplifies the chance of radiation exposure to large number of subjects. The exposed mass would include individuals with not only different genetic backgrounds but also different ages. In addition, some people may also have a previous history of sub-lethal radiation exposure. Therefore, the development of high-throughput biomarkers which are robust against differences in age and history of exposure is an essential component of countermeasures against ionizing radiation exposure.
DNA damage signatures, such as gamma-H2AX1 or changes in gene expression, such as XPC, CDKN1A, GADD45 and MDM22–4 have been proposed as biomarkers for ionizing radiation exposure. However, the effect of aging and a history of previous exposure on the robustness and dose-response of these signatures has not been studied in detail. The importance of examining this interaction is evident from the fact that aging and exposure to reactive oxygen species (ROS), which may occur due to previous exposure to ionization radiation, has been shown to modulate a number of pathways involved in DNA damage-repair5–7. In addition, many of these signatures, including the well established method for biodosimetry using the dicentric chromosome assay8, are either low-throughput or require invasive sample collection, which may compromise their utility in case of a mass exposure. On the other hand, recent studies have shown that ultra-performance liquid chromatography coupled with electrospray ionization and quadrupole time-of-flight (UPLC-ESI-QTOF) mass spectrometry-based metabolomic analysis of urine samples can efficiently identify exposed subjects. This approach is both noninvasive and high-throughput. A subset of these radiation biomarkers, primarily metabolites arising from DNA damage, were found to be common across exposed species with wide variation in genetic composition such as mouse9–11, rat12 and non-human primates13. However, the robustness of these markers against aging and exposure history has yet to be tested. The metabolome is the result of intertwined interactions between upstream molecular events and examination of these interactions may yield novel insights into molecular phenomenon involved in the modulation of DNA damage response with aging. This study investigates and compares the effect of aging and exposure history on metabolomic signatures of ionizing radiation exposure and their implication on biology of DNA damage-repair using mouse model.
MATERIALS AND METHODS
Chemicals
All compounds were obtained from Sigma-Aldrich (St. Louis, MO) and were of the highest grade available. HPLC grade solvents were purchased from Fisher Scientific (Hampton, NH).
Animals and Treatments
Twenty-four male 6-week-old wild-type C57BL/6 were purchased from Jackson Laboratories (Bar Harbor, ME) and acclimated to the animal facility for two weeks. Mice were fed a regular chow diet and water ad libitum throughout the experiment. Animals were randomly divided in to four groups (six mice each) at two months of age for radiation exposure. Mice from sham as well as repeated exposure groups were cohabited. The repeated radiation exposure protocol at a regular interval of two months starting at two months of age is schematically presented in Figure 1A. The groups, namely, Rad-1, Rad-2 and Rad-4 received one exposure (at 8 months), two exposures (at 6 and 8 months) or four exposures (at 2, 4, 6 and 8 months), respectively (Figure 1A). All mice were acclimated to the metabolic cages twice before the first urine collection at two months and once before subsequent collections. At every time point, mice were exposed to 3 Gy gamma radiation from a 137Cs source in a Mark I Model 68 small animal irradiator (J. L. Shepherd & Associates, San Fernando, CA) operating at 1.16 Gy/min. Urine samples were collected by placing mice 2 days prior to (D-2) or immediately after (D0) or 2 days after (D2) radiation exposure in Nalgene metabolic cages (Tecniplast USA, Inc., Exton, PA) for 24 hours as shown in Figure 1B. Urine volumes were measured and all samples were stored at −80°C until further analysis. Animals were regularly monitored for changes in body weight as well as any sign of deterioration of health. All animal studies were approved by the National Cancer Institute Animal Care and Use Committee.
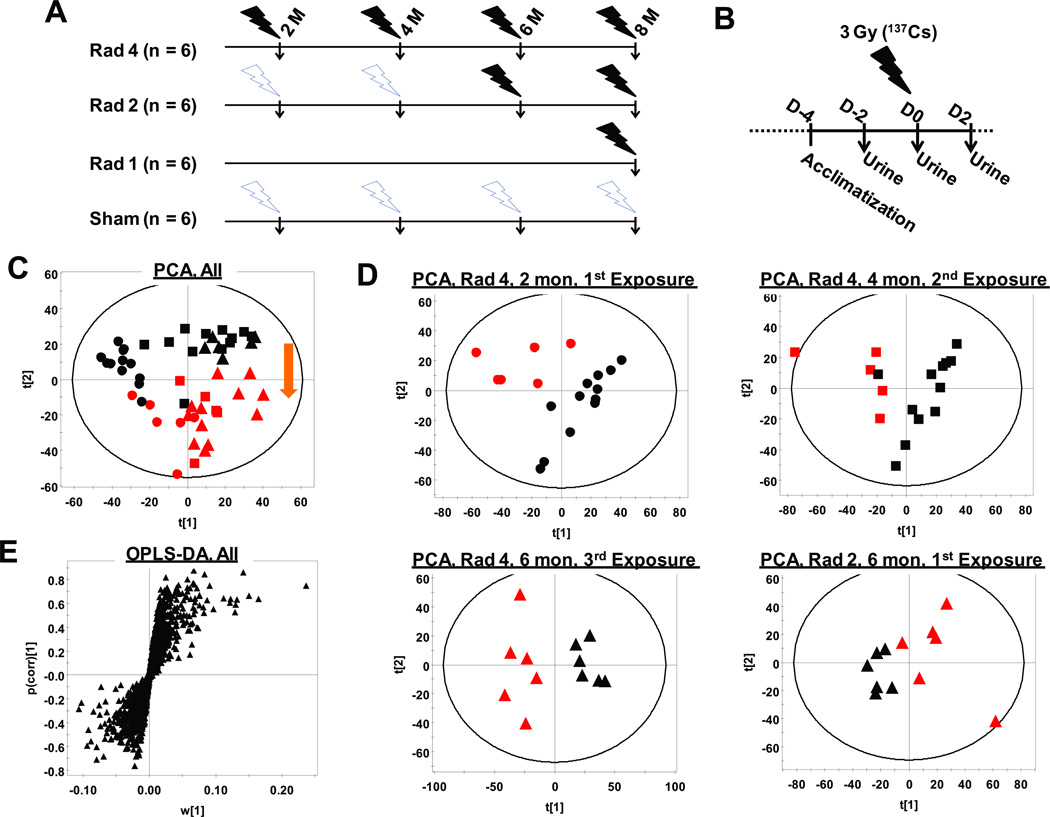
Figure 1. Metabolomic study design and multivariate data analysis.
(A) Rad-4 group of mice received four exposures (3 Gy; 1.16Gy/min from a 137Cs source) at 2, 4, 6, and 8 months of age. Rad-2 group of mice received two exposures at 6 month and 8 months of age. Rad-1 group of mice received one exposure at 8 months of age. (B) Mice were rested for 24 hours in their regular housing cages between 24-hour urine collections in metabowls on D-2, D0, and D2. At D0 mice were placed into metabolic cages immediately after exposure and urine collected for 24 hours. (C) Scores scatter plot generated from PCA of global metabolomic signatures of urine samples collected at D0 and recorded in ESI+ mode showing unsupervised clustering of sham (black) and irradiated (red) mice at 2 months (circle), 4 months (square), and 6 months (triangle) of age. The arrow shows the metabotype shift due to radiation exposure. (D) PCA scores scatter plots showing unsupervised segregation of the Rad-4 group of animals after the first (2 months), second (4 months), and third (6 months) exposures and that of Rad-2 mice after the first (6 months) exposure. (E) Loading S-plot generated from OPLS-DA including all irradiated and sham samples up to six months of age.
Preparation of Urine Samples and Untargeted UPLC-ESI-QTOFMS Analysis
Four volume of 50% acetonitrile containing 5µM chlorpropamide was added to one volume of urine sample, vortexed, centrifuged, and supernatant was transferred to 96-well sample plate (Waters Corp., Milford, MA). ESI-MS-based metabolite profiling was performed on a Waters ESI-QTOFMS (SYNAPT HDMS) coupled with an Acquity UPLC system (Milford, MA) in both positive and negative ionization mode as described earlier14. Briefly, a 5µL aliquot of the sample was injected into a BEH C18 column and chromatographic separation was achieved by an aqueous acetonitrile gradient containing 0.1% formic acid over a 10 min run with a flow rate of 0.5 ml/min. Column temperature was maintained at 40°C. Capillary and cone voltages used for ESI-Ms were 3 kV and 20 V, respectively. Source and desolvation temperatures were 120 °C and 350 °C, respectively. Nitrogen was used as both cone gas (50 L/h) and desolvation gas (600 L/h). The run also included intermittent injection of blanks as well as two different quality control samples and all samples were injected in randomized fashion. Mass chromatograms and mass spectral data (m/z = 50–850) were acquired using MassLynx software (Waters Corp.) in centroid format.
Multivariate Data Analysis
Chromatograms were manually inspected for consistency in retention time and peak shapes. MarkerLynx was used for feature identification, deconvolution of chromatogram, and integration. The area under the peak of each ion was normalized with respect to the total ion count (TIC) to generate a data matrix that consisted of the retention time, m/z value, and the normalized peak area. The multivariate data matrix was analyzed by SIMCA-P+ 12.0 software (Umetrics, Kinnelon, NJ). The unsupervised segregation of samples was checked by principal components analysis (PCA) using Pareto-scaled data15. The supervised orthogonal projection to latent structures discriminate analysis (OPLS-DA) model was used to generate loading S-plots for biomarker mining.
Identification of biomarkers
Putative candidates for biomarkers ions were searched using METLIN and HMDB16,17 databases and screened with the help of MS/MS fragmentation patterns. For MS/MS fragmentation of target ions, collision energies ranging from 10 to 40 eV were applied. The identities of ions were confirmed by comparison of retention times and fragmentation patterns with authentic standards. Authentic N1-acetylspermidine was synthesized as reported earlier12.
Quantitation of Urinary Metabolites
Quantitation of urinary metabolites was carried out using an AcquityÆ UPLC system coupled with a XEVO™ triple-quadrupole tandem mass spectrometer (Waters Corp.) by multiple reaction monitoring (MRM, see Table 1 for details). Standard calibration plots for quantitation were generated using authentic standards. Deproteinated urine samples containing 1.0 µM α-aminopimelic acid (176→112; ESI+), were analyzed in the same fashion as that of authentic compounds. The quantitative abundances were calculated from the normalized (with respect to internal standard) peak area with the help of the calibration plot.
Table 1.
List of aging and exposure history independent urinary radiation biomarkers
| RT (min)a |
m/z (ESI mode) |
Identity | Mass error (ppm) |
RT error (min) |
MRM (ionization mode)b |
Trendc | Metabolic pathway |
Sensitive to |
|---|---|---|---|---|---|---|---|---|
| 0.6 | 251.065+ | 2’-Deoxyuridine + Na+ | 4 | 0.0 | 227→ 184 (−) | ↑ | Pyrimidne metabolism | Aging |
| 0.3 | 188.175+ | N1-Acetylspermidine + H+ | 3 | 0.0 | 188→ 100 (+) | ↑ | Polyamine metabolism | Aging |
| 1.0 | 265.081+ | Thymidine + Na+ | 5 | 0.0 | 241→ 151 (−) | ↑ | Pyrimidne metabolism | Prior exposure |
| 0.3 | 228.097− | 2’-Deoxycytidine– H+ | 3 | 0.0 | 228→ 112 (−) | ↑ | Pyrimidne metabolism | Prior exposure |
| 0.4 | 151.026− | Xanthine – H+ | 0 | 0.1 | 151→ 107 (−) | ↑ | Purine metabolism | Aging |
| 0.5 | 227.066− | 2’-Deoxyuridine – H+ | 5 | 0.1 | 227→ 184 (−) | ↑ | Pyrimidne metabolism | Aging |
| 1.0 | 241.082− | Thymidine – H+ | 4 | 0.0 | 241→ 151 (−) | ↑ | Pyrimidne metabolism | Prior exposure |
| 0.8 | 283.066− | Xanthosine – H+ | 8 | 0.1 | 285→ 153 (+) | ↑ | Purine metabolism | Aging |
All RT (retention time) in reverse-phase global analysis.
Characteristic fragmentation reactions monitored for quantitation via multiple reaction monitoring (MRM).
All trends indicate statistically significant and consistent change after quantitation and normalization with respect to creatinine clearance.
Statistics
All values are presented as mean ± standard error of the mean (SEM). Two-way Mann-Whitney test was performed with 95% confidence interval to evaluate statistical significance of observed changes in metabolite excretion between sham and irradiated mice using GraphPad Prism 4 software and p < 0.05 was considered statistically significant. Logistic regression and ROC analysis for the predictive power of individual biomarkers were performed using level of urinary excretion of metabolites in sham and exposed animals at D0. All Logistic regression and ROC analysis was performed in STATA (Stata Statistical Software Release 11.2, College Station, TX).
RESULTS
Animal Performance
There was no significant change in the overall health and performance of mice irrespective of the exposure status during the study. The body weights (Figure S1A) and urine volumes (Figure S1B) of the sham and irradiated animals were also not significantly different.
Multivariate Data Analysis
There was no significant shift in retention times, and mass errors were found to be less than 10 ppm throughout the run. Unsupervised principal components analysis (PCA) showed a tight clustering of quality control and blank samples (data not shown) indicating consistent instrument response throughout the runs. PCA of deconvoluted, integrated, total ion count (TIC)-normalized and Pareto-scaled data from all urine samples collected up to 6 months, showed that the global urinary metabolomic signatures of the irradiated mice at D0 (24 h immediately after exposure) was distinct from the sham animals (Figure 1C). The shift in metabotype upon irradiation was similar (shown by arrow in Figure 1C) irrespective of age of the mice during irradiation as well as the history of previous exposures. The scores scatter plots for PCA analysis of urinary metabolomic signature from the Rad-4 group of mice at 2, 4, 6 months and that from the Rad-2 group of mice at 6 months also showed clear segregation from age-matched sham mice (Figure 1D). The OPLS-DA was performed to identify ions that contribute significantly to the discrimination of sham and irradiated animals. The loading S-plots for the discrimination of the Rad-4 group according to radiation exposure at 2, 4, 6 months and that for the Rad-2 group of animals at 6 months from age-matched sham mice are shown in Supplemental Figure S2A, B, C and D, respectively. Ions that contributed significantly to the separation of sham and irradiated mice in all these analysis were selected for further interrogation. OPLS-DA analysis was also performed on all sham and irradiated samples together to check the robustness of the contribution of ions in predicting exposure irrespective age of the animal and number of previous exposures. The loadings S-plot for the OPLS-DA model on all samples is shown in (Figure 1E). Finally, the ions that consistently contributed to the separation between sham and irradiated mice in all these models were selected as biomarkers for radiation exposure that could perform robustly against variation in age and exposure history.
Identification of Metabolites
The possible chemical formula of the ions were derived using the Seven Golden Rules18 and potential candidate metabolites were also identified using METLIN17 and HMDB16 databases using a maximum mass error of 10 ppm (Table 1). Finally, identities of the metabolites were confirmed by comparing retention time and fragmentation pattern against authentic standards. Thymidine was identified in both positive (as Na+-adduct) and negative (as deprotonated ion) mode. All others were identified as corresponding protonated or deprotonated ions in positive or negative mode, respectively, as shown in Table 1. The potential metabolic origins of these metabolites are also mentioned in the Table and show that purine and pyrimidine metabolism was most significantly affected along with arginine and polyamine metabolism following radiation exposure. All these metabolites were subsequently quantitated as described above along with putrescine that is the precursor in polyamine biosynthesis.
Effect of aging and repeated exposure on urinary radiation biomarker excretion
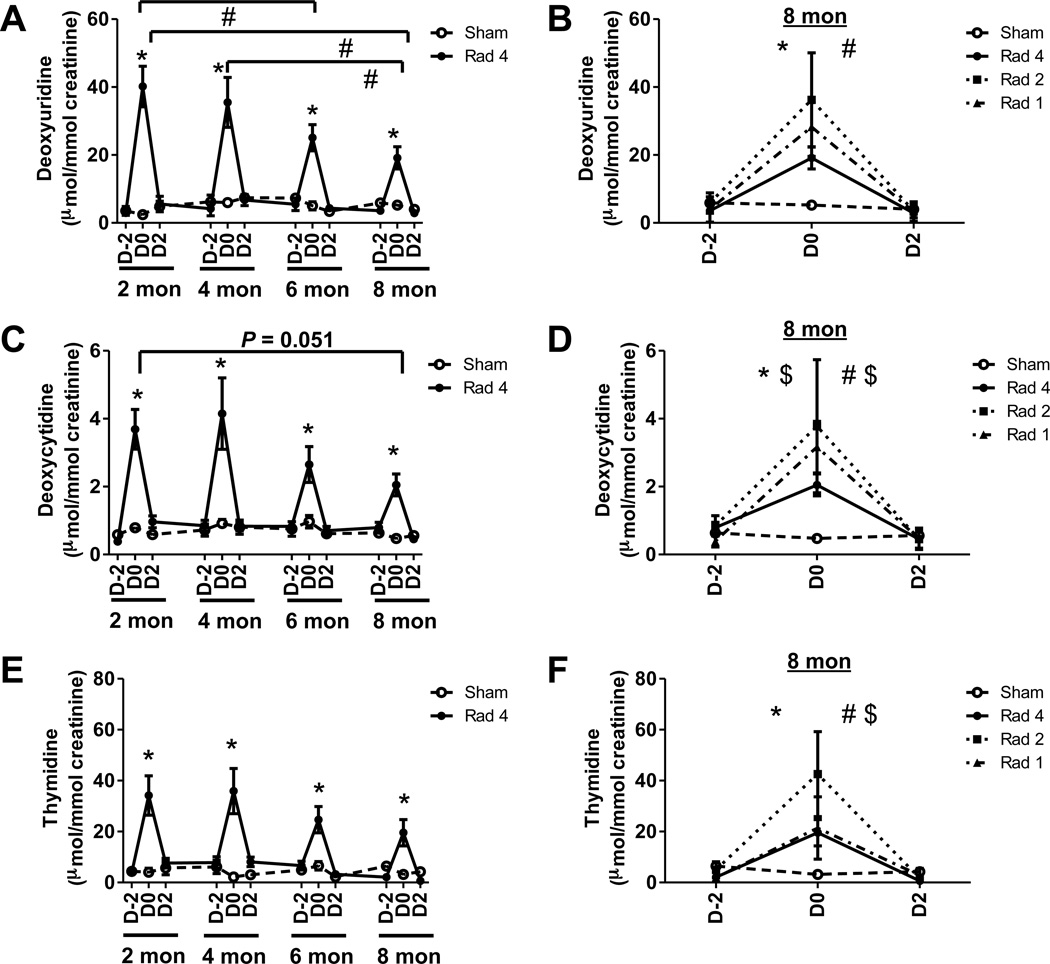
The urinary excretion of 2’-deoxyuridine was elevated by 16.6-fold (p < 0.005), 6-fold (p < 0.005), 5-fold (p < 0.005) and 3.7-fold (p < 0.005) after exposures (D0), respectively, at two, four, six and eight months of age in the Rad-4 group of mice compared to age-matched sham mice (Figure 2A). The urinary excretion of 2’-deoxyuridine on D0 at six and eight months was, respectively, 1.6-fold (p < 0.03) and 2.1-fold (p < 0.04) lower than that at two months of age. The 2-deoxyuridine excretion at D0 in eight months (p < 0.05) was also 1.9-fold lower than that at four months of age. The excretion of this metabolite in the Rad-2 and Rad-1 group of mice after exposure at eight months was 6.9-fold and 5.4 fold higher than that of the sham mice (Figure 2B). However, 2’-deoxyuridine excretion in the Rad-1 group, which was freshly exposed at eight months of age, was not significantly different from that in the age-matched Rad-2 or Rad-4 groups, which, respectively, received one or three previous exposures. These results indicated that aging, and not repeated exposure, contributes more significantly to the attenuation in elevation in excretion of 2’-deoxyuridine following radiation exposure.
Figure 2. Effect of aging and repeated radiation exposure on pyrimidine metabolites.
Temporal evolution of creatinine-normalized (A) 2’-deoxyuridine, (C) 2’-deoxycytidine and (E) thymidine excretion over the first (2 months), second (4 months), third (6 months), and fourth (8 months) exposures of the Rad-4 group (●, solid line) are shown along with sham (○, dotted line) mice. Although excretion levels between exposures (e.g., D2 at two month and D-2 at 4 months) are joined by straight lines no urine samples were collected during these rest periods. Significant elevations in metabolite excretion following radiation exposure (D0) compared to age-matched sham-mice are indicated (*). Significant decreases in excretion of the metabolite over time are indicated (#). The creatinine-normalized excretion of (B) 2’-deoxyuridine, (D) 2’-deoxycytidine and (F) thymidine from Rad-1 (▲) mice, the second exposure of Rad-2 (■) mice, fourth exposure Rad-4 (●) mice, and sham mice (○) at 8 months of age. Significant differences between D0 Rad-1 (*) and Rad-2 (#) and age-matched sham mice are indicated. In addition, $ next to * and #, respectively, indicates that excretion of the metabolite in Rad-1 and Rad-2 group was also significantly different than age-matched Rad-4 mice. Statistical significance for graphs (A), (C), and (E) was calculated by paired t test. The statistical significance for graphs (B), (D), and (F) was calculated by two-tailed Mann-Whitney test with 95% confidence interval. P value < 0.05 was considered significant.
2’-Deoxycytidine excretion was elevated by 4.7-fold (p < 0.005), 4.6-fold (p < 0.005), 2.6-fold (p < 0.02) and 4.4-fold (p < 0.005) after exposures (D0), respectively, at two, four, six and eight months of age in the Rad-4 group of mice compared to age-matched sham mice (Figure 2C). Although statistically insignificant, urinary excretions of 2’-deoxycytidine at D0 at eight months were 1.8-fold (p = 0.051) lower than that at two months of age. The urinary excretions of 2’-deoxycytidine in the Rad-2 and Rad-1 group of mice after exposure at eight months were 8-fold and 6.7-fold higher than the sham mice (Figure 2D). There was no significant difference between Rad-1 and Rad-2 groups with respect to urinary excretion of this metabolite following radiation exposure at eight months, in spite of Rad-2 receiving a previous exposure. However, the urinary excretion of 2’-deoxycytidine in the Rad-2 and Rad-1 groups were, respectively, 1.8-fold (p < 0.05) and 1.5-fold (p < 0.05) higher than that of the Rad-4 group. These results indicated that repeated exposure contributes more significantly to the attenuation in elevation of 2’-deoxycytidine excretion compared to aging.
The urinary excretion of thymidine was elevated by 8.4-fold (p < 0.005), 16.7-fold (p < 0.005), 3.8-fold (p < 0.005) and 6.1-fold (p < 0.01) after exposures (D0), respectively, at two, four, six and eight months of age in the Rad-4 group of mice compared to age-matched sham mice (Figure 2D). Although, thymidine excretions on D0 at six and eight months were lower than that at two and four months, the attenuation was not statistically significant. The urinary excretions of thymidine in the Rad-2 and Rad-1 group of mice after exposure at eight months were 13.4-fold (p < 0.005) and 6.7- fold (p < 0.01) higher than that of the sham mice (Figure 2E). Thymidine excretion in Rad-2 group was 2.2-fold higher (P < 0.05) than that in Rad-4 group at eight months. However, there was no significant difference between urinary excretion of this metabolite between the Rad-2 and Rad-1 group as well as between the Rad-4 and Rad-1 group at this age. These results indicated that repeated exposure contributes more significantly to the attenuation in elevation in excretion of thymidine compared to aging.
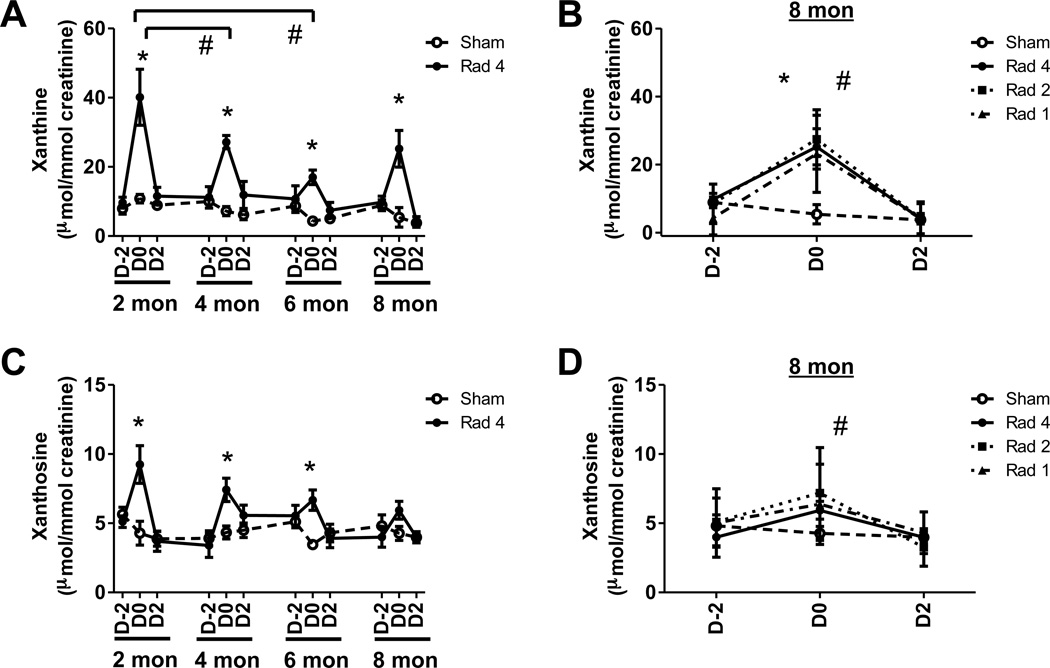
The urinary excretion of xanthine was elevated by 3.7-fold (p < 0.005), 3.8-fold (p < 0.005), 3.9-fold (p < 0.005) and 4.7-fold (p < 0.02) after exposures (D0), respectively, at two, four, six and eight months of age in Rad-4 group of mice compared to age-matched sham mice (Figure 3A). The urinary excretion of xanthine on D0 at six and eight months of age were, respectively, 1.6-fold (p <0.03) and 2.1-fold (p < 0.004) lower than that at two months of age. The urinary excretions of xanthine in the Rad-2 and Rad-1 group after exposure at eight months were, respectively, 6.1-fold (p < 0.008) and 4.3-fold (p <0.02) higher than that of sham mice (Figure 3B). However, there was no significant difference in excretion of xanthine among Rad-1, Rad-2 and Rad-4 groups that received their first, second and fourth exposure, respectively, at eight months. These results indicated that aging contributes more significantly to the attenuation of elevation in excretion of xanthine compared to repeated exposure.
Figure 3. Effect of aging and repeated radiation exposure on purine metabolites.
Temporal evolution of creatinine-normalized (A) xanthine and (C) xanthosine excretion during the first (2 months), second (4 months), third (6 months) and fourth (8 months) exposures of Rad-4 group (●, solid line) are shown along with sham (○, dotted line) mice. Although the excretion level between exposures (e.g., D2 at two month and D-2 at four month) are joined by straight lines no urine samples were collected during these rest periods. Significant elevations in metabolite excretion following radiation exposure (D0) compared to age-matched sham-mice are indicated (*). Significant decreases in excretion of the metabolite over time are indicated (#). The creatinine-normalized excretion of (B) xanthine and (D) xanthosine from Rad-1 (▲), the second exposure of Rad-2 (■), fourth exposure Rad-4 (●), and sham (○) at 8 months of age. Significant differences between D0 Rad-1 (*) and Rad-2 (#) and age-matched sham mice are indicated. Statistical significance for graphs (A) and (C) was calculated by paired t test. The statistical significance for graphs (B) and (D) was calculated by two-tailed Mann-Whitney test with 95% confidence interval. P value < 0.05 was considered significant.
The urinary excretion of xanthosine was elevated by 2.2-fold (p < 0.03), 1.7-fold (p < 0.01) and 1.9-fold (p < 0.005) after exposures (D0), respectively, at two, four and six months of age in the Rad-4 mice compared to age-matched sham mice (Figure 3C). However, the elevation in xanthosine excretion at eight months of age was insignificant in Rad-4 as well as Rad-1 group of mice (Figure 3D). Although Rad-2 group of mice showed 1.7-fold (p < 0.05) elevation in xanthosine excretion compared to sham mice at eight months, they were not significantly different from the Rad-1 and Rad-4 group. These results indicated that attenuation of elevation in excretion of xanthosine is mostly due to aging rather than repeated exposure.
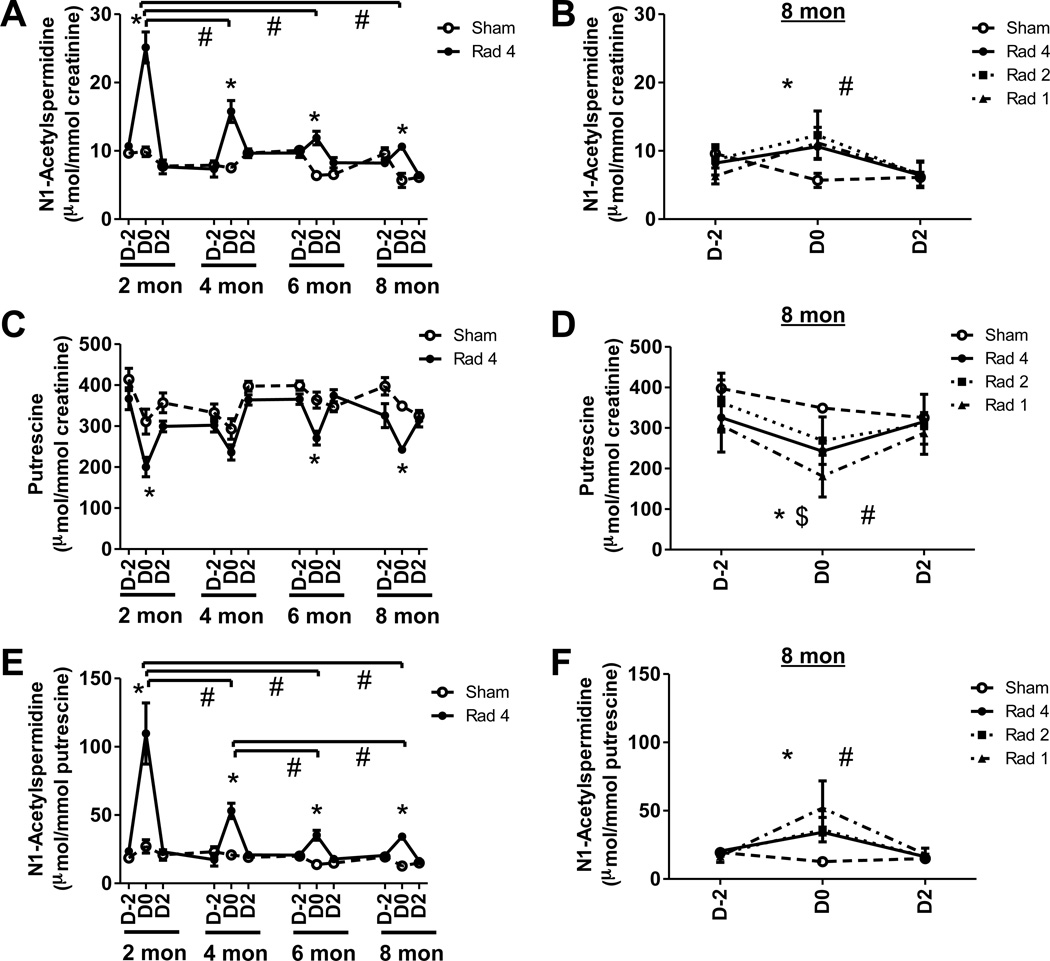
N1-acetylspermidine excretion was elevated by 2.5-fold (p < 0.005), 2.1-fold (p < 0.005), 1.9-fold (p < 0.005) and 1.9-fold (p < 0.02) after exposures (D0) at two, four, six and eight months of age in Rad-4 group of mice compared to age-matched sham mice (Figure 4A). The urinary excretion of N1-acetylspermidine on D0 at four, six and eight months were, respectively, 1.6-fold (p < 0.03), 2.1-fold (p < 0.001) and 2.4-fold (p <0.002) lower than that at two months of age. N1-acetylspermidine excretion in the Rad-2 and Rad-1 mice after exposure at eight months were 2.2-fold (p < 0.005) and 2-fold (p <0.02) higher than that of the sham mice (Figure 4B). Excretion of N1-acetylspermidine in Rad-1 mice at eight months was also 2.3-fold (p <0.005) lower than after the first exposure of the Rad-4 animals at two months. However, there was no significant difference in excretion of N1-acetylspermidine among Rad-1, Rad-2 and Rad-4 groups that received their first, second and fourth exposure, respectively, at eight months. These results indicated that the elevation of N1-acetylspermidine excretion is attenuated by aging.
Figure 4. Effect of aging and repeated radiation exposure on polyamine metabolites.
Temporal evolution of creatinine-normalized (A) N1-acetylspermidine, (C) putrescine excretion and (E) ratio of excretion of N1-acetylspermidine with respect to putrescine during the first (2 months), second (4 months), third (6 months) and fourth (8 months) exposures of Rad-4 group (●, solid line) are shown along with sham (○, dotted line) mice. Although excretion levels between exposures (e.g., D2 at two month and D-2 at four month) are joined by straight lines no urine samples were collected during these rest periods. Significant differences in metabolite excretion following radiation exposure (D0) compared to age-matched sham-mice are indicated (*). Significant decreases in excretion of the metabolite over time are indicated (#). The creatinine-normalized (B) N1-acetylspermidine, (D) putrescine excretion and (F) the ratio of N1-acetylspermidine excretion normalized to putrescine from Rad-1 (▲), second exposure of Rad-2 (■), fourth exposure of Rad-4 (●), and sham (○) mice at 8. Significant differences between D0 Rad-1 (*) and Rad-2 (#) and age-matched sham mice are indicated. In addition, $ next to * indicates that excretion of the metabolite in Rad-1 group was also significantly different than age-matched Rad-4 mice. The statistical significance of difference in metabolite excretion in different group of mice were calculated by two-tailed Mann-Whitney test with 95% confidence interval and the significance in difference in excretion of metabolites in mice from Rad-4 group during different exposures were calculated by paired t test. P value < 0.05 was considered significant. Statistical significance for graphs (A), (C), and (E) was calculated by paired t test. The statistical significance for graphs (B), (D), and (F) was calculated by two-tailed Mann-Whitney test with 95% confidence interval. P value < 0.05 was considered significant.
The urinary excretion of putrescine was depleted by 1.6-fold (p < 0.03), 1.3-fold (p < 0.01), and 1.4-fold (p < 0.005), respectively, after exposures (D0) at two, six and eight months of age in Rad-4 group of mice compared to age-matched sham mice (Figure 4C). Putrescine excretions in the Rad-2 and Rad-1 mice after exposure at eight months were 1.3-fold (p < 0.005) and 1.9-fold (p <0.005) lower than that of the sham mice (Figure 4D). In addition, the Rad-1 group was found to have 1.3-fold (p < 0.03) and 1.5-fold (p < 0.02) lower excretion of putrescine compared to the Rad-4 and Rad-2 groups, respectively. These results indicated that depletion in putrescine excretion may be affected by repeated exposure.
N1-Acetylspermidine is the downstream metabolite of putrescine in the polyamine metabolic pathway. The ratio of N1-acetylspermidine to putrescine in the urine of the Rad-4 group was, respectively, 4-fold (p < 0.005), 2.6-fold (p < 0.005), 2.5-fold (p < 0.005) and 2.7-fold (p < 0.005) higher than that of the sham mice on D0 at two, four, six and eight months (Figure 4E). The ratio of these two metabolites at four, six and eight months were, respectively, 2.1-fold (p <0.04), 3.1-fold (p <0.02), and 3.2-fold (p <0.03) lower than that at two months in the Rad-4 group. The ratios at six and eight months were also 1.5-fold (p < 0.01) and 1.6-fold (p < 0.02) lower than that at four months in this group. The ratios of these metabolites in Rad-2 and Rad-1 group of mice after exposure at eight months were, respectively, 2.9-fold (p < 0.005) and 4.1-fold (p <0.005) higher than that of the sham mice (Figure 4F). Post-exposure putrescine-normalized excretion of N1-acetylspermidine in Rad-1 mice at eight months was also 2.1-fold (p <0.02) lower than after first exposure of Rad-4 animals at two months. However, there was no significant difference between putrescine-normalized excretion of N1-acetylspermidine in Rad-1, Rad-2 and Rad-4 groups that received their first, second and fourth exposure, respectively, at eight months. These results indicated that the elevation in putrescine-normalized excretion of N1-acetylspermidine is attenuated by aging. Excretion of ornithine, dimethylarginines, creatine and pimelic acid did not show any consistent change following radiation exposure throughout the study.
These results showed that while elevation of 2’-deoxyuridine, xanthine, xanthosine, N1-acetylspermidine and the ratio of N1-acetylspermidine to putrescine were mainly attenuated by aging, repeated exposure contributed more significantly to the attenuation of 2’-deoxycytidine and thymidine. However, in spite of the attenuation, changes in all biomarkers were still significant in mice that were older as well as those receiving multiple exposures.
Predictive power of the urinary radiation biomarkers
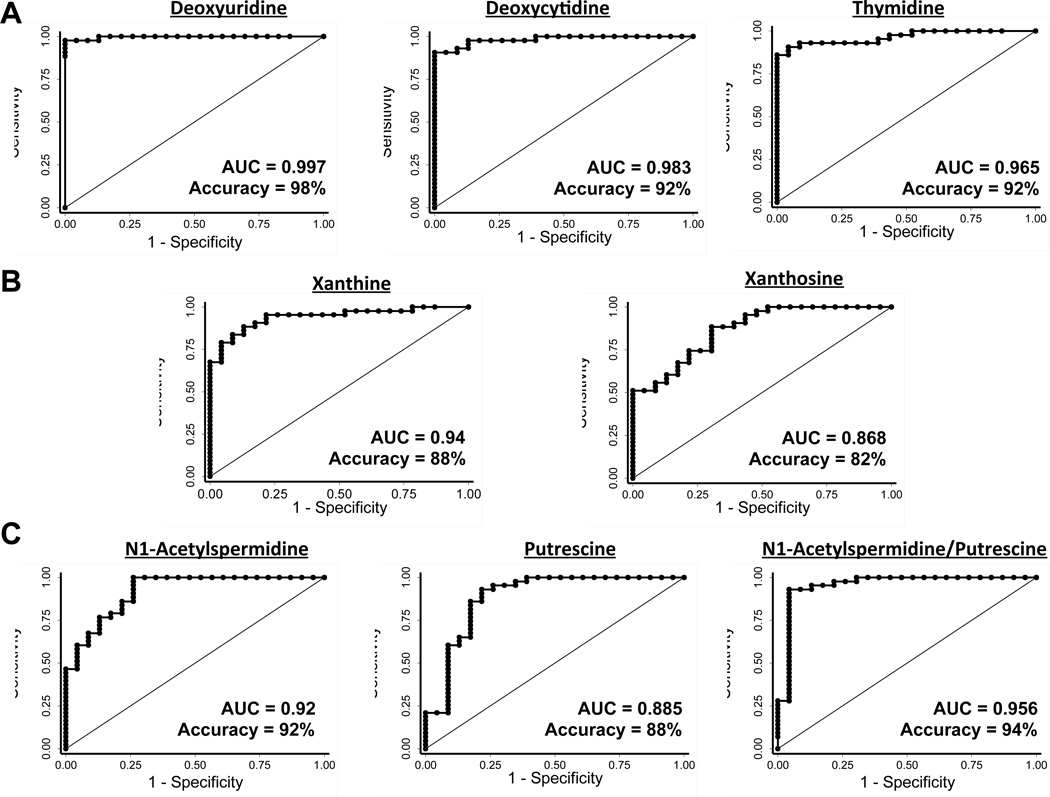
The ROC analysis was performed to assess the utility of individual metabolites in predicting radiation exposure status. The area under the ROC curve for 2’-deoxyuridine was found to be 1.0 at two (six sham and six R-4 animals), four (six sham and six R-4 animals), six (six sham, six R-2 and six R-4 animals) as well as eight months of age (six sham, six R-1, six R-2 and six R-4 animals) (Figure S3). The ROC analysis including sham and irradiated animals from all exposure and age groups showed an area under the curve of 0.997 with 98% accuracy of identifying radiation exposure using 2’-deoxyuridine as a noninvasive biomarker (Figure 5A). The area under the ROC curve for deoxycytidine was found to be 1.0, 0.943, 0.958 and 1.0 at two, four, six and eight months, respectively (Figure S3). The ROC analysis including sham and irradiated animals from all exposure and age groups showed an area under the curve of 0.983 with 92% accuracy of identifying radiation exposure using deoxycytidine as a noninvasive biomarker (Figure 5A). The area under the ROC curve for thymidine was found to be 1.0, 0.943, 0.986 and 0.983 at two, four, six and eight months, respectively (Figure S3). The ROC analysis including sham and irradiated animals from all exposure and age groups showed an area under the curve of 0.965 with 92% accuracy of identifying radiation exposure using thymidine as a noninvasive biomarker (Figure 5A). The area under the ROC curve for xanthine was found to be 1.0, 0.971, 0.931 and 0.944 at two, four, six and eight months, respectively (Figure S3). The ROC analysis including sham and irradiated animals from all exposure and age groups showed an area under the curve of 0.94 with 88% accuracy of identifying radiation exposure using xanthine as a noninvasive biomarker (Figure 5B). The area under the ROC curve for xanthosine was found to be 0.889, 0.971, 0.986 and 0.778 at two, four, six and eight months, respectively (Figure S3). The ROC analysis including sham and irradiated animals from all exposure and age groups showed an area under the curve of 0.868 with 82% accuracy of identifying radiation exposure using xanthosine as a noninvasive biomarker (Figure 5A). The area under the ROC curve for N1-acetylspermidine was found to be 1.0 at all time points except at six months (0.935) (Figure S3). The ROC analysis including sham and irradiated animals from all exposure and age groups showed an area under the curve of 0.92 with a 92% accuracy of identifying radiation exposure using N1-acetylspermidine as a noninvasive biomarker (Figure 5C). The area under the ROC curve for putrescine was found to be 0.889, 0.971, 0.986 and 0.778 at two, four, six and eight months, respectively (Figure S3). The ROC analysis including sham and irradiated animals from all exposure and age groups showed an area under the curve of 0.868 with 82% accuracy of identifying radiation exposure using putrescine as a noninvasive biomarker (Figure 5C). The area under the ROC curve for detection of radiation exposure using N1-acetylspermidine to putrescine ratio as a noninvasive signature was found to be 1.0 at all time points except at six months (0.943) (Figure S3). Consequently, the ROC analysis including sham and irradiated animals from all exposure and age groups showed an area under the curve of (0.956) and accuracy (94%), which was higher than that achieved using N1-acetylspermidine and putrescine as individual markers for identifying radiation exposure (Figure 5C).
Figure 5. Predictive power of noninvasive biomarkers.
Receiver operating characteristic (ROC) curves for evaluating the accuracy of radiation exposure prediction by logistic regression of the level of urinary excretion of (A) pyrimidine, (B) purine, and (C) polyamine metabolites. The analysis was performed including all post-exposure samples from sham, Rad-1, Rad-2, and Rad-4 groups irrespective of age and history of exposure. The area under the curve and accuracy of prediction is indicated on each ROC curve.
Correlation between changes in urinary radiation biomarker excretion
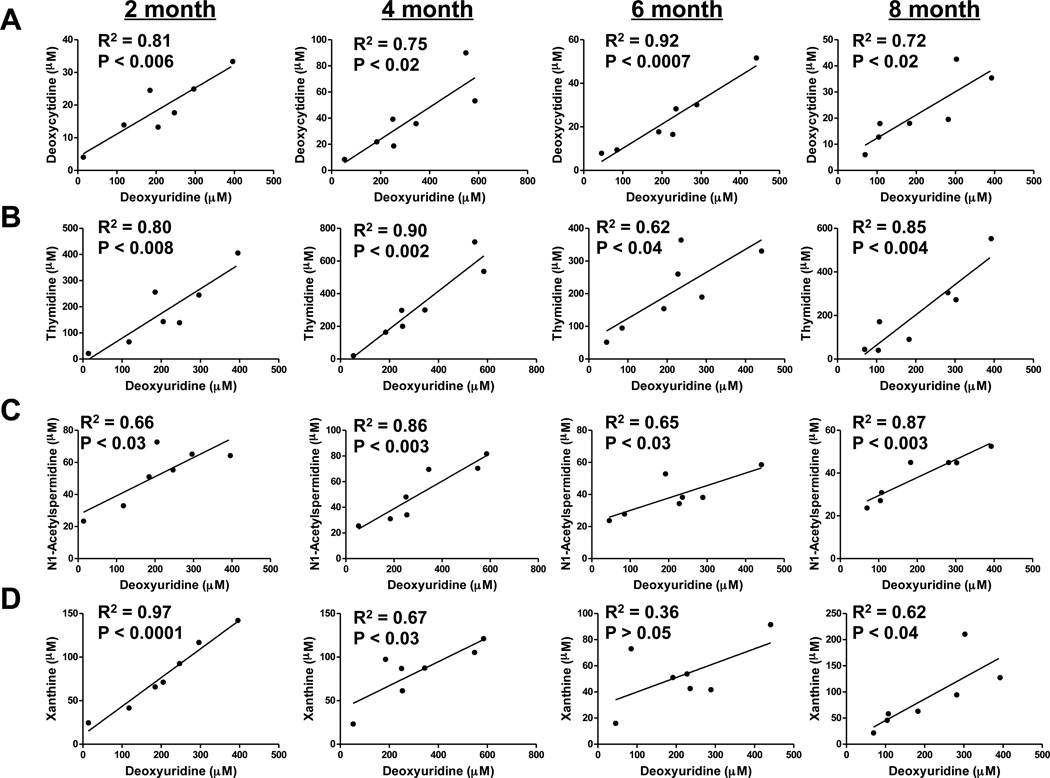
Although most of the metabolites showed a significant change in excretion upon irradiation and, consequently, very high ability to identify radiation exposure, there was considerable inter-individual variations in the elevation of urinary biomarkers among exposed mice. It was noted that this variation did not correlate with the body weight of the animals (data not shown) indicating that area of exposure is not the determining factor in the observed variation. In order to reveal possible collusion between metabolic events in response to radiation exposure in the context of aging, the correlation between changes in metabolites in same set of animals (Rad-4 group) at different ages were examined. Since elevation in 2’-deoxyuridine was found to be the most robust signature of radiation exposure, correlations between changes in other biomarkers and that of 2’-deoxyuridine were examined. The elevation of 2’-deoxycytidine excretion was highly correlated with that of 2’-deoxyuridine with correlation coefficients (R2) of 0.81 (p < 0.006), 0.75 (p < 0.002), 0.92 (p < 0.0007) and 0.72 (p < 0.02), respectively, at two, four, six and eight months (Figure 6A). The elevation of thymidine excretion was highly correlated with that of 2’-deoxyuridine with R2 of 0.80 (p < 0.008), 0.90 (p < 0.002), 0.62 (p < 0.04) and 0.85 (p < 0.004) at two, four, six and eight months, respectively (Figure 6B). The correlation coefficients between elevation of N1-acetylspermidine and deoxyuridine at two, four, six and eight months were 0.66 (p < 0.03), 0.86 (p < 0.003), 0.65 (p < 0.03), 0.87 (p < 0.003), respectively (Figure 6C). The correlation between elevation of xanthine and 2’-deoxyuridine were found to be very high at two months with R2 0.97 (p < 0.0001) (Figure 6D). However, the correlation was poorer, in general, for older mice with correlation coefficient of 0.67 (p < 0.03) and 0.62 (p < 0.04), respectively, at four and eight months and no significant correlation at six months. The correlation between xanthine and N1-acetylspermidine elevation was also relatively poorer, in general, and insignificant in older mice. The correlation coefficients between elevation of these two metabolites were found to be 0.60 (p < 0.05), 0.58 (p < 0.05) at two and four months, respectively. At six and eight months, this correlation was not statistically significant. There was no significant correlation between the excretion of xanthosine and other metabolites (data not shown).
Figure 6. Analysis of correlation between metabolomic response originating from different biochemical pathways.
Correlation between excretion of the most predictive metabolite 2’-deoxyuridine with that of other pyrimidine metabolites such as (A) 2’-deoxycytidine, (B) thymidine, the polyamine metabolite (C) N1-acetylspermidine and purine metabolite (D) xanthine is shown. The linearity between excretion of these biomarkers during consecutive exposure of Rad-4 animals at 2, 4, 6, and 8 months were examined. Pearson correlation coefficients at respective ages are indicated along with P values on the plots.
These results indicate that elevation in production of pyrimidine metabolites (2’-deoxyuridine, 2’-deoxycytidine and thymidine) are tightly coupled between themselves as well as with N1-acetylspermidine. On the other hand, elevation of the purine metabolites was either not correlated (xanthosine) or showed very poor correlation (xanthine) with elevation of pyrimidine metabolites and N1-acetylspermidine, particularly, in older mice.
DISCUSSION
In addition to the well-known γH2AX and dicentric chromosome assay, recent studies have revealed metabolome signatures of exposure to ionizing radiation in serum19,20. However, urine-based biomarkers are very attractive due to the ease and noninvasiveness of the collection procedure. In addition, the fact that these biomarkers originate from DNA damage, which is a well-established consequence of radiation exposure, and some of them were shown to be elevated in urine from non-human primates13, which are genetically very close to humans, as well as in human skin samples21 following irradiation, increases the likelihood of translation into viable biomarkers for human exposure. This study showed that elevation of these urinary metabolomic biomarkers are robust signatures of radiation exposure irrespective of aging and history of previous exposures in mice. However, it was found that the elevation of these biomarkers was significantly attenuated with aging. In addition, in the case of older mice (8 months) there was some significant attenuation in the elevation of thymidine and deoxycytidine in mice with a history of previous exposure compared to newly exposed mice. However, there was no difference in the elevation of xanthine, N1-acetylspermidine and the ratio of N1-acetylspermidine and putrescine in older mice due to previous exposure. This suggests that these three metabolites could be used to determine level of exposure if the age of the subject is known subject to establishment of an age-adjusted dose-response curve. In addition, a concomitant attenuation of thymidine and deoxycytidine excretion without any significant difference in level of xanthine, N1-acetylspermidine and ratio of N1-acetylspermidine to putrescine compared to other age-matched exposed subjects could yield unbiased information about previous exposure. These data are extremely important in the context of risk assessment and stratification for appropriate post-exposure intervention. Thus, these results indicate that detailed dose-response studies on the effect of aging and previous exposure on noninvasive metabolomic biomarkers of radiation exposure may help to develop methods for rapid identification of exposed subjects as well as for risk assessment according to age and exposure history. However, the overall attenuation in metabolic response due to repeated exposure at long intervals was not as drastic as that caused by aging. The non-uniformity of attenuation of metabolites also indicates that prior exposure-related tissue depletion may not be the reason for the observed changes.
Radiation exposure is well known to cause DNA damage22–25. In fact, DNA damage-related changes such as phosphorylation of H2AX at Ser-139 (γ-H2AX)1 and the dicentric chromosome assay8 have been established as the most reliable signatures of radiation exposure. Radiation was shown to cause DNA damage by production of ROS such as hydroxyl radical or by direct cleavage of covalent bonds in DNA molecule22–25. Modification of DNA bases and sugars are major ways of DNA damage due to radiation exposure and ROS production, in addition to single and double-strand breaks6,26,27. Under normal conditions, cells recognize such changes and activate a cascade of signaling events that either leads to apoptosis or cell cycle arrest28. During cell cycle arrest, a complex machinery of enzymes is recruited depending on the type of DNA damage. The damaged lesions are eventually repaired to restore the original sequence (except in the case of non-homologous end-joining) by removal of damaged DNA and joining broken strands27–29. The damaged DNA products such as bases or nucleosides are removed through base-excision repair or nucleoside excision repair28,30–32. It was shown that oxidatively modified DNA bases can be excreted in urine as 2’-deoxyribonucleosides during the DNA damage-repair process33. Therefore, it is not surprising to find that the origin of most of the urinary biomarkers reported in this study lies in nucleic acid metabolism. Apart from animal models, such markers that potentially arise from DNA damage have also been shown to be elevated after exposure of human skin tissue to low-dose gamma radiation21. The damaged nucleic acids are eventually metabolized and excreted. These indicate DNA damage to be the origin of urinary biomarkers such as 2’-deoxyuridine, 2’-deoxycytidine and thymidine.
Cytosine, which is frequently converted to uracil under oxidative stress, is one of the most sensitive bases to oxidative DNA damage29. Such modification can cause introduction of G→A mutations in DNA if replication continues without repair. The current as well as earlier radiation metabolomics studies has shown significant elevation in urinary excretion of the pyrimidine 2’-deoxyribonucleosides, such as thymidine, deoxyuridine and deoxycytidine10,13. It was noted that the elevation of 2’-deoxyuridine was highly correlated with that of 2’-deoxycytidine. As shown in Figure 7A, this may reflect possible conversion of 2’-deoxycytidine to 2’-deoxyuridine due to cytidine deaminase (EC 3.5.4.5) activity. 2’-Deoxyuridine could also be converted to thymidine via a rescue mechanism involving thymidine kinase (EC 2.7.1.21) and thymidilate synthase (EC 2.1.1.45). This could explain the strong correlation between thymidine and 2’-deoxyuridine excretion. The fact that the correlation between 2’-deoxyuridine, 2’-deoxycytidine and thymidine excretion was strong in spite of significant attenuation of the level of elevation, indeed, indicate to a biochemical link between production of these pyrimidine 2’-deoxynucleosides in response to exposure to ionizing radiation.
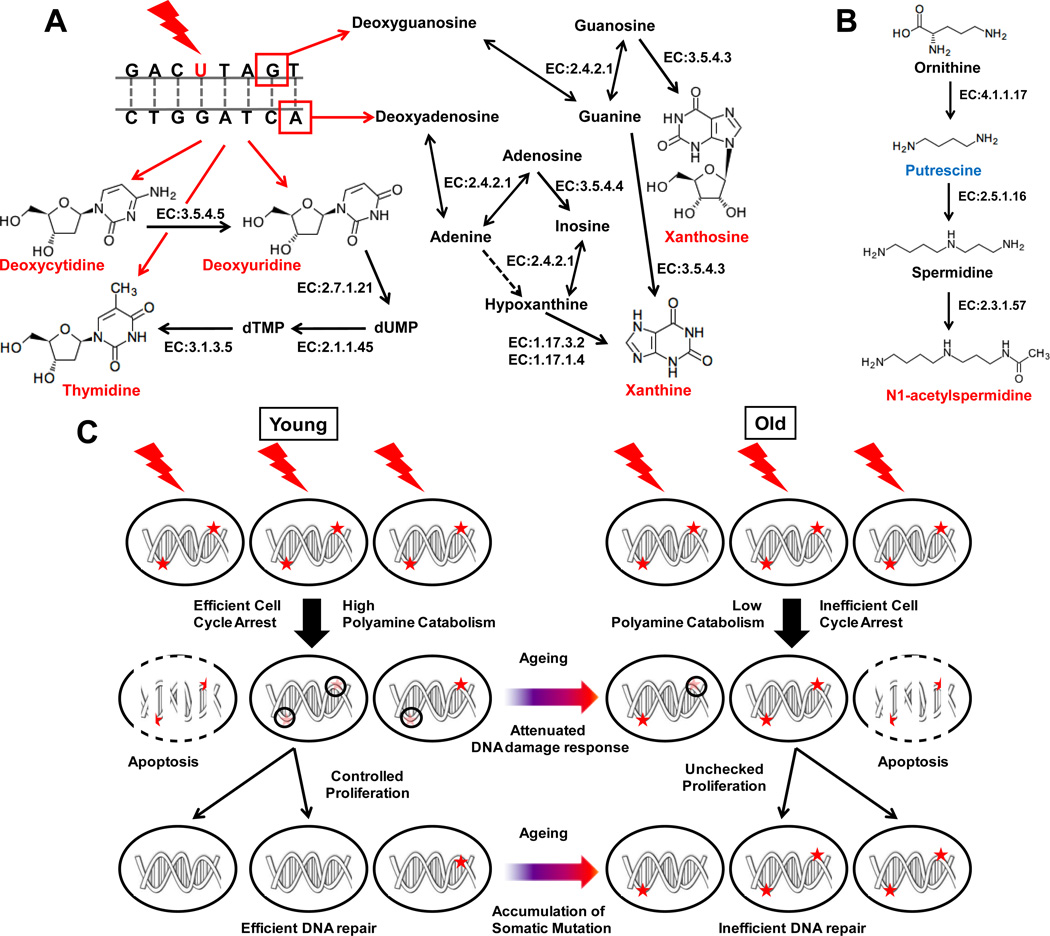
Figure 7. Biochemical pathway involved in production of radiation exposure biomarkers related to (A) nucleic acid metabolism and (B) polyamine metabolism.
Quantitatively elevated and depleted metabolites are shown in red and blue, respectively, while grey metabolites indicate no change or not measured in this study. Possible chemical modification of DNA contributing to production of these metabolites is shown with red bases indicating base modification (such as C→U) and red boxes indicating nucleotide excision sites following single-strand breaks. The enzymes involved in these metabolic pathways such as cytidine deaminase (EC 3.5.4.5), thymidine kinase (EC 2.7.1.21), thymidylate synthase (EC 2.1.1.45), 5',3'-nucleotidase (mitochondrial) (EC 3.1.3.5), purine-nucleoside phosphorylase (EC 2.4.2.1), guanine deaminase (EC 3.5.4.4), adenine deaminase (EC 3.5.4.3), xanthine dehydrogenase (EC 1.17.3.2), xanthine oxidase (EC 1.17.1.4), ornithine decarboxylase (EC 4.1.17), spermidine synthase (EC 2.5.1.16) and spermidine/spermine N1-acetyltransferase (EC 2.3.1.57) are indicated next to corresponding steps (shown by solid grey arrow). Lines with arrows in both directions for purine-nucleoside phosphorylase indicate its ability to catalyze ribosyltransferase activity in both directions. Dotted grey arrow indicates that the corresponding enzymatic activity has not been reported in mice. (C) Putative mechanism showing the role of polyamine catabolism in radiation-induced damage-repair through cell cycle arrest and the effect of aging in the process. Dark red asterisk indicate DNA damage lesion caused by radiation exposure that would cause mutation if replicated without repair during proliferation. Faint red asterisk encircled by black circles indicate recruitment of DNA-damage repair machinery during cell cycle arrest leading to successful DNA damage repair. Attenuation of polyamine catabolism in older mice may lead to inefficient arrest of cell cycle and, thus, impairment of DNA damage repair and propagation of mutation in proliferating somatic cells.
Putrescine clearance was found be attenuated after radiation exposure. This may be due to conversion putrescine to N1-acetylspermidine, which was elevated in the urine after exposure. N1-acetylspermidine is produced by the addition of a propylamine group onto putrescine by spermidine synthase (EC 2.5.1.16) followed by acetylation by spermine/spermidine acetyl transferase (EC 2.3.1.57) as shown in Figure 7B. It was observed that similar to the pyrimidine 2’-deoxynucleosides, N1-acetylspermidine elevation was attenuated with aging but remained highly correlated with that of deoxyuridine. This suggests a potential biochemical link between elevation of N1-acetylspermidine and pyrimidine 2’-deoxynucleosides generated during DNA damage-repair. It was also interesting to note that ROC analysis indicated that the ratio of excretion of N1-acetylspermidine and putrescine was a more robust signature of radiation exposure than the individual metabolites. This ratio essentially indicates the flux through polyamine biosynthesis towards production of acetylated polyamines. Whereas polyamines have been shown to promote proliferation, polyamine catabolism through acetylation has been shown to attenuate it34, 35. Cells need to halt proliferation in order to efficiently repair DNA damage caused by radiation exposure. Depletion of the polyamine pool would help cells to achieve lower proliferation rates and propagation of fewer mutations during subsequent replication. Thus, an increase in polyamine acetylation may be an integral part of effective cellular response to oxidative DNA damage, which is reflected by the urinary metabolomic signature. The fact that inter-individual variation in the elevation of 2’-deoxyuridine, a metabolic by-product of oxidative DNA-damage repair, was strongly correlated with that in elevation of polyamine acetylation, further supports this hypothesis. If this hypothesis is true, attenuation in elevation of polyamine acetylation would result in inefficient arrest of the cell cycle and, thereby, inefficient oxidative DNA damage repair. Consequently, one would expect a decline in clearance of metabolites resulting from oxidative DNA damage such as 2’-deoxyuridine. It was noted that elevation in polyamine acetylation was significantly attenuated with aging with a concomitant and correlated attenuation in excretion of 2’-deoxyuridine. It has been well documented that aging attenuates the ability to fight oxidative stress and reduces DNA damage repair efficiency6,36,37. In fact, similar to the observations reported in this study, DNA repair activity through base and nucleotide excision repair in human skin fibroblasts were affected by aging but not by chronic photoexposure37. Thus these results indicate that attenuation of polyamine metabolism may contribute to the decline of DNA damage-repair efficiency and accumulation of somatic mutation in older animals as shown in Figure 7C.
Radiation exposure also caused significant elevation of xanthine and xanthosine. Xanthine has been shown to be elevated in human skin tissue after low-dose ionizing radiation exposure21. Nucleotide excision repair can produce purine 2’-deoxyribonucleosides such as 2’-deoxyadenosine and 2’-deoxyguanosine. These 2’-deoxyribonucleosides can be deconjugated to their corresponding bases adenine and guanine by purine nucleoside phosphorylase (EC 2.4.2.1). Guanine can be converted to xanthine by guanine deaminase (EC 3.5.4.3). Adenine can be conjugated with ribose by purine nucleoside phosphorylase to produce adenosine that can be deaminated by adenine deaminase (EC 3.5.4.4) to inosine. Inosine can be converted to xanthine by the action of enzymes purine nucleoside phosphorylase and xanthine oxidase (EC 1.17.3.2) or xanthine dehydrogenase (EC 1.17.1.4) with production of hypoxanthine as an intermediate (Figure 7A). Both inosine and hypoxanthine was shown to be elevated in irradiated human skin tissue21. Guanine can also be conjugated to guanosine by purine nucleoside phosphorylase. It was shown earlier that guanine deaminase from human liver can directly convert guanosine to xanthosine38, which was found to be elevated after radiation exposure. The lack of correlation between xanthine and xanthosine excretion may be a reflection of the fact that a number of additional pathways contribute to xanthine production. Consequently, xanthine was found to be the major purine metabolite that was elevated in response to radiation exposure. Similar to pyrimidine metabolites, xanthine also showed a significant correlation with N1-acetylspermidine levels in younger mice, but not in older ones. The younger mice also showed a significant correlation between clearance of the major purine metabolite xanthine and that of modified pyrimidines such as 2’-deoxyuridine. However, this correlation was poor in older mice. In the absence of drastic changes in composition due to mutations, the purine and pyrimidine content of DNA is expected to remain constant throughout life. Assuming that the propensity of purine and pyrimidine nucleotides to undergo oxidative damage following radiation exposure, which is determined by their chemical reactivity, remain constant throughout life, the clearance of these metabolites should be determined by the efficiency of DNA damage repair. It is known that DNA damage repair ability is attenuated with aging. However, if the relative efficiency of repair of purine and pyrimidine modification is attenuated coherently (through the same mechanism) during aging, the elevation in xanthine and 2’-deoxyuridine would be attenuated but would remain correlated. However, the fact that their correlation was also poor in older mice, thus, indicates that the machinery involved in repair of purine and pyrimidine damage-repair is affected differently by aging. Such deficiency in repair of different types of DNA damages may further contribute to the accumulation of mismatches in older mice. Since, attenuation of polyamine catabolism with aging could also contribute to inefficient cell cycle arrest, which is essential for DNA damage repair; such mismatches would result in accumulation of somatic mutations in proliferating cells from older mice.
Thus, the results presented in this study shows that both attenuation of polyamine catabolism and loss of coherence between DNA damage repair mechanisms may contribute to attenuation in the ability of older mice to negotiate oxidative insults caused by radiation exposure and lead to the accumulation of somatic mutations. Accumulation of somatic mutation during aging due to chronic oxidative stress plays a major role in aberration cellular phenotype6,36,39,40. This eventually and stochastically results in pathogenesis of number of diseases including cancer. Therefore, the observations presented here invites detailed investigation into interaction of polyamine metabolism and DNA damage repair pathways. Future studies into this aspect may not only enrich our understanding of the chemical biology of DNA damage repair but also provide novel strategies to mitigate the challenges posed by an aging world population as well as increasing exposure to oxidative stress due to environmental and life-style-related factors.
Conclusion
This study showed that repeated exposure and, more importantly, aging, attenuates changes in the urinary metabolomic response to radiation exposure. Nevertheless, the metabolomic signature was robust enough to identify exposed animals irrespective of their age and history of previous exposure. In addition, comparative analysis of elevation in excretion of radiation biomarkers in age-matched subject may also help in extracting information about previous exposures. Thus, this study showed that metabolomic alterations could be used to noninvasively screen and triage exposed subjects from a heterogeneous population following an accidental radiation release. Finally, the analysis of the effect of aging on the metabolic fingerprint also indicated to a novel link between polyamine metabolism and DNA damage repair.
Supplementary Material
Acknowledgments
Funding
This work was supported by the National Cancer Institute Intramural Research Program, the Columbia University Center for Medical Countermeasures against Radiation (P.I. David Brenner) funded by NIH (NIAID) grant U19 AI067773-05/06 to JRI, and NIAID Radiation/Nuclear Medical Countermeasures Program, NIH Intramural Laboratory Collaboration Funding to FJG.
Abbreviations
- UPLC-ESI-QTOF-MS
ultra-performance liquid chromatography coupled with electrospray ionization time-of-flight mass spectrometry
- PCA
principal components analysis
- OPLS-DA
orthogonal projection to latent structures discriminant analysis
- ROC
receiver operating characteristic
- ROS
reactive oxygen species
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
S.K.M. designed the experiments and analyzed the results. S.K.M., K.W.K. and J.A.B. performed the experiments. S.K.M., J.A.B., J.R.I. and F.J.G. participated in data interpretation and manuscript preparation. All authors have given approval to the final version of the manuscript.
Contributor Information
Soumen K. Manna, Email: mannask@mail.nih.gov.
Kristopher W. Krausz, Email: krauszk@intra.nci.nih.gov.
Jessica A. Bonzo, Email: jessica.bonzo@lifetech.com.
Jeffrey R. Idle, Email: jeff.idle@ikp.unibe.ch.
Frank J. Gonzalez, Email: gonzalef@mail.nih.gov.
References
- 1.Mah LJ, El-Osta A, Karagiannis TC. gammaH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia. 24(4):679–686. doi: 10.1038/leu.2010.6. [DOI] [PubMed] [Google Scholar]
- 2.Amundson SA, Do KT, Fornace AJ., Jr Induction of stress genes by low doses of gamma rays. Radiat Res. 1999;152(3):225–231. [PubMed] [Google Scholar]
- 3.Amundson SA, Do KT, Shahab S, Bittner M, Meltzer P, Trent J, Fornace AJ., Jr Identification of potential mRNA biomarkers in peripheral blood lymphocytes for human exposure to ionizing radiation. Radiat Res. 2000;154(3):342–346. doi: 10.1667/0033-7587(2000)154[0342:iopmbi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 4.Amundson SA, Lee RA, Koch-Paiz CA, Bittner ML, Meltzer P, Trent JM, Fornace AJ., Jr Differential responses of stress genes to low dose-rate gamma irradiation. Mol Cancer Res. 2003;1(6):445–452. [PubMed] [Google Scholar]
- 5.Chen JH, Hales CN, Ozanne SE. DNA damage, cellular senescence and organismal : causal or correlative? Nucleic Acids Res. 2007;35(22):7417–7428. doi: 10.1093/nar/gkm681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 2003;17(10):1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- 7.Maynard S, Schurman SH, Harboe C, de Souza-Pinto NC, Bohr VA. Base excision repair of oxidative DNA damage and association with cancer and aging. Carcinogenesis. 2009;30(1):2–10. doi: 10.1093/carcin/bgn250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmann W, Schmitz-Feuerhake I. How radiation-specific is the dicentric assay? J Expo Anal Environ Epidemiol. 1999;9(2):113–133. doi: 10.1038/sj.jea.7500008. [DOI] [PubMed] [Google Scholar]
- 9.Lanz C, Patterson AD, Slavik J, Krausz KW, Ledermann M, Gonzalez FJ, Idle JR. Radiation metabolomics. 3. Biomarker discovery in the urine of gamma-irradiated rats using a simplified metabolomics protocol of gas chromatography-mass spectrometry combined with random forests machine learning algorithm. Radiat Res. 2009;172(2):198–212. doi: 10.1667/RR1796.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tyburski JB, Patterson AD, Krausz KW, Slavik J, Fornace AJ, Jr, Gonzalez FJ, Idle JR. Radiation metabolomics. 2. Dose- and time-dependent urinary excretion of deaminated purines and pyrimidines after sublethal gamma-radiation exposure in mice. Radiat Res. 2009;172(1):42–57. doi: 10.1667/RR1703.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tyburski JB, Patterson AD, Krausz KW, Slavik J, Fornace AJ, Jr, Gonzalez FJ, Idle JR. Radiation metabolomics. 1. Identification of minimally invasive urine biomarkers for gamma-radiation exposure in mice. Radiat Res. 2008;170(1):1–14. doi: 10.1667/RR1265.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson CH, Patterson AD, Krausz KW, Lanz C, Kang DW, Luecke H, Gonzalez FJ, Idle JR. Radiation metabolomics. 4. UPLC-ESI-QTOFMS-Based metabolomics for urinary biomarker discovery in gamma-irradiated rats. Radiat Res. 175(4):473–484. doi: 10.1667/RR2437.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson CH, Patterson AD, Krausz KW, Kalinich JF, Tyburski JB, Kang DW, Luecke H, Gonzalez FJ, Blakely WF, Idle JR. Radiation metabolomics. 5. Identification of urinary biomarkers of ionizing radiation exposure in nonhuman primates by mass spectrometry-based metabolomics. Radiat Res. 178(4):328–340. doi: 10.1667/rr2950.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manna SK, Patterson AD, Yang Q, Krausz KW, Idle JR, Fornace AJ, Gonzalez FJ. UPLC-MS-based urine metabolomics reveals indole-3-lactic acid and phenyllactic acid as conserved biomarkers for alcohol-induced liver disease in the Ppara-null mouse model. J Proteome Res. 10(9):4120–4133. doi: 10.1021/pr200310s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pearson K. On lines and planes of closest fit to systems of points in space. Philosophical Magazine. 1901;2(6):559–572. [Google Scholar]
- 16.Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E, Bouatra S, Sinelnikov I, Arndt D, Xia J, Liu P, Yallou F, Bjorndahl T, Perez-Pineiro R, Eisner R, Allen F, Neveu V, Greiner R, Scalbert A. HMDB 3.0--The Human Metabolome Database in 2013. Nucleic Acids Res. 41(D1):D801–D807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith CA, O'Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;27(6):747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 18.Kind T, Fiehn O. Seven Golden Rules for heuristic filtering of molecular formulas obtained by accurate mass spectrometry. BMC Bioinformatics. 2007;8:105. doi: 10.1186/1471-2105-8-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grison S, Martin JC, Grandcolas L, Banzet N, Blanchardon E, Tourlonias E, Defoort C, Fave G, Bott R, Dublineau I, Gourmelon P, Souidi M. The metabolomic approach identifies a biological signature of low-dose chronic exposure to cesium 137. J Radiat Res. 53(1):33–43. doi: 10.1269/jrr.11071. [DOI] [PubMed] [Google Scholar]
- 20.Liu H, Wang Z, Zhang X, Qiao Y, Wu S, Dong F, Chen Y. Selection of Candidate Radiation Biomarkers in the Serum of Rats Exposed to Gamma-Rays by Gc/Tofms-Based Metabolomics. Radiat Prot Dosimetry. doi: 10.1093/rpd/ncs138. [DOI] [PubMed] [Google Scholar]
- 21.Hu ZP, Kim YM, Sowa MB, Robinson RJ, Gao X, Metz TO, Morgan WF, Zhang Q. Metabolomic response of human skin tissue to low dose ionizing radiation. Mol Biosyst. 8(7):1979–1986. doi: 10.1039/c2mb25061f. [DOI] [PubMed] [Google Scholar]
- 22.Charles M. UNSCEAR report 2000: sources and effects of ionizing radiation. United Nations Scientific Comittee on the Effects of Atomic Radiation. J Radiol Prot. 2001;21(1):83–86. doi: 10.1088/0952-4746/21/1/609. [DOI] [PubMed] [Google Scholar]
- 23.Feldberg RS, Carew JA. Water radiolysis products and nucleotide damage in gamma-irradiated DNA. Int J Radiat Biol Relat Stud Phys Chem Med. 1981;40(1):11–17. [PubMed] [Google Scholar]
- 24.Spotheim-Maurizot M, Davidkova M. Radiation damage to DNA in DNA-protein complexes. Mutat Res. 711(1–2):41–48. doi: 10.1016/j.mrfmmm.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 25.Teoule R. Radiation-induced DNA damage and its repair. Int J Radiat Biol Relat Stud Phys Chem Med. 1987;51(4):573–589. doi: 10.1080/09553008414552111. [DOI] [PubMed] [Google Scholar]
- 26.Cadet J, Delatour T, Douki T, Gasparutto D, Pouget JP, Ravanat JL, Sauvaigo S. Hydroxyl radicals and DNA base damage. Mutat Res. 1999;424(1–2):9–21. doi: 10.1016/s0027-5107(99)00004-4. [DOI] [PubMed] [Google Scholar]
- 27.Evans MD, Dizdaroglu M, Cooke MS. Oxidative DNA damage and disease: induction, repair and significance. Mutat Res. 2004;567(1):1–61. doi: 10.1016/j.mrrev.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 28.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 29.Krokan HE, Drablos F, Slupphaug G. Uracil in DNA--occurrence, consequences and repair. Oncogene. 2002;21(58):8935–8948. doi: 10.1038/sj.onc.1205996. [DOI] [PubMed] [Google Scholar]
- 30.Fromme JC, Verdine GL. Base excision repair. Adv Protein Chem. 2004;69:1–41. doi: 10.1016/S0065-3233(04)69001-2. [DOI] [PubMed] [Google Scholar]
- 31.Robertson AB, Klungland A, Rognes T, Leiros I. DNA repair in mammalian cells: Base excision repair: the long and short of it. Cell Mol Life Sci. 2009;66(6):981–993. doi: 10.1007/s00018-009-8736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sancar A, Tang MS. Nucleotide excision repair. Photochem Photobiol. 1993;57(5):905–921. doi: 10.1111/j.1751-1097.1993.tb09233.x. [DOI] [PubMed] [Google Scholar]
- 33.Evans MD, Saparbaev M, Cooke MS. DNA repair and the origins of urinary oxidized 2'-deoxyribonucleosides. Mutagenesis. 25(5):433–442. doi: 10.1093/mutage/geq031. [DOI] [PubMed] [Google Scholar]
- 34.Li L, Rao JN, Guo X, Liu L, Santora R, Bass BL, Wang JY. Polyamine depletion stabilizes p53 resulting in inhibition of normal intestinal epithelial cell proliferation. Am J Physiol Cell Physiol. 2001;281(3):C941–C953. doi: 10.1152/ajpcell.2001.281.3.C941. [DOI] [PubMed] [Google Scholar]
- 35.Mandal S, Mandal A, Johansson HE, Orjalo AV, Park MH. Depletion of cellular polyamines, spermidine and spermine, causes a total arrest in translation and growth in mammalian cells. Proceedings of National academy of Science. doi: 10.1073/pnas.1219002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of. Nature. 2000;408(6809):239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 37.Sauvaigo S, Caillat S, Odin F, Nkengne A, Bertin C, Oddos T. Effect of aging on DNA excision/synthesis repair capacities of human skin fibroblasts. J Invest Dermatol. 130(6):1739–1741. doi: 10.1038/jid.2010.10. [DOI] [PubMed] [Google Scholar]
- 38.Kimm SW, Park JB, Lee IS. Purification and characterizationof guanine aminohydrolase from human liver. Korean J. Biochem. 1985;17:139–148. [Google Scholar]
- 39.Kregel KC, Zhang HJ. An integrated view of oxidative stress in aging: basic mechanisms, functional effects, and pathological considerations. Am J Physiol Regul Integr Comp Physiol. 2007;292(1):R18–R36. doi: 10.1152/ajpregu.00327.2006. [DOI] [PubMed] [Google Scholar]
- 40.Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, Morrow JD, Van Remmen H, Sedivy JM, Yamasoba T, Tanokura M, Weindruch R, Leeuwenburgh C, Prolla TA. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309(5733):481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.