Abstract
NF-κB (nuclear factor kappa B) family transcription factors are master regulators of immune and inflammatory processes in response to both injury and infection. In the latent state, NF-κBs are sequestered in the cytosol by their inhibitor IκB (inhibitor of NF-κB) proteins. Upon stimulations of innate immune receptors such as Toll-like receptors and cytokine receptors such as those in the TNF (tumor necrosis factor) receptor superfamily, a series of membrane proximal events lead to the activation of the IKK (IκB kinase). Phosphorylation of IκBs results in their proteasomal degradation and the release of NF-κB for nuclear translocation and activation of gene transcription. Here, we review the plethora of structural studies in these NF-κB activation pathways, including the TRAF (TNF receptor–associated factor) proteins, IKK, NF-κB, ubiquitin ligases, and deubiquitinating enzymes. Although these structures only provide snapshots of isolated processes, an emerging picture is that these signaling cascades coalesce into large oligomeric signaling complexes, or signalosomes, for signal propagation.
Keywords: TNF, Toll-like receptor, IKK, TRAF, ubiquitin, NEMO
INTRODUCTION
It has been over 25 years since David Baltimore discovered the first member of the nuclear factor-kappa B (NF-κB) protein family that bound selectively to the κ-light-chain enhancer in extracts of B-cell tumors (41, 43, 102). Since then, scientists have illuminated the role of NF-κB proteins in the orchestration of complex biological processes in many thousands of publications.
The eukaryotic NF-κB transcription factor family regulates the expression of a large variety of genes that are involved in a number of processes like inflammatory and immune responses of the cell, cell growth, and development. NF-κB transcription factors are activated as a response to a variety of signals, including cytokines, pathogens, injuries, and other stressful conditions. Activation of NF-κB proteins is tightly regulated, and inappropriate activation of the NF-κB signaling pathways has been linked to autoimmunity, chronic inflammation, and various cancers (4, 14, 111). In unstimulated cells, NF-κB is bound to an inhibitory protein, IκB. Binding to IκB masks the nuclear localization signal (NLS) of NF-κB, sequesters the NF-κB·IκB complex in the cytoplasm, and prevents NF-κB from binding to DNA.
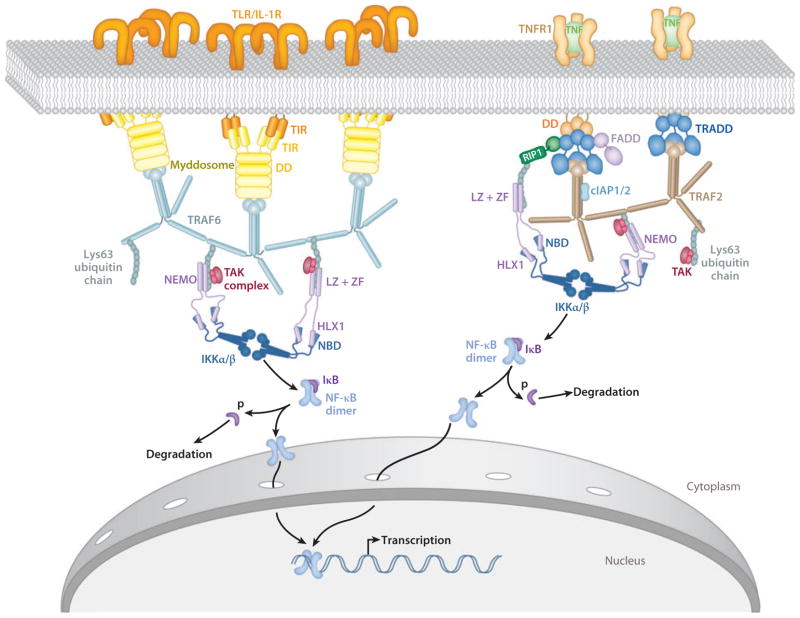
Activation of NF-κB signaling is initiated by extracellular stimuli. These stimuli are recognized by receptors and transmitted into the cell, where adaptor signaling proteins initiate a signaling cascade. These signaling cascades culminate in the activation of IκB kinase (IKK). IKK phosphorylates the inhibitory IκB subunit of the NF-κB·IκB complex in the cytoplasm. This phosphorylation marks IκB for degradation by the proteasome and releases NF-κB from the inhibitory complex (26, 41, 43, 74, 102). The freed NF-κB proteins are then transported into the nucleus where they bind to their target sequences and activate gene transcription. In this review, we focus on the molecular mechanisms of NF-κB signaling from receptor stimulation to activation of gene transcription (Figure 1).
Figure 1.
Simplified view of the TLR/IL-1R (left) and TNFR (right) pathways leading to activation of NF-κB. Known interactions between proteins are indicated and discussed in detail in the text. Abbreviations: IL-1R, interleukin-1 receptor; NF-κB, nuclear factor κB; TLR, Toll-like receptor; TNFR, tumor necrosis factor receptor.
NF-κB FAMILY
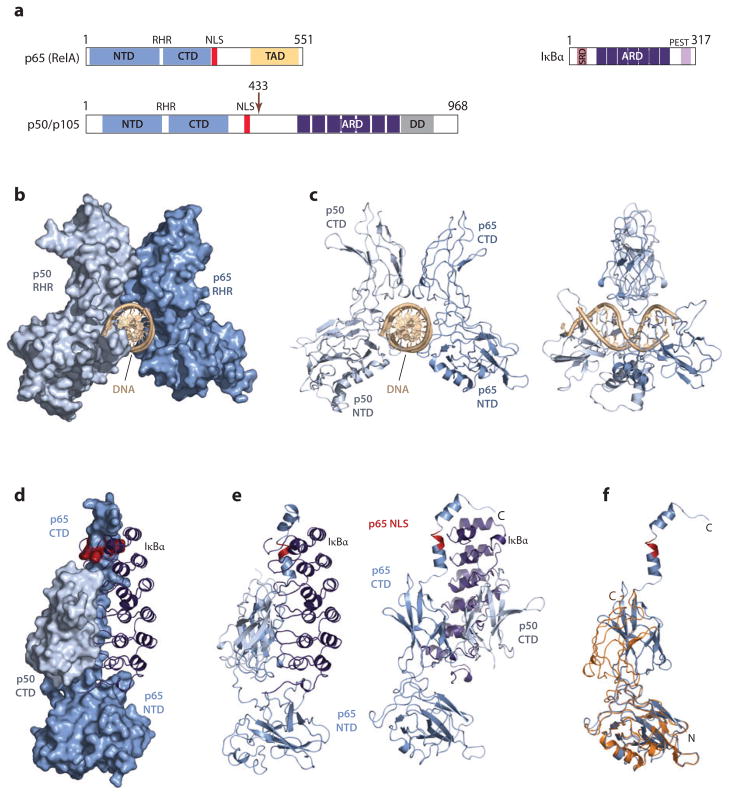
All members of the NF-κB protein family, RelA (p65), RelB, c-Rel, p50 (p105 precursor), p52 (p100 precursor), and Relish share a highly conserved DNA-binding and dimerization domain termed Rel homology region (RHR) that enables them to homo- or heterodimerize (4, 14, 27, 28, 39, 73, 108, 111) (Figure 2a). RelA (p65), RelB, and c-Rel contain a C-terminal transactivation domain (TAD) that allows them to activate target gene expression. p50 (p105 precursor), p52 (p100 precursor), and Relish contain a long, ankyrin repeat–containing domain (ARD) at their C terminus instead of the TAD domain and therefore cannot activate target gene expression as a homodimer.
Figure 2.
Structures of NF-κB and IκBs. (a) Domain organizations of representative members. (b) Space-filling model of the crystal structure of the p50/p65 heterocomplex bound to DNA. (c) The same structure shown in ribbon diagrams in two orientations. (d) Space-filling model of the crystal structure of p50/p65 heterocomplex bound to IκBα. NLS of p65 is shown in red. (e) The same structure shown in ribbon diagrams in two orientations. (f) Superposition of p65 in the IκBα-bound form (blue) and DNA-bound form (orange). Abbreviations: CTD, C-terminal domain; IκB, inhibitor of NF-κB; NF-κB, nuclear factor κB; NLS, nuclear localization signal; NTD, N-terminal domain.
Structures of NF-κB Bound to DNA
NF-κB proteins bind as a homo- or heterodimer to a 10-base-pairDNAsequence first identified in the enhancer of the immunoglobulin κ-light-chain gene in mature B cells (27, 38, 41, 43, 46, 102, 107). The first structure of an RHR was revealed from the structure of a p50 homodimer bound to an idealized κB target DNA sequence (4, 14, 25, 80, 111). To date, several other structures of NF-κB dimers bound to DNA have been solved and reveal a common mode of DNA binding and dimerization. The structures resemble a butterfly with the protein dimer forming the wings around a cylindrical body of DNA(25, 26, 54, 74, 80) (Figure 2b). The RHR domain of the NF-κB subunit forms into two distinct domains connected by a linker, the N-terminal domain (NTD) and the C-terminal domain (CTD) (Figure 2c). NF-κB uses both RHR domains to encircle the target DNA. The flexible region at the C-terminal end of the RHR contains the NLS. The core of the NTD and CTD folds into a β-sandwich structure. The CTD is solely responsible for dimerization and makes phosphate contacts with the DNA. In addition to nonspecific contacts with the sugar phosphate backbone of the DNA, the NTD also recognizes the target DNA sequence via specific interaction with DNA bases.
Analysis of the available structures has shown plasticity in the target sequence of NF-κB homo- and heterodimers. The canonical NF-κB p50·p65 heterodimer recognizes its target sequence via p50 binding to a 5′GGPyN half site and p65 binding to another 5′GGPyN site centered around an A:T base pair. However, flexibility in the short linker region and the modular architecture of the RHR allows NF-κB to recognize variations in κB DNA sequences by repositioning the RHR-NTD on the DNA and interacting with the DNA backbone.
NF-κB Bound to Inhibitory IκB
Proteins of the inhibitory κB family (IκB) serve as inhibitors and regulators of NF-κB activity. Members of the IκB family are the classical IκB proteins (IκBα, IκBβ, and IkBε), NF-κB precursor proteins (p100 and p105), and the nuclear IκBs (IκBζ, Bcl-3, and IκBNS). IκB proteins contain an N-terminal signal–receiving domain (SRD), a central ARD, and a C-terminal proline-, glutamate-, serine-, and threonine-rich (PEST) sequence (Figure 2a). IκBα was first discovered as a factor that dissociates preformed NF-κB·DNA complexes in vitro (1, 23, 27, 28, 39, 73, 108, 132). The transcription of IκBα was shown to be regulated by NF-κB; therefore, IκB in turn regulates activation and inactivation of NF-κB.
All IκBs recognize NF-κB via their ARD. The classical IκB subfamily has a preference for NF-κB dimers containing a p65 or c-Rel subunit, whereas the nuclear IκBs prefer p50 or p52 homodimer (40, 85). Structures of NF-κB bound to IκB have revealed a common mechanism of interaction (41, 43, 74). The NF-κB dimer is bound by one molecule of IκB (Figure 2d). The structure of the IκB ARD contains six ankyrin repeats, each of which consists of one β-loop and two α-helices. IκB folds into an elongated, barrel-like structure with a concave and a convex surface (Figure 2e). The concave surface of IκB faces the NF-κB dimerization domains. In the structure of the IκB-bound NF-κB heterodimer complex, p50·p65·IκBα, the ankyrin repeats 1 and 2 bind the NLS of p65; repeats 4 and 6 contact p50 at the interface of the paired dimerization domains; and repeats 5 and 6 contact the dimerization domain of p65 (41, 43). In the presence of IκB, the p65 subunit of the NF-κB heterodimer undergoes a conformational change and adopts a closed conformation when compared with the DNA-bound heterodimer (Figure 2f). The NTD of p65 rotates ~180° about its long axes and shifts by ~38Å toward its dimerization domain. This allows for allosteric inhibition of DNA binding of NF-κB. In addition, IκB contacts DNA-binding residues of NF-κB directly; ankyrin repeat 6 and the C-terminal PEST residues of IκB interact with the RHR-NTD and interfere with DNA binding.
UBIQUITINATION.
Ubiquitin is a small protein that can be attached to target proteins in a posttranslational process. During ubiquitination, the carboxy-terminal end of ubiquitin is covalently attached to a lysine in the target protein. This process requires several enzymes: an E1 ubiquitin-activating enzyme, an E2 ubiquitin-conjugating enzyme and an E3 ubiquitin ligase. After addition of the first ubiquitin to a target protein, further ubiquitin molecules can be attached to the first one, generating polyubiquitin chains.
Ubiquitin contains seven lysine residues, which can serve as attachment points for additional ubiquitin molecules. The classical polyubiquitin chains are the Lys48-linked chains, Ub1(Lys48)-Ub2(Met1), which target proteins for degradation by the proteasome (Figure 3a). Lys63-linked polyubiquitin chains are nondestructive and are often formed in signaling that triggers NF-kB activation. In these chains, the Lys63 of ubiquitin is used to attach to the next ubiquitin molecule, Ub1(Lys63)-Ub2(Met1) (Figure 3b). This results in a different conformation of the ubiquitin moieties with respect to each other. In addition, the N terminus of ubiquitin can be used as an attachment point, Ub1(Gly76)-Ub2(Met1) (Figure 3c). This results in the formation of a canonical peptide bond between the ubiquitin molecules and is termed a linear ubiquitin chain. Linear and Lys63-linked polyubiquitin chains are very similar in overall structure and differ mainly with respect to their isopeptide bonds.
Figure 3.
Structural overview of (a) Lys48-linked, (b) Lys63-linked, and (c) linear di-ubiquitin (di-Ub) showing the resulting different overall conformation. Lys48, Lys63, and Met1 are highlighted in red. Note that in linear di-Ub chains, no lysine residue is used for the linker.
In contrast to p50·p65·IκBα, the structure of the IκBβ-bound p65 NF-κB homodimer revealed that the NLS-containing region remains largely flexible and that the NTD of the p65 RHR does not contact IκBβ (27, 74). As a result, the crucial DNA-binding elements appear to be free to bind DNA even in the presence of IκBβ. These observations are supported by the subcellular distribution of these two complexes. Whereas the p50·p65·IκBα complex is exclusively localized in the cytoplasm in resting cells (27, 38, 39, 46, 73, 102, 107, 108), the p65·p65·IκBβ complex shuttles between cytoplasm and nucleus (25, 27, 38, 46, 54, 80, 107).
Although the classical IκBs are well understood, we are only beginning to shed light on the molecular mechanism of the nonclassical IκBs IκBζ, Bcl-3, and IκBNS. IκBζ was first identified as a gene induced in immune cells in response to bacterial LPS or IL-1 (1, 23, 54, 132), and IκBNS is expressed during negative selection in T-cells (23, 40, 85). Bcl-3 is a proto-oncogene and is often found to be translocated in B-cell leukemias (78, 85). The nuclear IκBs concentrate in the nucleus when expressed in cells (31, 78) and bind specifically to NF-κB p50 homodimers (16, 31, 77, 96, 119, 133).
THE IKK COMPLEX
A key step in the NF-κB signaling pathway is the regulation of the interaction between NF-κB and the inhibitory IκB. A majority of the signaling pathways leading to activation of NF-κB converge at a 700–900-kDa molecular mass complex containing a serin-specific IκB kinase, termed the IKK (16, 77, 96, 119, 133). The IKK complex consists of two catalytic subunits, IKKα and IKKβ, as well as a regulatory subunit, NEMO (IKKγ). The IKK complex consists of a homo- (α·α or β·β) or heterodimer (α·β) associated stoichiometrically with NEMO (50). The IKK complex is functionally pleiotropic and plays a major role in many biological processes, including inflammation, autophagy, insulin signaling, and DNA damage response. The complex is responsible for the phosphorylation of two serine residues in the SRD of IκB (76, 77, 97, 124, 133). Phosphorylation of IκB in turn leads to Lys48-linked polyubiquitination (Figure 3) (see sidebar, Ubiquitination), which targets IκB for degradation by the proteasome. Proteosomal degradation of IκB ultimately frees and activates NF-κB.
Structure of the Catalytic Subunit of the IKK Complex
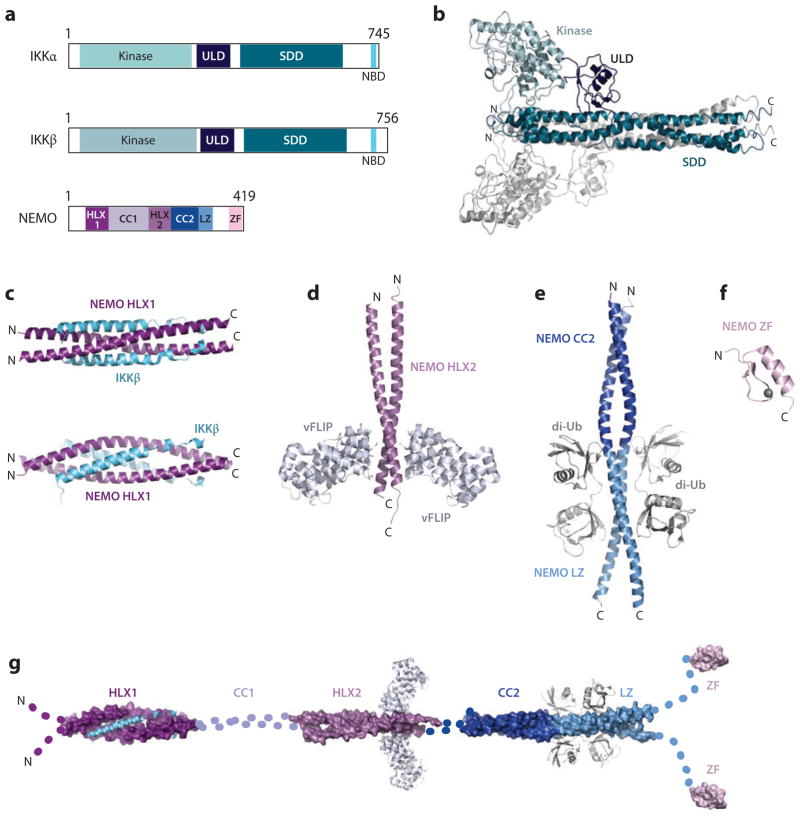
The catalytic subunits IKKα and IKKβ share ~50% sequence identity. The structure of IKKβ revealed a dimeric architecture containing an N-terminal kinase domain, followed by a ubiquitin-like domain (ULD), and a scaffold/dimerization domain (SDD), resembling the shape of a pair of scissors (122) (Figure 4a,b). The kinase and ULD domains form the “handle” of the scissors and the “blade” is formed by the SDD. All three domains interact mutually with each other. The C-terminal end contains the NEMO-binding domain (Figure 4a).
Figure 4.
Structures of the IKK complex. (a) Domain organizations. (b) Crystal structure of IKKβ dimer. (c) Crystal structure of IKKβ NBD in complex with the N-terminal kinase-binding domain (HLX1) of NEMO. (d) Crystal structure of NEMO HLX2 in complex with vFLIP. (e) Crystal structure of CC2-LZ of NEMO in complex with linear di-Ub. (f) Structure of the NEMO ZF domain. (g) A model of full-length NEMO dimer. Abbreviations: di-Ub, di-ubiquitin; IKK, IκB kinase; NBD, NEMO-binding domain; NEMO, NF-κB essential modulator; ZF, zinc finger.
The ULD is required for the catalytic activity of IKKβ and together with the SDD helps to bind, position, and orient IκBα with respect to the IKKβ catalytic pocket. Dimerization of IKKβ is mediated by the SDD domain, and full-length IKKβ has been shown to be a dimer in solution (122). However, in the IKKβ dimer, the kinase domains are not located close to each other and are unable to promote intradimer trans-autophosphorylation. This suggests a role for higher oligomerization in the activation of IKKβ. Indeed, IKKβ exists as a dimer of dimers in the observed crystal structure. In this conformation, the activation loops of the kinase domains are in a position close enough to activate the kinase of the neighboring dimer (26, 74, 122). This conformation may be transient and stabilized by interaction partners like NEMO and the ubiquitin chain network.
The regulatory subunit NEMO of the IKK complex is mainly helical and contains the helical domains termed HLX1, CC1, HLX2, and CC2, followed by a leucine-zipper domain and a C-terminal zinc-finger (ZF) region (27, 28, 39, 73, 76, 97, 98, 108, 124). The kinase-binding domain (KBD) contains HLX1 and part of CC1. There is no full-length structure of NEMO; however, several structures containing individual domains have been solved and enable us a view of an elongated, flexible coiled-coil (CC) NEMO model (Figure 4c–g).
The IKKβ-Binding Domain: Recognition of IKKβ by NEMO
The crystal structure of the KBD in complex with the C-terminal region of IKKβ reveals a heterotetramer in which the molecules pack as a parallel four-helix bundle (33, 60, 98, 105, 127) (Figure 4c). The two NEMO molecules assemble a head-to-head dimer with each molecule forming a crescent-shaped α-helix that interacts with the other NEMO molecule at the N and C termini. The NEMO dimer is stabilized by hydrophobic interactions at the N and C termini but leaves a slit-shaped aperture between them. The N-terminal dimerization patch is more extensive and is strengthened by additional electrostatic interactions.
The two IKKβ molecules do not contact each other but instead associate with each of the NEMO molecules respectively. Although the interaction between the NEMO and IKKβ molecule is not extensive at the N terminus, the IKKβ molecules are tightly wedged into the NEMO dimer at the C-terminal end. The IKKβ-binding pocket of NEMO mainly interacts with three, large hydrophobic side chains of IKKβ that are oriented toward the binding pocket on NEMO. This results in a specific, high-affinity interaction between NEMO and IKKβ.
NEMO and vFlip
During the constant battle between our immune system and pathogens, invading pathogens have learned to manipulate cellular processes to their advantage. Lymphogenic viruses, for instance, have been observed to increase cell survival and proliferation by activating NF-κB (22, 29, 33, 44, 52, 60, 105, 127). FLIP proteins are a group of cellular proteins identified as inhibitors of death receptor–induced apoptosis (42, 110). The viral FLIP (vFLIP) proteins are viral encoded proteins used to manipulate the host cells (11). In the case of the Kaposi’s sarcoma virus KSHV, vFLIP is produced in infected cells and has been shown to interact with and activate the IKK complex (2, 22, 29, 41–44, 102).
The crystal structure of the KSHV vFLIP in complex with its target region on NEMO has been solved (2, 4, 14, 111). The structure reveals two molecules of vFLIP bound to the C-terminal half of the head-to-head dimer of the NEMO HLX2 region (Figure 4d). The vFLIP monomers bind to NEMO in essentially identical fashion. They are oriented face-to-face but do not interact with each other. vFLIP consist of two death-effector domains, DED1 and DED2, of which DED1 is the main contributor to the extensive complex interface. The interface shows a high degree of structural complementarity and is characterized by two vertical clefts on the surface of vFLIP. Each of the clefts interacts with one molecule of the NEMO dimer. However, one of the clefts, cleft 1, is more extensive and comprises an asymmetric pocket. The upper compartment of this pocket is deep and hydrophobic, whereas the lower part of the cleft is more open and mediates hydrophilic interactions.
Interestingly, the mutationNEMOL227P is found in patients with the chronic genetic disorder anhidrotic ectodermal dysplasia with immunodeficiency and is located in the HLX2 region of NEMO(2, 14, 26, 41, 43, 74, 102). The L227P mutation may therefore change the helical folding tendency of NEMO and destabilize the NEMO dimer in the cell (2, 4, 14, 18, 27, 28, 39, 73, 108, 111, 120). It is assumed that stabilization of NEMO dimerization, either by receptor signaling or viral intervention, activates NEMO. Therefore, the stabilization or destabilization of NEMO may be an important point of regulation in the NF-κB signaling pathway.
NEMO and Di-Ubiquitin
Although NEMO does not have a catalytic activity, it is essential for the NF-κB pathway. NEMO can specifically recognize linear and Lys63-linked polyubiquitin chains (Figure 3). Many of the proteins in the NF-κB signaling cascade, including NEMO itself and TRAF6, are polyubiquitinated. Binding of NEMO to polyubiquitin is crucial for IKK recruitment and subsequent NF-κB activation (12, 13, 18, 27, 38, 41, 43, 46, 68, 102, 107, 120, 131). It was postulated that polyubiquitin chains serve as an extended scaffold for recruitment of signaling molecules and enhance higher oligomerization (21).
Secondary structure analysis and several X-ray structures showed that, with the exception of the carboxy-terminal ZF, NEMO is essentially an elongated, α-helical protein. Whereas the N-terminal region interacts with the catalytic IKK subunits, the C-terminal region is responsible for ubiquitin chain binding (122).
The crystal structures of the NEMO CC2-LZ region alone and in complex with linear or Lys63-linked di-ubiquitin (di-Ub) have been determined (4, 12–14, 25, 68, 80, 111, 131). NEMO forms a head-to head dimer spanning ~110Å (Figure 4e). The dimer observes a pseudo-twofold symmetry, and each molecule of the NEMO dimer forms a continuous helix (25, 26, 54, 68, 74, 80). In contrast to classical CC proteins, the two NEMO molecules use hydrophobic residues and aliphatic portions of charged residues to pack against each other.
The NEMO CC2-LZ dimer provides a composite binding surface with contributions from both NEMO molecules for ubiquitin. NEMO CC2-LZ preferentially binds linear di-Ub and has a modest affinity toward Lys63-linked di-Ub. Lys48-linked di-Ub was found not to bind to the NEMO CC2-LZ. In linear di-Ub binding, NEMO binds to a conserved hydrophobic surface patch and the C-terminal tail of the distal ubiquitin while recognizing an adjacent surface on the proximal ubiquitin (1, 23, 27, 28, 39, 68, 73, 108, 131, 132). In the case of Lys63-linked di-Ub binding, only one ubiquitin contacts each NEMO dimer, which explains the observed lower affinity (5, 40, 85, 131). Therefore, even though linear and Lys63-linked ubiquitin chains share an extended, open structural architecture (56), the subtle differences in the linkages and the conformations underlie their distinct functions in NF-κB activation.
NEMO ZF
The very C-terminal end of NEMO contains a ZF that has been shown to bind ubiquitin. Mutations in the ZF have been linked to human anhidrotic ectodermal dysplasia with immunodeficiency and incontinentia pigmenti (24, 41, 43, 74). Nuclear magnetic resonance studies revealed that the NEMOZF folds into a canonical β-β-α fold (Figure 4f). Interestingly, a mutant missing the last zinc-chelating residue (C417F) retains the ability to bind zinc with a native-like affinity (13, 41, 43). However, analysis of the ZF surface and biochemical experiments identifies two putative protein interaction sites that are destabilized in the mutant and may explain the signaling defects of the C417F mutant. Furthermore, the NEMOZF was shown to be a ubiquitin-binding domain, which binds ubiquitin with a 1:1 stoichiometry and submillimolar affinity (12, 27, 74). The interaction between the NEMOZF and ubiquitin resembles previously solved interactions between α-helical ubiquitin-binding domains and ubiquitin; the amphipathic α-helix of the NEMO ZF binds to a hydrophobic patch centered around residue Ile44 of ubiquitin (12, 27, 38, 39, 46, 73, 102, 107, 108).
SIGNALING VIA THE TNF RECEPTOR SUPERFAMILY
In the canonical NF-κB pathway, proinflammatory signals like cytokines, pathogen-associated molecular patterns (PAMPs), and danger-associated molecular patterns activate a signaling cascade that leads to the activation of IKK, which ultimately frees NF-κB from its inhibitory complex. Several receptor-induced activation pathways of IKK have been elucidated for the tumor necrosis factor receptor (TNFR) superfamily and the Toll-like receptor/interleukin-1 receptor (TLR/IL-1R) superfamily.
Signaling cascades triggered by the TNFR superfamily are diverse and can induce either the NF-κB pathway or apoptosis (3, 5, 25, 27, 38, 46, 54, 80, 107). The signaling outcome is determined by the proteins recruited to the intracellular domain of the TNFRs. Upon ligand-dependent trimerization of the TNFR, TNFR-associated factors (TRAFs) are recruited to the receptor either directly or via adaptor proteins (1, 3, 23, 54, 75, 92, 132). For instance, after binding of TNF-α, TNFR1 recruits the adaptor protein TRADD through death domain (DD) interaction, which in turn recruits TRAF2 (Figure 1).
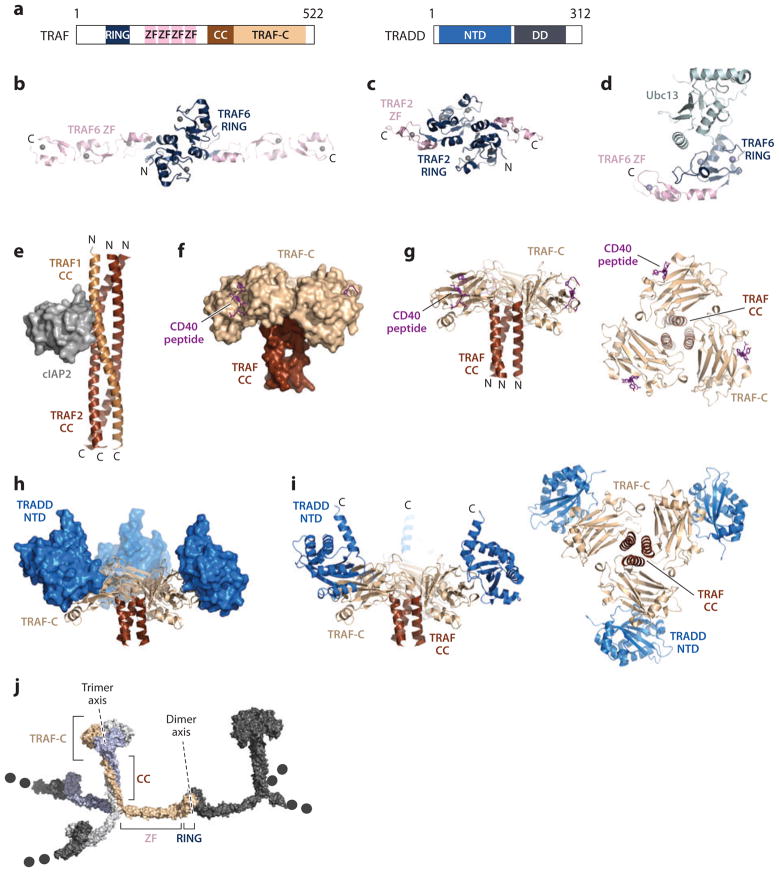
TRAF proteins contain an N-terminal RING domain, followed by several ZFs and a CTD containing a CC and the TRAF-C region (5, 23, 34, 35, 40, 75, 85, 92) (Figure 5a). The C-terminal part of TRAFs facilitates trimerization and is involved in sequence-specific interactions with the TNF receptor and adaptor proteins. The N-terminal region of TRAFs mediates dimerization and in the case of TRAF6, functions as a ubiquitin ligase (E3) for Lys63-linked polyubiquitination (5, 34, 35, 70, 78, 85, 115, 134). The difference in the symmetry of the N-terminal region and the C-terminal region likely results in alternating dimerization and trimerization, leading to infinite TRAF aggregation (129). Formation of high-order assemblies is required for TRAF6-mediated polyubiquitination and NF-κB activation (129).
Figure 5.
Structures of TRAFs. (a) Domain organizations. (b) Crystal structure of the dimeric RING and ZF domains (ZF1-3) of TRAF6. (c) Crystal structure of the dimeric RING and ZF domain (ZF1) of TRAF2. (d) Crystal structure of TRAF6 RING and ZF domain (ZF1) in complex with Ubc13. (e) Crystal structure of TRAF2 CC in complex with cIAP2 BIR1 domain. (f, g) Crystal structure of the TRAF domain of TRAF2 in complex with a peptide from the CD40 receptor in space-filling and ribbon diagrams, respectively. (h, i) Crystal structure of the TRAF domain of TRAF2 in complex with the NTD of TRADD in space-filling and ribbon diagrams, respectively. (j) An infinite aggregation model of full-length TRAF proteins by alternating dimerization and trimerization. Abbreviations: CC, coiled coil; NTD, N-terminal domain; TRAF, tumor necrosis factor (TNF) receptor–associated factor.
The CC region of the TRAF2 trimer recruits cIAP1/2 (5, 7, 31, 34, 35, 70, 78, 115, 134) and the RING domains of cIAP1/2 nucleate polyubiquitination of target proteins, including RIP1 kinase, NIK, TRAF2, and cIAP1/2 (5, 7, 16, 30, 31, 34, 35, 69, 77, 95, 96, 119, 133). Ubiquitinated RIP1 serves as a platform for the assembly of downstream proteins like transforming growth factor (TGF)-β-activated kinase 1 (TAK1), TAK1-binding protein (TAB) 2 and TAB3, and NEMO, as well as other ubiquitin ligases such as the linear ubiquitin chain assembly complex (7, 16, 30, 69, 77, 95, 96, 119, 129, 133). Binding of NEMO to ubiquitin chains promotes IKK activation, either by induction of conformational changes or by placing IKK in a context where it is susceptible to phosphorylation by upstream kinases like TAK1 or trans-autophosphorylation. Ubiquitination of NIK by cIAP1/2 leads to its proteasomal degradation and suppression of the noncanonical NF-κB pathway.
Structure of TRAF RING-ZF and Interaction with Ubc13
TRAF6 is an E3 ubiquitin ligase crucial for TNFR-, TLR-, and IL-1R-induced NF-κB activation and AP-1 signaling pathways (50, 121). It was shown that dimerization of TRAF6 is crucial for E3 ligase activity in vitro and activation of NF-κB in vivo (76, 77, 97, 124, 129, 133). TRAF6 mutants incapable of forming dimers are compromised in polyubiquitin chain assembly and promotion of IκB phosphorylation (122, 129).
The structures of the N-terminal region of TRAF2 and TRAF6 contain the RING domain and part of the ZF region and have revealed that both proteins form homodimers (122, 128, 129) (Figure 5b,c). Homodimerization is mediated by the RING domain and adjacent linker region and has been confirmed in solution (122, 128, 129). The structure of TRAF6 can be described as a golf club–like formation with the ZFs forming the shaft and the RING domain forming the club head (76, 97, 98, 124, 128, 129). The RING domain folds into a canonical cross-brace fold but contains an Asp instead of a more classical zinc-coordinating residue. The hydrophobic dimer interface is formed by stacked aromatic and aliphatic side chains, which are conserved among TRAF proteins. The three ZFs form the canonical β-β-α-fold and exhibit conserved orientations with respect to each other.
The N-terminal region of TRAF6 was shown to function as an E3 ubiquitin ligase (33, 60, 98, 105, 121, 127). The complex structure between TRAF6 and its cognate E2 Ubc13 resembles other RING·E2 structures. The core of the complex is formed by hydrophobic interactions of the RING domain near the first zinc-binding site, whereas residues N-terminal to the RING domain form additional charged interactions (Figure 5d). The first ZF of the region is required for the interaction of TRAF6 with Ubc13, although ZF does not directly contact Ubc13. The ZF1 forms a three-stranded, antiparallel β-sheet with residues N-terminal to the RING domain, thereby locking these residues into a conformation suitable for Ubc13 binding.
Interestingly, superposition of theTRAF2RING-ZFonto theTRAF6·Ubc13 structure reveals steric clashes between TRAF2 and Ubc13 (22, 29, 33, 44, 52, 60, 63, 75, 81, 92, 105, 126–128), and purified TRAF2 fails to interact with UBC13 in gel-shift assays in vitro (128). Residues critical for Ubc13 binding in TRAF6 are not conserved in TRAF2, and mutations of these residues to their TRAF2 counterparts abolish Ubc13 interaction as well as E3 activity of TRAF6. Therefore, TRAF2 requires interaction with cIAP to induce polyubiquitination (115, 134).
TRAF CC Domain and Interaction with cIAP
TRAF2 and cIAP form an E3 complex and are essential components of the TNF-α-induced canonical and noncanonical NF-κB pathways (70, 71, 115,134). Upon binding to TNF-α,TNFR1 recruits the adaptor protein TRADD, which in turn recruits TRAF2 and RIP1. Lys63-linked polyubiquitination of RIP1 is essential for IKK activation (18) and dependent on binding of TRAF2 to cIAP1/2 (115, 134).
The TRAF2-CC domain forms a continuous homotrimer both alone and in complex with the BIR1 domain of cIAP2 (70, 134) (Figure 5e). Strikingly, each TRAF-CC trimer interacts with only one cIAP molecule in the crystal structure as well as in solution. Upon binding of cIAP the TRAF2-CC trimer undergoes subtle conformational changes, which result in an enhanced binding site while at the same time prohibiting binding of further cIAP molecules to the other two potential binding sites on the TRAF-CC trimer.
In contrast to TRAF2, the role of TRAF1 in NF-κB activation is still unclear. TRAF1 has been suggested to act as both a negative and a positive regulator, possibly in a cell type–dependent manner. Little is known about the exact mechanism of regulation. However, in the presence of TRAF1-CC, a heterotrimer consisting of one TRAF1-CC molecule and two TRAF2-CC molecules (TRAF1·[TRAF2]2) is preferentially formed (134). Interestingly, biochemical analysis of this interaction showed that the TRAF1·[TRAF2]2 heterotrimer has a higher affinity to cIAP2 than the TRAF2 homotrimer. Inspection of the TRAF2-CC homotrimer and the TRAF1·[TRAF2]2 heterotrimer structures display a high degree of similarity. However, upon superpositioning of the cIAP2 molecules, a relative rotation of ~9° between the two trimers is observed. Additionally, the interaction with cIAP2 is mediated mostly by TRAF1 and exhibits higher hydrophobicity and better shape complementarity than that observed in the homotrimer.
TRAF1 was shown to rescue a cIAP2-binding-defective mutant of TRAF2 and was able to inhibit TNFR2-induced proteosomal degradation of TRAF2 (17, 134). Therefore, the differences in the binding affinity and mode of cIAP to TRAF1·[TRAF2]2 and TRAF2 trimer provide a first explanation for these regulatory functions of TRAF1 in NF-κB activation.
TRAF CTD and Interactions with Peptides and TRADD
The activated TNFR recruits signaling adaptors to their intracellular domains. The TNFR superfamily can be divided into two subgroups: TNFRs called death receptors that contain an intracellular DD, and TNFRs without a DD. Those without a DD are able to directly recruit members of the TRAFs. Death receptors related to TNFR1 require the adaptor protein TRADD to recruit TRAF2. The DD of TRADD binds to the receptor, and the NTD of TRADD facilitates interaction with TRAF2 (92, 93).
Several complexes betweenTRAF2 and receptor peptides have been solved (63, 75, 81, 92, 126). Overall, these complexes resemble a mushroom-like shape with the TRAF2-CC domain as the stalk and the β-sandwich domain as the cap of the mushroom (Figure 5f,g). In all receptor·TRAF2 structures, the receptor peptides have an extended main chain conformation and form a β-strand. This β-strand extends the β-sandwich of TRAF2 at the edge of the mushroom cap. The TRAF2-binding motifs are defined by specific side chain interactions between the receptor peptides and TRAF2 (75, 92, 126).
The crystal structure of the CTD (CC and TRAF-C) of TRAF2 and the NTD of TRADD (TRADD-N) reveal that the mushroom-like shape of TRAD2 is decorated by three molecules of TRADD bound to the top side of the mushroom cap (93) (Figure 5h,i). Each of the TRADD molecules exclusively interacts with the β-sandwich fold of one TRAF2 subunit. TRADD-N folds into an α-β-sandwich with a four-stranded β-sheet and six α-helices (Figure 5h,i). The interface is divided into two regions. Region 1 is largely hydrophobic and formed by a surface protrusion on TRAF2 and an exposed shallow face of the β-sheet of TRADD. Region II is formed by a highly charged ridge on TRADD and a surface depression of TRAF2 and features a hydrophilic interface with many hydrogen bonds and salt bridges.
The C terminus of TRADD-N projects away from theTRADD·TRAF2 complex and indicates a possible location of the C-terminal DD of TRADD, which directly binds the intracellular DD of TNFR1 close to the cell membrane. The DD domain of TRADD is also a central platform for the recruitment of intracellular signaling molecules such as FADD for caspase-8 binding or RIP1 for NF-κB activation. In this conformation, the trimeric CC regions are protruding toward the center of the cell, poised to interact with downstream signaling proteins like cIAPs. Interestingly, the binding region of TRADD on TRAF2 is similar to the surface region occupied by receptor peptides (71, 75, 92, 115, 126) and highlights the competitive nature of direct and indirect recruitment of TRAFs by the TNFR family.
TRAF Higher-Order Oligomerization
Although the N-terminal part of TRAF proteins forms dimers, the CTDs form trimers. This apparent symmetry discrepancy within the same protein can be rectified with a higher-order oligomerization network. Indeed, spontaneous aggregation of endogenous TRAF6 has been observed as a response to receptor stimulation in vivo (129). Furthermore, fluorescence resonance energy transfer experiments have shown that TRAF6 can form higher-order oligomers. The capability to oligomerize can be abolished by a mutant incapable of dimerization. Taking all the available structural information together, a model of activated, full-length TRAF6 was proposed (88, 106, 129) (Figure 5j). The TRAF6 model reveals the potential for infinite oligomerization into a two-dimensional lattice via the dimerization and trimerization interfaces. This network of interactions would lead to an increase in local concentration of all associated signaling proteins by providing a docking platform and promoting autoubiquitination, polyubiquitination, and downstream signaling.
SIGNALING CASCADES TRIGGERED BY THE TLR/IL-1R SUPERFAMILY
The TLR/IL-1R superfamily belongs to the pattern recognition receptors, with each of the TLRs recognizing a specific PAMP via their extracellular domain. PAMPs include lipopolysaccharides of gram-negative bacteria, peptidoglycan and lipoteichoic acid of gram-positive bacteria, RNAs indicative of virus presence, and various forms of stress signals.
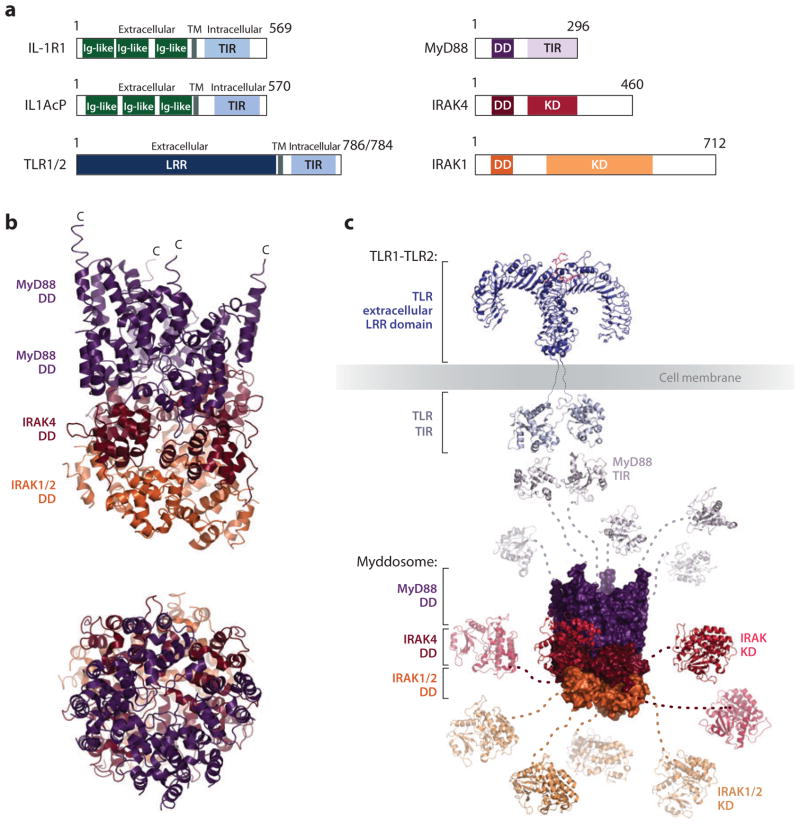
The TLRs are related to the IL-1R subfamily of membrane receptors. They share a common cytoplasmic TIR domain and consequently utilize overlapping components for downstream signaling. TLRs are evolutionarily conserved from Caenorhabditis elegans to mammals, and to date, a total of 13 mammalian TLRs have been identified. TLRs are composed of three domains: an N-terminal extracellular leucine-rich repeat region (LRR) that senses extracellular pathogens and signals generated during tissue injury, a single transmembrane domain, and a C-terminal intracellular Toll/IL-1R (TIR) domain (88, 106, 112) (Figure 6a). Upon ligand-induced activation, TLRs and IL-1Rs form dimers (62, 112) or alter their preexisting conformation (45, 49, 62, 67, 90, 130).
Figure 6.
Structures of the Myddosome and other membrane-proximal interactions in the TLR/IL-1R pathway. (a) Domain organizations. (b) Crystal structure of the DD complex of the Myddosome in two orientations, showing a structure with 6 MyD88, 4 IRAK4, and 4 IRAK1 or IRAK2 molecules. (c) A structural model of the membrane-proximal events in TLR signaling. Abbreviations: DD, death domain; IL-1R, interleukin-1 receptor; IRAK, interleukin-1 receptor (IL-1R)-associated kinase; MyD88, myeloid differentiation primary-response gene 88; TLR, Toll-like receptor.
Membrane Proximal Interactions
To date, several structures of the extracellular domain of TLRs bound to a ligand have been solved and reveal a common overall structure (45, 49, 67, 90, 130). The extracellular LRR domains of the dimeric TLRs resemble an m-like shape with the two N termini extending in opposite directions and the two C termini converging in the middle region. However, each of the different ligands is bound to distinct surfaces on the TLR dimers.
Receptors of the IL-1R family contain three immunoglobulin-like domains in their extracellular portion (Figure 6a). Upon initial binding to IL-1R1, association with IL-1RAcP is required for tight binding of the cytokine and downstream signaling (57, 87, 102). The structures of the IL-1-bound IL-1R1·IL-1RAcP and the IL-1R1·IL-1RII heterodimer reveal similar architecture and show that the Ig-like domains of the receptor are brought in close proximity upon dimerization (26, 109, 114, 117).
The TLRs and IL-1R family of membrane receptors share a common cytoplasmic TIR domain and consequently utilize overlapping components for downstream signaling (28, 86). It was suggested that higher-order oligomers of receptors may be critical for intracellular signal initiation (79, 102). The TIR domain forms a small, globular, parallel β-sheet surrounded by α-helices (25, 80, 123) (Figure 6c). It is thought that upon binding of the ligand to the extracellular domain, the intracellular TIR domains come in close proximity to each other and can engage in homotypic interaction. This creates a platform from which oligomerization of TIR-domain-containing adaptor molecules can be nucleated (1, 88, 106, 132). To date, five adaptor molecules have been identified: myeloid differentiation primary-response gene 88 (MyD88), MyD88-adaptor-like protein, TIR-domain-containing adaptor protein–inducing IFNβ (TRIF), TRIF-related adaptor molecule, and sterile α- and armadillo-motif-containing protein (40, 88). Several structures of TIR-domains are known. However, due to the difficulty in reconstituting stable TIR domain oligomers, the nature of TIR domain oligomerization still remains elusive. And several proposed models based on mutagenesis studies, crystal structures, or docking studies still await conformation (41, 43, 51, 74, 83, 84, 113).
Oligomeric DD Interactions and Activation of IRAK: The Myddosome
It was suggested that higher-order oligomers of receptors may be critical for intracellular signal initiation (79, 102). Upon binding of the ligand to the receptor, the intracellular TIR domains come in close proximity to each other and can engage in homotypic interaction. This creates an initial TIR platform at which other TIR-domain-containing adaptor molecules can assemble and oligomerize (88, 106, 132).
The primary signaling adaptor protein for TLR and IL-1R signaling is MyD88. In addition to its TIR domain, it contains a C-terminal DD (Figure 6a). MyD88 is a member of the DD superfamily, which also includes pyrin, caspase recruitment, and death effector domains (91). The DD fold contains six antiparallel α-helices arranged in a Greek key bundle (37, 104). The DD fold is characterized by a low sequence homology that generates diverse, specific interaction surfaces. Within each subfamily, members form conserved homotypic interactions and facilitate the assembly of oligomeric signaling complexes, which are crucial components of inflammatory and apoptotic signaling pathways.
In the TLR- and IL-1R-induced pathways, MyD88 associates with members of the IL-1R-associated kinase (IRAK) family. IRAK proteins contain an N-terminal DD followed by a C-terminal kinase domain (Figure 6a). The DDs of MyD88 and IRAKs assemble into a large oligomeric complex termed a Myddosome (79). The structure of this Myddosome has been solved and reveals a tower shape consisting of four layers; a layer of IRAK4 is sandwiched between two top layers of MyD88 and a bottom layer of IRAK2 (66) (Figure 6b). The DDs of MyD88, IRAK4, and IRAK2 assemble into a left-handed, helical structure that is stabilized by three types of conserved DD interactions (20). Surface shape and charge complementarity between the adjacent layers and the fact that the IRAK4DDis monomeric whereas the DD of MyD88 is prone to oligomerization in solution suggests a sequential assembly order of the entire complex (66). Therefore, a strict hierarchical assembly mechanism can be easily envisioned. First, binding of a ligand to the receptor brings the intracellular receptor TIR domains into close proximity, which leads to recruitment of MyD88. Second, the DD of MyD88 oligomerizes and nucleates IRAK4 binding and oligomerization. Third, after four IRAK4 molecules have assembled a layer of the Myddosome, IRAK2 and IRAK1 molecules can bind to the Myddosome.
IRAK4 was shown to phosphorylate the activation loop of IRAK1 and autophosphorylate itself in vitro (64), whereas MyD88 was found to promote phosphorylation of IRAK1 and IRAK4 (9). This suggests that the Myddosome provides a platform at which phosphorylation is initiated by IRAK4 and propagated downstream to IRAK1 and IRAK2. Phosphorylated IRAK1 and IRAK2 in turn recruit TRAF6 to the membrane (10, 94, 125) (Figure 6c).
Activation of the TAK1 Complex
The TAK complex is involved in signaling pathways induced by various cytokines, including TGF-β, TNF-α, IL-1, and even LPS (61), and is required to activate IKK. The TAK complex consists of a kinase, TAK1, and TAB1, TAB2, and TAB3 (Figure 7a). TAK1 is a member of the MAPKKK family (82). TAK1 requires TAB1 for its kinase activity (53, 103). The structure of the TAK1·TAB1 complex shows that the interacting α-helix of TAB1 is bound in an extensive pocket on the surface of the C-terminal TAK1 kinase lobe (8) (Figure 7b). This close interaction promotes autophosphorylation of the TAK1 kinase activation lobe, likely through an allosteric mechanism (8, 89, 100).
Figure 7.
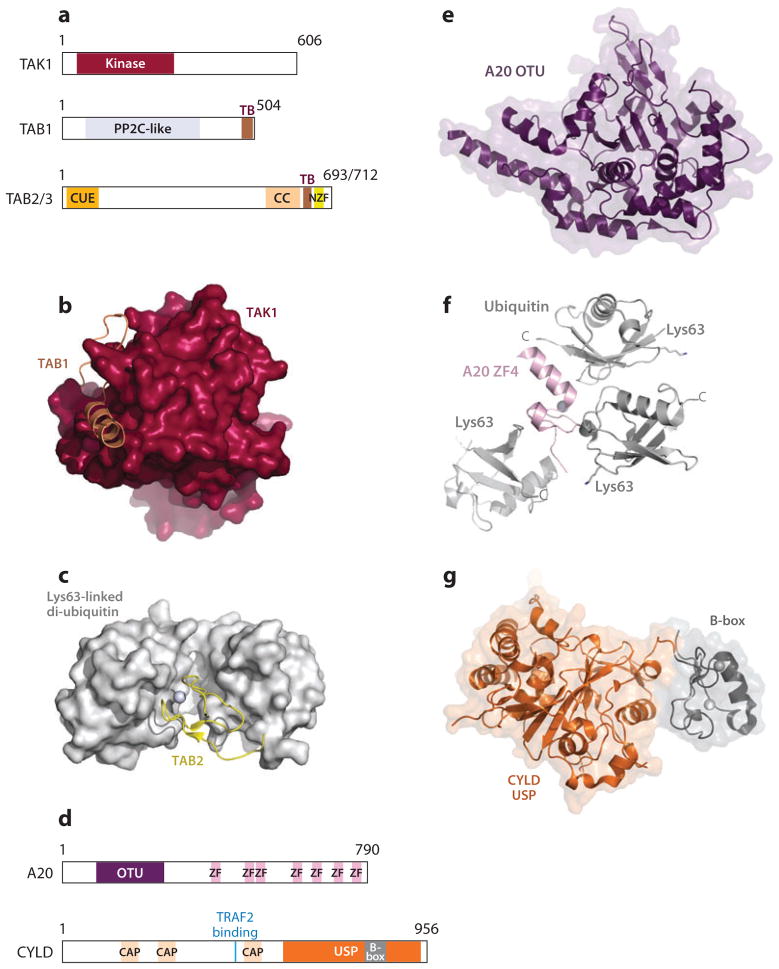
The TAK1 complex and deubiquitinases in downregulating NF-κB signaling. (a) Domain organizations of TAK1 complex components. (b) Crystal structure of TAK1 kinase domain in complex with an activating peptide from TAB1. (c) Crystal structure of TAB2 NZF in complex with Lys63-linked di-ubiquitin. (d) Domain organizations of deubiquitinases A20 and CYLD. (e) Crystal structure of the OTU domain of A20. (f) Crystal structure of the fourth ZF domain of A20 in complex with three ubiquitin molecules. (g) Crystal structure of the USP domain of CYLD. The B-box is a small zinc-binding domain similar to RING domains. Abbreviations: NZF, Npl4 zinc finger; OTU, ovarian tumor domain; USP, ubiquitin-specific protease; ZF, zinc finger.
The TAK1 complex is regulated by Lys63-linked polyubiquitination (116). The ZF domains of TAB2 and TAB3 were shown to specifically recognize Lys63-linked polyubiquitin chains that are either unanchored or anchored to substrate proteins such as RIP1, TRAF6, or NEMO; mutations preventing polyubiquitin binding abolish TAK1 or IKK complex activation (48, 116). Secondary structure analysis shows a similar domain structure for TAB2 and TAB3: an N-terminal ubiquitin binding (CUE) domain and a CC region, followed by a TAK1-binding (TB) domain and a C-terminal Npl4 ZF (NZF) (48). The sequences of the TAB2 and TAB3 NZF domains are ~80% identical, and crystal structures of both in complex with Lys63-linked polyubiquitin have been solved and are practically identical (59, 101). The TABNZF folds into a pair of antiparallel β-sheets followed by a long loop and binds to both ubiquitin moieties simultaneously (Figure 7c). TAB does not recognize the Lys63-linked isopeptide bond but interacts with hydrophobic patches centered around Ile44 on adjacent ubiquitin moieties. Specificity for Lys63-linked ubiquitin is achieved by conformational constraints. The distal and proximal ubiquitin-binding sites of the TAB NZF are organized to optimize simultaneous binding to adjacent ubiquitin moieties in a configuration specific to Lys63-linked ubiquitin chains (56). The conformational constraints imposed by TAB on the Lys63-linked di-Ub cannot be adopted by a linear linkage (59).
Multiple TAB NZF domains could bind to long Lys63-linked ubiquitin chains. Recruitment of multiple TAK1 complexes to the TRAF6-polyubiquitination scaffold would bring the kinase domains of TAK1 in close proximity to each other. This would promote autophosphorylation and activation of the TAK1 kinase, which in turn can activate the IKK complex.
NEGATIVE REGULATION BY DEUBIQUITINASES
Polyubiquitin chains play a crucial role in the activation of the NF-κB pathway by proving an assembly platform for signaling molecules. Therefore, the removal of the polyubiquitin chains by deubiquitinases (DUBs) presents one way of terminating NF-κB activation. Similar to E3 ligases, DUBs can have specificity for certain polyubiquitin linkage types. The specificity can be mediated by selectivity of the catalytic core, ubiquitin-binding domains of the DUB, or adaptor proteins. Several DUBs have been found to regulate the NF-κB pathway. The DUBs, A20 (TNF-α-induced protein 3), Cezanne, Usp21, and the ubiquitin carboxyl-terminal hydrolase CYLD have been shown to be important for negative feedback regulation of the NF-κB pathway. A20, CYLD, Cezanne, and Usp21 differ in their temporal activation and ubiquitin linkage specificity. However, all four DUBs have been implicated in the removal of ubiquitin chains from RIP1 in TNF signaling.
A20 belongs to the ovarian tumor superfamily of DUBs (72). It contains an N-terminal ovarian tumor domain (OTU) and seven ZF domains (Figure 7d). A20 is a ubiquitin-editing enzyme (19). It mediates deubiquitination of Lys63-linked polyubiquitin but can also generate Lys48-linked polyubiquitin chains via the E3 ligase activity of the fourth ZF motif (15, 19, 118). Both activities are required for NF-κB inhibition (32).
The fourth ZF (ZF4) of A20 was also shown to selectively bind Lys63-linked ubiquitin chains (6). A crystal structure of ZF4 bound to monoubiquitin in addition to solution-phase characterization revealed a three-interface-binding site for Lys63-linked ubiquitin.
The OTU domain of A20 folds into wedge-like shape (Figure 7e). A highly conserved, negatively charged surface patch close to the active site of A20 was suggested as the ubiquitin-binding region (65). The putative binding site is located at a position similar to ubiquitin-binding sites of two other DUBs, Yuh1 and HAUSP (36, 47). A20 was shown to remove the entire Lys63-linked polyubiquitin chain from TRAF6 in one step, thereby effectively turning off the NF-κB pathway (65). It was suggested that the ZF domain recruits A20 to Lys63-linked polyubiquitin chains on activated signaling complexes. The A20 OTU disassembles the polyubiquitin chains, which in turn could facilitate the synthesis of degradative Lys48-linked ubiquitin chains on the substrates (Figure 7f).
CYLD is a member of the ubiquitin-specific protease (USP) family. It contains three cytoskeletal-associated protein (CAP)–glycine-conserved repeats in its N-terminal portion and a USP domain at the C terminus (Figure 6d). CYLD contains a TRAF2-binding sequence, and the third CAP was shown to interact with a Pro-rich sequence in NEMO (58, 99). The USP domain mediates disassembly of Lys63-linked polyubiquitin chains. Unlike other USPs, CYLD is able to hydrolyze polyubiquitin chains internally, resulting in an efficient inhibition of the NF-κB pathway. The structure of the CYLD USP domain revealed a proximal ubiquitin-binding site near the active site and a Lys63-specific set of conserved residues near the catalytic center, explaining the specificity for Lys63 (Figure 7g).
CONCLUDING REMARKS
Structural studies of NF-κB signaling have helped illustrate the underlying molecular mechanisms in many aspects of the signaling cascades from adaptor recruitment to activation of ubiquitin ligases and kinases, culminating in transcriptional control of immune and inflammatory responses. Several central themes are emerging from these studies. First, the conventional view that signal transduction is composed of linear strings of events is being challenged in that large multimeric assemblies, or signalosomes, are formed in these signaling processes. Second, at least three types of multimerization scaffolds have been identified from these studies: the infinite high-order oligomerization of TRAF6, the ubiquitin chain network, and the helical assembly in the DD superfamily. Third, oligomerization appears to occur at all levels of the signaling cascade in which multiple signaling oligomers combine to form gigantic signalosomes to perform multiple reactions simultaneously and efficiently, such as ubiquitination and phosphorylation. The intrinsic cooperativity in the formation of multimeric signalosomes may predict a digital threshold response in NF-κB signaling, a property that may be quite general in many other biological processes.
SUMMARY POINTS.
NF-κB family transcription factors are master regulators of immune and inflammatory responses.
NF-κB is sequestered in the cytosol by IκB proteins and released through specific signaling cascades that lead to activation of IKK, phosphorylation of IκB, and degradation of IκB.
NF-κBs bind to DNA as homo- or heterodimers.
The IKK complex contains a catalytic subunit and the regulatory subunit NEMO capable of multiple interactions.
IKK signaling is regulated by ubiquitin ligases and DUBs in ubiquitin-chain networks.
TRAF proteins may form infinite assemblies through alternating dimerization and trimerization.
DD proteins in NF-κB signaling assemble into large signalosomes using helical symmetry.
Formation of large signaling assemblies using various structural scaffolds may be a general principle in signaling to NF-κB and other biological processes.
Acknowledgments
We apologize for incomplete citations due to space limitations and acknowledge support from the National Institutes of Health (grants R01AI50872, R01AI045937, and R01AI079260).
Glossary
- NF-κB
nuclear factor κB
- IκB
inhibitor of NF-κB
- IKK
IκB kinase
- Kinase
enzyme that transfers phosphate groups from high-energy donor molecules, such as ATP, to specific substrates
- NEMO
NF-κB essential modulator; also known as IKKγ
- TRAF
tumor necrosis factor (TNF) receptor–associated factor
- TLR
Toll-like receptor
- TNF
tumor necrosis factor
- Ubiquitin ligase
protein that in combination with an ubiquitin-conjugating enzyme causes the attachment of ubiquitin to a lysine on a target protein via an isopeptide bond
- cIAP
cellular inhibitor of apoptosis protein
- MyD88
myeloid differentiation primary response protein 88
- IRAK
interleukin-1 receptor (IL-1R)-associated kinase
- Signalosomes
large multimeric signaling complexes
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Baeuerle PA, Baltimore D. I kappa B: a specific inhibitor of the NF-kappa B transcription factor. Science. 1988;242(4878):540–46. doi: 10.1126/science.3140380. [DOI] [PubMed] [Google Scholar]
- 2.Bagnéris C, Ageichik AV, Cronin N, Wallace B, Collins M, et al. Crystal structure of a vFlip-IKKγ complex: insights into viral activation of the IKK signalosome. Mol Cell. 2008;30(5):620–31. doi: 10.1016/j.molcel.2008.04.029. [DOI] [PubMed] [Google Scholar]
- 3.Banner DW, D’Arcy A, Janes W, Gentz R, Schoenfeld HJ, et al. Crystal structure of the soluble human 55 kd TNF receptor–human TNFβ complex: implications for TNF receptor activation. Cell. 1993;73(3):431–45. doi: 10.1016/0092-8674(93)90132-a. [DOI] [PubMed] [Google Scholar]
- 4.Bassères DS, Baldwin AS. Nuclear factor-κB and inhibitor of κB kinase pathways in oncogenic initiation and progression. Oncogene. 2006;25(51):6817–30. doi: 10.1038/sj.onc.1209942. [DOI] [PubMed] [Google Scholar]
- 5.Bianchi K, Meier P. A tangled web of ubiquitin chains: breaking news in TNF-R1 signaling. Mol Cell. 2009;36(5):736–42. doi: 10.1016/j.molcel.2009.11.029. [DOI] [PubMed] [Google Scholar]
- 6.Bosanac I, Wertz IE, Pan B, Yu C, Kusam S, et al. Ubiquitin binding to A20 ZnF4 is required for modulation of NF-κB signaling. Mol Cell. 2010;40(4):548–57. doi: 10.1016/j.molcel.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 7.Broemer M, Meier P. Ubiquitin-mediated regulation of apoptosis. Trends Cell Biol. 2009;19(3):130–40. doi: 10.1016/j.tcb.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 8.Brown K, Vial SCM, Dedi N, Long JM, Dunster NJ, Cheetham GMT. Structural basis for the interaction of TAK1 kinase with its activating protein TAB1. J Mol Biol. 2005;354(5):1013–20. doi: 10.1016/j.jmb.2005.09.098. [DOI] [PubMed] [Google Scholar]
- 9.Burns K, Janssens S, Brissoni B, Olivos N, Beyaert R, Tschopp J. Inhibition of interleukin 1 receptor/Toll-like receptor signaling through the alternatively spliced, short form of MyD88 is due to its failure to recruit IRAK-4. J Exp Med. 2003;197(2):263–68. doi: 10.1084/jem.20021790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao Z, Henzel WJ, Gao X. IRAK: a kinase associated with the interleukin-1 receptor. Science. 1996;271(5252):1128–31. doi: 10.1126/science.271.5252.1128. [DOI] [PubMed] [Google Scholar]
- 11.Chaudhary PM, Jasmin A, Eby MT, Hood L. Modulation of the NF-κB pathway by virally encoded death effector domains–containing proteins. Oncogene. 1999;18(42):5738–46. doi: 10.1038/sj.onc.1202976. [DOI] [PubMed] [Google Scholar]
- 12.Cordier F, Grubisha O, Traincard F, Véron M, Delepierre M, Agou F. The zinc finger of NEMO is a functional ubiquitin-binding domain. J Biol Chem. 2009;284(5):2902–7. doi: 10.1074/jbc.M806655200. [DOI] [PubMed] [Google Scholar]
- 13.Cordier F, Vinolo E, Véron M, Delepierre M, Agou F. Solution structure of NEMO zinc finger and impact of an anhidrotic ectodermal dysplasia with immunodeficiency-related point mutation. J Mol Biol. 2008;377(5):1419–32. doi: 10.1016/j.jmb.2008.01.048. [DOI] [PubMed] [Google Scholar]
- 14.Courtois G, Gilmore TD. Mutations in the NF-κB signaling pathway: implications for human disease. Oncogene. 2006;25(51):6831–43. doi: 10.1038/sj.onc.1209939. [DOI] [PubMed] [Google Scholar]
- 15.De Valck D, Heyninck K, Van Criekinge W, Contreras R, Beyaert R, Fiers W. A20, an inhibitor of cell death, self-associates by its zinc finger domain. FEBS Lett. 1996;384(1):61–64. doi: 10.1016/0014-5793(96)00283-9. [DOI] [PubMed] [Google Scholar]
- 16.DiDonato JA, Hayakawa M, Rothwarf DM, Zandi E, Karin M. A cytokine-responsive IκB kinase that activates the transcription factor NF-κB. Nature. 1997;388(6642):548–54. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 17.Dupoux A, Cartier J, Cathelin S, Filomenko R, Solary E, Dubrez-Daloz L. cIAP1-dependent TRAF2 degradation regulates the differentiation of monocytes into macrophages and their response to CD40 ligand. Blood. 2009;113(1):175–85. doi: 10.1182/blood-2008-02-137919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ea C-K, Deng L, Xia Z-P, Pineda G, Chen ZJ. Activation of IKK by TNFα requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22(2):245–57. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 19.Evans PC, Smith TS, Lai M-J, Williams MG, Burke DF, et al. A novel type of deubiquitinating enzyme. J Biol Chem. 2003;278(25):23180–86. doi: 10.1074/jbc.M301863200. [DOI] [PubMed] [Google Scholar]
- 20.Ferrao R, Wu H. Helical assembly in the death domain (DD) superfamily. Curr Opin Struct Biol. 2012;22(2):241–47. doi: 10.1016/j.sbi.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferrao R, Li J, Bergamin E, Wu H. Structural insights into the assembly of large oligomeric signalosomes in the Toll-like receptor-interleukin-1 receptor superfamily. Sci Signal. 2012;5:re3. doi: 10.1126/scisignal.2003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Field N, Low W, Daniels M, Howell S, Daviet L, et al. KSHV vFLIP binds to IKK-γ to activate IKK. J Cell Sci. 2003;116(Pt. 18):3721–28. doi: 10.1242/jcs.00691. [DOI] [PubMed] [Google Scholar]
- 23.Fiorini E, Schmitz I, Marissen WE, Osborn SL, Touma M, et al. Peptide-induced negative selection of thymocytes activates transcription of an NF-κB inhibitor. Mol Cell. 2002;9(3):637–48. doi: 10.1016/s1097-2765(02)00469-0. [DOI] [PubMed] [Google Scholar]
- 24.Fusco F, Pescatore A, Bal E, Ghoul A, Paciolla M, et al. Alterations of the IKBKG locus and diseases: an update and a report of 13 novel mutations. Hum Mutat. 2008;29(5):595–604. doi: 10.1002/humu.20739. [DOI] [PubMed] [Google Scholar]
- 25.Ghosh G, van Duyne G, Ghosh S, Sigler PB. Structure of NF-κB p50 homodimer bound to a κB site. Nature. 1995;373(6512):303–10. doi: 10.1038/373303a0. [DOI] [PubMed] [Google Scholar]
- 26.Ghosh S, Baltimore D. Activation in vitro of NF-κB by phosphorylation of its inhibitor IκB. Nature. 1990;344(6267):678–82. doi: 10.1038/344678a0. [DOI] [PubMed] [Google Scholar]
- 27.Ghosh S, Karin M. Missing pieces in the NF-κB puzzle. Cell. 2002;109(Suppl):S81–96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- 28.Gilmore TD. NF-κB, KBF1, dorsal, and related matters. Cell. 1990;62(5):841–43. doi: 10.1016/0092-8674(90)90257-f. [DOI] [PubMed] [Google Scholar]
- 29.Guasparri I, Keller SA, Cesarman E. KSHVvFLIP is essential for the survival of infected lymphoma cells. J Exp Med. 2004;199(7):993–1003. doi: 10.1084/jem.20031467. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Haas TL, Emmerich CH, Gerlach B, Schmukle AC, Cordier SM, et al. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol Cell. 2009;36(5):831–44. doi: 10.1016/j.molcel.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 31.Hatada EN, Nieters A, Wulczyn FG, Naumann M, Meyer R, et al. The ankyrin repeat domains of the NF-κB precursor p105 and the protooncogene bcl-3 act as specific inhibitors of NF-κB DNA binding. Proc Natl Acad Sci USA. 1992;89(6):2489–93. doi: 10.1073/pnas.89.6.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heyninck K, Beyaert R. A20 inhibits NF-κB activation by dual ubiquitin-editing functions. Trends Biochem Sci. 2005;30(1):1–4. doi: 10.1016/j.tibs.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 33.Hiscott J, Nguyen T-LA, Arguello M, Nakhaei P, Paz S. Manipulation of the nuclear factor-κB pathway and the innate immune response by viruses. Oncogene. 2006;25(51):6844–67. doi: 10.1038/sj.onc.1209941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsu H, Huang J, Shu HB, Baichwal V, Goeddel DV. TNF-dependent recruitment of the protein kinase RIP to the TNF receptor-1 signaling complex. Immunity. 1996;4(4):387–96. doi: 10.1016/s1074-7613(00)80252-6. [DOI] [PubMed] [Google Scholar]
- 35.Hsu H, Shu HB, Pan MG, Goeddel DV. TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell. 1996;84(2):299–308. doi: 10.1016/s0092-8674(00)80984-8. [DOI] [PubMed] [Google Scholar]
- 36.Hu M, Li P, Li M, Li W, Yao T, et al. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell. 2002;111(7):1041–54. doi: 10.1016/s0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- 37.Huang B, Eberstadt M, Olejniczak ET, Meadows RP, Fesik SW. NMR structure and mutagenesis of the Fas (APO-1/CD95) death domain. Nature. 1996;384(6610):638–41. doi: 10.1038/384638a0. [DOI] [PubMed] [Google Scholar]
- 38.Huang TT, Kudo N, Yoshida M, Miyamoto S. A nuclear export signal in the N-terminal regulatory domain of IκBα controls cytoplasmic localization of inactive NF-κB/IκBα complexes. Proc Natl Acad Sci USA. 2000;97(3):1014–19. doi: 10.1073/pnas.97.3.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang TT, Miyamoto S. Postrepression activation of NF-κB requires the amino-terminal nuclear export signal specific to IκBα. Mol Cell Biol. 2001;21(14):4737–47. doi: 10.1128/MCB.21.14.4737-4747.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huxford T, Ghosh G. A structural guide to proteins of the NF-κB signaling module. Cold Spring Harb Perspect Biol. 2009;1(3):a000075. doi: 10.1101/cshperspect.a000075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huxford T, Huang DB, Malek S, Ghosh G. The crystal structure of the IκBα/NF-κB complex reveals mechanisms of NF-κB inactivation. Cell. 1998;95(6):759–70. doi: 10.1016/s0092-8674(00)81699-2. Describes the mechanism of inhibition of NF-κB by IκB (see also Reference 43) [DOI] [PubMed] [Google Scholar]
- 42.Irmler M, Thome M, Hahne M, Schneider P, Hofmann K, et al. Inhibition of death receptor signals by cellular FLIP. Nature. 1997;388(6638):190–95. doi: 10.1038/40657. [DOI] [PubMed] [Google Scholar]
- 43.Jacobs MD, Harrison SC. Structure of an IκBα/NF-κB complex. Cell. 1998;95(6):749–58. doi: 10.1016/s0092-8674(00)81698-0. [DOI] [PubMed] [Google Scholar]
- 44.Jenner RG, Albà MM, Boshoff C, Kellam P. Kaposi’s sarcoma–associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J Virol. 2001;75(2):891–902. doi: 10.1128/JVI.75.2.891-902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jin MS, Kim SE, Heo JY, Lee ME, Kim HM, et al. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell. 2007;130(6):1071–82. doi: 10.1016/j.cell.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 46.Johnson C, Van Antwerp D, Hope TJ. An N-terminal nuclear export signal is required for the nucleocytoplasmic shuttling of IκBα. EMBO J. 1999;18(23):6682–93. doi: 10.1093/emboj/18.23.6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnston SC, Riddle SM, Cohen RE, Hill CP. Structural basis for the specificity of ubiquitin C-terminal hydrolases. EMBO J. 1999;18(14):3877–87. doi: 10.1093/emboj/18.14.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kanayama A, Seth RB, Sun L, Ea C-K, Hong M, et al. TAB2 and TAB3 activate the NF-κB pathway through binding to polyubiquitin chains. Mol Cell. 2004;15(4):535–48. doi: 10.1016/j.molcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 49.Kang JY, Nan X, Jin MS, Youn S-J, Ryu YH, et al. Recognition of lipopeptide patterns by Toll-like receptor 2–Toll-like receptor 6 heterodimer. Immunity. 2009;31(6):873–84. doi: 10.1016/j.immuni.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 50.Karin M. How NF-κB is activated: the role of the IκB kinase (IKK) complex. Oncogene. 1999;18(49):6867–74. doi: 10.1038/sj.onc.1203219. [DOI] [PubMed] [Google Scholar]
- 51.Khan JA, Brint EK, O’Neill LAJ, Tong L. Crystal structure of the Toll/interleukin-1 receptor domain of human IL-1RAPL. J Biol Chem. 2004;279(30):31664–70. doi: 10.1074/jbc.M403434200. [DOI] [PubMed] [Google Scholar]
- 52.Kilger E, Kieser A, Baumann M, Hammerschmidt W. Epstein-Barr virus-mediated B-cell proliferation is dependent upon latent membrane protein 1, which simulates an activated CD40 receptor. EMBO J. 1998;17(6):1700–9. doi: 10.1093/emboj/17.6.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kishimoto K, Matsumoto K, Ninomiya-Tsuji J. TAK1 mitogen-activated protein kinase kinase kinase is activated by autophosphorylation within its activation loop. J Biol Chem. 2000;275(10):7359–64. doi: 10.1074/jbc.275.10.7359. [DOI] [PubMed] [Google Scholar]
- 54.Kitamura H, Kanehira K, Okita K, Morimatsu M, Saito M. MAIL, a novel nuclear IκB protein that potentiates LPS-induced IL-6 production. FEBS Lett. 2000;485(1):53–56. doi: 10.1016/s0014-5793(00)02185-2. [DOI] [PubMed] [Google Scholar]
- 55.Komander D, Lord CJ, Scheel H, Swift S, Hofmann K, et al. The structure of the CYLD USP domain explains its specificity for Lys63-linked polyubiquitin and reveals a B box module. Mol Cell. 2008;29(4):451–64. doi: 10.1016/j.molcel.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 56.Komander D, Reyes-Turcu F, Licchesi JDF, Odenwaelder P, Wilkinson KD, Barford D. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 2009;10(5):466–73. doi: 10.1038/embor.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Korherr C, Hofmeister R, Wesche H, Falk W. A critical role for interleukin-1 receptor accessory protein in interleukin-1 signaling. Eur J Immunol. 1997;27(1):262–67. doi: 10.1002/eji.1830270139. [DOI] [PubMed] [Google Scholar]
- 58.Kovalenko A, Chable-Bessia C, Cantarella G, Israël A, Wallach D, Courtois G. The tumour suppressor CYLD negatively regulates NF-κB signalling by deubiquitination. Nature. 2003;424(6950):801–5. doi: 10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- 59.Kulathu Y, Akutsu M, Bremm A, Hofmann K, Komander D. Two-sided ubiquitin binding explains specificity of the TAB2 NZF domain. Nat Struct Mol Biol. 2009;16(12):1328–30. doi: 10.1038/nsmb.1731. [DOI] [PubMed] [Google Scholar]
- 60.Laherty CD, Hu HM, Opipari AW, Wang F, Dixit VM. The Epstein-Barr virus LMP1 gene product induces A20 zinc finger protein expression by activating nuclear factor κB. J Biol Chem. 1992;267(34):24157–60. [PubMed] [Google Scholar]
- 61.Landström M. The TAK1–TRAF6 signalling pathway. Int J Biochem Cell Biol. 2010;42(5):585–89. doi: 10.1016/j.biocel.2009.12.023. [DOI] [PubMed] [Google Scholar]
- 62.Latz E, Verma A, Visintin A, Gong M, Sirois CM, et al. Ligand-induced conformational changes allosterically activate Toll-like receptor 9. Nat Immunol. 2007;8(7):772–79. doi: 10.1038/ni1479. [DOI] [PubMed] [Google Scholar]
- 63.Li C, Norris PS, Ni C-Z, Havert ML, Chiong EM, et al. Structurally distinct recognition motifs in lymphotoxin-β receptor and CD40 for tumor necrosis factor receptor-associated factor (TRAF)-mediated signaling. J Biol Chem. 2003;278(50):50523–29. doi: 10.1074/jbc.M309381200. [DOI] [PubMed] [Google Scholar]
- 64.Li S, Strelow A, Fontana EJ, Wesche H. IRAK-4: a novel member of the IRAK family with the properties of an IRAK-kinase. Proc Natl Acad Sci USA. 2002;99(8):5567–72. doi: 10.1073/pnas.082100399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin S-C, Chung JY, Lamothe B, Rajashankar K, Lu M, et al. Molecular basis for the unique deubiquitinating activity of the NF-κB inhibitor A20. J Mol Biol. 2008;376(2):526–40. doi: 10.1016/j.jmb.2007.11.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin S-C, Lo Y-C, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465(7300):885–90. doi: 10.1038/nature09121. Describes the helical oligomerization mechanism of proteins in TLR signaling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu L, Botos I, Wang Y, Leonard JN, Shiloach J, et al. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science. 2008;320(5874):379–81. doi: 10.1126/science.1155406. Describes the structure of the extracellular domain of a TLR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lo Y-C, Lin S-C, Rospigliosi CC, Conze DB, Wu C-J, et al. Structural basis for recognition of diubiquitins by NEMO. Mol Cell. 2009;33(5):602–15. doi: 10.1016/j.molcel.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lu M, Lin S-C, Huang Y, Kang YJ, Rich R, et al. XIAP induces NF-κB activation via the BIR1/TAB1 interaction and BIR1 dimerization. Mol Cell. 2007;26(5):689–702. doi: 10.1016/j.molcel.2007.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mace PD, Smits C, Vaux DL, Silke J, Day CL. Asymmetric recruitment of cIAPs by TRAF2. J Mol Biol. 2010;400(1):8–15. doi: 10.1016/j.jmb.2010.04.055. [DOI] [PubMed] [Google Scholar]
- 71.Mahoney DJ, Cheung HH, Mrad RL, Plenchette S, Simard C, et al. Both cIAP1 and cIAP2 regulate TNFα-mediated NF-κB activation. Proc Natl Acad Sci USA. 2008;105(33):11778–83. doi: 10.1073/pnas.0711122105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Makarova KS, Aravind L, Koonin EV. A novel superfamily of predicted cysteine proteases from eukaryotes, viruses and Chlamydia pneumoniae. Trends Biochem Sci. 2000;25(2):50–52. doi: 10.1016/s0968-0004(99)01530-3. [DOI] [PubMed] [Google Scholar]
- 73.Malek S, Chen Y, Huxford T, Ghosh G. IκBβ, but not IκBα, functions as a classical cytoplasmic inhibitor of NF-κB dimers by masking both NF-κB nuclear localization sequences in resting cells. J Biol Chem. 2001;276(48):45225–35. doi: 10.1074/jbc.M105865200. [DOI] [PubMed] [Google Scholar]
- 74.Malek S, Huang DB, Huxford T, Ghosh S, Ghosh G. X-ray crystal structure of an IκBβŸNF-κB p65 homodimer complex. J Biol Chem. 2003;278(25):23094–100. doi: 10.1074/jbc.M301022200. [DOI] [PubMed] [Google Scholar]
- 75.McWhirter SM, Pullen SS, Holton JM, Crute JJ, Kehry MR, Alber T. Crystallographic analysis of CD40 recognition and signaling by human TRAF2. Proc Natl Acad Sci USA. 1999;96(15):8408–13. doi: 10.1073/pnas.96.15.8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mercurio F, Murray BW, Shevchenko A, Bennett BL, Young DB, et al. IκBkinase (IKK)-associated protein 1, a common component of the heterogeneous IKK complex. Mol Cell Biol. 1999;19(2):1526–38. doi: 10.1128/mcb.19.2.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mercurio F, Zhu H, Murray BW, Shevchenko A, Bennett BL, et al. IKK-1 and IKK-2: cytokine-activated IκB kinases essential for NF-κB activation. Science. 1997;278(5339):860–66. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 78.Michel F, Soler-Lopez M, Petosa C, Cramer P, Siebenlist U, Müller CW. Crystal structure of the ankyrin repeat domain of Bcl-3: a unique member of the IκB protein family. EMBO J. 2001;20(22):6180–90. doi: 10.1093/emboj/20.22.6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Motshwene PG, Moncrieffe MC, Grossmann JG, Kao C, Ayaluru M, et al. An oligomeric signaling platform formed by the Toll-like receptor signal transducers MyD88 and IRAK-4. J Biol Chem. 2009;284(37):25404–11. doi: 10.1074/jbc.M109.022392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Müller CW, Rey FA, Sodeoka M, Verdine GL, Harrison SC. Structure of the NF-κB p50 homodimer bound to DNA. Nature. 1995;373(6512):311–17. doi: 10.1038/373311a0. [DOI] [PubMed] [Google Scholar]
- 81.Ni CZ, Welsh K, Leo E, Chiou CK, Wu H, et al. Molecular basis for CD40 signaling mediated by TRAF3. Proc Natl Acad Sci USA. 2000;97(19):10395–99. doi: 10.1073/pnas.97.19.10395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ninomiya-Tsuji J, Kajino T, Ono K, Ohtomo T, Matsumoto M, et al. A resorcylic acid lactone, 5Z-7-oxozeaenol, prevents inflammation by inhibiting the catalytic activity of TAK1 MAPK kinase kinase. J Biol Chem. 2003;278(20):18485–90. doi: 10.1074/jbc.M207453200. [DOI] [PubMed] [Google Scholar]
- 83.Nyman T, Stenmark P, Flodin S, Johansson I, Hammarström M, Nordlund P. The crystal structure of the human toll-like receptor 10 cytoplasmic domain reveals a putative signaling dimer. J Biol Chem. 2008;283(18):11861–65. doi: 10.1074/jbc.C800001200. [DOI] [PubMed] [Google Scholar]
- 84.Ohnishi H, Tochio H, Kato Z, Orii KE, Li A, et al. Structural basis for the multiple interactions of the MyD88 TIR domain in TLR4 signaling. Proc Natl Acad Sci USA. 2009;106(25):10260–65. doi: 10.1073/pnas.0812956106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ohno H, Takimoto G, McKeithan TW. The candidate proto-oncogene bcl-3 is related to genes implicated in cell lineage determination and cell cycle control. Cell. 1990;60(6):991–97. doi: 10.1016/0092-8674(90)90347-h. [DOI] [PubMed] [Google Scholar]
- 86.O’Neill L. The Toll/interleukin-1 receptor domain: a molecular switch for inflammation and host defence. Biochem Soc Trans. 2000;28(5):557–63. doi: 10.1042/bst0280557. [DOI] [PubMed] [Google Scholar]
- 87.O’Neill LAJ. The interleukin-1 receptor/Toll-like receptor superfamily: 10 years of progress. Immunol Rev. 2008;226:10–18. doi: 10.1111/j.1600-065X.2008.00701.x. [DOI] [PubMed] [Google Scholar]
- 88.O’Neill LAJ, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. 2007;7(5):353–64. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- 89.Ono K, Ohtomo T, Sato S, Sugamata Y, Suzuki M, et al. An evolutionarily conserved motif in the TAB1 C-terminal region is necessary for interaction with and activation of TAK1 MAPKKK. J Biol Chem. 2001;276(26):24396–400. doi: 10.1074/jbc.M102631200. [DOI] [PubMed] [Google Scholar]
- 90.Park BS, Song DH, Kim HM, Choi B-S, Lee H, Lee J-O. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458(7242):1191–95. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 91.Park HH, Lo Y-C, Lin S-C, Wang L, Yang JK, Wu H. The death domain superfamily in intracellular signaling of apoptosis and inflammation. Annu Rev Immunol. 2007;25:561–86. doi: 10.1146/annurev.immunol.25.022106.141656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Park YC, Burkitt V, Villa AR, Tong L, Wu H. Structural basis for self-association and receptor recognition of human TRAF2. Nature. 1999;398(6727):533–38. doi: 10.1038/19110. Describes the structure of a TRAF protein and its complex with a receptor tail. [DOI] [PubMed] [Google Scholar]
- 93.Park YC, Ye H, Hsia C, Segal D, Rich RL, et al. A novel mechanism of TRAF signaling revealed by structural and functional analyses of the TRADD-TRAF2 interaction. Cell. 2000;101:777–87. doi: 10.1016/s0092-8674(00)80889-2. [DOI] [PubMed] [Google Scholar]
- 94.Qian Y, Commane M, Ninomiya-Tsuji J, Matsumoto K, Li X. IRAK-mediated translocation of TRAF6 and TAB2 in the interleukin-1-induced activation of NFκB. J Biol Chem. 2001;276(45):41661–67. doi: 10.1074/jbc.M102262200. [DOI] [PubMed] [Google Scholar]
- 95.Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-κB activation. Cell. 2009;136(6):1098–109. doi: 10.1016/j.cell.2009.03.007. Describes the mode of interaction between NEMO and linear di-Ub. [DOI] [PubMed] [Google Scholar]
- 96.Régnier CH, Song HY, Gao X, Goeddel DV, Cao Z, Rothe M. Identification and characterization of an IκB kinase. Cell. 1997;90(2):373–83. doi: 10.1016/s0092-8674(00)80344-x. [DOI] [PubMed] [Google Scholar]
- 97.Rothwarf DM, Zandi E, Natoli G, Karin M. IKK-γ is an essential regulatory subunit of the IκB kinase complex. Nature. 1998;395(6699):297–300. doi: 10.1038/26261. [DOI] [PubMed] [Google Scholar]
- 98.Rushe M, Silvian L, Bixler S, Chen LL, Cheung A, et al. Structure of a NEMO/IKK-associating domain reveals architecture of the interaction site. Structure. 2008;16(5):798–808. doi: 10.1016/j.str.2008.02.012. Describes the interaction between NEMO and the catalytic subunit of IKK. [DOI] [PubMed] [Google Scholar]
- 99.Saito K, Kigawa T, Koshiba S, Sato K, Matsuo Y, et al. The CAP-Gly domain of CYLD associates with the proline-rich sequence in NEMO/IKKγ. Structure. 2004;12(9):1719–28. doi: 10.1016/j.str.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 100.Sakurai H, Miyoshi H, Mizukami J, Sugita T. Phosphorylation-dependent activation of TAK1 mitogen-activated protein kinase kinase kinase by TAB1. FEBS Lett. 2000;474(2–3):141–45. doi: 10.1016/s0014-5793(00)01588-x. [DOI] [PubMed] [Google Scholar]
- 101.Sato Y, Yoshikawa A, Yamashita M, Yamagata A, Fukai S. Structural basis for specific recognition of Lys 63–linked polyubiquitin chains by NZF domains of TAB2 and TAB3. EMBO J. 2009;28(24):3903–9. doi: 10.1038/emboj.2009.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sen R, Baltimore D. Inducibility of κ immunoglobulin enhancer-binding protein NF-κB by a posttranslational mechanism. Cell. 1986;47(6):921–28. doi: 10.1016/0092-8674(86)90807-x. [DOI] [PubMed] [Google Scholar]
- 103.Shibuya H, Yamaguchi K, Shirakabe K, Tonegawa A, Gotoh Y, et al. TAB1: an activator of the TAK1 MAPKKK in TGF-β signal transduction. Science. 1996;272(5265):1179–82. doi: 10.1126/science.272.5265.1179. [DOI] [PubMed] [Google Scholar]
- 104.Steward A, McDowell GS, Clarke J. Topology is the principal determinant in the folding of a complex all-α Greek key death domain from human FADD. J Mol Biol. 2009;389(2):425–37. doi: 10.1016/j.jmb.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Suzuki T, Hirai H, Fujisawa J, Fujita T, Yoshida M. A transactivator Tax of human T-cell leukemia virus type 1 binds to NF-κB p50 and serum response factor (SRF) and associates with enhancer DNAs of the NF-κB site and CArG box. Oncogene. 1993;8(9):2391–97. [PubMed] [Google Scholar]
- 106.Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–76. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 107.Tam WF, Lee LH, Davis L, Sen R. Cytoplasmic sequestration of Rel proteins by IκBα requires CRM1-dependent nuclear export. Mol Cell Biol. 2000;20(6):2269–84. doi: 10.1128/mcb.20.6.2269-2284.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tam WF, Sen R. IκB family members function by different mechanisms. J Biol Chem. 2001;276(11):7701–4. doi: 10.1074/jbc.C000916200. [DOI] [PubMed] [Google Scholar]
- 109.Thomas C, Bazan JF, Garcia KC. Structure of the activating IL-1 receptor signaling complex. Nat Struct Mol Biol. 2012;19(4):455–57. doi: 10.1038/nsmb.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Thome M, Schneider P, Hofmann K, Fickenscher H, Meinl E, et al. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature. 1997;386(6624):517–21. doi: 10.1038/386517a0. [DOI] [PubMed] [Google Scholar]
- 111.Toubi E, Shoenfeld Y. Toll-like receptors and their role in the development of autoimmune diseases. Autoimmunity. 2004;37(3):183–88. doi: 10.1080/08916930410001704944. [DOI] [PubMed] [Google Scholar]
- 112.Tsukamoto H, Fukudome K, Takao S, Tsuneyoshi N, Kimoto M. Lipopolysaccharide-binding protein-mediated Toll-like receptor 4 dimerization enables rapid signal transduction against lipopolysaccharide stimulation on membrane-associated CD14-expressing cells. Int Immunol. 2010;22(4):271–80. doi: 10.1093/intimm/dxq005. [DOI] [PubMed] [Google Scholar]
- 113.Valkov E, Stamp A, Dimaio F, Baker D, Verstak B, et al. Crystal structure of Toll-like receptor adaptor MAL/TIRAP reveals the molecular basis for signal transduction and disease protection. Proc Natl Acad Sci USA. 2011;108(36):14879–84. doi: 10.1073/pnas.1104780108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Vigers GP, Anderson LJ, Caffes P, Brandhuber BJ. Crystal structure of the type-I interleukin-1 receptor complexed with interleukin-1β. Nature. 1997;386(6621):190–94. doi: 10.1038/386190a0. [DOI] [PubMed] [Google Scholar]
- 115.Vince JE, Pantaki D, Feltham R, Mace PD, Cordier SM, et al. TRAF2 must bind to cellular inhibitors of apoptosis for tumor necrosis factor (TNF) to efficiently activate NF-κB and to prevent TNF-induced apoptosis. J Biol Chem. 2009;284(51):35906–15. doi: 10.1074/jbc.M109.072256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412(6844):346–51. doi: 10.1038/35085597. Describes the dependence of ubiquitin in NF-κB signaling. [DOI] [PubMed] [Google Scholar]
- 117.Wang D, Zhang S, Li L, Liu X, Mei K, Wang X. Structural insights into the assembly and activation of IL-1β with its receptors. Nat Immunol. 2010;11(10):905–11. doi: 10.1038/ni.1925. [DOI] [PubMed] [Google Scholar]
- 118.Wertz IE, O’Rourke KM, Zhou H, Eby M, Aravind L, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature. 2004;430(7000):694–99. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 119.Woronicz JD, Gao X, Cao Z, Rothe M, Goeddel DV. IκB kinase-β:NF-κB activation and complex formation with IκB kinase-α and NIK. Science. 1997;278(5339):866–69. doi: 10.1126/science.278.5339.866. [DOI] [PubMed] [Google Scholar]
- 120.Wu C-J, Conze DB, Li T, Srinivasula SM, Ashwell JD. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-κB activation [corrected] Nat Cell Biol. 2006;8(4):398–406. doi: 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]
- 121.Wu H, Arron JR. TRAF6, a molecular bridge spanning adaptive immunity, innate immunity and osteoimmunology. Bioessays. 2003;25(11):1096–105. doi: 10.1002/bies.10352. [DOI] [PubMed] [Google Scholar]
- 122.Xu G, Lo Y-C, Li Q, Napolitano G, Wu X, et al. Crystal structure of inhibitor of κB kinase β. Nature. 2011;472(7343):325–30. doi: 10.1038/nature09853. Describes the structure of IKKβ. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xu Y, Tao X, Shen B, Horng T, Medzhitov R, et al. Structural basis for signal transduction by the Toll/interleukin-1 receptor domains. Nature. 2000;408(6808):111–15. doi: 10.1038/35040600. [DOI] [PubMed] [Google Scholar]
- 124.Yamaoka S, Courtois G, Bessia C, Whiteside ST, Weil R, et al. Complementation cloning of NEMO, a component of the IκB kinase complex essential for NF-κB activation. Cell. 1998;93(7):1231–40. doi: 10.1016/s0092-8674(00)81466-x. [DOI] [PubMed] [Google Scholar]
- 125.Ye H, Arron JR, Lamothe B, Cirilli M, Kobayashi T, et al. Distinct molecular mechanism for initiating TRAF6 signalling. Nature. 2002;418(6896):443–47. doi: 10.1038/nature00888. Describes the structure of TRAF6 in complex with receptor peptides, showing the different specificity from TRAF2. [DOI] [PubMed] [Google Scholar]
- 126.Ye H, Park YC, Kreishman M, Kieff E, Wu H. The structural basis for the recognition of diverse receptor sequences by TRAF2. Mol Cell. 1999;4(3):321–30. doi: 10.1016/s1097-2765(00)80334-2. [DOI] [PubMed] [Google Scholar]
- 127.Yin MJ, Christerson LB, Yamamoto Y, Kwak YT, Xu S, et al. HTLV-I Tax protein binds to MEKK1 to stimulate IκB kinase activity and NF-κB activation. Cell. 1998;93(5):875–84. doi: 10.1016/s0092-8674(00)81447-6. [DOI] [PubMed] [Google Scholar]
- 128.Yin Q, Lamothe B, Darnay BG, Wu H. Structural basis for the lack of E2 interaction in the RING domain of TRAF2. Biochemistry. 2009;48(44):10558–67. doi: 10.1021/bi901462e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yin Q, Lin S-C, Lamothe B, Lu M, Lo Y-C, et al. E2 interaction and dimerization in the crystal structure of TRAF6. Nat Struct Mol Biol. 2009;16(6):658–66. doi: 10.1038/nsmb.1605. Describes the dimeric ubiquitin ligase domain of TRAF6 and proposes the model of infinite multimerization of TRAF6 through alternating dimerization and trimerization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yoon S-I, Kurnasov O, Natarajan V, Hong M, Gudkov AV, et al. Structural basis of TLR5-flagellin recognition and signaling. Science. 2012;335(6070):859–64. doi: 10.1126/science.1215584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yoshikawa A, Sato Y, Yamashita M, Mimura H, Yamagata A, Fukai S. Crystal structure of the NEMO ubiquitin-binding domain in complex with Lys 63–linked di-ubiquitin. FEBS Lett. 2009;583(20):3317–22. doi: 10.1016/j.febslet.2009.09.028. [DOI] [PubMed] [Google Scholar]
- 132.Zabel U, Baeuerle PA. Purified human IκB can rapidly dissociate the complex of the NF-κB transcription factor with its cognate DNA. Cell. 1990;61(2):255–65. doi: 10.1016/0092-8674(90)90806-p. [DOI] [PubMed] [Google Scholar]
- 133.Zandi E, Rothwarf DM, Delhase M, Hayakawa M, Karin M. The IκB kinase complex (IKK) contains two kinase subunits, IKKα and IKKβ, necessary for IκB phosphorylation and NF-κB activation. Cell. 1997;91(2):243–52. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]
- 134.Zheng C, Kabaleeswaran V, Wang Y, Cheng G, Wu H. Crystal structures of the TRAF2: cIAP2 and the TRAF1: TRAF2: cIAP2 complexes: affinity, specificity, and regulation. Mol Cell. 2010;38(1):101–13. doi: 10.1016/j.molcel.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]