Preface
New technologies have widened our view of “complex diseases”--diseases with both genetic and environmental risk factors. Here, we explore recent genetic and virologic evidence implicating host-virus interactions in three diseases—type 1 diabetes, inflammatory bowel disease, and asthma. In these examples, the viruses implicated in disease are mucosal infections that affect most of the population and that are asymptomatic or mild in many hosts. These findings place a new emphasis on common viral infections as an important environmental factor in complex disease pathogenesis, and they compel us to pursue a better understanding of host interactions with the human virome.
Introduction
Mucosal surfaces provide portals of entry into the body—for substances essential to survival, such as food, water, and air—and for many pathogens, including viruses. Over evolutionary time, hosts have developed a battery of anti-viral immune mechanisms to combat these attacks. When a virus encounters a mucosal surface, the interplay between viral invasion and host antiviral defense strategies ultimately determines the severity and outcome of the infection.
Like all traits, the host anti-viral response is dictated in part by genetics and can vary with genetic polymorphisms across the population. Similarly, other host proteins important for viral infection, such as cell surface receptors utilized by viruses, can vary across the host population. The significance of these variations may not be clear when studying the pathogenesis of a highly virulent virus in susceptible hosts. However, recent advances in viral detection are beginning to paint a new picture of mucosal viral infections in humans and other hosts—a picture in which viruses are frequently present, often in the absence of a clear-cut disease association. In the context of frequent, less virulent infections, variations in the host anti-viral response and host viral targets may take center stage in determining the physiological impact of a viral infection.
Here, we will discuss how genetic and genomic technologies have widened our view of mucosal virus infections. In contrast to a one pathogen-one disease model, we describe a model of the human virome in which we are nearly continually exposed to viruses, which may or may not cause symptoms. In this context, the virome is an important component of the environment that can interact with host genetic traits to contribute to the pathogenesis of complex diseases. We highlight several recent examples in which interactions between host genetics and viral infections have been implicated in autoimmune and inflammatory diseases.
Advances in viral detection: the expanding virome
PCR and other genetic methods have revolutionized our ability to detect viruses in samples from infected hosts. Since the 1970s, viruses have been routinely detected in clinical and research laboratories using viral culture in combination with various antibody-based techniques1. While these methods can be powerful, they are also labor-intensive, can require multiple days for a result, and can fail to detect many virus types. Since the invention of PCR2, nucleic acid based techniques have been increasingly applied to the detection of viral genetic material within host fluids and tissues. In comparison to viral culture, a PCR-based test is fast, requiring only several hours to complete, and is extremely sensitive. With the development of many reagents and platforms over the past decade, nucleic acid amplification techniques have become widely available and accessible; they are now the primary method for detecting viruses in research and are increasingly being adopted in clinical settings3.
As one might expect, the introduction of sensitive, easily-performed tests has led to a marked increase in virus detection in general. The largest application has been to improve pathogen detection in the setting of symptomatic illness. Classic epidemiologic studies of the 1950s and 1960s sought to identify agents of common respiratory and gastrointestinal infections. These studies were incredibly informative, yet had low virus detection rates by modern standards. For example, the Tecumseh Michigan study from 1965–69 tested 1419 secretions from symptomatic respiratory illnesses and isolated a pathogen from only 25%4. The most frequently detected agent in these studies, human rhinovirus, was isolated in only 140 specimens (10%). In contrast, a 1998 study employing PCR identified pathogens in 69% of adult colds, the majority of which were rhinoviruses (in 50% of illnesses)5. Furthermore, in addition to improving detection of known viruses, genetic methods have helped reveal previously-unknown viral pathogens underlying symptomatic illnesses. A number of disease-associated viruses have been discovered in recent years using techniques such as PCR, viral gene arrays, and most recently, deep pyrosequencing6–9. Deep pyrosequencing technology can read thousands to millions of short sequences in parallel from a single sample, allowing rare sequences to be detected. This powerful approach has been used to discover previously-unidentified pathogens in patient samples, such as a novel picornavirus found in stool from patients with gastroenteritis6,10. New genetic techniques have only just begun to be applied to pathogen discovery, and they hold great promise for further elucidating the members of the human virome.
In addition to improving detection of disease-causing viruses, genetic methods have highlighted the frequent presence of viruses in healthy individuals. For mucosal surfaces, this is well-exemplified by two virus groups within the Picornavirus family: rhinoviruses and gastrointestinal enteroviruses. In cross-sectional studies, rhinoviruses are quite prevalent in the respiratory tract by PCR. Asymptomatic detection rates have averaged 15% across multiple studies, often with higher rates in young children11. Rhinoviruses were already considered prevalent with isolation rates in routine respiratory samples of 2.3% (the Seattle Virus Watch, 1965–69)12; it is now clear that they are present at an additional order of magnitude. Similarly, classic studies employing viral culture found a significant prevalence of gastrointestinal enteroviruses in healthy children13. Recent PCR-based studies extend these results. For example, two large studies detected enterovirus in stool samples from 11% of asymptomatic infants14,15. Further analysis showed that the same enterovirus serotype could persist in sequential stool samples from healthy infants for 2-, 3-, or 4-month periods16. These initial observations may represent the “tip of the iceberg”, since comprehensive surveys of mucosal viruses in asymptomatic subjects have not yet been done.
The significance of a positive PCR test in the absence of symptoms is a subject of current investigation. Longitudinal studies of respiratory viruses suggest that a positive test can be associated with an imminent or antecedent illness17–19. In addition, there is evidence for infections that begin, persist, and terminate in the absence of symptoms. For example, Winther et al. followed 15 children with weekly nasal sampling and documented 8 examples of PCR-positive picornavirus infections that appeared, persisted for 2–3 weeks, and disappeared without any symptom development17. Episodes of asymptomatic respiratory infection were also seen in a molecular epidemiology study in Finland, which tracked family members of rhinovirus positive index children for three weeks with twice weekly nasal sampling20. One hundred percent of siblings and 50% of parents became rhinovirus PCR positive during the study, but many of the parents had asymptomatic infections. Interestingly, family members sometimes became infected with the same strain as the index case, but many became positive for distinct strains, highlighting the great diversity of rhinoviruses circulating in a given season. These observations reveal that viruses often visit our mucosal surfaces “under the radar” of symptoms, and indicate that interactions between viruses and our antiviral immune system may be occurring even when we are unaware of an infection.
In the remainder of this review, we will discuss new evidence that common mucosal virus infections are important players in the pathogenesis of complex disease. Even prior to the current age of gene-based pathogen detection, mucosal viral infections were known to be quite frequent. In the Cleveland Family Study from 1948–57, common respiratory illnesses were documented at average rates of 8 per year for children ages 1–4, 6 per year for children ages 5–9, 5 per year for children ages 10–19, and 3–4 per year for adults21. When surveyed by PCR, picornavirus infections alone can occur in the respiratory tract at rates as high as once per month in young children (on average once every two months)17. If we consider symptomatic plus asymptomatic infections, a view emerges in which viruses are very frequently if not continually present at our mucosal surfaces. In our model, this constant albeit changing barrage of mucosal infections constitutes a virome, which is frequently interacting with host targets and host antiviral responses. This model brings genetic polymorphisms in these host pathways into focus as we consider complex diseases.
In the digestive tract: host genes, viruses, and autoimmune disease
For many years, investigators have sought infectious causes for autoimmune diseases such as Type 1 diabetes. Autoimmune diseases, like infections, involve activation of the immune response and inflammation. However, no straightforward causal relationship between these diseases and particular pathogens has been uncovered. Recent evidence puts this search in a new context. In the last several years, autoimmune diseases have been investigated extensively in genome-wide association studies (GWAS). With a variety of approaches, investigators have sought to learn how the “hits” in GWAS relate to disease pathogenesis. For both Type I Diabetes and inflammatory bowel disease, new evidence links disease development to regions of the host genome involved in host-virus interactions. Here, we present an overview of the relevant findings and refer readers to the included references for more detailed reviews of GWAS results.
Type 1 Diabetes
Type 1 diabetes (T1D) is an autoimmune disease with both genetic and environmental components. At its onset, usually during childhood, the host immune system attacks the insulin-producing β cells of the pancreatic islets. This leads to life-long dependence on exogenous insulin and many long-term health consequences. While there is a tendency for the disease to be inherited in families, identical twins are not 100% concordant22–25. Genetic loci linked to Type 1 diabetes have been identified through various methods, most recently through GWAS.
Studies employing genome-wide association methods have confirmed some previously-known T1D associations and revealed new ones26. For example, GWAS confirmed the association between T1D and polymorphisms in the HLA-DR, -DQ region on chromosome 6. This region encodes MHC Class II molecules, which present antigenic peptides to CD4+ T cells. This association fits with a large body of evidence that adaptive immune responses contribute to T1D pathogenesis27. GWAS have also revealed new associations, often in genes that affect immune function. In 2006, Todd and colleagues found a significant association between T1D and single-nucleotide polymorphisms (SNPs) in the IFI1H gene28. IFI1H, also known as MDA5, is a cytoplasmic helicase that acts as an innate immune sensor of virus infection29. MDA-5 is one of the RIG-I-like receptors (RLRs) expressed in the cytosol. Upon sensing viral RNA, activated MDA-5 signals the cell to produce Type I interferons (IFN), initiating the anti-viral immune response. Interestingly, mda5 alleles present in the general population are associated with disease, whereas rare variants that result in loss of function or reduced expression confer disease protection30–32. Consistent with these findings, a large population-based study showed that MDA5 is expressed at higher levels in leukocytes from individuals with TID-susceptible alleles than those with resistant alleles33. These results suggest a link between a robust anti-viral immune response and T1D pathogenesis.
Recently, an innovative genomic analysis took GWAS data further to reveal a more extensive association between T1D and the antiviral immune response. In interpreting GWAS studies for complex diseases, a major limitation is the low strength of association between individual polymorphisms and the disease state. To overcome this, investigators looked for networks of genes co-regulated by transcription factors, then looked for associations between an identified gene network and GWAS data.
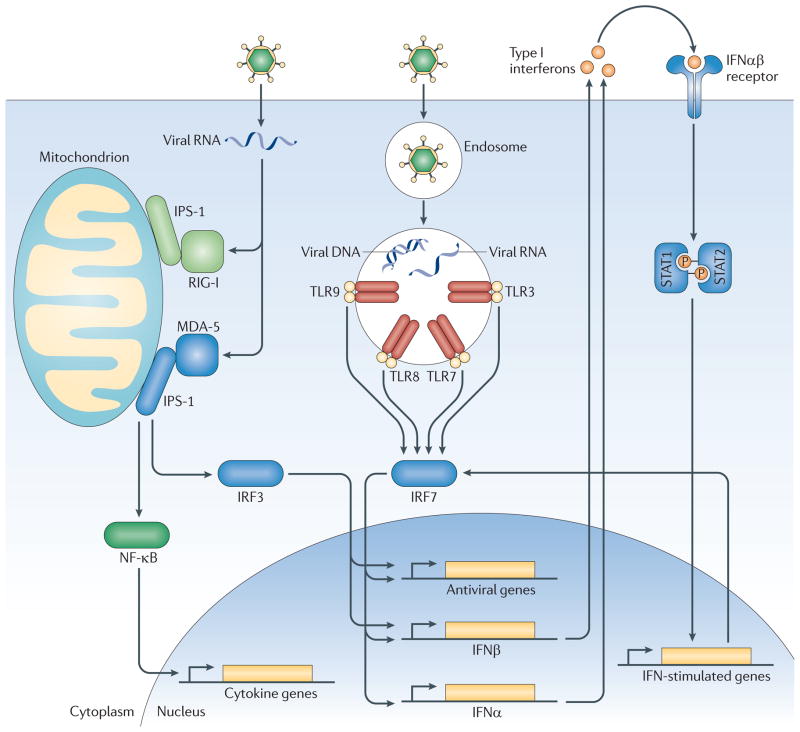
Heinig et al. focused on a gene expression network driven by the immune system transcription factor, IRF-7 (Ref 34). Using expression data from inbred rat strains, a network of genes co-regulated by IRF-7 was found. IRF-7 is a master regulator of Type I interferon genes35. In plasmacytoid dendritic cells, IRF-7 is constitutively expressed and transmits the signal from endosomal toll-like receptors (TLR)-7 and TLR-9 to the nucleus, to mediate transcription of type I IFN genes. In most other cell types, IRF-7 expression is induced by IFNαβ receptor signaling. By forming a homodimer or heterodimer with IRF-3, IRF-7 triggers transcription of IFN genes. Innate recognition of viral nucleic acids by endosomal TLRs and cytosolic RLRs and DNA sensors all results in the activation of type I IFN genes. Secreted IFNs, in turn, bind IFNαβ receptor on cell surfaces and can induce the expression of ~300 IFN-stimulated genes, some of which have antiviral functions. Having identified a genomic locus that regulates the expression of the IRF-7-driven network in rats, the investigators sought to translate these findings to human disease by using human gene expression data from two large studies. The investigators compared SNPs close to the IRF-driven network genes with T1D GWAS “hits”. Interestingly, a significant association was found. Furthermore, they examined the human orthologue of the locus that regulates IRF-7 network expression in rats. This region in humans, 13q23, showed a significant association with T1D risk. In addition, they studied how SNPs in this region would be expected to modulate IRF-7 network expression levels. Of note, SNPs associated with T1D were predicted to increase expression of the IRF-7 driven network. These results demonstrate a link between T1D and not only IFI1H/mda-5, but an entire network of IFN response genes (Figure 1).
Figure 1. Components of the anti-viral immune response genetically linked to Type I Diabetes.
Viruses are recognized in general by two separate pathways. Intrinsic recognition occurs through detection of viral nucleic acids by cytosolic RLRs and other nucleic acid sensors in the infected cells. MDA-5/IFI1H is a cytosolic RLR that can recognize Picornavirus genomes. Activated MDA-5 activates the transcription factors IRF-3 and NFκB. In contrast, extrinsic recognition of virus occurs through Toll-like receptors (TLR) 3, 7, 8 and 9, which can recognize viral DNA and RNA within endosomes and activate transcription factors IRF-7 and NF-κb. IRF-3 and IRF-7 function in homo- or heterodimers to initiate transcription of Type I interferons and other anti-viral genes, as shown. Via NF-κB, both pathways induce the expression of pro-inflammatory cytokines. Secreted Type I interferons bind to the IFNαβR on the cell surface, which signals via STAT1 and STAT2 to induce expression of ~300 interferon-stimulated genes (ISGs) including MDA-5 and IRF-7. Type 1 diabetes is linked to genetic polymorphisms in MDA-5/IFI1H, IRF-7, and an IRF-7 driven network of 305 genes (likely the depicted network of IFN response genes.) Interestingly, T1D is predicted to correlate with robust responses through these pathways. For MDA-5, polymorphisms that result greater expression correlate with disease, whereas rare alleles that result in loss of or reduced function are protective. Similarly, T1D-associated polymorphisms in the genomic locus regulating the IRF-7 driven network predict increased expression of this network in T1D.
If T1D development does involve the host anti-viral response, which virus(es) are inducing this response? While many pathogens have been investigated, recent studies of T1D risk have focused on gastrointestinal enteroviruses such as coxsackie B virus. A variety of observations have implicated enteroviruses in T1D. In 1979, investigators isolated a coxsackie B4 virus from the post-mortem pancreas of a diabetic child. Re-inoculation of this virus into mice led to pancreatic islet inflammation with β cell death36. Subsequently, other enteroviruses and related viruses have been shown to induce diabetes in model hosts37. Epidemiological studies have also implicated enteroviruses, particularly recent longitudinal studies showing that a positive enterovirus PCR test in the blood correlates with subsequent development of T1D in high-risk individuals37–39. Additional evidence includes post-mortem exams showing enteroviruses in the islets of diabetic but not control pancreases and in vitro studies of β cells showing enterovirus tropism and destructive effects37,38. One of the most intriguing clues invoking enteroviruses in T1D is the genetic evidence of a role for MDA-5, as discussed above. MDA5 specifically senses members of the picornavirus family (which includes enteroviruses), whereas RIG-I detects other virus families40. Despite these numerous associations, the role of enteroviruses in TID pathogenesis has not yet been clearly defined.
Many pathogenic mechanisms are possible. One scenario would be the infection of β cells by an enterovirus, followed by β cell destruction—either due to the virus itself, due to immune responses that kill virus-infected cells, or due to “autoimmune” sequelae of such an infection. Detailed discussions of possible mechanisms have been published recently27,37,38,41. Whatever the mechanism, in this scenario the host immune system genotype is likely to be critical. In mouse models, a “diabetogenic” picornavirus, the EMCV M variant, causes β cell destruction and diabetes, but only in some genetic backgrounds42,43. Also, enterovirus infections are quite common and usually do not lead to diabetes, invoking additional host (or viral) genetic factors. In addition, it is noteworthy that in a typical infectious disease, the infectious agent is more pathogenic in the setting of a diminished immune response (eg., immunosuppression.) However, the genetic studies described above predict that a robust anti-viral response correlates with the predisposition for T1D. This makes sense if the host immune response to the inciting virus is more pathogenic than the virus itself. Consistent with this idea, mice genetically engineered to produce IFN alpha in pancreatic β cells develop islet inflammation and diabetes44. Thus, type 1 diabetes may yet turn out to be a virus-induced disease, in the setting of a detrimental host response dictated partly by genotype, and partly by other environmental factors (for example, the commensal microbiota45). Clearly, more investigation will be required to delineate the role of viruses in T1D. At this time, we can only state that converging evidence implicates a frequently-encountered mucosal infection (in this case, enterovirus) as a key environmental factor in TID pathogenesis.
Crohn’s disease
As in Type I diabetes, genetic studies have offered many clues to the pathogenesis of Crohn’s disease (CD), a major subtype of inflammatory bowel disease. Crohn’s disease usually presents in young adults, with a second, smaller peak of onset in older adults, and involves transmural inflammation of the intestinal wall46. Inflammation usually occurs in the bacteria-dense distal ileum and colon, although multiple discontinuous regions of the intestine can be affected. Patients typically experience episodic symptoms including fever, abdominal pain, and diarrhea, and may also experience more serious gastrointestinal problems. CD risk factors are both genetic and environmental, with monozygotic twin concordance rates around 60%47,48. In the late 1990s, genome-wide studies revealed links between several chromosomal regions and Crohn’s disease49. The first CD gene identified in one of these regions was NOD-2, which encodes an innate immune receptor that recognizes bacterial cell wall components50,51. Subsequently, genetic studies have implicated a number of genes in CD, most of which have roles in immunological pathways or epithelial barrier functions. These findings fit with a current hypothesis that CD pathogenesis involves disruption of the normal homeostatic interactions between the host and the intestinal bacterial microbiome. In addition, several studies have linked CD to members of the autophagy pathway, ATG16L1 and IRGM54–57. Autophagy is an ancient cellular process important for removing unwanted debris from the cytosol, and may impact CD in a number of ways. During autophagy, a double membrane vesicle surrounds cytosolic waste or in some cases pathogens. This autophagosome ultimately fuses with a lysosome leading to digestion and recycling of the vesicle contents. In immune defense, autophagy can be important for both pathogen removal and pathogen recognition58–60. The role of autophagy in CD pathogenesis is an area of active investigation.
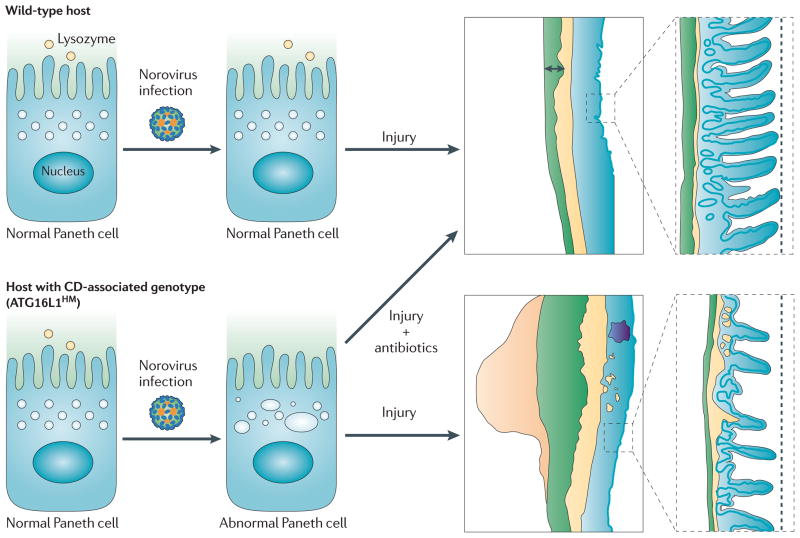
Recent studies of the autophagy protein ATG16L1 reveal two distinct roles, either or both of which may be important in CD. First, ATG16L1 can interact with innate immune receptors NOD-1 and NOD-2 to mediate autophagy of bacteria61,62. Second, ATG16L1 appears to play a role in the biology of Paneth cells. Paneth cells are specialized secretory cells within the intestinal crypts that release antimicrobial compounds and other substances that may affect the gut microbiota. One group studied the function of ATG16L1 by creating gene-trap mutant mice that express reduced levels of ATG16L1 (ATG16L1 “hypomorphs”)63. In these mice, Paneth cells display structural abnormalities in their secretory granules and an altered gene expression profile. This model likely parallels human biology, since the histological abnormalities observed mouse Paneth cells were also observed in intestinal biopsies from CD patients with the disease-associated ATG16L1 allele. Interestingly, further exploration of this Paneth cell phenotype uncovered an unexpected role for viruses in CD pathogenesis. In a follow-up study, the same group described how intestinal noroviruses are an essential ingredient for the abnormal Paneth cell phenotype of ATG16L1 hypomorphic mice64. In this study, Virgin and colleagues re-derived ATG16L1 hypomorphic mice in an enhanced barrier facility to better understand the contributions of microbes in this disease model. Surprisingly, when they analyzed these hosts, the Paneth cells were indistinguishable from those of wild type mice. These investigators had previously identified murine norovirus from their conventional mouse facility65. Therefore, they tested whether norovirus infection had any effect on enhanced barrier facility-derived hosts. Strikingly, norovirus infection restored the Paneth cell phenotype that these investigators had documented previously in conventionally-raised mice. This result was strain specific; a non-persisting norovirus strain did not induce the disease phenotype, but a persistent virus strain did. To further model Crohn’s disease, mice were exposed to -dextran sulphate sodium (DSS), an intestinal injury agent that produces an inflammatory bowel disease-like phenotype when ingested66. Virus-infected (but not uninfected) ATG16L1 hypomorphic mice displayed an abnormal response to intestinal injury, with pathology resembling CD (Figure 2). This phenotype was virus-strain specific and also required intestinal bacteria, as antibiotic-treated mice were protected. The mechanism whereby norovirus infection influences Paneth cell biology awaits further study; the ATG16L1 mutation did not result in increased viral titers and the virus was not detected within Paneth cells themselves. Nonetheless, these results provide a fascinating experimental demonstration of how a viral infection can profoundly influence the expression of a complex disease. It is noteworthy that intestinal noroviruses are common pathogens, which have frequently been found in mouse colonies worldwide and are not symptomatic in wild-type hosts65,67,68. These results prompt us to ask whether virus-host interactions are covertly influencing other disease phenotypes in model organisms raised in conventional facilities. Similarly, these results urge us to search for unrecognized effects of common viral infections in the pathogenesis of CD and other complex human diseases.
Figure 2. Inflammatory bowel disease requires multiple “hits”, including norovirus infection.
A mouse model of Crohn’s disease (CD) requires both a genetic predisposition and a viral infection for the expression of the disease phenotype. In this model, Virgin and colleagues compared intestinal physiology in wild-type hosts and in hosts with reduced expression of a Crohn’s disease-associated autophagy gene, ATG16L1. Wild-type hosts display the predicted intestinal histology during recovery from an intestinal injury. Similarly, hosts deficient in ATG16L1 have normal-appearing intestinal Paneth cells and the usual response to intestinal injury if norovirus infection is absent. However, if ATG16L1-mutant hosts are infected with a persistent strain of norovirus, they develop abnormal Paneth cells and have an abnormal response to intestinal injury with histopathology resembling Crohn’s disease. As illustrated, the injured colon in these mice colon displays lymphoid aggregates, abnormal thickening of the muscularis propria, and submucosa inflammation. In parallel, the intestinal injury agent induced abnormal histology in the ileum of mice with the predisposing gene + norovirus infection, but not in wild-type norovirus infected mice. Of note, CD-like intestinal histopathology does not develop if genetically-predisposed, virus infected mice are treated with antibiotics, indicating that the intestinal microbiota is also an important environmental factor for disease expression.
In the respiratory tract: respiratory viruses and asthma
Asthma is an important inflammatory disease of the lower airways. Although both genetic and environmental risk factors have been defined, many mysteries remain about asthma pathogenesis. Worldwide, asthma affects approximately 300 million people, including >10% of the population in the U.S. and the U.K.69. The disease is usually characterized by asymptomatic periods punctuated with episodes of wheezing and respiratory distress (“exacerbations”) accompanying acute lung inflammation. Over the long term, findings can include chronic inflammation and changes in lung structure and function. Asthma often occurs in association with allergies, and classic studies in both patients and animal models have demonstrated a role for allergic inflammation in asthma pathogenesis70–72. However, asthma is increasingly recognized to be a heterogeneous disorder, since allergic mechanisms do not explain many findings in human patients73.
An intriguing development in asthma research has been the growing evidence linking asthma to respiratory viruses, especially human rhinovirus. Inhaled allergens can trigger asthma exacerbations, and this phenomenon has been well studied in model organisms. However, in the 1990s, longitudinal studies showed that most exacerbations in asthma patients are associated with respiratory infections (80–85%)74,75. Multiple studies since that time have confirmed these findings, using PCR-based virus detection methods76. While a variety of different respiratory infections have been associated with asthma exacerbations, the most frequent pathogen found is human rhinovirus, representing 60% or more of those pathogens identified74–78. Recent studies have suggested that the rhinovirus-asthma association may be even stronger: for example, Bizzintino et al. detected rhinovirus in nearly 90% of children with severe asthma exacerbations using sensitive detection techniques79. Interestingly, the results of this study and others suggest that the newly-discovered rhinovirus group C may be particularly important in asthma, although additional studies will be needed to clarify the roles of rhinovirus subtypes80. . In addition to viral detection studies, the epidemiology of childhood asthma implicates rhinoviruses: the timing of childhood asthma hospitalizations closely mirrors the timing of the annual rhinovirus epidemic, peaking dramatically 2 weeks after school return each year81,82. Further evidence for the role of viruses in asthma stems from a recent studies on cohorts of high-risk children. In the Wisconsin COAST study83, 259 children were followed from birth and analyzed for many asthma risk factors, including episodes of severe respiratory infection with wheezing. Children who had a severe rhinovirus infection prior to the age of three developed asthma by age six in almost 90% of cases, representing an odds ratio of 25.6. In contrast, the odds ratio for developing asthma after early aeroallergen sensitization was only 3.4. In another recent cohort study, Kusel et al. also found a positive association between early rhinovirus wheezing illness and subsequent development of asthma84. The mounting evidence for a rhinovirus-asthma association suggests that it is an important clue to asthma pathogenesis.
Several studies have sought to determine whether asthmatic subjects have increased susceptibility to rhinovirus infections. The Wisconsin COAST results raise three possibilities: (1) that asthmatics (or those with the predisposition to asthma) have enhanced susceptibility to rhinovirus, (2) that rhinovirus infections affect the lung and contribute to asthma development, or (3) that both processes occur, possibly synergistically85–87. A longitudinal study of domestic partners showed that both asthmatics and non-asthmatics encountered rhinovirus at the same rate, but that asthmatics had more prolonged and severe symptoms88. In addition, experimental rhinovirus infections induce more lower airway symptoms in asthmatic than control subjects89,90. Also, ex vivo studies have demonstrated increased virus replication and diminished rhinovirus-induced interferon responses in airway epithelial cells from asthmatic vs. control subjects90–93, consistent with diminished interferon responses in asthma patients in vivo90,94. These results support the idea that asthma patients have an altered response to rhinovirus infection with more virus-induced lower airway pathology. However, these observations do not address whether this response is a consequence of the disease or part of the underlying cause.
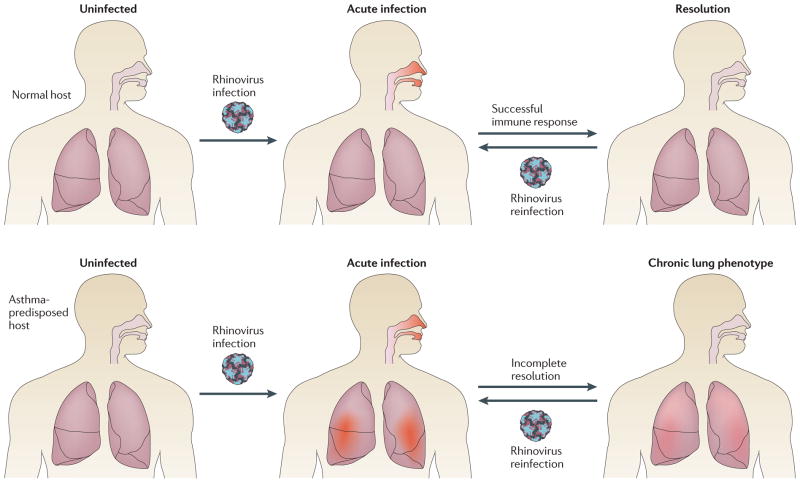
If asthma patients have an abnormal response to rhinovirus, it is quite likely that this aberrant host-virus interaction is central to asthma pathogenesis. Almost all children encounter rhinovirus in early childhood, with seroconversion rates of 90% by the age of two years95. As described above, childhood infections occur at least every few months. Furthermore, new rhinovirus epidemics occur every year, likely due to great virus diversity and continual emergence of new viral variants96–98. With this perpetual exposure to virus, it is easy to imagine how an individual with a deleterious response to rhinovirus infection could sustain acute and/or chronic lung injury over time (Figure 3). Since many features of asthma originate early in life99, sequelae of childhood infections could be particularly important. In the example of the Crohn’s disease model above, it would be incorrect to consider norovirus infection the “cause” of this multifactorial disease, but it would also be incorrect to consider norovirus an exacerbating factor in an established disease. Norovirus is an essential contributor to disease pathogenesis, along with other genetic and environmental factors. In an analogous fashion, we propose that rhinovirus infections are fundamental for the expression of asthma, at least in many patients with this heterogeneous disease. In fact, it would be surprising if rhinovirus-host interactions did not contribute to asthma pathogenesis if asthma patients truly have an enhanced inflammatory response to this very common infection.
Figure 3. Possible role of rhinovirus in asthma pathogenesis.
PCR-based studies have revealed a high incidence and prevalence of rhinovirus respiratory infections. At the same time, rhinovirus has been identified as a major precipitant of asthma exacerbations, and growing evidence indicates that asthmatic subjects develop more severe symptoms upon rhinovirus infection than healthy controls. This diagram illustrates how genetic polymorphisms affecting the host rhinovirus response could contribute to asthma pathogenesis. In this simplified scenario, all hosts experience frequent, repeated rhinovirus infections throughout their lifetimes (at least once every few months as children.) These infections may be asymptomatic, result in mild symptoms of the common cold, or result in upper and lower airway inflammation with wheezing and respiratory distress. In normal hosts, rhinovirus infections affect only the upper airway, and are mild or asymptomatic. Each infection fully resolves and leaves airways intact. In asthma-predisposed hosts, genetic polymorphisms in the antiviral immune response result in more prolonged and severe rhinovirus infections, with both upper and lower airway inflammation. In addition to acute episodes, repeated more severe infections lead to chronic inflammation and airway remodeling over time. This example illustrates how a common and frequent viral infection, in the setting of a detrimental host response, could contribute to the pathogenesis of asthma.
Thus far, patient observations provide the main evidence for an abnormal rhinovirus response in asthma. Is there any genetic evidence linking asthma to variations in the host antiviral response, as in type 1 diabetes? At this point, there are many possibilities. A number of genetic polymorphisms have been identified in asthma GWAS, and many of these occur in immunological pathways that could affect host-virus interactions (eg., polymorphisms in toll-like receptors)86,100,101. Recent studies using siRNA knockdown have implicated several innate immune pathways in host-rhinovirus interactions, including the TLR3 and RIG-I-like receptor pathways102,103 It will be interesting to see whether genetic polymorphisms that affect rhinovirus-triggered immune pathways influence rhinovirus susceptibility and/or correlate with asthma. Another intriguing finding is the observation that severe rhinovirus-induced symptoms correlate, to some extent, with aeroallergen sensitization98,104,105. One possible explanation is that rhinovirus lower respiratory infection promotes aeroallergen sensitization, or vice versa. However, an alternate explanation is that a common genetic polymorphism predisposes the host to both phenotypes. For example, a toll-like receptor polymorphism could conceivably bias the immune system towards allergic sensitization and also promote enhanced rhinovirus-induced inflammation. 98. While many questions in asthma pathogenesis remain, a growing body of evidence indicates that host-virus interactions will be an important part of the story. In addition to genetic and clinical studies, studies in model organisms have great potential to help elucidate the role of respiratory infections in asthma, such as a recent study paper showing an asthma-like phenotype in mice following viral infection106. The recent developments in asthma research illustrate how mainstream views of complex disease are being altered by a growing awareness of the role of mucosal viral infections.
Chronic infections and endogenous viruses: additional members of the virome
Although an extensive discussion is beyond the scope of this review, it is important to mention that the human virome encompasses more than frequent mucosal infections. Genetic techniques have demonstrated a high burden of chronic infections in the human population, many of which have unknown consequences, as recently reviewed by Virgin et al.107 Furthermore, genome sequencing has revealed multiple endogenous viruses in animal genomes, reflecting a “historical record” of viral infections with gene integration over evolutionary time108–110. In humans, endogenous retroviruses comprise approximately 8% of the genome108. Interestingly, while endogenous human viruses are generally defective in that they cannot reactivate to form complete viral particles, viral genes can be expressed at the RNA and even protein levels111. This leaves open questions about whether and to what extent endogenous viruses can activate the anti-viral immune response or interact with host targets, as suggested by recent studies112–114. Recent findings about chronic infections and endogenous viruses further demonstrate how genetic and genomic techniques have expanded our knowledge of the human virome, and they focus our attention on how host variation in the anti-viral immune response may influence complex disease pathogenesis.
Host-virome interactions in homeostasis and disease
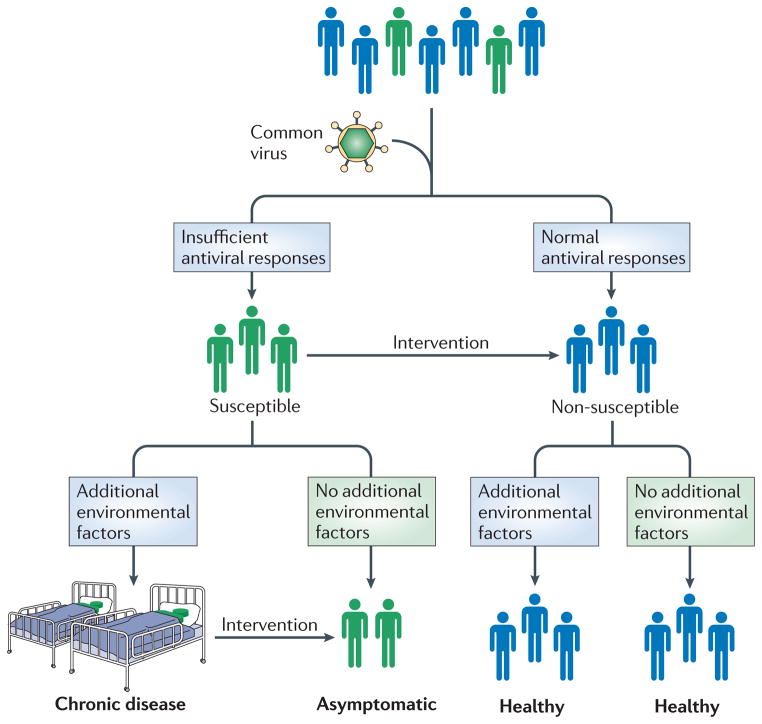
In a traditional view, mucosal virus infections are a deviation from host homeostasis, leading to disease followed by resolution of disease. However, emerging evidence suggests a different view, in which virus infections comprise a more constant feature of our environment. In contrast to studies of the human bacterial microbiome, relatively few studies to date have examined the presence of mucosal viruses in healthy subjects using modern techniques. However, those studies that have been done have found prevalent viral infections in the respiratory and gastrointestinal tract, even in the absence of symptoms. These results suggest a model in which mucosal infections are not a deviation, but rather “the norm”, comprising a frequent or even constant-albeit-changing virome. In this context, genetic variations in host viral targets and antiviral responses have the potential to dramatically affect host physiology, in some cases leading to complex disease (Figure 4). This model compels us to further investigate the nature and composition of the human mucosal virome and to identify the host proteins that interact with prevalent mucosal infections. Although projects have been proposed115, no study has yet characterized the virome of human pathogens in a comprehensive and systematic way. More detailed information about the human mucosal virome, particularly in healthy subjects, would be very relevant to the issues discussed here and hopefully such studies will be pursued as technology advances.
Figure 4. A model for the role of common infections in complex disease.
Converging evidence suggests that variations in the host antiviral response are important features of complex disease pathogenesis, in the context of a constant-albeit-changing virome of common infections. In this model, all hosts are exposed to certain highly prevalent viruses. Those with a non-susceptible antiviral response do not get the disease. Those with an altered antiviral response get the disease, but only if certain other key environmental factors are present. Avoiding these key environmental factors offers one avenue for circumventing complex diseases. This model also suggests a new mode for intervention in complex disease pathogenesis. If we understood the mechanisms whereby the altered antiviral response led to disease, we could potentially design interventions to shift this response to that of a non-predisposed host. Such interventions might include vaccines, well-timed virus exposures, or other immune modulating therapeutics. This model suggests that to tackle complex diseases, it is critical to learn more about the human virome and factors that influence host-virome interactions.
If we understood the mechanisms whereby host-virus interactions contribute to complex disease, we might be able to develop new ways of tackling these diseases. Autoimmune and inflammatory diseases, such as those discussed here, are increasing in prevalence in Western countries116–118. Understanding the role of the virome may help elucidate the reason for this epidemiological trend and indicate ways to intervene. Furthermore, if we could identify an altered host-virus interaction that promotes complex disease pathogenesis, it might be possible to design therapies to convert that interaction to the response of non-susceptible individuals, thereby protecting susceptible subjects (Figure 4). Future investigations of host-virome interactions hold great promise for providing new approaches to complex disease.
References
- 1.Leland DS, Ginocchio CC. Role of cell culture for virus detection in the age of technology. Clin Microbiol Rev. 2007;20:49–78. doi: 10.1128/CMR.00002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mullis KB, Faloona FA. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Meth Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- 3.Mackay IM, Mackay JF, Nissen MD, Sloots TP. In: Real-time PCR in Microbiology: From Diagnosis to Characterisation. Mackay IM, editor. Caister Academic Press; Norfolk, U. K: 2007. pp. 1–40. [Google Scholar]
- 4.Monto AS, Cavallaro JJ. The Tecumseh study of respiratory illness. II Patterns of occurrence of infection with respiratory pathogens, 1965–1969. Am J Epidemiol. 1971;94:280–289. doi: 10.1093/oxfordjournals.aje.a121321. [DOI] [PubMed] [Google Scholar]
- 5.Makela MJ, et al. Viruses and bacteria in the etiology of the common cold. J Clin Microbiol. 1998;36:539–542. doi: 10.1128/jcm.36.2.539-542.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiu C, Miller S. Microarrays and deep sequencing in clinical microbiology. Microbe. 2011;6:13–20. This review summarizes the recent technological advances in viral detection and describes examples of their use. [Google Scholar]
- 7.Wang D, et al. Viral discovery and sequence recovery using DNA microarrays. PLoS Biol. 2003;1:e2. doi: 10.1371/journal.pbio.0000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kahn J. Newly discovered respiratory viruses: significance and implications. Curr Opin Pharmacol. 2007;7:478–483. doi: 10.1016/j.coph.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee WM, et al. A diverse group of previously unrecognized human rhinoviruses are common causes of respiratory illnesses in infants. PLoS ONE. 2007;2:e966. doi: 10.1371/journal.pone.0000966. This paper describes how genetic approaches facilitated the discovery of a new group of rhinoviruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greninger AL, et al. The complete genome of klassevirus - a novel picornavirus in pediatric stool. Virol J. 2009;6:82. doi: 10.1186/1743-422X-6-82. This paper provides an example of how deep pyrosequencing can enable the detection of previously unknown pathogens — in this case, a virus in children with gastroenteritis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jartti T, Jartti L, Peltola V, Waris M, Ruuskanen O. Identification of respiratory viruses in asymptomatic subjects: asymptomatic respiratory viral infections. Pediatr Infect Dis J. 2008;27:1103–1107. doi: 10.1097/INF.0b013e31817e695d. [DOI] [PubMed] [Google Scholar]
- 12.Fox JP, Cooney MK, Hall CE. The Seattle Virus Watch. V Epidemiologic observations of rhinovirus infections, 1965–1969, in families with young children. Am J Epidemiol. 1975;101:122–143. doi: 10.1093/oxfordjournals.aje.a112078. [DOI] [PubMed] [Google Scholar]
- 13.Gelfand HM, Holguin AH, Marchetti GE, Feorino PM. A Continuing surveillance of enterovirus infections in healthy children in six United States Cities. i viruses isolated during 1960 and 1961. Am J Hyg. 1963;78:358–375. doi: 10.1093/oxfordjournals.aje.a120355. [DOI] [PubMed] [Google Scholar]
- 14.Cinek O, et al. Longitudinal observation of enterovirus and adenovirus in stool samples from Norwegian infants with the highest genetic risk of type 1 diabetes. J Clin Virol. 2006;35:33–40. doi: 10.1016/j.jcv.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 15.Witso E, et al. Predictors of sub-clinical enterovirus infections in infants: a prospective cohort study. Int J Epidemiol. 2010;39:459–468. doi: 10.1093/ije/dyp333. [DOI] [PubMed] [Google Scholar]
- 16.Witso E, et al. High prevalence of human enterovirus a infections in natural circulation of human enteroviruses. J Clin Microbiol. 2006;44:4095–4100. doi: 10.1128/JCM.00653-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winther B, Hayden FG, Hendley JO. Picornavirus infections in children diagnosed by RT-PCR during longitudinal surveillance with weekly sampling: association with symptomatic illness and effect of season. J Med Virol. 2006;78:644–650. doi: 10.1002/jmv.20588. This longitudinal study follows healthy children over three seasons and sheds light on the importance of the positive PCR tests that have been observed in cross-sectional studies in asymptomatic subjects. [DOI] [PubMed] [Google Scholar]
- 18.Wright PF, et al. Comparison of results of detection of rhinovirus by PCR and viral culture in human nasal wash specimens from subjects with and without clinical symptoms of respiratory illness. J Clin Microbiol. 2007;45:2126–2129. doi: 10.1128/JCM.02553-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jartti T, Lehtinen P, Vuorinen T, Koskenvuo M, Ruuskanen O. Persistence of rhinovirus and enterovirus RNA after acute respiratory illness in children. J Med Virol. 2004;72:695–699. doi: 10.1002/jmv.20027. [DOI] [PubMed] [Google Scholar]
- 20.Peltola V, et al. Rhinovirus transmission within families with children: incidence of symptomatic and asymptomatic infections. J Infect Dis. 2008;197:382–389. doi: 10.1086/525542. This paper combines longitudinal surveillance with PCR-based testing and viral genotyping to reveal the high prevalence and spread of rhinoviruses within families. [DOI] [PubMed] [Google Scholar]
- 21.Monto AS. Studies of the community and family: acute respiratory illness and infection. Epidemiol Rev. 1994;16:351–373. doi: 10.1093/oxfordjournals.epirev.a036158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barnett AH, Eff C, Leslie RD, Pyke DA. Diabetes in identical twins. A study of 200 pairs. Diabetologia. 1981;20:87–93. doi: 10.1007/BF00262007. [DOI] [PubMed] [Google Scholar]
- 23.Hyttinen V, Kaprio J, Kinnunen L, Koskenvuo M, Tuomilehto J. Genetic liability of type 1 diabetes and the onset age among 22, 650 young Finnish twin pairs: a nationwide follow-up study. Diabetes. 2003;52:1052–1055. doi: 10.2337/diabetes.52.4.1052. [DOI] [PubMed] [Google Scholar]
- 24.Redondo MJ, et al. Heterogeneity of type I diabetes: analysis of monozygotic twins in Great Britain and the United States. Diabetologia. 2001;44:354–362. doi: 10.1007/s001250051626. [DOI] [PubMed] [Google Scholar]
- 25.Redondo MJ, Jeffrey J, Fain PR, Eisenbarth GS, Orban T. Concordance for islet autoimmunity among monozygotic twins. N Engl J Med. 2008;359:2849–2850. doi: 10.1056/NEJMc0805398. [DOI] [PubMed] [Google Scholar]
- 26.Concannon P, Rich SS, Nepom GT. Genetics of type IA diabetes. N Engl J Med. 2009;360:1646–1654. doi: 10.1056/NEJMra0808284. [DOI] [PubMed] [Google Scholar]
- 27.Lehuen A, Diana J, Zaccone P, Cooke A. Immune cell crosstalk in type 1 diabetes. Nature Rev Immunol. 2010;10:501–513. doi: 10.1038/nri2787. [DOI] [PubMed] [Google Scholar]
- 28.Smyth DJ, et al. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nature Genet. 2006;38:617–619. doi: 10.1038/ng1800. [DOI] [PubMed] [Google Scholar]
- 29.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 30.Nejentsev S, Walker N, Riches D, Egholm M, Todd JA. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324:387–389. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shigemoto T, et al. Identification of loss of function mutations in human genes encoding RIG-I and MDA5: implications for resistance to type I diabetes. J Biol Chem. 2009;284:13348–13354. doi: 10.1074/jbc.M809449200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Downes K, et al. Reduced expression of IFIH1 is protective for type 1 diabetes. PLoS ONE. 2010;5:e12646. doi: 10.1371/journal.pone.0012646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu S, et al. IFIH1 polymorphisms are significantly associated with type 1 diabetes and IFIH1 gene expression in peripheral blood mononuclear cells. Hum Mol Genet. 2009;18:358–365. doi: 10.1093/hmg/ddn342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heinig M, et al. A trans-acting locus regulates an anti-viral expression network and type 1 diabetes risk. Nature. 2010;467:460–464. doi: 10.1038/nature09386. This study combines GWA study data and gene expression analysis to reveal a link between T1D and a network of co-regulated immune system genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Honda K, Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nature Rev Immunol. 2006;6:644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- 36.Yoon JW, Austin M, Onodera T, Notkins AL. Isolation of a virus from the pancreas of a child with diabetic ketoacidosis. N Engl J Med. 1979;300:1173–1179. doi: 10.1056/NEJM197905243002102. [DOI] [PubMed] [Google Scholar]
- 37.Jaïdane H, et al. Enteroviruses and type 1 diabetes: towards a better understanding of the relationship. Rev Med Virol. 2010;20:265–280. doi: 10.1002/rmv.647. [DOI] [PubMed] [Google Scholar]
- 38.Tauriainen S, Oikarinen S, Oikarinen M, Hyöty H. Enteroviruses in the pathogenesis of type 1 diabetes. Semin Immunopathol. 2010;33:45–55. doi: 10.1007/s00281-010-0207-y. [DOI] [PubMed] [Google Scholar]
- 39.Stene LC, et al. Enterovirus Infection and progression from islet autoimmunity to type 1 diabetes: the diabetes and autoimmunity study in the young (DAISY) Diabetes. 2010;59:3174–3180. doi: 10.2337/db10-0866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 41.von Herrath M. A virus-gene collaboration. Nature. 2009;459:518–519. doi: 10.1038/459518a. [DOI] [PubMed] [Google Scholar]
- 42.Ross ME, Onodera T, Brown KS, Notkins AL. Virus-induced diabetes mellitus. IV Genetic and environmental factors influencing the development of diabetes after infection with the M variant of encephalomyocarditis virus. Diabetes. 1976;25:190–197. doi: 10.2337/diab.25.3.190. [DOI] [PubMed] [Google Scholar]
- 43.Kruppenbacher JP, Mertens T, Muntefering H, Eggers HJ. Encephalomyocarditis virus and diabetes mellitus: studies on virus mutants in susceptible and non-susceptible mice. J Gen Virol. 1985;66:727–732. doi: 10.1099/0022-1317-66-4-727. [DOI] [PubMed] [Google Scholar]
- 44.Stewart TA, et al. Induction of type I diabetes by interferon-α in transgenic mice. Science. 1993;260:1942–1946. doi: 10.1126/science.8100367. [DOI] [PubMed] [Google Scholar]
- 45.Chervonsky AV. Influence of microbial environment on autoimmunity. Nature Immunol. 2010;11:28–35. doi: 10.1038/ni.1801. [DOI] [PubMed] [Google Scholar]
- 46.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 47.Tysk C, Lindberg E, Jarnerot G, Floderus-Myrhed B. Ulcerative colitis and Crohn’s disease in an unselected population of monozygotic and dizygotic twins. A study of heritability and the influence of smoking. Gut. 1988;29:990–996. doi: 10.1136/gut.29.7.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Orholm M, Binder V, Sorensen TI, Rasmussen LP, Kyvik KO. Concordance of inflammatory bowel disease among Danish twins. Results of a nationwide study. Scand J Gastroenterol. 2000;35:1075–1081. doi: 10.1080/003655200451207. [DOI] [PubMed] [Google Scholar]
- 49.Cho JH. The Nod2 gene in Crohn’s disease: implications for future research into the genetics and immunology of Crohn’s disease. Inflamm Bowel Dis. 2001;7:271–275. doi: 10.1097/00054725-200108000-00014. [DOI] [PubMed] [Google Scholar]
- 50.Ogura Y, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 51.Hugot JP, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 52.Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med. 2009;361:2066–2078. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parkes M. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nature Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hampe J, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nature Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 55.Rioux JD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nature Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parkes M, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nature Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McCarroll SA, et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn’s disease. Nature Genet. 2008;40:1107–1112. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- 59.Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nature Rev Immunol. 2007;7:767–777. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Virgin HW, Levine B. Autophagy genes in immunity. Nature Immunol. 2009;10:461–470. doi: 10.1038/ni.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cooney R, et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nature Med. 2010;16:90–97. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 62.Travassos LH, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nature Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 63.Cadwell K, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cadwell K, et al. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. This paper shows how a viral infection can contribute to complex disease pathogenesis in the context of a genetically predisposed host. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Karst SM, Wobus CE, Lay M, Davidson J, Virgin HWt. STAT1-dependent innate immunity to a Norwalk-like virus. Science. 2003;299:1575–1578. doi: 10.1126/science.1077905. [DOI] [PubMed] [Google Scholar]
- 66.Okayasu I, et al. A novel method in the induction of reliable experimental acute and chronic ulcerative colitis in mice. Gastroenterology. 1990;98:694–702. doi: 10.1016/0016-5085(90)90290-h. [DOI] [PubMed] [Google Scholar]
- 67.Hsu CC, Wobus CE, Steffen EK, Riley LK, Livingston RS. Development of a microsphere-based serologic multiplexed fluorescent immunoassay and a reverse transcriptase PCR assay to detect murine norovirus 1 infection in mice. Clin Diagn Lab Immunol. 2005;12:1145–1151. doi: 10.1128/CDLI.12.10.1145-1151.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goto K, et al. Molecular detection of murine norovirus from experimentally and spontaneously infected mice. Exp Anim. 2009;58:135–140. doi: 10.1538/expanim.58.135. [DOI] [PubMed] [Google Scholar]
- 69.WHO. Global surveillance, prevention and control of chronic respiratory diseases: a comprehensive approach. WHO; Geneva: 2007. [Google Scholar]
- 70.Robinson DS, et al. Predominant TH2-like bronchoalveolar T-lymphocyte population in atopic asthma. N Engl J Med. 1992;326:298–304. doi: 10.1056/NEJM199201303260504. [DOI] [PubMed] [Google Scholar]
- 71.Cohn L, Homer RJ, Marinov A, Rankin J, Bottomly K. Induction of airway mucus production By T helper 2 (Th2) cells: a critical role for interleukin 4 in cell recruitment but not mucus production. J Exp Med. 1997;186:1737–1747. doi: 10.1084/jem.186.10.1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Barrett NA, Austen KF. Innate cells and T helper 2 cell immunity in airway inflammation. Immunity. 2009;31:425–437. doi: 10.1016/j.immuni.2009.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Holgate ST, Davies DE. Rethinking the pathogenesis of Asthma. Immunity. 2009;31:362–367. doi: 10.1016/j.immuni.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 74.Johnston SL, et al. Community study of role of viral infections in exacerbations of asthma in 9–11 year old children. BMJ. 1995;310:1225–1229. doi: 10.1136/bmj.310.6989.1225. This longitudinal study provides important evidence for a role for viruses in asthma. The investigators follow children with asthma for >1 year and test respiratory specimens collected during each asthma exacerbation; 80–85% of children [Au:OK? or specimens collected?] were positive for viruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nicholson KG, Kent J, Ireland DC. Respiratory viruses and exacerbations of asthma in adults. BMJ. 1993;307:982–986. doi: 10.1136/bmj.307.6910.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gern JE. The ABCs of rhinoviruses, wheezing, and asthma. J Virol. 2010;84:7418–7426. doi: 10.1128/JVI.02290-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Leung TF, et al. Multiplex molecular detection of respiratory pathogens in children with asthma exacerbation. Chest. 2009;137:348–354. doi: 10.1378/chest.09-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Khetsuriani N, et al. Prevalence of viral respiratory tract infections in children with asthma. J Allergy Clin Immunol. 2007;119:314–321. doi: 10.1016/j.jaci.2006.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bizzintino J, et al. Association between human rhinovirus C and severity of acute asthma in children. Eur Respir J. 2010 Aug 6; doi: 10.1183/09031936.00092410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miller EK. New human rhinovirus species and their significance in asthma exacerbation and airway remodeling. Immunol Allergy Clin North Am. 2010;30:541–552. doi: 10.1016/j.iac.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Johnston SL, et al. The relationship between upper respiratory infections and hospital admissions for asthma: a time-trend analysis. Am J Respir Crit Care Med. 1996;154:654–660. doi: 10.1164/ajrccm.154.3.8810601. [DOI] [PubMed] [Google Scholar]
- 82.Sears MR, Johnston NW. Understanding the September asthma epidemic. J Allergy Clin Immunol. 2007;120:526–529. doi: 10.1016/j.jaci.2007.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jackson DJ, et al. Wheezing rhinovirus illnesses in early life predict asthma development in high-risk children. Am J Respir Crit Care Med. 2008;178:667–672. doi: 10.1164/rccm.200802-309OC. This report details the results of a prospective study of high-risk children from birth to age 6 years, with an analysis of the correlation between early childhood risk factors and subsequent diagnosis of asthma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kusel M, et al. Early-life respiratory viral infections, atopic sensitization, and risk of subsequent development of persistent asthma. J Allergy Clin Immunol. 2007;119:1105–1110. doi: 10.1016/j.jaci.2006.12.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lemanske RF., Jr The childhood origins of asthma (COAST) study. Pediatr Allergy Immunol. 2002;13(Suppl 15):38–43. doi: 10.1034/j.1399-3038.13.s.15.8.x. [DOI] [PubMed] [Google Scholar]
- 86.Bartlett NW, McLean GR, Chang YS, Johnston SL. Genetics and epidemiology: asthma and infection. Curr Opin Allergy Clin Immunol. 2009;9:395–400. doi: 10.1097/ACI.0b013e32833066fa. [DOI] [PubMed] [Google Scholar]
- 87.Gern JE. Rhinovirus and the initiation of asthma. Curr Opin Allergy Clin Immunol. 2009;9:73–78. doi: 10.1097/ACI.0b013e32831f8f1b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Corne J, et al. Frequency, severity, and duration of rhinovirus infections in asthmatic and non-asthmatic individuals: a longitudinal cohort study. Lancet. 2002;359:831–834. doi: 10.1016/S0140-6736(02)07953-9. [DOI] [PubMed] [Google Scholar]
- 89.Message SD, et al. Rhinovirus-induced lower respiratory illness is increased in asthma and related to virus load and Th1/2 cytokine and IL-10 production. Proc Natl Acad Sci. 2008;105:13562–13567. doi: 10.1073/pnas.0804181105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Contoli M, et al. Role of deficient type III interferon-λ production in asthma exacerbations. Nature Med. 2006;12:1023–1026. doi: 10.1038/nm1462. [DOI] [PubMed] [Google Scholar]
- 91.Wark PAB. Asthmatic bronchial epithelial cells have a deficient innate immune response to infection with rhinovirus. J Exp Med. 2005;201:937–947. doi: 10.1084/jem.20041901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wark PAB, Grissell T, Davies B, See H, Gibson PG. Diversity in the bronchial epithelial cell response to infection with different rhinovirus strains. Respirology. 2009;14:180–186. doi: 10.1111/j.1440-1843.2009.01480.x. [DOI] [PubMed] [Google Scholar]
- 93.Uller L, et al. Double-stranded RNA induces disproportionate expression of thymic stromal lymphopoietin versus interferon-β in bronchial epithelial cells from donors with asthma. Thorax. 2010;65:626–632. doi: 10.1136/thx.2009.125930. [DOI] [PubMed] [Google Scholar]
- 94.Bosco A, Ehteshami S, Stern DA, Martinez FD. Decreased activation of inflammatory networks during acute asthma exacerbations is associated with chronic airflow obstruction. Mucosal Immunol. 2010;3:399–409. doi: 10.1038/mi.2010.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Blomqvist S, Roivainen M, Puhakka T, Kleemola M, Hovi T. Virological and serological analysis of rhinovirus infections during the first two years of life in a cohort of children. J Med Virol. 2002;66:263–268. doi: 10.1002/jmv.2140. [DOI] [PubMed] [Google Scholar]
- 96.Savolainen C, Mulders MN, Hovi T. Phylogenetic analysis of rhinovirus isolates collected during successive epidemic seasons. Virus Res. 2002;85:41–46. doi: 10.1016/s0168-1702(02)00016-3. [DOI] [PubMed] [Google Scholar]
- 97.Kistler A, et al. Pan-viral screening of respiratory tract infections in adults with and without asthma reveals unexpected human coronavirus and human rhinovirus diversity. J Infect Dis. 2007;196:817–825. doi: 10.1086/520816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Olenec JP, et al. Weekly monitoring of children with asthma for infections and illness during common cold seasons. J Allergy Clin Immunol. 2010;125:1001–1006. e1. doi: 10.1016/j.jaci.2010.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Martinez FD. The origins of asthma and chronic obstructive pulmonary disease in early life. Proc Am Thorac Soc. 2009;6:272–277. doi: 10.1513/pats.200808-092RM. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Le Souef PN. Gene-environmental interaction in the development of atopic asthma: new developments. Curr Opin Allergy Clin Immunol. 2009;9:123–127. doi: 10.1097/ACI.0b013e3283292283. [DOI] [PubMed] [Google Scholar]
- 101.Weiss ST, Raby BA, Rogers A. Asthma genetics and genomics 2009. Curr Opin Genet Dev. 2009;19:279–282. doi: 10.1016/j.gde.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 102.Wang Q, et al. Role of double-stranded RNA pattern recognition receptors in rhinovirus-induced airway epithelial cell responses. J Immunol. 2009;183:6989–6997. doi: 10.4049/jimmunol.0901386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Slater L, et al. Co-ordinated role of TLR3, RIG-I and MDA5 in the innate response to rhinovirus in bronchial epithelium. PLoS Pathog. 2010;6:e1001178. doi: 10.1371/journal.ppat.1001178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jartti T, et al. Allergic sensitization is associated with rhinovirus-, but not other virus-, induced wheezing in children. Pediatr Allergy Immunol. 2010;21:1008–1014. doi: 10.1111/j.1399-3038.2010.01059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kloepfer KM, Gern JE. Virus/allergen interactions and exacerbations of asthma. Immunol Allergy Clin North Am. 2010;30:553–563. doi: 10.1016/j.iac.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kim EY, et al. Persistent activation of an innate immune response translates respiratory viral infection into chronic lung disease. Nature Med. 2008;14:633–640. doi: 10.1038/nm1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Virgin HW, Wherry EJ, Ahmed R. Redefining chronic viral infection. Cell. 2009;138:30–50. doi: 10.1016/j.cell.2009.06.036. [DOI] [PubMed] [Google Scholar]
- 108.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 109.Horie M, et al. Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature. 2010;463:84–87. doi: 10.1038/nature08695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Taylor DJ, Leach RW, Bruenn J. Filoviruses are ancient and integrated into mammalian genomes. BMC Evol Biol. 2010;10:193. doi: 10.1186/1471-2148-10-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.de Parseval N, Heidmann T. Human endogenous retroviruses: from infectious elements to human genes. Cytogenet Genome Res. 2005;110:318–332. doi: 10.1159/000084964. [DOI] [PubMed] [Google Scholar]
- 112.Colmegna I, Garry RF. Role of endogenous retroviruses in autoimmune diseases. Infect Dis Clin North Am. 2006;20:913–929. doi: 10.1016/j.idc.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 113.Rolland A, et al. The envelope protein of a human endogenous retrovirus-W family activates innate immunity through CD14/TLR4 and promotes Th1-like responses. J Immunol. 2006;176:7636–7644. doi: 10.4049/jimmunol.176.12.7636. [DOI] [PubMed] [Google Scholar]
- 114.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Anderson NG, Gerin JL, Anderson NL. Global screening for human viral pathogens. Emerg Infect Dis. 2003;9:768–774. doi: 10.3201/eid0907.030004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vehik K, Dabelea D. The changing epidemiology of type 1 diabetes: why is it going through the roof? Diabetes Metab Res Rev. 2010 doi: 10.1002/dmrr.1141. [DOI] [PubMed] [Google Scholar]
- 117.Loftus EV, Jr, Schoenfeld P, Sandborn WJ. The epidemiology and natural history of Crohn’s disease in population-based patient cohorts from North America: a systematic review. Aliment Pharmacol Ther. 2002;16 doi: 10.1046/j.1365-2036.2002.01140.x. [DOI] [PubMed] [Google Scholar]
- 118.Eder W, Ege MJ, von Mutius E. The asthma epidemic. N Engl J Med. 2006;355:2226–2235. doi: 10.1056/NEJMra054308. [DOI] [PubMed] [Google Scholar]