Synopsis
Preservation of genome integrity via the DNA damage response is critical to prevent disease. Ataxia-telangiectasia mutated and Rad3-related (ATR) is essential for life and functions as a master regulator of the DNA damage response, especially during DNA replication. ATR controls and coordinates DNA replication origin firing, replication fork stability, cell cycle checkpoints, and DNA repair. Since its identification 15 years ago, a model of ATR activation and signaling has emerged that involves localization to sites of DNA damage and activation through protein-protein interactions. Recent research has added an increasingly detailed understanding of the canonical ATR pathway, and an appreciation that the canonical model does not fully capture the complexity of ATR regulation. Here we review the ATR signaling process, focusing on mechanistic findings garnered from the identification of new ATR interacting proteins and substrates. We discuss how to incorporate these new insights into a model of ATR regulation, and point out the significant gaps in our understanding of this essential genome maintenance pathway.
Keywords: ATR, ATRIP, checkpoint, DNA damage response, DNA replication
Introduction
Safeguarding against unfavorable changes in the genome during cell division is vital. Although packaging DNA in chromatin may protect it, genes need to be transcribed, and chromosomes need to be duplicated and segregated. Thus, even for normal DNA metabolism, DNA is precariously unwound, nicked, copied, broken, and recombined. The cell also produces reactive metabolites and oxidation products that damage DNA. Finally, exogenous sources of DNA damage like radiation and other genotoxins are prevalent in the environment. This combined assault on DNA yields tens of thousands of DNA lesions per day in every human cell.
In response, the cell mounts an evolutionarily conserved DNA damage response (DDR) that coordinates cell cycle progression, DNA repair, DNA replication, DNA transcription, and even cell death to promote genome maintenance. Mutation or deletion of many DDR genes results in lethality, cancer susceptibility syndromes, neurodegenerative disorders, and premature aging syndromes. Therefore, genome maintenance via the DDR is essential to prevent disease.
At the apex of the DDR are three related protein kinases, Ataxia-telangiectasia Mutated (ATM), ATM and Rad3-related (ATR), and DNA-dependent protein kinase (DNA-PK). These kinases belong to the phosphoinositide-3-like kinase kinase (PIKK) family and share similar domain architecture and several modes of regulation [1-3]. While DNA double-strand breaks (DSBs) activate ATM and DNA-PK, many types of DNA damage activate ATR, including DSBs, base adducts, and crosslinks [4]. Once activated, these kinases preferentially phosphorylate serines and threonines followed by a glutamine (S/TQ) in hundreds of protein substrates. In some cases, substrates contain multiple phosphorylated S/TQs within a small region of the primary sequence —S/TQ cluster domains [5, 6].
The most common signal for ATR activation likely involves replication stress —interference of replication fork progression caused by DNA damage, lack of sufficient deoxynucleotides, and even difficult to replicate DNA sequences. In response to replication stress, ATR regulates replisome stability, origin firing, and prevents premature mitotic entry [4]. ATR, unlike ATM and DNA-PK, is an essential gene in replicating cells [7-9]. This probably is due to ATR activation by replication stress in every S phase and perhaps regulation of specific aspects of DNA replication such as origin firing or nucleotide production. Failure to resolve stalled replication forks results in unreplicated DNA, single-stranded DNA replication intermediates, and DSBs. Single-stranded DNA (ssDNA) and DSBs are highly recombinogenic, and aberrant recombination of these structures yields chromatid- and chromosome-type errors [10, 11].
Homozygous loss of function mutations in ATR are not compatible with mammalian cell viability [7-9]. However, hypomorphic mutations in ATR that cause reduced ATR function are found in a few patients with the rare Seckel Syndrome, which is characterized by microcephaly and growth retardation [12]. In certain genetic backgrounds ATR is a haploinsufficient tumor suppressor [13, 14], and ATR mutations in microsatellite instability tumors are associated with reduced overall survival and disease-free survival [15, 16]. Oncogene-induced replication stress activates the ATR pathway explaining high levels of DDR activation in many neoplasias [17-19]. Therefore, the ATR pathway and the DDR potentially constitute a barrier to cancer [20]. Bypass of this barrier may be a double-edged sword for the cancer cell, as cancer cells may exhibit an increased dependency on the ATR pathway, analogous to oncogene addiction, to continue to replicate in the presence of oncogene-induced replication stress. Many current therapies are DNA damaging agents that increase the signaling burden on the ATR pathway, and ATR pathway proteins may be good drug targets in cancers containing elevated levels of replicative stress.
Canonical ATR signaling
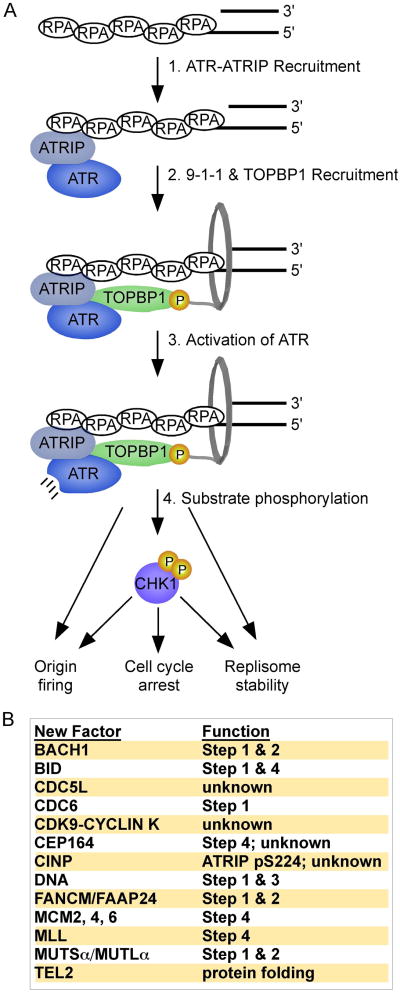
Replication stress often results in the generation of excess ssDNA through uncoupling of enzymatic activities at the replication fork. For example, many DNA lesions stall the DNA polymerase but not the replicative helicase [21]. While lesions, such as interstrand crosslinks, block both the polymerase and helicase, enzymatic remodeling of the blocked replication fork by helicases and nucleases also creates ssDNA. Similarly, nuclease-mediated resection of DNA DSBs produces ssDNA [22]. ssDNA is bound by the ssDNA binding protein Replication Protein A (RPA). In the canonical ATR signaling pathway, RPA-ssDNA is the ligand that recruits ATR and other ATR signaling components to sites of replication stress. Once RPA-ssDNA appears, ATR signaling canonically requires: 1. Recruitment of ATR via its obligate partner ATR Interacting Protein (ATRIP) to RPA-ssDNA; 2. Independent recruitment of a checkpoint clamp, containing RAD9-HUS1-RAD1 proteins, and the ATR activator Topoisomerase Binding Protein 1 (TOPBP1) to RPA-ssDNA; 3. Activation of ATR by TOPBP1; and 4 Phosphorylation of ATR substrates (Figure 1). Cases where ATR activation occurs independently of steps 1-4 and RPA-ssDNA are defined as non-canonical for the purposes of this review. For a more detailed description of the canonical pathway we refer the readers to a recent review [4]. Here we will review new advances in understanding each of these steps in ATR signaling, emphasizing novel ATR regulatory factors, ATR targets, and non-canonical pathways of ATR regulation.
Figure 1.
Canonical ATR signaling pathway and functions of newly identified regulators.
(A) ATR signaling is activated in response to single-stranded DNA gaps in the genome. Independent recruitment of several checkpoint proteins leads to TOPBP1-dependent activation of the kinase and phosphorylation of numerous substrates including CHK1 to regulate cellular responses to DNA damage and replication stress. (B) The table lists newly identified regulators of this pathway and, when known, describes their position in the pathway. See text for details.
The ATR-ATRIP Complex
Of all the identified ATR-interacting proteins, perhaps only ATRIP is obligatory for all known ATR functions. Deletion of ATRIP in yeast and silencing in mammalian systems phenocopies loss of ATR--resulting in lethality, DNA damage sensitivity, and abrogation of ATR substrate phosphorylation [7, 23-25]. ATRIP has at least three well-defined functions in ATR signaling —regulating ATR stability, localization, and kinase activation. The stability of ATR and ATRIP are mutually dependent [7], consistent with the formation of a stoichiometric complex. Indeed, ATR and ATRIP stably associate independent of DNA damage [26, 27]. ATRIP binds directly to RPA to promote accumulation of ATR into damage-induced foci [26, 28]. Finally, TOPBP1 activates ATR through a direct interaction with ATRIP [29, 30]. In addition to these clearly elucidated functions, there are several additional ATR signaling regulators that may depend on ATRIP for their effects.
TEL2
ATR and other PIKKs, including ATM and DNA-PK, associate with TEL2 [31]. Initial results suggested that TEL2 regulates the stability of these kinases. However, unlike ATRIP, TEL2 does not co-purify stoichiometrically with ATR, suggesting a more transient interaction. Recently, several groups determined that TEL2 functions as part of multi-protein complexes that include protein chaperone activities [32-35]. TEL2 binds TTI1 and TTI2 forming a Triple T complex, which also contains HSP90 [33, 34]. TEL2 preferentially associates with newly synthesized ATR within one hour of synthesis and not after. In the absence of TripleT/HSP90, newly synthesized ATR is unable to co-immunoprecipitate with ATRIP. Thus, it is likely that the TripleT/HSP90 complex promotes the correct folding of ATR and other PIKKs to allow their assembly into proper protein complexes. TEL2 also facilitates PIKK maturation through the R2TP/prefoldin-like complex [32]. Thus, in the absence of TEL2, newly synthesized ATR is not properly folded and assembled with ATRIP explaining the checkpoint defects associated with TEL2 silencing. The PIKKs are very large proteins with extensive HEAT repeats. HEAT repeats typically do not form globular domains. Instead, they create extended surfaces that mediate protein and DNA interactions. This architecture may explain the need for these chaperone complexes for proper folding and assembly.
CINP and CEP164
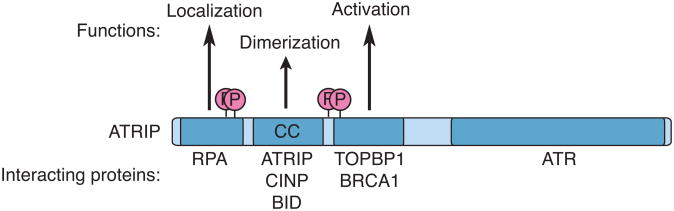
Although all eukaryotic organisms have an ATRIP protein, the amino acid sequence is highly divergent to the extent that the vertebrate ATRIP could not be identified through homology to lower eukaryotic proteins. Despite this poor sequence conservation, the overall architecture of the protein is conserved with an N-terminal RPA binding domain followed by a coiled-coil domain and a C-terminal tail that includes the ATR binding region (Figure 2). The ATRIP coiled-coil domain is required for ATRIP dimerization and stable association with ATR [36, 37]. Disruption of the coiled-coil domain perturbs ATRIP localization to foci and ATR signaling to CHK1 in mammalian cells. Replacement of the ATRIP coiled-coil domain with a heterologous coiled-coiled domain restores some but not all ATRIP functions [36]. This data suggested that the ATRIP coiled-coil domain might mediate a protein-protein interaction important to regulate ATR signaling. A search for proteins that interact with ATRIP through the coiled-coil identified CDK2-interacting protein (CINP) [38]. The function of CINP in ATR-ATRIP signaling is not yet clear, but CINP silencing causes modest defects in ATR-dependent CHK1 and SMC1 phosphorylation, hydroxyurea (HU) sensitivity, and loss of the G2 checkpoint. Since CINP does not appear to form foci with ATR-ATRIP or alter the in vitro activity of ATR, it may function through a transient protein-protein interaction that facilitates modifications of the ATR-ATRIP complex in cells. G2 checkpoint defects in CINP silenced cells may be due to reduced CDK2-dependent ATRIP S224 phosphorylation, which is required for G2 checkpoint maintenance [39]. However, mutation of S224 does not fully phenocopy CINP silencing.
Figure 2.
Functions of ATRIP in ATR regulation.
ATRIP has several functional domains that regulate localization, stability, dimerization, and activation of ATR. These include an N-terminal RPA binding domain, a coiled-coil domain (CC), and a C-terminal domain that interacts with TOPBP1 and ATR.
Interestingly, CINP also complexes with CEP152 [40]. Like ATR, CEP152 mutations cause Seckel Syndrome [12, 40]. Furthermore, evidence has linked mutations in PERICENTRIN and CENPJ to Seckel Syndrome as well [41, 42]. CEP152, CENPJ, and PERICENTRIN all function at centrosomes, suggesting that one underlying cause of Seckel Syndrome is a defect in centrosome function or duplication. In this model, ATR and CINP may participate in centrosome regulation. A large-scale screen for ATM and ATR substrates identified centrosome components, lending support to this idea [43]. Furthermore, new data indicate another centrosomal protein CEP164 functions in ATR signaling [44]. CEP164 was originally identified as a centriole appendage protein required for primary cilium function [45]. CEP164 exists in both ATR and ATM complexes as well as nucleotide excision repair protein complexes [46]. Silencing CEP164 results in a G2 checkpoint defect as well as CHK1 and RPA phosphorylation defects following UV damage. While the interaction with ATR is not DNA damage regulated, ATR phosphorylates CEP164, and CEP164 localizes to nuclear foci in response to DNA damage. These linkages between ATR signaling and centrosome biology, though preliminary, warrant further investigation to understand both the mechanistic relationship and how defects in this pathway may cause Seckel Syndrome.
BID
The pro-apoptotic BH3 only protein BID also functions in the DDR and is an ATR and ATM substrate. Metaphase spreads from BID-deficient cells exhibit an increase in chromatid-type errors following mitomycin C treatment and increased sensitivity to DNA damage [47, 48]. A new study determined that BID silencing results in less ATR-ATRIP association on chromatin following HU treatment and reduced CHK1 phosphorylation [49]. Helix4 of BID interacts with the coiled-coil domain of ATRIP, suggesting that BID may regulate ATR signaling via ATRIP. However, BID is not an essential gene, and its deletion phenotypes are not as severe as ATR or ATRIP deficiency. Therefore, its contribution to ATRIP function might be context dependent.
ATR Recruitment to Sites of DNA Damage
In addition to ATR stability, ATRIP is required for ATR recruitment to single-stranded DNA (ssDNA) gaps in the genome. Recognition of ssDNA by ATR-ATRIP occurs via an interaction between the acidic checkpoint recruitment domain (CRD) of ATRIP and a basic cleft in the ssDNA binding protein RPA70 [50]. Mutation of the ATRIP CRD as well as the basic cleft of RPA70 prevents ATR-ATRIP from binding to ssDNA [50, 51]. However, an ATRIP mutant unable to bind RPA70 does not exhibit severe checkpoint signaling defects [26, 50]. Mutation of the RPA70 basic cleft also does not fully compromise checkpoint signaling [52, 53]. This suggests there may be alternative, cooperating, or semi-redundant mechanisms to localize ATR-ATRIP to DNA. Several new ATR interacting proteins affect ATR recruitment to chromatin, and ATR can bind DNA directly. In this section we describe how these interactions may cooperate with the canonical RPA-dependent mechanism of ATR-ATRIP recruitment.
CDC6
CDC6 is an evolutionarily conserved DNA replication factor required to load the MCM2-7 (minichromosome maintenance) protein complex onto replication origins [54]. In addition to replication initiation, evidence for CDC6 function in the DNA damage response comes from several experimental systems. Mutations in S. pombe CDC18CDC6 causes a mitotic checkpoint defect [55]. Xenopus CDC6 is required for CHK1 phosphorylation in response to replication stress agents [56]. In human cells, CDC6 overexpression in G2 results in a mitotic block that correlates with CHK1 activation [57], and silencing CDC6 in S phase cells causes premature mitotic entry [58].
A challenge in interpreting many of these results is separating the function of CDC6 in replication initiation from its checkpoint signaling activity. After all, lack of initiation would lead to less checkpoint activation by agents that cause replication fork stalling. Two recent papers now provide some mechanistic insight into how CDC6 contributes to the DNA damage response via retention of ATR to chromatin.
In S. pombe, CDC18CDC6 and RAD3ATR increase on chromatin in response to replication stress [59]. The increase of RAD3 on chromatin is lost in CDC18-deficient cells. Furthermore, RAD3 and CDC18 reside in the same complex that also contains RAD26ATRIP in cells treated with HU. Two hybrid analysis and experiments with recombinant proteins produced in bacteria indicate a direct interaction between CDC18 and RAD26. Furthermore, RAD26 mutation abrogates CDC18 recruitment to chromatin and the CDC18-RAD3 interaction. Experiments in human cells confirmed that CDC6, ATR, and ATRIP exist in a complex [60]. Furthermore, chromatin retention of Xenopus ATR decreases with CDC6 depletion from extracts [60]. It will be important to determine the molecular requirements for CDC6-mediated retention of ATR to chromatin. Ultimate proof that CDC6 functions upstream of ATR independently of its origin initiation activity requires the production of separation of function mutants.
FANCM/FAAP24
FANCM is a gene mutated in patients with Fanconi Anemia (FA) and helps tether a FA core complex to chromatin to regulate repair of DNA interstrand crosslinks (ICL). However, unlike other FA proteins, FANCM is highly evolutionarily conserved with orthologues found in all eukaryotes and even archae. FANCM contains a conserved helicase domain with DNA translocase activity [61-63]. It associates with FA-associated protein 24 (FAAP24), which possesses DNA fork-structure binding activities [64, 65]. FANCM/FAAP24 localizes to chromatin and regulates fork progression and stability independently of the FA core complex [66-69].
FANCM/FAAP24 silencing abrogates the G2 checkpoint and reduces ATR signaling in response to many types of DNA lesions. FANCM potentially functions in ATR signaling at multiple steps in response to different damaging agents. For instance, in human cells FANCM silencing reduced the amount of RPA-ssDNA induced by DNA ICLs [70]. This is in contrast to HU, UV, and camptothecin-induced RPA-ssDNA, which is actually increased in FANCM silenced cells. Unlike HU and UV, DNA ICLs do not uncouple the polyermase and helicase to generate ssDNA, so FANCM/FAAP24 could be required to expose ssDNA in these contexts, placing the complex at the node of RPA-ssDNA generation. While FANCM helicase activity is required for full checkpoint signaling in response to ICLs, it is not required for RPA-ssDNA formation as detected by RPA foci. Rather, FAAP24 DNA binding activity is needed. It is still unclear how FAAP24 DNA binding mediates RPA-ssDNA formation. In response to other types of DNA damage, FANCM may function at the level of TOPBP1 recruitment to chromatin [66] since FANCM deletion causes reduced TOPBP1 binding following camptothecin treatment. Thus, depending on the context, FANCM may be important at several steps in the ATR pathway.
MUTS and MUTL
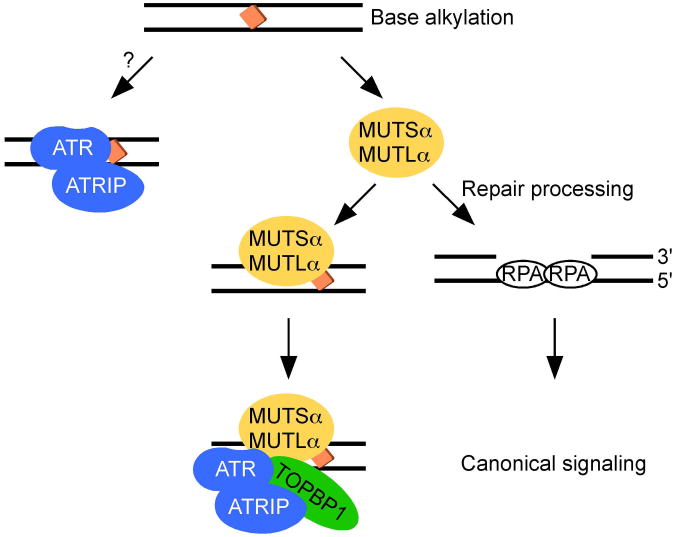
Activation of the ATR pathway depends on mismatch repair proteins in cells treated with base alkylation-inducing agents like N-methyl-N′-nitro-N-nitrosoguanidine but not replication fork stalling agents like HU [71]. There are two primary mechanistic explanations for the function of mismatch repair in ATR activation. Mismatch repair-dependent processing of the DNA may create RPA-ssDNA that then mediates ATR activation. Alternatively, there might be a direct function of specific mismatch repair proteins in ATR signaling mediated by direct protein-protein interactions. For example, the mismatch repair proteins could serve as a lesion recognition platform on which the signaling apparatus assembles (Figure 3).
Figure 3.
ATR recruitment to some DNA base lesions proceeds through a mismatch repair-dependent mechanism.
Mismatch repair proteins may contribute directly and indirectly to ATR recognition of some times of DNA damage. Direct interactions between the ATR-ATRIP complex and MUTSα and MUTLα proteins may promote activation of ATR independently of RPA and single-stranded DNA. Alternatively, mismatch repair-dependent DNA processing may promote the RPA-dependent canonical ATR signaling pathway.
Accumulating evidence supports the second model. MUTSα associates with ATR, CHK1, and TOPBP1 [71-74], and MUTLα interacts with TOPBP1 [74]. Some of these interactions may be direct, although domain-mapping studies are incomplete, and interaction deficient mutants have not been isolated. MUTSα and MUTLα regulate recruitment of ATR, CHK1, and TOPBP1 to chromatin in response to base damage [74]. Importantly, RPA, RAD17, and RAD9 were not enriched on chromatin in these experiments and were not found to interact with mismatch repair proteins. Deletion of MSH2 also reduces ATR recruitment to damage sites after N-methyl-N-nitrosourea treatment, supporting a recruitment function for mismatch repair proteins in ATR activation [75].
These results suggest an RPA-independent mode of ATR activation in response to specific types of lesions detected by mismatch repair proteins. These findings fit with observations that ATR can be activated in the absence of a direct RPA-ATRIP interaction [26, 50]. This model makes several testable predictions. First, the ATRIP-CRD mutation, which cannot interact with RPA [50], should have no effect on ATR chromatin association following base damage. Second, RAD17 or RAD9 silencing would not impact ATR signaling and recruitment of TOPBP1 and CHK1 after base damage. Finally, it should be possible to identify separation of function mutations in ATR or more likely ATRIP that block ATR activation by N-methyl-N′-nitro-N-nitrosoguanidine but not activation by HU.
DNA
Flag-ATR purified from mammalian cells can interact with duplex DNA and ultraviolet radiation (UV)-damaged DNA [76]. Additionally, linearized plasmid stimulates ATR activity in vitro, and this stimulation increases when the DNA is damaged by UV or benzo[a]pyrene diol epoxide [76, 77]. This implies that ATR binding to damaged duplex DNA occurs in a manner distinct from the RPA-ssDNA binding mode. Yet ATR lacks a known DNA binding domain, and DNA binding activity has not been localized to any specific ATR fragment. A clue may come from research on the Bacillus cereus AlkD glycosylase. Rubinson et al. recently reported this glycosylase binds alkylated and abasic DNA via its HEAT repeats [78]. The HEAT repeats create a concave surface lined with positively charged residues that mediate electrostatic interactions with the phosphoribose DNA backbone. The lesion itself is not recognized directly. Instead, AlkD recognizes helical distortion created when the damaged base is flipped out into solution. These findings suggest the possibility that ATR binds damaged DNA via its HEAT repeats. This mechanism of DNA binding would explain how multiple types of DNA lesions could be recognized by ATR. Structural studies of DNA-PK support the idea that its HEAT repeats can bind DNA [79, 80]. Unfortunately testing this speculative model will be difficult given the slow progress in creating soluble recombinant forms of ATR in sufficient quantities for structural studies.
ATR Activation
Recruitment of ATR-ATRIP to DNA lesions or stalled forks is not sufficient for checkpoint signaling. The ATR activation process is not well understood, but it requires the ATR activator TOPBP1 that is recruited independently of ATR-ATRIP localization. A region of TOPBP1 termed the ATR Activating Domain (AAD) binds surfaces on both ATR and ATRIP [30], and this domain is sufficient to stimulate ATR kinase activity both in vitro and in vivo[29]. Mapping of the AAD binding surface on ATR delineated a regulatory domain called the PIKK Regulatory Domain (PRD) [30]. The PRD resides between the ATR kinase domain and the FATC domain, and mutation of this region results in loss of ATR activation by TOPBP1 as well as checkpoint and viability defects. Interestingly, this region of the other PIKKs, including ATM and DNA-PK, is also important for their activation [30, 81]. The PRD of ATM contains a critical lysine and cysteine residue that are acetylated or oxidized respectively [81, 82]. Acetylation by TIP60 activates ATM, and oxidation provides a mechanism of regulating ATM by oxidative stress independently of double-strand breaks. Mutations in the PRD of DNA-PK also interfere with its activation [30]. It should be noted that the PRD region is not the same as the FATC domain. The FATC is defined by sequence similarity among the PIKK family of kinases. In contrast, the PRD of each PIKK is unique, which may allow different inputs to activate each PIKK [3].
TOPBP1-mediated activation of ATR is conserved to budding yeast. S. cerevisiae DPB11TOPBP1 stimulates MEC1ATR kinase activity in vitro[83, 84]. Similar to TOPBP1, a discrete region of the C-terminus of DPB11 interacts with MEC1ATR and DDC2ATRIP and is sufficient to activate MEC1. Additional proteins, such as the checkpoint clamp protein DDC1, can also serve as direct ATR activators in the budding yeast system [85, 86]. Whether this mechanism is conserved in other organisms is unknown.
A thorough understanding of ATR activation mechanisms will require both structural studies and reconstitution of the activation steps in vitro. The Sancar group made an important advance in this area when they partially reconstituted ATR activation using purified RPA, ATR-ATRIP, and TOPBP1 proteins along with DNA [87]. Interestingly, they reported that only native purified ATR-ATRIP and not overexpressed versions of these proteins were a good source for their assays. This might imply that overexpressed protein lacks some regulatory partners or post-translational modifications necessary for the RPA-ssDNA and TOPBP1-dependent regulation of ATR.
TOPBP1 recruitment
It is becoming increasingly clear that like ATR-ATRIP recruitment, TOPBP1 recruitment may occur through multiple mechanisms possibly depending on the DNA damage context. One mechanism of TOPBP1 recruitment involves the checkpoint clamp complex RAD9-HUS1-RAD1 (9-1-1). The RFC-like clamp loader containing RAD17 loads the 9-1-1 complex onto the junction of double and single-stranded DNA [88-90]. This loading is specific to a 5′ recessed junction perhaps due to the interaction of RAD9 with the 70N domain of RPA [53]. Interaction between the phosphorylated tail of RAD9 and BRCA1 C-terminal repeats (BRCT) 1-2 of TOPBP1 [91-93] position TOPBP1 for ATR activation. Importantly 9-1-1 and TOPBP1 recruitment occurs independently of ATR-ATRIP [94, 95]. The independent recruitment of ATR and its protein activator TOPBP1 builds in an important regulatory feature recently described as the “two-man rule” for ATR activation [4]. Requiring multiple complexes to separately sense a DNA lesion or stalled fork, prevents precocious activation of the checkpoint.
The Michael group recently reported that TOPBP1 can be recruited to chromatin prior to RAD9 in Xenopus extracts [96]. Furthermore, RAD9 recruitment required TOPBP1. One possible model to explain these results would be that, at least under some circumstances, the RAD9-TOPBP1 interaction is a mechanism of signal amplification instead of initiation. A critical unanswered question, however, is how TOPBP1 recognizes damaged DNA and stalled replication forks without RAD9.
TOPBP1 can bind ssDNA directly in reconstituted in vitro systems. However, if RPA is pre-loaded onto the ssDNA, TOPBP1 association with RPA-ssDNA requires the presence of ATRIP [87]. Given that RPA has a much higher affinity for ssDNA than TOPBP1, it seems unlikely that a TOPBP1-ssDNA interaction would be sufficient for recruitment. An interaction between the TOPBP1 AAD and ATRIP mediates ATR activation [1], but the recruitment of TOPBP1 to chromatin requires different regions of TOPBP1 and not its AAD [97, 98]. TOPBP1 has multiple additional binding partners including TRESLIN, CtIP, BACH1 and the MRN complex [99-103]. Each of these may contribute to TOPBP1 and ATR signaling regulation in specific circumstances. For example, a TOPBP1-BACH1 interaction contributes to the exposure of ssDNA at stalled replication forks [103]. However, further research is necessary to understand how TOPBP1 initially recognizes the damaged fork.
TOPBP1-mediated activation of ATR and post-translational modifications
Mechanistic information on how TOPBP1 stimulates ATR kinase activity is lacking. It is postulated that TOPBP1 binding to ATR induces a conformational change that increases substrate affinity for the kinase. This is based on the observation that TOPBP1 greatly reduces the Km of ATR for its substrates [1]. In this model, ATR would only be active when directly bound by TOPBP1. This is consistent with an inability to purify an active ATR protein from damaged cells. Such purification attempts may fail because TOPBP1 does not remain bound to the complex. This model would also imply that ATR activation may not be accompanied by a post-translational modification that is sufficient to stabilize the activated form of the kinase.
ATR is phosphorylated in cells and can autophosphorylate in vitro. An antibody to one phosphorylation site (S428) is commercially available, and some reports have used it as a marker of ATR activation. However, this site is unlikely to be an autophosphorylation site, and it is not clear whether mutation of the site to alanine has any impact on ATR activation or function. Thus, it is questionable whether antibodies to this site provide a meaningful description of ATR activity. Currently, the best measure of ATR activation is CHK1 phosphorylation on ATR-dependent S317 and S345 residues [104, 105]. Of course, this is at best an indirect measure of ATR activity. CHK1 is one of several hundreds of ATR substrates. Unlike other ATR substrates, its phosphorylation by ATR has unique requirements for other proteins including CLASPIN [106]. Thus, identifying a regulatory ATR autophosphorylation site would provide a much-needed tool for ATR studies and could be used as a potential biomarker in clinical development of ATR inhibitors for cancer therapy.
ATR Signaling
Upon activation, ATR coordinates cell cycle checkpoints, replication fork stability and restart, and origin firing. However, few studies have elucidated specific mechanisms through which ATR elicits these responses. The best-characterized mechanisms are the checkpoint responses mediated by ATR-activation of CHK1. These include the G2 checkpoint that prevents entry into mitosis in the presence of damaged DNA via regulation of the CDC25 phosphatase. Since the CHK1-dependent ATR pathway is the subject of many recent reviews [4, 107], we will focus the following discussion on other ATR signaling activities.
ATR-dependent regulation of replication origin activity
One of the well-established activities of ATR is to regulate origin firing. Evidence exists for both positive and negative regulation of origin firing by ATR. For example, delaying late origin firing at least partly mediates the S-phase checkpoint [108]. However, replication-associated DNA damage that stalls replication forks also can induce nearby replication origins to fire in an apparent attempt to complete DNA synthesis independently of the damaged fork [109-111]. A recent report in S. cerevisiae suggests MEC1ATR also has a function during normal replication to promote replication initiation [112]. The mechanisms underlying these ATR activities are complex and may differ between organisms. However, we are beginning to obtain some important insights into these processes.
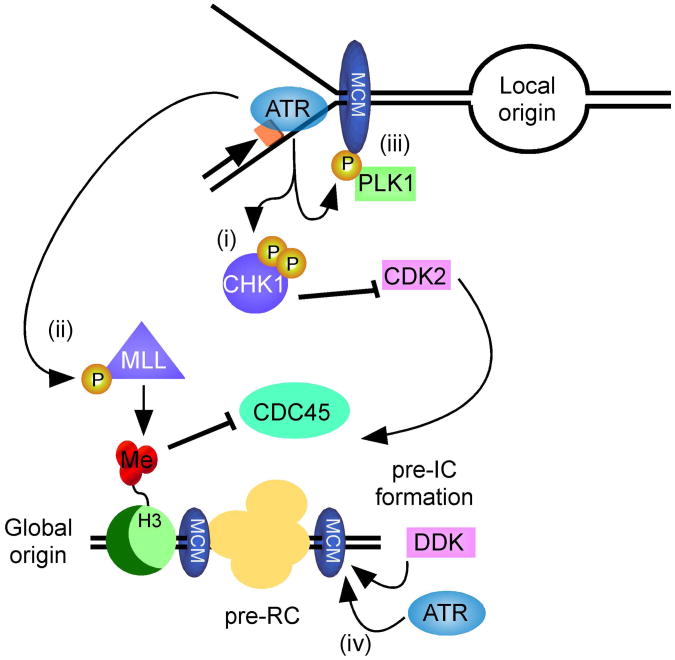
ATR represses origin firing in vertebrate cells at least partly by regulating chromatin structure around origins. Specifically, ATR regulates the lysine methyl transferase MLL [113]. MLL methylates histone H3 on lysine 4 (H3K4) and functions in embryogenesis and cell fate determination, in addition to cell cycle regulation [114]. MLL deficient cells exhibit chromatid-type errors and S-phase checkpoint defects. MLL activity is tightly controlled by its abundance. It is degraded in S and M phase by the SCFSKP2 and APCCDC20 complexes respectively [115], and overexpression of MLL perturbs S phase progression. ATR phosphorylates MLL in response to DNA damage in S-phase [113]. This phosphorylation prevents MLL from interacting with SKP2, thereby stabilizing it. Increased MLL abundance leads to increased H3 methylation at origins. Importantly, the replication initiation protein CDC45 binds directly to H3 but not methylated H3K4me3. Thus, in response to damage during S-phase, ATR phosphorylates and stabilizes MLL, which prevents CDC45 loading onto origins by methylating H3K4 (Figure 4).
Figure 4.
ATR signaling at stalled forks regulates origin firing through multiple mechanisms.
ATR negatively regulates origin firing by regulating S-phase kinases via CHK1 activation (i) and histone methylation (ii). These activities prevent pre-initiation complex (pre-IC) formation. ATR also positively promotes local origin firing via MCM2 phosphorylation and recruitment of PLK1 (iii). Finally, at least in budding yeast, it can also act as a priming kinase for DDK-dependent MCM phosphorylation (iv). See text for details.
Paradoxically, ATR also promotes origin firing in some circumstances. First, there are many more licensed replication origins than are used in each round of replication. The extra origins provide a resource to use to complete DNA syntheses when there is stalling of elongating replication forks [109-111]. ATR-dependent phosphorylation of MCM2 promotes the recruitment of PLK1 [116-118]. At least locally, this has the effect of allowing origins to fire perhaps through localized differences in CHK1 signaling. Evidence now indicates that these localized differences specifically inhibit activation of new replication factories while allowing dormant origins to fire within preexisting factories in an ATR-CHK1 dependent manner [119]. A second activity of ATR in promoting replication initiation also occurs through MCM protein phosphorylation. Phosphorylation of the MCM proteins by the DBF4-CDC7 (DDK) replication promoting kinase initiates origin firing [120]. However, the DDK kinase often requires a priming phosphorylation for it to recognize its substrates. The S. cerevisiae ATR protein catalyzes some of these priming phosphorylation events [112].
The discovery of these ATR-dependent activities to regulate origin firing provides exciting insights into ATR function. However, they also raise many new questions. How can ATR both promote and inhibit origin firing? What determines the local vs. global response to ATR activation? Why would a checkpoint kinase known to slow DNA synthesis and cell cycle progression be involved in priming events on pre-replication complexes to promote initiation? Are these mechanisms of ATR function conserved in all eukaryotic organisms or has evolution made use of the ATR kinase in different ways in different organisms? Based on the rapid pace of research in this area, hopefully, we can anticipate answers to many of these questions in the near future.
Crosstalk between transcription and the replication checkpoint
Transcription and replication are intimately linked if for no other reason than they both must make use of the same DNA template. Coordination of transcription (and additional associated RNA processing activities) and replication is necessary to prevent fork stalling at active transcription units. Indeed, silencing many RNA processing enzymes by RNAi causes DNA damage partly due to formation of R-loop intermediates where RNA is bound to the DNA duplex [121]. These may be processed directly to cause DNA breakage or may serve as obstacles for elongating DNA polymerases. In the most highly transcribed regions of DNA and in ribosomal gene clusters, programmed origins and replication pause sites assure that replication and transcription machinery do not make head-on collisions [122]. We will not attempt to review all the literature on how RNA processing activities crosstalk with the ATR-dependent replication checkpoint. However, we offer two examples to illustrate that the crosstalk may be more direct than simply collisions of separate RNA and DNA processing machineries.
CDK9 regulates transcription by phosphorylating the C-terminal tail of RNA polymerase II. It performs this activity as part of the pTEFb complex containing the regulatory CYCLIN T1 or T2 [123, 124]. Interestingly, CDK9 regulates the ATR-dependent response to stalled replication forks independently of its pTEFb activity [125]. In this case, CDK9 complexes with CYCLIN K, and this complex is essential for the resumption of DNA synthesis following a challenge with replication fork stalling agents like HU or aphidicolin. CDK9 can be found in complexes with ATR-ATRIP but does not appear to act upstream of ATR, since ATR substrate phosphorylation is not perturbed in CDK9- or CYCLIN K-silenced cells. These data suggest that CDK9-CYCLIN K may act downstream of ATR to regulate replication fork stability. Its activities in replication require an intact CDK9 kinase domain; therefore, identifying the substrates of CDK9-CYCLIN K will be essential to understand its function. Notably, this activity in the DDR appears to be evolutionarily conserved, since yeast CDK9-cyclin proteins also participate in the DDR [126].
A second example of ATR crosstalk with RNA processing machinery is the splicing factor CDC5L. CDC5L is part of a conserved mRNA splicing complex, which contains PSO4/PRP19, SPF27, and PLRG1 [127]. Silencing CDC5L causes sensitivity to replication stress agents and defects in ATR-dependent checkpoint signaling [128]. The simplest explanation would be an indirect effect of splicing problems on production of ATR or its regulatory partners. Surprisingly, further experiments suggested a more direct function when CDC5L was found to interact with ATR. As yet, the data are too preliminary to conclude how CDC5L actually regulates ATR signaling. While deleting an ATR interaction domain on CDC5L causes DDR defects, it is not clear whether this deletion actually separates its DDR and RNA processing functions. Nonetheless, these data are intriguing especially in light of the identification of multiple RNA processing proteins in genetic and biochemical screens looking for new DDR factors [38, 43, 121].
Concluding Remarks
Fifteen years of ATR research since the gene was first cloned has established the biological importance of ATR in DNA replication and the DDR. A mechanistic understanding of ATR activation has progressed dramatically in this time, but much is still unclear. Regulation of ATR recruitment to sites of DNA damage and the mechanism of activation defy simple linear models. The discovery of multiple mechanisms of ATR regulation that do not fit neatly into the canonical signaling pathway suggests context of the activation signal is important. Some of these mechanisms may function in response to specific lesion types while others may be more general modes of regulation that potentiate or amplify signaling without being essential for all ATR function.
The number of ATR interacting proteins and substrates is steadily increasing. Now that ATR signaling has been partially reconstituted in vitro, systematic examination of these new factors biochemically is feasible. TOPBP1-mediated activation of ATR is perhaps the most important step in the regulatory pathway, but the lack of high-resolution ATR structural information stymies mechanistic understanding of this activation step. Finally, clues found in the past couple of years hint at a broad and complex function for ATR in many aspects of cellular physiology. Much remains to be learned about the mechanisms through which ATR signaling regulates genome maintenance.
Acknowledgments
Funding: The authors' work on ATR is supported by the National Institutes of Health [grant number CA102729 (to D.C.)].
Abbreviations used
- ATR
Ataxia-telangiectasia mutated and Rad3-related
- ATRIP
ATR-interacting protein
- ATM
Ataxia-telangiectasia mutated
- DDR
DNA damage response
- DNA-PK
DNA-dependent protein kinase
- PIKK
phosphoinositide-3-like kinase kinase
- DSB
double-strand break
- RPA
replication protein A
- PRD
PIKK regulatory domain
- ssDNA
single-stranded DNA
- RNAi
RNA interference
- BRCT
Brca1 C-terminal
- 9-1-1
RAD9-HUS1-RAD1
- HU
hydroxyurea
- DDK
Dbf4-Cdc7
- CINP
CDK2-interacting protein
- ICL
interstrand crosslink
References
- 1.Mordes DA, Cortez D. Activation of ATR and related PIKKs. Cell Cycle. 2008;7:2809–2812. doi: 10.4161/cc.7.18.6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lempiainen H, Halazonetis TD. Emerging common themes in regulation of PIKKs and PI3Ks. The EMBO journal. 2009;28:3067–3073. doi: 10.1038/emboj.2009.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lovejoy CA, Cortez D. Common mechanisms of PIKK regulation. DNA Repair (Amst) 2009;8:1004–1008. doi: 10.1016/j.dnarep.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cortez D, Wang Y, Qin J, Elledge SJ. Science. Vol. 286. New York, N.Y.: 1999. Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks; pp. 1162–1166. [DOI] [PubMed] [Google Scholar]
- 6.Traven A, Heierhorst J. SQ/TQ cluster domains: concentrated ATM/ATR kinase phosphorylation site regions in DNA-damage-response proteins. Bioessays. 2005;27:397–407. doi: 10.1002/bies.20204. [DOI] [PubMed] [Google Scholar]
- 7.Cortez D, Guntuku S, Qin J, Elledge SJ. Science. Vol. 294. New York, N.Y.: 2001. ATR and ATRIP: partners in checkpoint signaling; pp. 1713–1716. [DOI] [PubMed] [Google Scholar]
- 8.Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes & development. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- 9.de Klein A, Muijtjens M, van Os R, Verhoeven Y, Smit B, Carr AM, Lehmann AR, Hoeijmakers JH. Targeted disruption of the cell-cycle checkpoint gene ATR leads to early embryonic lethality in mice. Curr Biol. 2000;10:479–482. doi: 10.1016/s0960-9822(00)00447-4. [DOI] [PubMed] [Google Scholar]
- 10.Aguilera A, Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet. 2008;9:204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- 11.Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol. 2010;11:208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- 12.O'Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat Genet. 2003;33:497–501. doi: 10.1038/ng1129. [DOI] [PubMed] [Google Scholar]
- 13.Fang Y, Tsao CC, Goodman BK, Furumai R, Tirado CA, Abraham RT, Wang XF. ATR functions as a gene dosage-dependent tumor suppressor on a mismatch repair-deficient background. The EMBO journal. 2004;23:3164–3174. doi: 10.1038/sj.emboj.7600315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis KA, Mullany S, Thomas B, Chien J, Loewen R, Shridhar V, Cliby WA. Heterozygous ATR mutations in mismatch repair-deficient cancer cells have functional significance. Cancer Res. 2005;65:7091–7095. doi: 10.1158/0008-5472.CAN-05-1019. [DOI] [PubMed] [Google Scholar]
- 15.Zighelboim I, Schmidt AP, Gao F, Thaker PH, Powell MA, Rader JS, Gibb RK, Mutch DG, Goodfellow PJ. ATR mutation in endometrioid endometrial cancer is associated with poor clinical outcomes. J Clin Oncol. 2009;27:3091–3096. doi: 10.1200/JCO.2008.19.9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Menoyo A, Alazzouzi H, Espin E, Armengol M, Yamamoto H, Schwartz S., Jr Somatic mutations in the DNA damage-response genes ATR and CHK1 in sporadic stomach tumors with microsatellite instability. Cancer Res. 2001;61:7727–7730. [PubMed] [Google Scholar]
- 17.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, Orntoft T, Lukas J, Bartek J. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 18.Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA, Jr, Kastrinakis NG, Levy B, Kletsas D, Yoneta A, Herlyn M, Kittas C, Halazonetis TD. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 19.Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, Takaoka M, Nakagawa H, Tort F, Fugger K, Johansson F, Sehested M, Andersen CL, Dyrskjot L, Orntoft T, Lukas J, Kittas C, Helleday T, Halazonetis TD, Bartek J, Gorgoulis VG. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 20.Halazonetis TD, Gorgoulis VG, Bartek J. Science. Vol. 319. New York, N.Y.: 2008. An oncogene-induced DNA damage model for cancer development; pp. 1352–1355. [DOI] [PubMed] [Google Scholar]
- 21.Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes & development. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huertas P. DNA resection in eukaryotes: deciding how to fix the break. Nat Struct Mol Biol. 2010;17:11–16. doi: 10.1038/nsmb.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paciotti V, Clerici M, Lucchini G, Longhese MP. The checkpoint protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes & development. 2000;14:2046–2059. [PMC free article] [PubMed] [Google Scholar]
- 24.Rouse J, Jackson SP. LCD1: an essential gene involved in checkpoint control and regulation of the MEC1 signalling pathway in Saccharomyces cerevisiae. The EMBO journal. 2000;19:5801–5812. doi: 10.1093/emboj/19.21.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edwards RJ, Bentley NJ, Carr AM. A Rad3-Rad26 complex responds to DNA damage independently of other checkpoint proteins. Nature cell biology. 1999;1:393–398. doi: 10.1038/15623. [DOI] [PubMed] [Google Scholar]
- 26.Ball HL, Myers JS, Cortez D. ATRIP Binding to RPA-ssDNA Promotes ATR-ATRIP Localization but Is Dispensable for Chk1 Phosphorylation. Mol Biol Cell. 2005;16:2372–2381. doi: 10.1091/mbc.E04-11-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Unsal-Kacmaz K, Sancar A. Quaternary structure of ATR and effects of ATRIP and replication protein A on its DNA binding and kinase activities. Molecular and cellular biology. 2004;24:1292–1300. doi: 10.1128/MCB.24.3.1292-1300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zou L, Elledge SJ. Science. Vol. 300. New York, N.Y.: 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes; pp. 1542–1548. [DOI] [PubMed] [Google Scholar]
- 29.Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 30.Mordes DA, Glick GG, Zhao R, Cortez D. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes & development. 2008;22:1478–1489. doi: 10.1101/gad.1666208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takai H, Wang RC, Takai KK, Yang H, de Lange T. Tel2 regulates the stability of PI3K-related protein kinases. Cell. 2007;131:1248–1259. doi: 10.1016/j.cell.2007.10.052. [DOI] [PubMed] [Google Scholar]
- 32.Horejsi Z, Takai H, Adelman CA, Collis SJ, Flynn H, Maslen S, Skehel JM, de Lange T, Boulton SJ. CK2 phospho-dependent binding of R2TP complex to TEL2 is essential for mTOR and SMG1 stability. Molecular cell. 2010;39:839–850. doi: 10.1016/j.molcel.2010.08.037. [DOI] [PubMed] [Google Scholar]
- 33.Hurov KE, Cotta-Ramusino C, Elledge SJ. A genetic screen identifies the Triple T complex required for DNA damage signaling and ATM and ATR stability. Genes & development. 2010;24:1939–1950. doi: 10.1101/gad.1934210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takai H, Xie Y, de Lange T, Pavletich NP. Tel2 structure and function in the Hsp90-dependent maturation of mTOR and ATR complexes. Genes & development. 2010;24:2019–2030. doi: 10.1101/gad.1956410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaizuka T, Hara T, Oshiro N, Kikkawa U, Yonezawa K, Takehana K, Iemura S, Natsume T, Mizushima N. Tti1 and Tel2 are critical factors in mammalian target of rapamycin complex assembly. The Journal of biological chemistry. 2010;285:20109–20116. doi: 10.1074/jbc.M110.121699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ball HL, Cortez D. ATRIP oligomerization is required for ATR-dependent checkpoint signaling. The Journal of biological chemistry. 2005;280:31390–31396. doi: 10.1074/jbc.M504961200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Itakura E, Sawada I, Matsuura A. Dimerization of the ATRIP Protein through the Coiled-Coil Motif and Its Implication to the Maintenance of Stalled Replication Forks. Mol Biol Cell. 2005;16:5551–5562. doi: 10.1091/mbc.E05-05-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lovejoy CA, Xu X, Bansbach CE, Glick GG, Zhao R, Ye F, Sirbu BM, Titus LC, Shyr Y, Cortez D. Functional genomic screens identify CINP as a genome maintenance protein. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:19304–19309. doi: 10.1073/pnas.0909345106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Myers JS, Zhao R, Xu X, Ham AJ, Cortez D. Cyclin-dependent kinase 2 dependent phosphorylation of ATRIP regulates the G2-M checkpoint response to DNA damage. Cancer Res. 2007;67:6685–6690. doi: 10.1158/0008-5472.CAN-07-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalay E, Yigit G, Aslan Y, Brown KE, Pohl E, Bicknell LS, Kayserili H, Li Y, Tuysuz B, Nurnberg G, Kiess W, Koegl M, Baessmann I, Buruk K, Toraman B, Kayipmaz S, Kul S, Ikbal M, Turner DJ, Taylor MS, Aerts J, Scott C, Milstein K, Dollfus H, Wieczorek D, Brunner HG, Hurles M, Jackson AP, Rauch A, Nurnberg P, Karaguzel A, Wollnik B. CEP152 is a genome maintenance protein disrupted in Seckel syndrome. Nat Genet. 2010 doi: 10.1038/ng.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Griffith E, Walker S, Martin CA, Vagnarelli P, Stiff T, Vernay B, Al Sanna N, Saggar A, Hamel B, Earnshaw WC, Jeggo PA, Jackson AP, O'Driscoll M. Mutations in pericentrin cause Seckel syndrome with defective ATR-dependent DNA damage signaling. Nat Genet. 2008;40:232–236. doi: 10.1038/ng.2007.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Al-Dosari MS, Shaheen R, Colak D, Alkuraya FS. Novel CENPJ mutation causes Seckel syndrome. J Med Genet. 2010;47:411–414. doi: 10.1136/jmg.2009.076646. [DOI] [PubMed] [Google Scholar]
- 43.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. Science. Vol. 316. New York, N.Y.: 2007. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage; pp. 1160–1166. [DOI] [PubMed] [Google Scholar]
- 44.Sivasubramaniam S, Sun X, Pan YR, Wang S, Lee EY. Cep164 is a mediator protein required for the maintenance of genomic stability through modulation of MDC1, RPA, and CHK1. Genes & development. 2008;22:587–600. doi: 10.1101/gad.1627708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Graser S, Stierhof YD, Lavoie SB, Gassner OS, Lamla S, Le Clech M, Nigg EA. Cep164, a novel centriole appendage protein required for primary cilium formation. The Journal of cell biology. 2007;179:321–330. doi: 10.1083/jcb.200707181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pan YR, Lee EY. UV-dependent interaction between Cep164 and XPA mediates localization of Cep164 at sites of DNA damage and UV sensitivity. Cell Cycle. 2009;8:655–664. doi: 10.4161/cc.8.4.7844. [DOI] [PubMed] [Google Scholar]
- 47.Zinkel SS, Hurov KE, Gross A. Bid plays a role in the DNA damage response. Cell. 2007;130:9–10. doi: 10.1016/j.cell.2007.06.035. author reply 10-11. [DOI] [PubMed] [Google Scholar]
- 48.Kamer I, Sarig R, Zaltsman Y, Niv H, Oberkovitz G, Regev L, Haimovich G, Lerenthal Y, Marcellus RC, Gross A. Proapoptotic BID is an ATM effector in the DNA-damage response. Cell. 2005;122:593–603. doi: 10.1016/j.cell.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 49.Liu Y, Bertram CC, Shi Q, Zinkel SS. Proapoptotic Bid mediates the Atr-directed DNA damage response to replicative stress. Cell Death Differ. 2010 doi: 10.1038/cdd.2010.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ball HL, Ehrhardt MR, Mordes DA, Glick GG, Chazin WJ, Cortez D. Function of a conserved checkpoint recruitment domain in ATRIP proteins. Molecular and cellular biology. 2007;27:3367–3377. doi: 10.1128/MCB.02238-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mohni KN, Livingston CM, Cortez D, Weller SK. ATR and ATRIP are recruited to herpes simplex virus type 1 replication compartments even though ATR signaling is disabled. J Virol. 2010;84:12152–12164. doi: 10.1128/JVI.01643-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Umezu K, Sugawara N, Chen C, Haber JE, Kolodner RD. Genetic analysis of yeast RPA1 reveals its multiple functions in DNA metabolism. Genetics. 1998;148:989–1005. doi: 10.1093/genetics/148.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu X, Vaithiyalingam S, Glick GG, Mordes DA, Chazin WJ, Cortez D. The basic cleft of RPA70N binds multiple checkpoint proteins including RAD9 to regulate ATR signaling. Molecular and cellular biology. 2008;28:7345–7353. doi: 10.1128/MCB.01079-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Remus D, Diffley JF. Eukaryotic DNA replication control: lock and load, then fire. Current opinion in cell biology. 2009;21:771–777. doi: 10.1016/j.ceb.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 55.Murakami H, Yanow SK, Griffiths D, Nakanishi M, Nurse P. Maintenance of replication forks and the S-phase checkpoint by Cdc18p and Orp1p. Nature cell biology. 2002;4:384–388. doi: 10.1038/ncb789. [DOI] [PubMed] [Google Scholar]
- 56.Oehlmann M, Score AJ, Blow JJ. The role of Cdc6 in ensuring complete genome licensing and S phase checkpoint activation. The Journal of cell biology. 2004;165:181–190. doi: 10.1083/jcb.200311044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clay-Farrace L, Pelizon C, Santamaria D, Pines J, Laskey RA. Human replication protein Cdc6 prevents mitosis through a checkpoint mechanism that implicates Chk1. The EMBO journal. 2003;22:704–712. doi: 10.1093/emboj/cdg046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lau E, Zhu C, Abraham RT, Jiang W. The functional role of Cdc6 in S-G2/M in mammalian cells. EMBO Rep. 2006;7:425–430. doi: 10.1038/sj.embor.7400624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hermand D, Nurse P. Cdc18 enforces long-term maintenance of the S phase checkpoint by anchoring the Rad3-Rad26 complex to chromatin. Molecular cell. 2007;26:553–563. doi: 10.1016/j.molcel.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 60.Yoshida K, Sugimoto N, Iwahori S, Yugawa T, Narisawa-Saito M, Kiyono T, Fujita M. CDC6 interaction with ATR regulates activation of a replication checkpoint in higher eukaryotic cells. J Cell Sci. 2010;123:225–235. doi: 10.1242/jcs.058693. [DOI] [PubMed] [Google Scholar]
- 61.Meetei AR, Medhurst AL, Ling C, Xue Y, Singh TR, Bier P, Steltenpool J, Stone S, Dokal I, Mathew CG, Hoatlin M, Joenje H, de Winter JP, Wang W. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat Genet. 2005;37:958–963. doi: 10.1038/ng1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mosedale G, Niedzwiedz W, Alpi A, Perrina F, Pereira-Leal JB, Johnson M, Langevin F, Pace P, Patel KJ. The vertebrate Hef ortholog is a component of the Fanconi anemia tumor-suppressor pathway. Nat Struct Mol Biol. 2005;12:763–771. doi: 10.1038/nsmb981. [DOI] [PubMed] [Google Scholar]
- 63.Collis SJ, Ciccia A, Deans AJ, Horejsi Z, Martin JS, Maslen SL, Skehel JM, Elledge SJ, West SC, Boulton SJ. FANCM and FAAP24 function in ATR-mediated checkpoint signaling independently of the Fanconi anemia core complex. Molecular cell. 2008;32:313–324. doi: 10.1016/j.molcel.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 64.Ciccia A, Ling C, Coulthard R, Yan Z, Xue Y, Meetei AR, Laghmani el H, Joenje H, McDonald N, de Winter JP, Wang W, West SC. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Molecular cell. 2007;25:331–343. doi: 10.1016/j.molcel.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 65.Kim JM, Kee Y, Gurtan A, D'Andrea AD. Cell cycle-dependent chromatin loading of the Fanconi anemia core complex by FANCM/FAAP24. Blood. 2008;111:5215–5222. doi: 10.1182/blood-2007-09-113092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schwab RA, Blackford AN, Niedzwiedz W. ATR activation and replication fork restart are defective in FANCM-deficient cells. The EMBO journal. 2010;29:806–818. doi: 10.1038/emboj.2009.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Luke-Glaser S, Luke B, Grossi S, Constantinou A. FANCM regulates DNA chain elongation and is stabilized by S-phase checkpoint signalling. The EMBO journal. 2010;29:795–805. doi: 10.1038/emboj.2009.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Singh TR, Saro D, Ali AM, Zheng XF, Du CH, Killen MW, Sachpatzidis A, Wahengbam K, Pierce AJ, Xiong Y, Sung P, Meetei AR. MHF1-MHF2, a histone-fold-containing protein complex, participates in the Fanconi anemia pathway via FANCM. Molecular cell. 2010;37:879–886. doi: 10.1016/j.molcel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yan Z, Delannoy M, Ling C, Daee D, Osman F, Muniandy PA, Shen X, Oostra AB, Du H, Steltenpool J, Lin T, Schuster B, Decaillet C, Stasiak A, Stasiak AZ, Stone S, Hoatlin ME, Schindler D, Woodcock CL, Joenje H, Sen R, de Winter JP, Li L, Seidman MM, Whitby MC, Myung K, Constantinou A, Wang W. A histone-fold complex and FANCM form a conserved DNA-remodeling complex to maintain genome stability. Molecular cell. 2010;37:865–878. doi: 10.1016/j.molcel.2010.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang M, Kim JM, Shiotani B, Yang K, Zou L, D'Andrea AD. The FANCM/FAAP24 complex is required for the DNA interstrand crosslink-induced checkpoint response. Molecular cell. 2010;39:259–268. doi: 10.1016/j.molcel.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stojic L, Mojas N, Cejka P, Di Pietro M, Ferrari S, Marra G, Jiricny J. Mismatch repair-dependent G2 checkpoint induced by low doses of SN1 type methylating agents requires the ATR kinase. Genes & development. 2004;18:1331–1344. doi: 10.1101/gad.294404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yoshioka K, Yoshioka Y, Hsieh P. ATR kinase activation mediated by MutSalpha and MutLalpha in response to cytotoxic O6-methylguanine adducts. Molecular cell. 2006;22:501–510. doi: 10.1016/j.molcel.2006.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Y, Qin J. MSH2 and ATR form a signaling module and regulate two branches of the damage response to DNA methylation. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:15387–15392. doi: 10.1073/pnas.2536810100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu Y, Fang Y, Shao H, Lindsey-Boltz L, Sancar A, Modrich P. Interactions of human mismatch repair proteins MutSalpha and MutLalpha with proteins of the ATR-Chk1 pathway. The Journal of biological chemistry. 2010;285:5974–5982. doi: 10.1074/jbc.M109.076109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pabla N, Ma Z, McIlhatton MA, Fishel R, Dong Z. hMSH2 recruits ATR to DNA damage sites for activation during DNA damage-induced apoptosis. The Journal of biological chemistry. 2011 doi: 10.1074/jbc.M110.210989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Unsal-Kacmaz K, Makhov AM, Griffith JD, Sancar A. Preferential binding of ATR protein to UV-damaged DNA. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:6673–6678. doi: 10.1073/pnas.102167799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Choi JH, Lindsey-Boltz LA, Sancar A. Cooperative activation of the ATR checkpoint kinase by TopBP1 and damaged DNA. Nucleic acids research. 2009;37:1501–1509. doi: 10.1093/nar/gkn1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rubinson EH, Gowda AS, Spratt TE, Gold B, Eichman BF. An unprecedented nucleic acid capture mechanism for excision of DNA damage. Nature. 2010;468:406–411. doi: 10.1038/nature09428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Williams DR, Lee KJ, Shi J, Chen DJ, Stewart PL. Cryo-EM Structure of the DNA-Dependent Protein Kinase Catalytic Subunit at Subnanometer Resolution Reveals alpha Helices and Insight into DNA Binding. Structure. 2008;16:468–477. doi: 10.1016/j.str.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sibanda BL, Chirgadze DY, Blundell TL. Crystal structure of DNA-PKcs reveals a large open-ring cradle comprised of HEAT repeats. Nature. 2010;463:118–121. doi: 10.1038/nature08648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sun Y, Xu Y, Roy K, Price BD. DNA damage induced acetylation of lysine 3016 of ATM activates ATM kinase activity. Molecular and cellular biology. 2007;24:8502–8509. doi: 10.1128/MCB.01382-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guo Z, Kozlov S, Lavin MF, Person MD, Paull TT. Science. Vol. 330. New York, N.Y.: 2010. ATM activation by oxidative stress; pp. 517–521. [DOI] [PubMed] [Google Scholar]
- 83.Mordes DA, Nam EA, Cortez D. Dpb11 activates the Mec1-Ddc2 complex. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:18730–18734. doi: 10.1073/pnas.0806621105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Navadgi-Patil VM, Burgers PM. Yeast DNA replication protein Dpb11 activates the Mec1/ATR checkpoint kinase. The Journal of biological chemistry. 2008;283:35853–35859. doi: 10.1074/jbc.M807435200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Navadgi-Patil VM, Burgers PM. The unstructured C-terminal tail of the 9-1-1 clamp subunit Ddc1 activates Mec1/ATR via two distinct mechanisms. Molecular cell. 2009;36:743–753. doi: 10.1016/j.molcel.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Majka J, Niedziela-Majka A, Burgers PM. The Checkpoint Clamp Activates Mec1 Kinase during Initiation of the DNA Damage Checkpoint. Molecular cell. 2006;24:891–901. doi: 10.1016/j.molcel.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Choi JH, Lindsey-Boltz LA, Kemp M, Mason AC, Wold MS, Sancar A. Reconstitution of RPA-covered single-stranded DNA-activated ATR-Chk1 signaling. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:13660–13665. doi: 10.1073/pnas.1007856107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ellison V, Stillman B. Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5′ recessed DNA. PLoS Biol. 2003;1:E33. doi: 10.1371/journal.pbio.0000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bermudez VP, Lindsey-Boltz LA, Cesare AJ, Maniwa Y, Griffith JD, Hurwitz J, Sancar A. Loading of the human 9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17-replication factor C complex in vitro. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:1633–1638. doi: 10.1073/pnas.0437927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zou L, Cortez D, Elledge SJ. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes & development. 2002;16:198–208. doi: 10.1101/gad.950302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Furuya K, Poitelea M, Guo L, Caspari T, Carr AM. Chk1 activation requires Rad9 S/TQ-site phosphorylation to promote association with C-terminal BRCT domains of Rad4TOPBP1. Genes & development. 2004;18:1154–1164. doi: 10.1101/gad.291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes & development. 2007;21:1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lee J, Kumagai A, Dunphy WG. The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. The Journal of biological chemistry. 2007;282:28036–28044. doi: 10.1074/jbc.M704635200. [DOI] [PubMed] [Google Scholar]
- 94.Kondo T, Wakayama T, Naiki T, Matsumoto K, Sugimoto K. Science. Vol. 294. New York, N.Y.: 2001. Recruitment of Mec1 and Ddc1 checkpoint proteins to double-strand breaks through distinct mechanisms; pp. 867–870. [DOI] [PubMed] [Google Scholar]
- 95.Melo JA, Cohen J, Toczyski DP. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes & development. 2001;15:2809–2821. doi: 10.1101/gad.903501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yan S, Michael WM. TopBP1 and DNA polymerase-alpha directly recruit the 9-1-1 complex to stalled DNA replication forks. The Journal of cell biology. 2009;184:793–804. doi: 10.1083/jcb.200810185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cescutti R, Negrini S, Kohzaki M, Halazonetis TD. TopBP1 functions with 53BP1 in the G1 DNA damage checkpoint. The EMBO journal. 2010;29:3723–3732. doi: 10.1038/emboj.2010.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yamane K, Wu X, Chen J. A DNA damage-regulated BRCT-containing protein, TopBP1, is required for cell survival. Molecular and cellular biology. 2002;22:555–566. doi: 10.1128/MCB.22.2.555-566.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yoo HY, Kumagai A, Shevchenko A, Dunphy WG. The Mre11-Rad50-Nbs1 complex mediates activation of TopBP1 by ATM. Mol Biol Cell. 2009;20:2351–2360. doi: 10.1091/mbc.E08-12-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kumagai A, Shevchenko A, Dunphy WG. Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell. 2010;140:349–359. doi: 10.1016/j.cell.2009.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ramirez-Lugo JS, Yoo HY, Yoon SJ, Dunphy WG. CtIP interacts with TopBP1 and Nbs1 in the response to double-stranded DNA breaks (DSBs) in Xenopus egg extracts. Cell Cycle. 2011;10:469–480. doi: 10.4161/cc.10.3.14711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Leung CC, Gong Z, Chen J, Glover JN. Molecular Basis of BACH1/FANCJ Recognition by TopBP1 in DNA Replication Checkpoint Control. The Journal of biological chemistry. 2011;286:4292–4301. doi: 10.1074/jbc.M110.189555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gong Z, Kim JE, Leung CC, Glover JN, Chen J. BACH1/FANCJ acts with TopBP1 and participates early in DNA replication checkpoint control. Molecular cell. 2010;37:438–446. doi: 10.1016/j.molcel.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K, Luo G, Carattini-Rivera S, DeMayo F, Bradley A, Donehower LA, Elledge SJ. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes & development. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 105.Zhao H, Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Molecular and cellular biology. 2001;21:4129–4139. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kumagai A, Kim SM, Dunphy WG. Claspin and the activated form of ATR-ATRIP collaborate in the activation of Chk1. The Journal of biological chemistry. 2004 doi: 10.1074/jbc.M408353200. [DOI] [PubMed] [Google Scholar]
- 107.Stracker TH, Usui T, Petrini JH. Taking the time to make important decisions: the checkpoint effector kinases Chk1 and Chk2 and the DNA damage response. DNA Repair (Amst) 2009;8:1047–1054. doi: 10.1016/j.dnarep.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tercero JA, Diffley JF. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature. 2001;412:553–557. doi: 10.1038/35087607. [DOI] [PubMed] [Google Scholar]
- 109.Woodward AM, Gohler T, Luciani MG, Oehlmann M, Ge X, Gartner A, Jackson DA, Blow JJ. Excess Mcm2-7 license dormant origins of replication that can be used under conditions of replicative stress. The Journal of cell biology. 2006;173:673–683. doi: 10.1083/jcb.200602108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ge XQ, Jackson DA, Blow JJ. Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes & development. 2007;21:3331–3341. doi: 10.1101/gad.457807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ibarra A, Schwob E, Mendez J. Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proceedings of the National Academy of Sciences. 2008;105:8956–8961. doi: 10.1073/pnas.0803978105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Randell JC, Fan A, Chan C, Francis LI, Heller RC, Galani K, Bell SP. Mec1 is one of multiple kinases that prime the Mcm2-7 helicase for phosphorylation by Cdc7. Molecular cell. 2010;40:353–363. doi: 10.1016/j.molcel.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liu H, Takeda S, Kumar R, Westergard TD, Brown EJ, Pandita TK, Cheng EH, Hsieh JJ. Phosphorylation of MLL by ATR is required for execution of mammalian S-phase checkpoint. Nature. 2010;467:343–346. doi: 10.1038/nature09350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liu H, Takeda S, Cheng EH, Hsieh JJ. Biphasic MLL takes helm at cell cycle control: implications in human mixed lineage leukemia. Cell Cycle. 2008;7:428–435. doi: 10.4161/cc.7.4.5426. [DOI] [PubMed] [Google Scholar]
- 115.Liu H, Cheng EH, Hsieh JJ. Bimodal degradation of MLL by SCFSkp2 and APCCdc20 assures cell cycle execution: a critical regulatory circuit lost in leukemogenic MLL fusions. Genes & development. 2007;21:2385–2398. doi: 10.1101/gad.1574507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yoo HY, Shevchenko A, Dunphy WG. Mcm2 is a direct substrate of ATM and ATR during DNA damage and DNA replication checkpoint responses. The Journal of biological chemistry. 2004;279:53353–53364. doi: 10.1074/jbc.M408026200. [DOI] [PubMed] [Google Scholar]
- 117.Cortez D, Glick G, Elledge SJ. Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proceedings of National Academy of Sciences of the United States of America. 2004;101:10078–10083. doi: 10.1073/pnas.0403410101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Trenz K, Errico A, Costanzo V. Plx1 is required for chromosomal DNA replication under stressful conditions. The EMBO journal. 2008;27:876–885. doi: 10.1038/emboj.2008.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ge XQ, Blow JJ. Chk1 inhibits replication factory activation but allows dormant origin firing in existing factories. The Journal of cell biology. 2010 doi: 10.1083/jcb.201007074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Labib K. How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes & development. 2010;24:1208–1219. doi: 10.1101/gad.1933010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero DE, Meyer T, Cimprich KA. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Molecular cell. 2009;35:228–239. doi: 10.1016/j.molcel.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Labib K, Hodgson B. Replication fork barriers: pausing for a break or stalling for time? EMBO Rep. 2007;8:346–353. doi: 10.1038/sj.embor.7400940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Romano G, Giordano A. Role of the cyclin-dependent kinase 9-related pathway in mammalian gene expression and human diseases. Cell Cycle. 2008;7:3664–3668. doi: 10.4161/cc.7.23.7122. [DOI] [PubMed] [Google Scholar]
- 124.Pirngruber J, Shchebet A, Johnsen SA. Insights into the function of the human P-TEFb component CDK9 in the regulation of chromatin modifications and co-transcriptional mRNA processing. Cell Cycle. 2009;8:3636–3642. doi: 10.4161/cc.8.22.9890. [DOI] [PubMed] [Google Scholar]
- 125.Yu DS, Zhao R, Hsu EL, Cayer J, Ye F, Guo Y, Shyr Y, Cortez D. Cyclin-dependent kinase 9-cyclin K functions in the replication stress response. EMBO Rep. 2010;11:876–882. doi: 10.1038/embor.2010.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Clausing E, Mayer A, Chanarat S, Muller B, Germann SM, Cramer P, Lisby M, Strasser K. The transcription elongation factor Bur1-Bur2 interacts with replication protein A and maintains genome stability during replication stress. The Journal of biological chemistry. 2010 doi: 10.1074/jbc.M110.193292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Grote M, Wolf E, Will CL, Lemm I, Agafonov DE, Schomburg A, Fischle W, Urlaub H, Luhrmann R. Molecular architecture of the human Prp19/CDC5L complex. Molecular and cellular biology. 2010;30:2105–2119. doi: 10.1128/MCB.01505-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhang N, Kaur R, Akhter S, Legerski RJ. Cdc5L interacts with ATR and is required for the S-phase cell-cycle checkpoint. EMBO Rep. 2009;10:1029–1035. doi: 10.1038/embor.2009.122. [DOI] [PMC free article] [PubMed] [Google Scholar]