Summary
Assembly of the endosomal sorting complex required for transport (ESCRT)-III executes the formation of intralumenal vesicles (ILVs) at endosomes. Repeated cycles of ESCRT-III function requires disassembly of the complex by Vps4, an ATPase with a microtubule interaction and trafficking (MIT) domain that binds MIT-interacting motifs (MIM1 or MIM2) in ESCRT-III subunits. We identified a putative MIT domain at the N-terminus of Doa4, which is the ubiquitin (Ub) hydrolase in Saccharomyces cerevisiae that deubiquitinates ILV cargo proteins. The Doa4 N-terminus is predicted to have the α-helical structure common to MIT domains, and it binds directly to a MIM1-like sequence in the Vps20 subunit of ESCRT-III. Disrupting this interaction does not prevent endosomal localization of Doa4 but enhances the defect in ILV cargo protein deubiquitination observed in cells lacking Bro1, which is an ESCRT-III effector protein that stimulates Doa4 catalytic activity. Deletion of the BRO1 gene (bro1Δ) blocks ILV budding, but ILV budding was rescued upon disrupting the interaction between Vps20 and Doa4. This rescue in ILV biogenesis requires Doa4 expression but is independent of its Ub hydrolase activity. Thus, binding of Vps20 to the Doa4 N-terminus inhibits a non-catalytic function of Doa4 that promotes ILV formation.
Key words: Multivesicular body, Vesicle budding, Deubiquitination
Introduction
The endosomal sorting complexes required for transport (ESCRTs) sort ubiquitinated transmembrane proteins at endosomes into intralumenal vesicles (ILVs) that are subsequently degraded in the hydrolytic interior of lysosomes upon endolysosomal fusion. ILV cargo recognition is mediated by ESCRT-0, -I, and -II, which bind directly to ubiquitin (Ub) conjugates on the cytosolic domains of transmembrane proteins targeted for destruction (reviewed in Henne et al., 2011). Based on in vitro studies that reconstituted ILV budding at synthetic membranes, ESCRT-I and -II induce the formation of ILV buds that detach as free ILVs upon membrane scission catalyzed by ESCRT-III (Wollert and Hurley, 2010; Wollert et al., 2009). How the activities of ESCRTs are regulated in vivo to drive the ILV budding reaction is poorly understood.
ESCRT-III subunits exist as soluble inactive monomers and polymerize into the active complex only on membranes (Babst et al., 2002; Shim et al., 2007), which is a prerequisite to execute membrane scission (Wollert et al., 2009). Saccharomyces cerevisiae has seven ESCRT-III subunits, four of which (Vps20, Snf7, Vps24 and Vps2) are thought to comprise the core of the complex and assemble in the above order (Teis et al., 2008). Vps20 initiates ESCRT-III assembly by stimulating homopolymerization of Snf7, the most abundant subunit of the complex (Saksena et al., 2009; Teis et al., 2008). Vps24 and Vps2 terminate ESCRT-III assembly by capping the Snf7 polymer (Teis et al., 2008). The other ESCRT-III subunits (Did2, Ist1 and Vps60) regulate disassembly of the complex but are not strictly required for ILV budding (Dimaano et al., 2008; Nickerson et al., 2006; Rue et al., 2008).
Vps4 catalyzes disassembly and dissociation of ESCRT-III subunits from the membrane, which is essential to recycle subunits for subsequent rounds of complex assembly. Vps4 belongs to the diverse family of AAA+ ATPases that unfold proteins and/or disassemble protein complexes (Babst et al., 1998). Direct contact between Vps4 and ESCRT-III is mediated by the microtubule interacting and trafficking (MIT) domain at the N-terminus of Vps4, which binds two distinct MIT-interacting motifs (MIM1 or MIM2) at or near the C termini of ESCRT-III subunits (Kieffer et al., 2008; Obita et al., 2007; Stuchell-Brereton et al., 2007). The MIT domain of Vps4 binds every ESCRT-III subunit through either its MIM1 (in Vps24, Vps2, Did2 and Ist1) or MIM2 (in Vps20, Snf7 and Ist1). However, other ESCRT-III effector proteins that contain MIT domains bind more selectively to a single subunit or subset of subunits (reviewed in Hurley, 2010), including AMSH and UBPY, two Ub hydrolases in humans that bind a distinct but overlapping subset of ESCRT-III proteins (Agromayor and Martin-Serrano, 2006; Row et al., 2007).
In yeast, two MIT domains exist in Vta1 (Xiao et al., 2008), which is a Vps4 cofactor that stimulates its ATPase activity (Azmi et al., 2006; Azmi et al., 2008; Lottridge et al., 2006), but no other yeast proteins that associate with ESCRT-III are known to have an MIT domain. We show that the yeast Doa4 Ub hydrolase contains a candidate MIT domain at its N-terminus that interacts specifically with Vps20. Binding to the Doa4 N-terminus requires conserved amino acids within a MIM1-like sequence in helix α6 at the C terminus of Vps20. This site, which we refer to as MIMα6, is spatially separate from the MIM2 sequence in Vps20 that binds the MIT domain of Vps4 (Kieffer et al., 2008; Shestakova et al., 2010). Mutation of Vps20 MIMα6 (vps20ΔMIMα6) alone did not impair Doa4 function, but the vps20ΔMIMα6 mutation caused a strong synthetic inhibition of deubiquitination when combined with deletion of the BRO1 gene, which encodes an ESCRT-III-associated protein that stimulates Doa4 Ub hydrolase activity (Richter et al., 2007). Surprisingly, the vps20ΔMIMα6 mutation rescued ILV budding in the absence of Bro1, and this rescue required Doa4 expression but was independent of Doa4 Ub hydrolase activity. These results reveal an unexpected non-catalytic role for Doa4 in ILV budding that is inhibited through its interaction with the Vps20 subunit of ESCRT-III.
Results
The N-terminus of Doa4 binds a MIM1-like sequence in Vps20 helix α6
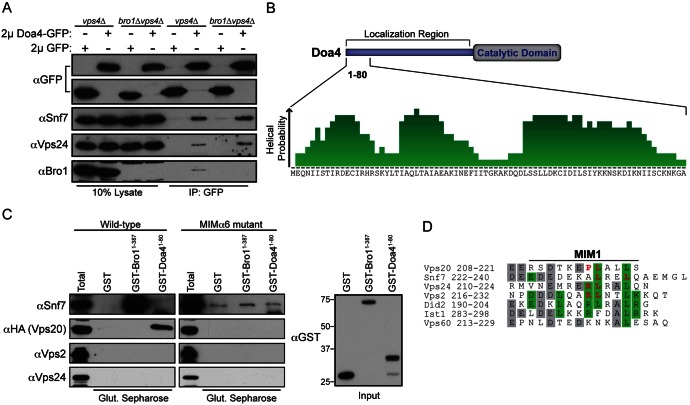
The Snf7 subunit of ESCRT-III in yeast binds Bro1 (Kim et al., 2005; Odorizzi et al., 2003), which is an auxiliary protein that stabilizes ESCRT-III assembly (Wemmer et al., 2011). Bro1 also functions as an ESCRT-III effector by promoting the deubiquitination of ILV cargoes through its recruitment and activation of Doa4 (Luhtala and Odorizzi, 2004; Richter et al., 2007). Deubiquitination by Doa4 is required for cargoes to be sorted into ILVs (Nikko and André, 2007), and the need for Bro1 in Doa4 function can be bypassed by overexpressing the DOA4 gene (Amerik et al., 2006; Luhtala and Odorizzi, 2004). Deleting BRO1 (bro1Δ) in tandem with other ESCRT genes revealed that each of the core ESCRT-III subunits (Snf7, Vps20, Vps2 or Vps24) is required for overexpressed Doa4-GFP to localize to endosomes in the absence of Bro1 (Fig. 1). In addition, ESCRT-III subunits co-immunoprecipitated with Doa4-GFP overexpressed from a high-copy (2µ) plasmid regardless of whether BRO1 was deleted (Fig. 2A), indicating that Doa4 associates with ESCRT-III independently of Bro1, provided that the ESCRT-III core complex is intact.
Fig. 1.
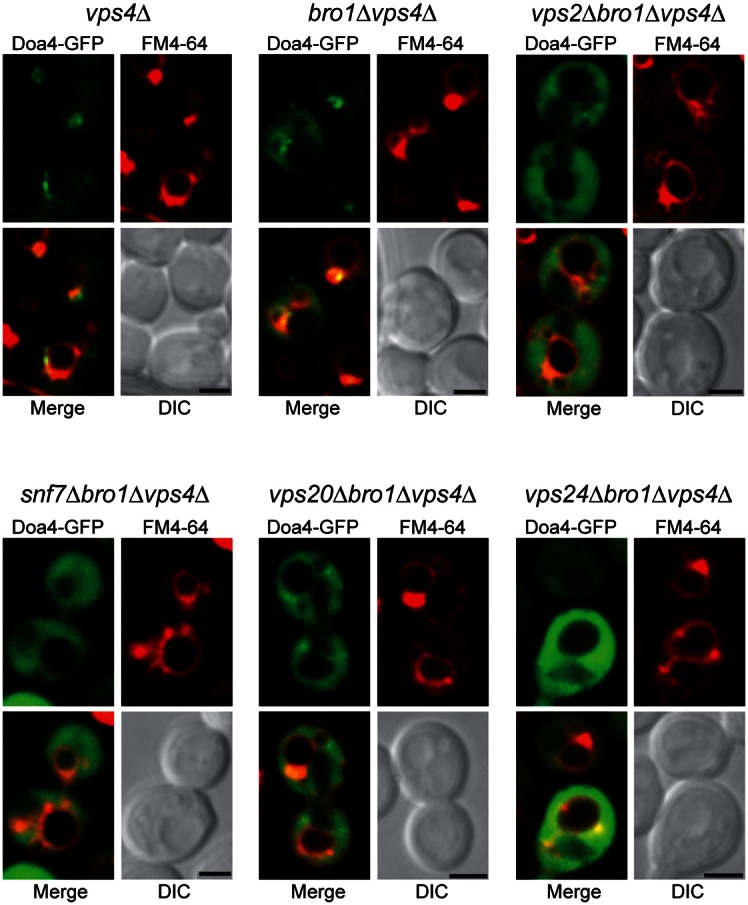
ESCRT-III is required for Bro1-independent localization of overexpressed Doa4 to endosomes. Fluorescence and differential interference contrast (DIC) microscopy of 2μ Doa4-GFP and FM4-64, a fluorescent dye that labels class E compartments and vacuolar membranes (Vida and Emr, 1995). Scale bars: 2 µm.
Fig. 2.
The N-terminal domain of Doa4 binds to a MIM1-like sequence in Vps20 helix α6. (A) In vivo αGFP immunoprecipitation (IP) of 2μ GFP or Doa4-GFP and immunoblot with αGFP, αSnf7, αVps24 and αBro1. The experiment was performed in vps4Δ cells to stabilize ESCRT-III, which is largely disassembled at steady state in the presence of VPS4. (B) PredictProtein (Rost et al., 2004) helical probability plot for amino acids 1-80 of Doa4. (C) In vitro glutathione (Glut.)–Sepharose pulldowns of purified GST, GST-Bro11-387, or GST-Doa41-80 mixed with E. coli lysates expressing wild-type or MIMα6-mutant Snf7, Vps20-HA, Vps2, or Vps24. GST-Bro11-387 served as a positive control for binding to the α6 helix of Snf7. For unknown reasons, the MIMα6 mutant of Snf7 had a higher propensity for non-specific binding than wild-type Snf7. Immunoblots were probed with αSnf7, αHA, αVps2, αVps24 or αGST. (D) Sequence alignment of the α6 helices of S. cerevisiae ESCRT-III subunits using the PredictProtein online server (Rost et al., 2004), which contain MIM1 or MIM1-like sequences. Grey boxes indicate conserved residues and green boxes indicate residues that conform to the MIM1 consensus sequence (D/E)xxLxxRLxxL(K/R). Red lettering designates the positions at which the endogenous residues were substituted with aspartic acid in the MIMα6 mutants in C.
Amino acids 1-80 of Doa4 are predicted to fold into three α helices (Fig. 2B), which is similar to the size and secondary structure of MIT domains in Vps4 and Vta1 (Obita et al., 2007; Xiao et al., 2008). Therefore, we hypothesized Doa4 residues 1-80 comprise an MIT-like domain lacking sequence homology to the known MIT domains that mediate binding of Vps4 and Vta1 to ESCRT-III subunits (Kieffer et al., 2008; Obita et al., 2007; Stuchell-Brereton et al., 2007; Xiao et al., 2008). In support of this hypothesis, purified recombinant GST-Doa41-80 bound HA-tagged Vps20 (Fig. 2C), and this interaction was abolished by mutation of conserved residues in Vps20 helix α6 that are similar to the MIM1 consensus sequence that binds the Vps4 MIT domain (Fig. 2D; Obita et al., 2007). Like the MIT domains of human AMSH and UBPY (Agromayor and Martin-Serrano, 2006; Row et al., 2007), the N-terminus of Doa4 exhibits binding specificity because GST-Doa41-80 did not bind other core ESCRT-III subunits, Vps2, Snf7 or Vps24 (Fig. 2C).
Vps20 MIMα6 is not required for Doa4 endosomal recruitment
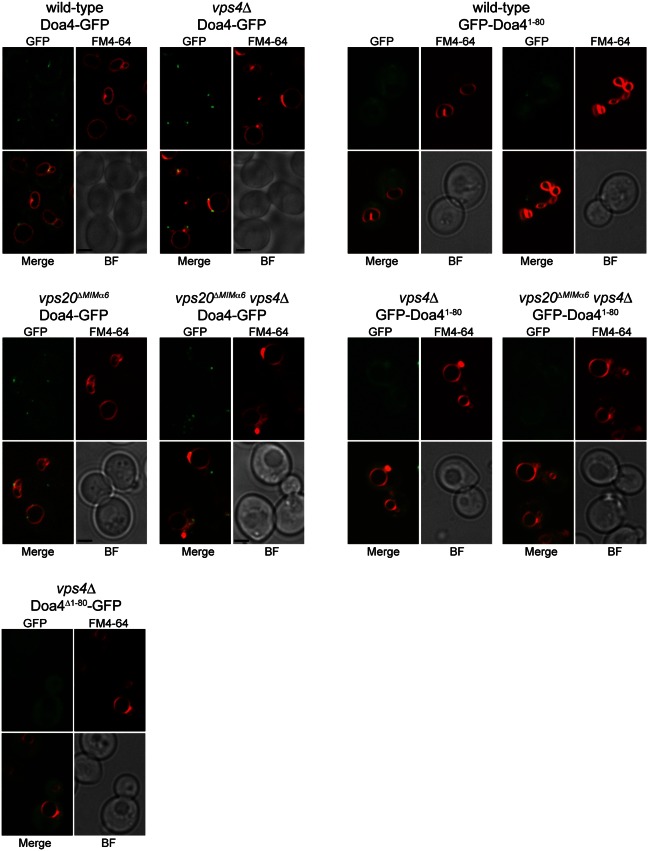
Endosomal localization of Doa4 is disabled by mutations within residues 1-80 (Amerik et al., 2006), raising the possibility that endosomal recruitment of Doa4 is mediated by its interaction with the MIM1-like sequence in Vps20 helix α6 (hereafter referred to as Vps20 MIMα6). However, unlike full-length Doa4-GFP (Luhtala and Odorizzi, 2004; Fig. 3), GFP-Doa41-80 exhibited a predominantly cytosolic distribution rather than the punctate localization characteristic of endosomes (Fig. 3). GFP-Doa41-80 was similarly cytosolic in vps4Δ cells (Fig. 3), which provided more compelling evidence that the N-terminus of Doa4 cannot autonomously mediate endosomal recruitment because the absence of Vps4 traps wild-type Doa4 at aberrant endosomal structures known as ‘class E compartments’ formed in ESCRT-mutant strains (Luhtala and Odorizzi, 2004; Fig. 3). Curiously, GFP-Doa41-80 was present occasionally within the vacuole lumen of wild-type cells (Fig. 3), suggesting it had been packaged into ILVs bound for vacuolar delivery because of transient association with ESCRT-III.
Fig. 3.
Vps20 MIMα6 does not recruit Doa4 to endomembranes. Fluorescence and brightfield (BF) microscopy of Doa4-GFP, GFP-Doa41-80, or Doa4Δ1-80-GFP and FM4-64. Scale bars: 2 µm.
Further evidence that endosomal recruitment of Doa4 is not mediated by interaction of its N-terminus with Vps20 came from our analysis of cells expressing the vps20ΔMIMα6 allele, in which the conserved residues essential for binding of Vps20 to Doa41-80 were mutated (Fig. 2D). The vps20ΔMIMα6 mutation did not disable localization of full-length Doa4-GFP to endomembranes either in wild-type or vps4Δ cells (Fig. 3). Nonetheless, the Doa4 N-terminus might be required in the context of a larger localization determinant because deletion of residues 1-80 blocked the accumulation of Doa4-GFP at class E compartments in vps4Δ cells (Fig. 3).
Mutation of Vps20 MIMα6 causes dominant-synthetic inhibition of deubiquitination
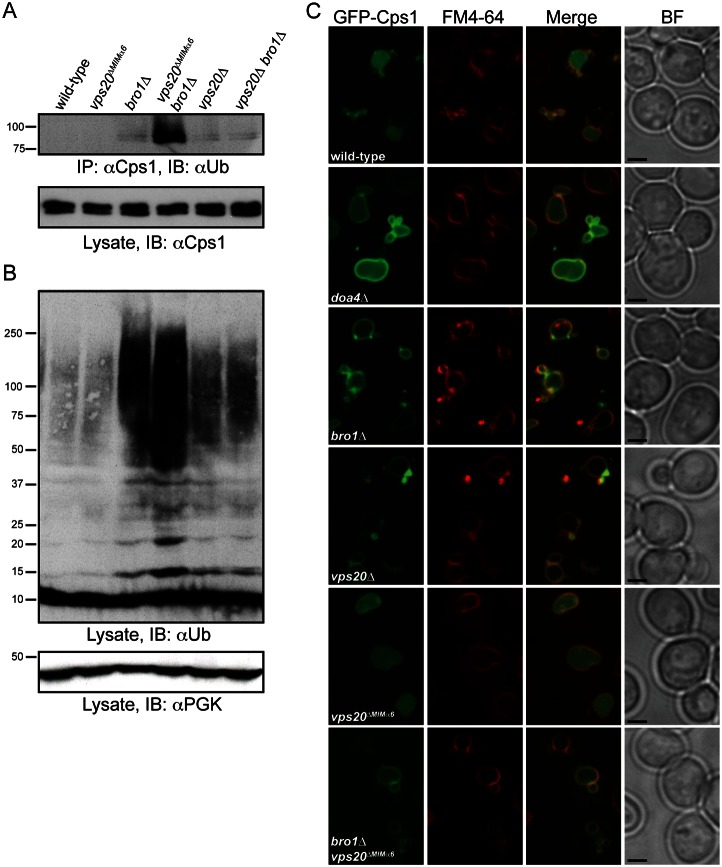
The ATPase activity of Vps4 is stimulated by interaction of its MIT domain with MIMs in ESCRT-III subunits (Merrill and Hanson, 2010; Obita et al., 2007; Stuchell-Brereton et al., 2007). We investigated whether the Ub hydrolase activity of Doa4 is similarly stimulated by interaction of its N-terminal domain with MIMα6 in Vps20. Direct in vitro assay of Doa4 catalytic activity in response to binding of its N-terminus was complicated by difficulties in purifying full-length recombinant Doa4 (data not shown). However, Doa4 activity can be monitored in vivo by assaying the abundance of ubiquitinated Cps1, which is a transmembrane protein deubiquitinated by Doa4 prior to its enclosure within ILVs (Dupré and Haguenauer-Tsapis, 2001; Katzmann et al., 2001; Reggiori and Pelham, 2001). Because Bro1 stimulates Doa4 catalytic activity, Cps1 accumulates in its ubiquitinated form (Ub-Cps1) in bro1Δ cells (Richter et al., 2007; Fig. 4A). Introduction of the vps20ΔMIMα6 mutation in bro1Δ cells further increased the accumulation of Ub-Cps1, but cells harboring the vps20ΔMIMα6 mutation alone had no apparent defect in Cps1 deubiquitination (Fig. 4A). Enhancement of the Cps1 deubiquitination defect in bro1Δ cells did not occur upon simultaneous deletion of the entire VPS20 gene. Instead, the abundance of Ub-Cps1 both in vps20Δ cells and in vps20Δ bro1Δ cells was similar to that in bro1Δ cells (Fig. 4A), consistent with the epistatic relationship Vps20 has in initiating ESCRT-III assembly for subsequent recruitment of Bro1 and Doa4 (Saksena et al., 2009; Teis et al., 2008). The dominant-synthetic inhibition of Cps1 deubiquitination seen when the vps20ΔMIMα6 and bro1Δ mutations were combined was mirrored in the analysis of total polyubiquitin conjugates from cell extracts, which accumulated to a greater extent in bro1Δ cells when the vps20ΔMIMα6 mutation was introduced (Fig. 4B). That the accumulation of total polyubiquitin conjugates in vps20ΔMIMα6 bro1Δ was less pronounced than the accumulation of Ub-Cps1 might be due to the abundance of cellular proteins that do not rely on Doa4 for deubiquitination, whereas Doa4 is the primary Ub hydrolase for Cps1.
Fig. 4.
Mutation of Vps20 MIMα6 exacerbates deubiquitination defects in bro1Δ cells. (A) Upper panel: Cps1 immunoprecipitations (IP) followed by αUb immunoblotting (IB). Lower panel: αCps1 immunoblotting of whole-cell lysates. (B) Immunoblot of whole-cell lysates with αUb and αPGK. (C) Fluorescence and brightfield (BF) microscopy of GFP-Cps1 and FM4-64. Scale bars: 2 µm.
The results described above suggest that Vps20 functions in parallel with Bro1 to activate Doa4 catalytic activity. However, Bro1 must have the predominant role in this regard since the vps20ΔMIMα6 mutation alone caused no apparent loss of Doa4 Ub hydrolase activity in vivo. This conclusion was further supported by the correct localization of GFP-Cps1 in the vacuole lumen in vps20ΔMIMα6 mutant cells, which contrasted with the mislocalization of GFP-Cps1 to the vacuole membrane that occurs upon loss of Doa4 function (Katzmann et al., 2001; Fig. 4C). In bro1Δ cells and vps20Δ cells, GFP-Cps1 was mislocalized both to the vacuole membrane and class E compartments (Odorizzi et al., 2003; Fig. 4C). GFP-Cps1 was similarly mislocalized to the vacuole membrane in vps20ΔMIMα6 bro1Δ cells, but class E compartments were less prominent in this strain (Fig. 4C), suggesting this aberrant endosomal morphology in bro1Δ cells is suppressed by the vps20ΔMIMα6 mutation.
Mutation of Vps20 MIMα6 rescues ILV formation in bro1Δ cells
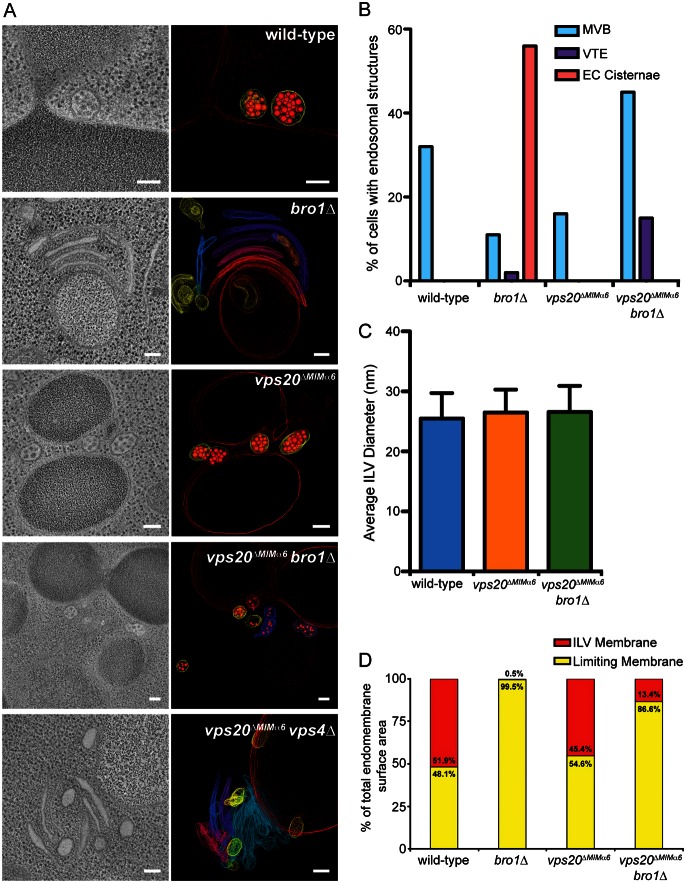
The class E compartments formed upon deleting BRO1 or other ESCRT genes are tubular and cisternal endosomes in which ILVs are largely absent (Odorizzi et al., 2003; Fig. 5A). However, overexpression of the DOA4 gene in bro1Δ cells suppresses class E compartment formation and restores the normal MVB morphology of endosomes (Luhtala and Odorizzi, 2004), raising the possibility that Doa4 promotes ILV formation in a manner that compensates for the absence of Bro1. We speculated such a role for Doa4 might involve Vps20 MIMα6 based on the apparent lack of class E compartment puncta in vps20ΔMIMα6 bro1Δ cells observed by fluorescence microscopy (Fig. 4C). Therefore, we used electron tomography to determine if the vps20ΔMIMα6 mutation has an effect on endosomal morphology.
Fig. 5.
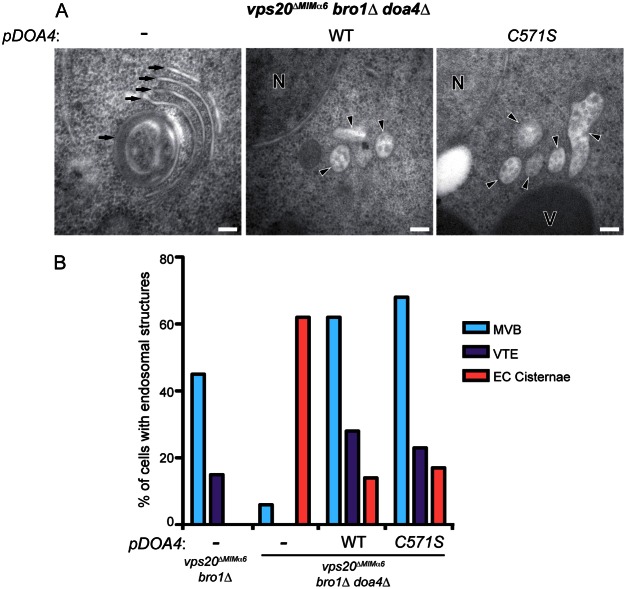
Mutation of Vps20 MIMα6 rescues ILV budding defects in bro1Δ cells. (A) Electron tomograms and corresponding models of the indicated strains. In models, ILVs are represented with small red spherical membranes and surrounded by yellow limiting membrane of spherical MVBs. Limiting membranes of tubular MVBs and individual cisternae of the class E compartments are shown in various colors; vacuoles are shown as large red membranes. Scale bars: 100 nm. (B) Quantification of the frequency of three endosomal morphologies: multivesicular body (MVB), vesicular tubular endosome (VTE), and class E compartment (EC) cisternae from random planes of 100 cells using transmission electron microcopy (TEM) of 80-nm thin sections of each indicated strain. (C) Quantification of average ILV diameters in each indicated strain. Error bars represent ±s.d. (D) Quantification of the relative percentages of membrane surface areas within ILVs and limiting membranes in each indicated strain. vps20ΔMIMα6 vps4Δ cells exclusively contained class E compartment cisternae without ILVs so were not included in the quantifications in (B–D).
Mutation of Vps20 MIMα6 alone did not inhibit ILV budding because vps20ΔMIMα6 cells had MVBs morphologically similar to those in wild-type yeast (Fig. 5A). However, the class E compartment morphology in bro1Δ cells (Fig. 5A,B) was strongly suppressed by introduction of the vps20ΔMIMα6 mutation: class E compartments were largely absent in vps20ΔMIMα6 bro1Δ cells, while spherical MVBs and vesicular tubular endosomes (VTEs) were abundant (Fig. 5A,B). The ILVs present in both vps20ΔMIMα6 and vps20ΔMIMα6 bro1Δ cells had average diameters indistinguishable from those of wild-type cells (Fig. 5C). The vps20ΔMIMα6 mutation did not generally suppress class E compartment formation and rescue MVB morphology because vps20ΔMIMα6 vps4Δ cells had class E compartments and lacked MVBs (Fig. 5A). Despite the rescue of MVB morphology in vps20ΔMIMα6 bro1Δ cells, ILVs sparsely filled the lumenal space (Fig. 5A), comprising only ∼13% of the total endosomal membrane surface area (Fig. 5D). In contrast, ILVs in MVBs of vps20ΔMIMα6 cells accounted for 45% of endosomal membrane surface area, which is similar to that observed in wild-type yeast (Fig. 5D) and is consistent with Bro1 being required for maximal ILV budding efficiency (Wemmer et al., 2011).
Although the modest recovery in ILV budding in bro1Δ cells upon introduction of the vps20ΔMIMα6 mutation might explain why GFP-Cps1 mislocalized to the vacuole membrane under these conditions (Fig. 4C), Cps1 deubiquitination is a prerequisite for the sorting of Cps1 into ILVs (Nikko and André, 2007) and is potently inhibited in vps20ΔMIMα6 bro1Δ cells (Fig. 4A). Therefore, we also examined the localization of Sna3, which is an ILV cargo that does not require Doa4 to be sorted into ILVs (Reggiori and Pelham, 2001). Like GFP-Cps1, however, Sna3-GFP failed to be sorted into the vacuole lumen in vps20ΔMIMα6 bro1Δ cells (Fig. 6), indicating that the recovery in ILV budding in bro1Δ cells upon introduction of the vps20ΔMIMα6 mutation is not sufficient to rescue the sorting of ILV cargoes to the vacuole lumen, regardless of whether these cargoes require deubiquitination to be sorted into ILVs.
Fig. 6.
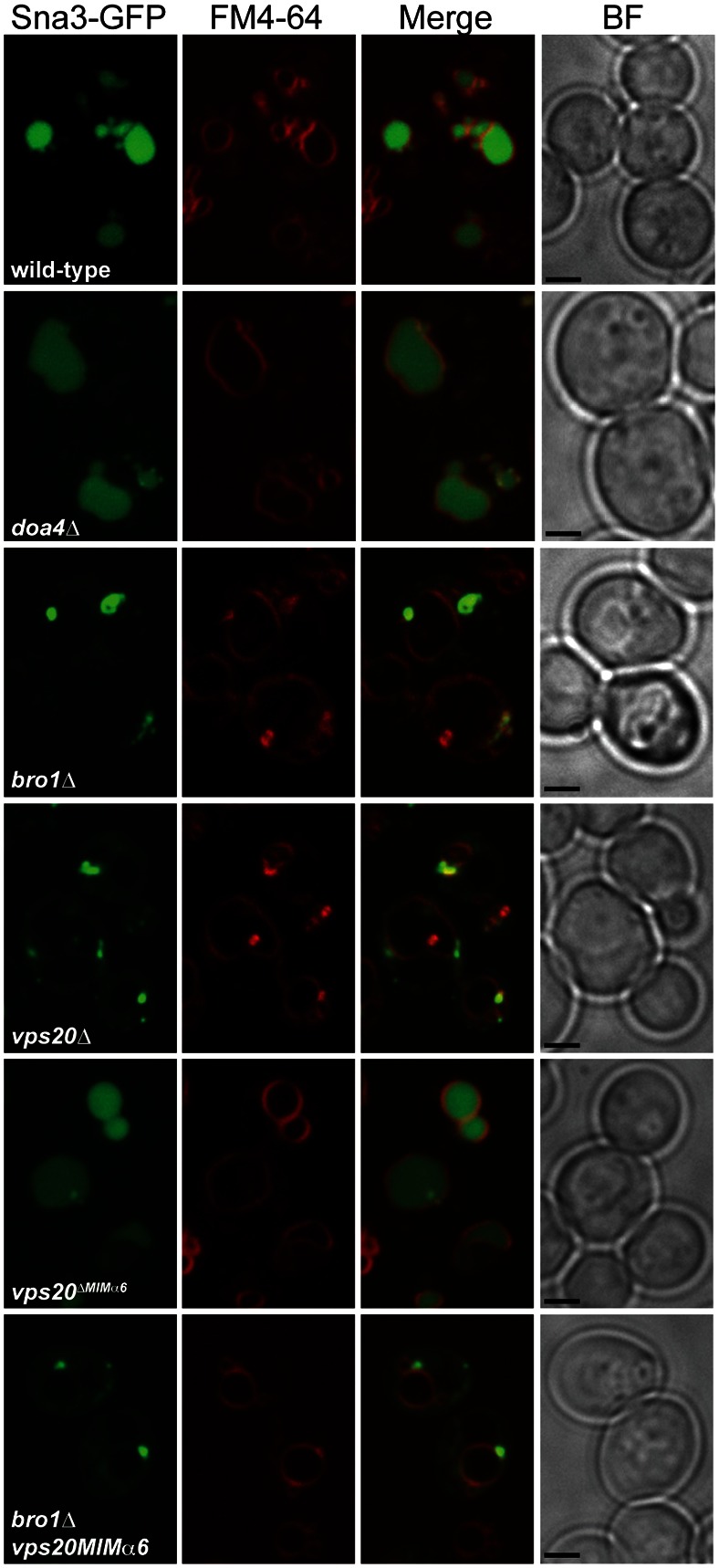
Mutation of Vps20 MIMα6 does not restore ILV sorting of Sna3-GFP. Fluorescence and brightfield (BF) microscopy of Sna3-GFP and FM4-64. Scale bars: 2 µm.
Doa4 is required for the rescue of ILV budding in vps20ΔMIMα6 bro1Δ cells
The suppression of class E compartment formation and rescue of MVB morphology in bro1Δ cells upon mutation of the Doa4-binding site in Vps20 (Fig. 5A) or upon overexpression of the DOA4 gene (Luhtala and Odorizzi, 2004) suggested that Vps20 MIMα6 inhibits an unknown function for Doa4 in promoting ILV budding in the absence of Bro1. Indeed, the deletion of DOA4 in vps20ΔMIMα6 bro1Δ cells abrogated MVB biogenesis and resulted almost exclusively in class E compartments (Fig. 7A,B). Expression of wild-type DOA4 from a low-copy plasmid in vps20ΔMIMα6 bro1Δ doa4Δ cells restored MVBs and suppressed class E compartments (Fig. 7A,B), confirming that Doa4 is required for the vps20ΔMIMα6 mutation to rescue ILV budding in bro1Δ cells. Surprisingly, the Doa4-dependent rescue of ILV budding did not require its Ub hydrolase activity, as plasmid-borne expression of the catalytically inactive doa4C571S allele also rescued MVB biogenesis in vps20ΔMIMα6 bro1Δ doa4Δ cells (Fig. 7A,B). Thus, the interaction of Vps20 MIMα6 with the N-terminal domain of Doa4 inhibits a non-catalytic function for Doa4 in promoting ILV budding.
Fig. 7.
Rescue of ILV budding in vps20ΔMIMα6 bro1Δ cells requires DOA4 expression. (A) 80-nm thin section transmission electron micrographs (TEM) of the indicated strains with or without a plasmid expressing wild-type DOA4 or doa4C571S. Arrows and arrowheads indicate class E compartment cisternae and multivesicular bodies, respectively. N, nucleus; V, vacuole. Scale bars: 100 nm. (B) Quantification of the frequency of three endosomal morphologies: multivesicular body (MVB), vesicular tubular endosome (VTE), and class E compartment (EC) cisternae from random planes of 100 cells using TEM as in A.
Discussion
Disassembly of ESCRT-III relies on Vps4 and its cofactor, Vta1, both of which contain MIT domains that interact with MIMs at or near the C termini of ESCRT-III subunits (Kieffer et al., 2008; Obita et al., 2007; Stuchell-Brereton et al., 2007). Beyond the Vps4–Vta1 complex, however, no MIT-containing ESCRT-III-associated proteins have been discovered in yeast, despite the abundance of human MIT proteins that are essential for ESCRT-III function in MVB sorting, retroviral budding, and cytokinesis (Kieffer et al., 2008; Renvoisé et al., 2010; Row et al., 2007). The N-terminus of the Doa4 Ub hydrolase in yeast is predicted to have the α-helical structure common to MIT domains, and we found that this region binds directly to a MIM1-like motif in helix α6 (MIMα6) of the Vps20 subunit of ESCRT-III. Although we speculate that the Doa4 N-terminus comprises an MIT domain, structural studies will be required to test this hypothesis.
The most surprising result from our study is that the vps20ΔMIMα6 mutation that disables binding of Vps20 to the Doa4 N-terminus rescued ILV budding to a modest extent in bro1Δ cells. That ILV biogenesis in vps20ΔMIMα6 bro1Δ cells was entirely dependent upon DOA4 expression argues that ILV budding was rescued specifically because Vps20 could not bind Doa4. While it is possible that another protein might also bind Vps20 MIMα6, mutation of this site alone had no apparent effect on endosome morphology or ILV cargo sorting. The requirement for DOA4 expression to rescue ILV budding in vps20ΔMIMα6 bro1Δ cells is also inconsistent with a model in which Doa4 antagonizes Vps20 activation of ESCRT-III assembly (Saksena et al., 2009). These findings suggest, instead, that Doa4 has a positive role in ILV budding that is inhibited by its interaction with Vps20. The Doa4–Vps20 interaction might, therefore, constitute a checkpoint that governs the timing of ILV scission in coordination with release of Doa4 from its inhibitory interaction with Vps20. Such a function for Doa4 is unexpected given that, by itself, deletion of the DOA4 gene does not block ILV biogenesis (Richter et al., 2007). The function of Doa4 that promotes ILV budding must, therefore, be redundant to some extent.
That expression of the catalytically inactive doa4C571S allele rescued MVB biogenesis as effectively as did expression of wild-type DOA4 in vps20ΔMIMα6 bro1Δ cells indicates that the function of Doa4 that promotes ILV budding does not involve Doa4 Ub hydrolase activity. Current models of ILV budding posit that Vps20 seeds the polymerization of Snf7 into fibrils that are remodeled by Vps24 and Vps2 into helices that promote membrane deformation and scission (Henne et al., 2012; Saksena et al., 2009; Teis et al., 2008). In this light, a mechanism by which Doa4 promotes ILV budding might be through Doa4 contributing to the assembly or remodeling of ESCRT-III oligomers into a scission-competent state. Alternatively, Doa4 might regulate ESCRT-III association with Vps4 to influence the timing of complex disassembly, an idea supported by observation that the MIT domain of AMSH in humans competes with VPS4 for binding to CHMP1B (Agromayor and Martin-Serrano, 2006).
The rescue of MVB biogenesis by the vps20ΔMIMα6 mutation in bro1Δ but not in vps4Δ cells indicates that the liberation of Doa4 from its interaction with Vps20 cannot rescue ILV budding in the face of general ESCRT dysfunction. Moreover, the paucity of ILVs in vps20ΔMIMα6 bro1Δ cells signifies that, despite recovery of the ILV budding mechanism, Bro1 is required for this process to achieve maximal efficiency. Binding of Bro1 to the Snf7 subunit of ESCRT-III stabilizes the complex by preventing its disassembly, which affects the efficiency with which ILV scission occurs (Wemmer et al., 2011). However, it is unclear whether Doa4 can substitute for Bro1 in this capacity. We detected no direct interaction between the Doa4 N-terminal domain and Snf7 in vitro, but the possibility that another region of Doa4 is capable of binding Snf7 is supported by the interaction between full-length Doa4 and Snf7 in a yeast two-hybrid assay (Bowers et al., 2004).
The ATPase activity of human VPS4A is stimulated in vitro by its interactions with ESCRT-III subunits, which is predicted to induce a structural rearrangement in VPS4A that prevents the MIT domain and nearby linker region from auto-inhibiting the active site (Merrill and Hanson, 2010). Based on our finding that mutation of the Doa4 binding site in Vps20 exacerbates the deubiquitination defect observed in cells that lack Bro1, Vps20 might have a role in the activation of Doa4. Such a function for Vps20, however, must be secondary to the direct activation of Doa4 Ub hydrolase activity by Bro1 (Richter et al., 2007) because deubiquitination of Cps1, a Doa4 substrate, was unaffected in vps20ΔMIMα6 cells that express Bro1.
MIT domains in two human Ub hydrolases, UBPY and AMSH, mediate recruitment to sites of ESCRT-III assembly (Row et al., 2007; Solomons et al., 2011). However, we found no evidence that the MIT-like N-terminal domain of Doa4 serves the same role because this region alone was not efficiently recruited to endomembranes, and mutation of its binding site in Vps20 did not disable endosomal localization of wild-type Doa4. Nonetheless, we found the N-terminal domain is required for Doa4 endomembrane recruitment, which is consistent with earlier work showing that mutations within this region disrupted Doa4 localization (Amerik et al., 2006) and suggests that amino acids 1-80 are part of a larger endosomal localization domain. Although the N-terminal domain is insufficient for robust endosomal localization of Doa4, the presence of GFP-Doa41-80 in the vacuole lumen of wild-type cells suggests that this domain, when expressed alone, transiently associates with ESCRTs but lacks the ability to be released back into the cytoplasm before its enclosure within an ILV.
That the binding of its N-terminus to Vps20 does not localize Doa4 to endosomes but, instead, inhibits Doa4 function in the formation of ILVs suggests that the Vps20–Doa4 interaction serves as a checkpoint in the ILV budding pathway. Further study is needed to validate the concept of a checkpoint involving this interaction, to establish the time at which Vps20 inhibits Doa4 relative to ESCRT-III assembly, and to identify the mechanism that normally relieves this inhibition.
Materials and Methods
Yeast strains and plasmid construction
Standard techniques were used for growth and genetic manipulation of S. cerevisiae. Yeast strains created for this study (Table 1) were constructed using integration cassettes described in Longtine et al. (Longtine et al., 1998). Construction of plasmids expressing GST-Bro11-387 (Kim et al., 2005), 2μ GFP, 2μ GFP-Cps1 (Odorizzi et al., 1998), DOA4, and doa4C571S (Richter et al., 2007) have been described previously. To construct 2μ DOA4-GFP, a PCR product corresponding to the DOA4-GFP locus from GOY74 (Luhtala and Odorizzi, 2004) was subcloned into pRS426, resulting in pCR142. To construct 2μ GFP-DOA41-80 and 2μ DOA4Δ1-80, PCR products corresponding to DOA4 codons 1-80 or DOA4 codons 81-926, respectively, were subcloned into pGO35 (Odorizzi et al., 1998), resulting in pGO640 and pGO704, respectively. To construct GST-DOA41-80, a PCR product corresponding to DOA4 codons 1-80 was cloned into pCR2.1, yielding pCR149, and the BamHI/XhoI fragment of pCR149 was subcloned into pGEX-4T1, yielding pCR152. A PCR product corresponding to VPS2 was cloned into pCR2.1, yielding pGO480. Site-directed mutagenesis was used to create SnaBI sites flanking the VPS2 intron, yielding pGO497, which was then digested with SnaBI and re-ligated to make pGO503. The XbaI/BamHI fragment of pGO503 was subcloned into the bacterial expression vector pST39 (Tan, 2001) to create pGO516. To construct SNF7 in pST39, pGO465 (Kim et al., 2005) was digested with BspEI and MluI to remove BRO1, treated with T4 polymerase to generate blunt ends, and re-ligated, yielding pGO547. A PCR product corresponding to VPS20-HA was digested with SacI/KpnI and subcloned into pST39, yielding pGO569. To create MIM mutants, Site directed mutagenesis was used on templates pGO516, pGO547, pGO569, and pDN63 (Nickerson et al., 2006) to generate vps2R224D,L225D (pCR160), snf7L231D,L234D (pGO560), vps20P218D,L219D-HA (pCR162), and His6-vps24R218D,L219D (pCR161), respectively.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Reference |
| SEY6210 | MAT-alpha leu2-3,112 ura3-52 his3Δ200 trp1-Δ901 lys2-Δ801 suc2-Δ9 | Robinson et al., 1988 |
| GOY74 | SEY6210; DOA4-GFP::HIS3MX6 | Luhtala and Odorizzi, 2004 |
| GOY75 | SEY6210; DOA4-GFP::HIS3MX6 vps4Δ::TRP1 | Luhtala and Odorizzi, 2004 |
| GOY23 | SEY6210; pep4Δ::LEU2 prb1Δ::LEU2 | Luhtala and Odorizzi, 2004 |
| DBY5 | SEY6210; doa4Δ::HIS3 | Richter et al., 2007 |
| GOY65 | SEY6210; bro1Δ::HIS3 | Luhtala and Odorizzi, 2004 |
| MBY3 | SEY6210; vps4Δ::TRP1 | Babst et al., 1998 |
| DBY19 | GOY23; vps4Δ::TRP1 | Nickerson et al., 2006 |
| DBY43 | GOY23; bro1Δ::HIS3 | Odorizzi et al., 2003 |
| DBY44 | GOY23; bro1Δ::HIS3 vps4Δ::TRP1 | Odorizzi et al., 2003 |
| GOY248 | SEY6210; vps20ΔMIMα6::KANMX6 | This study |
| GOY249 | GOY248; vps4Δ::TRP1 | This study |
| GOY250 | GOY65; vps20ΔMIMα6::KANMX6 | This study |
| GOY307 | SEY6210; vps20ΔMIMα6::KANMX6 bro1Δ::TRP1 doa4Δ::HIS3MX6 | This study |
| GOY312 | GOY23; vps20ΔMIMα6::KANMX6 | This study |
| GOY311 | DBY43; vps20ΔMIMα6::KANMX6 | This study |
| EEY2-1 | SEY6210; vps20Δ::HIS3 | Babst et al., 2002 |
| GOY314 | GOY23; vps20Δ::HIS3MX6 | This study |
| GOY315 | DBY43; vps20ΔKANMX6 | This study |
Fluorescence microscopy
Strains were grown to logarithmic phase at 30°C before observation at room temperature using a Nikon TE2000-U inverted fluorescence microscope equipped with a Yokogawa spinning disc confocal unit (CSU-Xm2; Nikon Instruments, Inc.) and a 100× oil objective with a numerical aperture of 1.4. Fluorescence and brightfield images were acquired with a Photometrics Cascade II EM-CCD camera using MetaMorph (v7.0) software, then processed with ImageJ and Photoshop CS4 software (Adobe). Endosomal membranes were stained with FM4-64 (Molecular Probes, Inc.) using a 20 minute pulse and 90 minute chase (Odorizzi et al., 2003).
Immunoprecipitations and western blotting
For native immunoprecipitations of Doa4-GFP, 20 A600 equivalents of logarithmic phase cells were converted to spheroplasts and osmotically lysed and homogenized on ice in 1 ml of lysis buffer (200 mM sorbitol, 50 mM potassium acetate, 20 mM Hepes, pH 7.2, 2 mM EDTA, supplemented with a protease inhibitor cocktail; Roche). Triton X-100 was added to a final concentration of 0.5% and lysates were spun at 16,000 g for 10 min at 4°C to remove insoluble material. One A600 equivalent of detergent-soluble lysate was precipitated by the addition of 10% TCA to generate total lysate samples. 10 A600 equivalents were incubated with mouse anti-GFP monoclonal antibody (Roche) and Protein G–Sepharose beads (GE Healthcare) for 2 hours at 4°C, after which the beads were collected by centrifugation, washed thrice in lysis buffer, and boiled in Laemmli buffer to elute bound material. Five A600 equivalents of immunoprecipitates and 0.5 A600 equivalents of total lysate were resolved by SDS-PAGE, transferred to nitrocellulose, and analyzed by western blot using rabbit anti-Bro1, anti-Vps24 and anti-Snf7 polyclonal antiserum (Odorizzi et al., 2003) and mouse anti-GFP (Roche) monoclonal antibody.
Denatured immunoprecipitations to detect Ub-Cps1 were performed as described previously (Katzmann et al., 2001). Twenty A600 equivalents of logarithmic phase cells were precipitated by the addition of 10% TCA containing 5 mM N-ethylmaleimide (NEM), and whole-cell lysates were generated by glass-bead disruption in lysis buffer (6 M urea, 1% SDS, 50 mM Tris pH 7.5, 1 mM EDTA, 5 mM NEM). Lysates were diluted 10-fold in immunoprecipitation buffer (50 mM Tris pH 7.5, 150 mM NaCl, 0.5% Tween, 1 mM EDTA, 5 mM NEM), insoluble material was cleared by centrifugation at 16,000 g, and lysates were immunoprecipitated with anti-Cps1 polyclonal antiserum (Cowles et al., 1997). Five A600 equivalents of immunoprecipitates and 0.5 A600 equivalents of total lysate were resolved by SDS-PAGE, transferred to nitrocellulose, and analyzed by western blot using anti-Ub (Invitrogen), anti-phosphoglycerate kinase (PGK), and anti-GFP (Roche) monoclonal antibodies (Invitrogen).
Affinity purification of recombinant proteins and in vitro binding studies
GST-Doa41-80, GST-Bro11-387, and GST were expressed in E. coli BL21-CodonPlus (DE3) cells (Stratagene) by induction with 0.5 mM isopropyl-β-D-thiogalactoside (IPTG) at 20°C for 18 hours and purified using glutathione–Sepharose (GE Healthcare). Liquid cultures (100 ml) of BL21(DE3) transformed with Vps2, Vps20-HA, His6-Vps24, and Snf7 expression plasmids were grown and lysed as described previously (McNatt et al., 2007). Lysates were clarified of cell debris by centrifugation at 100,000 g for 30 min at 4°C. 10 µg of purified GST, GST-Bro11-387, or GST-Doa41-80 was added to 1/3 of each lysate and incubated at 4°C for 2 hours with glutathione–Sepharose (GE Healthcare). Sepharose was washed 4 times with GST wash buffer [phosphate-buffered saline (PBS), 0.5% Triton X-100] and once with PBS. Samples were resolved by SDS-PAGE, transferred to nitrocellulose, and analyzed by western blot using anti-Vps2, anti-Vps24, and anti-Snf7 polyclonal antiserum (Odorizzi et al., 2003) as well as anti-HA (Covance) and anti-GST (Invitrogen) monoclonal antibodies.
Electron microscopy and tomography
Yeast cells were harvested at log phase, vacuum-filtered on 0.45 micron Millipore paper, loaded into 0.25-mm aluminum planchettes, and high-pressure frozen in a Balzers Bal-Tec HPM 010 (Boeckeler Instruments) as previously described (Wemmer et al., 2011). A Leica AFS (Automated Freeze Substitution, Vienna, Austria) was used for freeze-substitution preparation of 0.1% uranyl acetate and 0.25% glutaraldehyde in anhydrous acetone (Giddings, 2003). Samples were then washed in pure acetone, embedded in Lowicryl HM20 resin (Polysciences, Warrington, PA), and polymerized at −60°C. A Leica Ultra-Microtome was used to cut 80-nm serial thin sections and 250-nm serial semi-thick sections, which were collected onto 1% formvar films, adhered to rhodium-plated copper grids (Electron Microscopy Sciences). A Phillips CM10 (Mahwah, NJ) transmission electron microscope was used to image 80-nm sections at 80 kV to quantify the frequency of MVBs, VTEs, and class E compartments. For tomography, grids were labeled on both sides with fiduciary 15-nm colloidal gold (British Biocell International). Typically, Z-shrinkage of semi-thick sections was 20 percent volume and corrected in final models and measurements. Dual-axis tilt series were collected from ±60° with 1° increments at 200 kV using a Tecnai F20 (FEI-Company, Eindhoven, the Netherlands and Hillsboro, OR) at a magnification of 29,000× using SerialEM (Mastronarde, 2005). 2× binning on the recording 4K × 4K CCD camera (Gatan, Inc., Abingdon, UK) creates a 2K × 2K image with a pixel size of 0.764 nm. Dual-axis electron tomograms (Mastronarde, 1997) of endosomes and ILVs required the IMOD package (Kremer et al., 1996) for tomogram construction and modeling (3DMOD 4.0.11). Manually assigned contours of the endosomal limiting membrane at the inner leaflet were used to measure the surface of the bilayers periodically every 3.85 nm and calculated using imodmesh. Best-fit sphere models were used to measure the diameters of nearly spherical lumenal vesicles from the outer leaflet of the membrane bilayers (O'Toole et al., 2002). IMODINFO provided surface area and volume data of contour models. Data were sorted, analyzed, and graphed using Microsoft Excel and Prism 5.
Acknowledgments
We thank Tess Shideler (University of Colorado) for assistance with fluorescence microscopy and James Hurley (National Institutes of Health) for helpful discussions.
Footnotes
Author contributions
M.W. performed electron microscopy and tomography experiments. All other experiments were performed by C.R.. G.O. and C.R. designed the project, and C.R. wrote the manuscript with comments from co-authors.
Funding
This work was funded by the National Institutes of Health [grant numbers R01GM-065505 and T32GM-08759]. Deposited in PMC for release after 12 months.
References
- Agromayor M., Martin-Serrano J. (2006). Interaction of AMSH with ESCRT-III and deubiquitination of endosomal cargo. J. Biol. Chem. 281, 23083–23091 10.1074/jbc.M513803200 [DOI] [PubMed] [Google Scholar]
- Amerik A., Sindhi N., Hochstrasser M. (2006). A conserved late endosome-targeting signal required for Doa4 deubiquitylating enzyme function. J. Cell Biol. 175, 825–835 10.1083/jcb.200605134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azmi I., Davies B., Dimaano C., Payne J., Eckert D., Babst M., Katzmann D. J. (2006). Recycling of ESCRTs by the AAA-ATPase Vps4 is regulated by a conserved VSL region in Vta1. J. Cell Biol. 172, 705–717 10.1083/jcb.200508166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azmi I. F., Davies B. A., Xiao J., Babst M., Xu Z., Katzmann D. J. (2008). ESCRT-III family members stimulate Vps4 ATPase activity directly or via Vta1. Dev. Cell 14, 50–61 10.1016/j.devcel.2007.10.021 [DOI] [PubMed] [Google Scholar]
- Babst M., Wendland B., Estepa E. J., Emr S. D. (1998). The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17, 2982–2993 10.1093/emboj/17.11.2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M., Katzmann D. J., Estepa-Sabal E. J., Meerloo T., Emr S. D. (2002). Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell 3, 271–282 10.1016/S1534-5807(02)00220-4 [DOI] [PubMed] [Google Scholar]
- Bowers K., Lottridge J., Helliwell S. B., Goldthwaite L. M., Luzio J. P., Stevens T. H. (2004). Protein-protein interactions of ESCRT complexes in the yeast Saccharomyces cerevisiae. Traffic 5, 194–210 10.1111/j.1600-0854.2004.00169.x [DOI] [PubMed] [Google Scholar]
- Cowles C. R., Snyder W. B., Burd C. G., Emr S. D. (1997). Novel Golgi to vacuole delivery pathway in yeast: identification of a sorting determinant and required transport component. EMBO J. 16, 2769–2782 10.1093/emboj/16.10.2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimaano C., Jones C. B., Hanono A., Curtiss M., Babst M. (2008). Ist1 regulates Vps4 localization and assembly. Mol. Biol. Cell 19, 465–474 10.1091/mbc.E07-08-0747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupré S., Haguenauer-Tsapis R. (2001). Deubiquitination step in the endocytic pathway of yeast plasma membrane proteins: crucial role of Doa4p ubiquitin isopeptidase. Mol. Cell. Biol. 21, 4482–4494 10.1128/MCB.21.14.4482-4494.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giddings T. H. (2003). Freeze-substitution protocols for improved visualization of membranes in high-pressure frozen samples. J. Microsc. 212, 53–61 10.1046/j.1365-2818.2003.01228.x [DOI] [PubMed] [Google Scholar]
- Henne W. M., Buchkovich N. J., Emr S. D. (2011). The ESCRT pathway. Dev. Cell 21, 77–91 10.1016/j.devcel.2011.05.015 [DOI] [PubMed] [Google Scholar]
- Henne W. M., Buchkovich N. J., Zhao Y., Emr S. D. (2012). The endosomal sorting complex ESCRT-II mediates the assembly and architecture of ESCRT-III helices. Cell 151, 356–371 10.1016/j.cell.2012.08.039 [DOI] [PubMed] [Google Scholar]
- Hurley J. H. (2010). The ESCRT complexes. Crit. Rev. Biochem. Mol. Biol. 45, 463–487 10.3109/10409238.2010.502516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann D. J., Babst M., Emr S. D. (2001). Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106, 145–155 10.1016/S0092-8674(01)00434-2 [DOI] [PubMed] [Google Scholar]
- Kieffer C., Skalicky J. J., Morita E., De Domenico I., Ward D. M., Kaplan J., Sundquist W. I. (2008). Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding. Dev. Cell 15, 62–73 10.1016/j.devcel.2008.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Kim J., Sitaraman S., Hierro A., Beach B. M., Odorizzi G., Hurley J. H. (2005). Structural basis for endosomal targeting by the Bro1 domain. Dev. Cell 8, 937–947 10.1016/j.devcel.2005.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer J. R., Mastronarde D. N., McIntosh J. R. (1996). Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 10.1006/jsbi.1996.0013 [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- Lottridge J. M., Flannery A. R., Vincelli J. L., Stevens T. H. (2006). Vta1p and Vps46p regulate the membrane association and ATPase activity of Vps4p at the yeast multivesicular body. Proc. Natl. Acad. Sci. USA 103, 6202–6207 10.1073/pnas.0601712103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luhtala N., Odorizzi G. (2004). Bro1 coordinates deubiquitination in the multivesicular body pathway by recruiting Doa4 to endosomes. J. Cell Biol. 166, 717–729 10.1083/jcb.200403139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde D. N. (1997). Dual-axis tomography: an approach with alignment methods that preserve resolution. J. Struct. Biol. 120, 343–352 10.1006/jsbi.1997.3919 [DOI] [PubMed] [Google Scholar]
- Mastronarde D. N. (2005). Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 10.1016/j.jsb.2005.07.007 [DOI] [PubMed] [Google Scholar]
- McNatt M. W., McKittrick I., West M., Odorizzi G. (2007). Direct binding to Rsp5 mediates ubiquitin-independent sorting of Sna3 via the multivesicular body pathway. Mol. Biol. Cell 18, 697–706 10.1091/mbc.E06-08-0663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill S. A., Hanson P. I. (2010). Activation of human VPS4A by ESCRT-III proteins reveals ability of substrates to relieve enzyme autoinhibition. J. Biol. Chem. 285, 35428–35438 10.1074/jbc.M110.126318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson D. P., West M., Odorizzi G. (2006). Did2 coordinates Vps4-mediated dissociation of ESCRT-III from endosomes. J. Cell Biol. 175, 715–720 10.1083/jcb.200606113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikko E., André B. (2007). Evidence for a direct role of the Doa4 deubiquitinating enzyme in protein sorting into the MVB pathway. Traffic 8, 566–581 10.1111/j.1600-0854.2007.00553.x [DOI] [PubMed] [Google Scholar]
- O'Toole E. T., Winey M., McIntosh J. R., Mastronarde D. N. (2002). Electron tomography of yeast cells. Methods Enzymol. 351, 81–95 10.1016/S0076-6879(02)51842-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obita T., Saksena S., Ghazi-Tabatabai S., Gill D. J., Perisic O., Emr S. D., Williams R. L. (2007). Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature 449, 735–739 10.1038/nature06171 [DOI] [PubMed] [Google Scholar]
- Odorizzi G., Babst M., Emr S. D. (1998). Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell 95, 847–858 10.1016/S0092-8674(00)81707-9 [DOI] [PubMed] [Google Scholar]
- Odorizzi G., Katzmann D. J., Babst M., Audhya A., Emr S. D. (2003). Bro1 is an endosome-associated protein that functions in the MVB pathway in Saccharomyces cerevisiae. J. Cell Sci. 116, 1893–1903 10.1242/jcs.00395 [DOI] [PubMed] [Google Scholar]
- Reggiori F., Pelham H. R. (2001). Sorting of proteins into multivesicular bodies: ubiquitin-dependent and -independent targeting. EMBO J. 20, 5176–5186 10.1093/emboj/20.18.5176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renvoisé B., Parker R. L., Yang D., Bakowska J. C., Hurley J. H., Blackstone C. (2010). SPG20 protein spartin is recruited to midbodies by ESCRT-III protein Ist1 and participates in cytokinesis. Mol. Biol. Cell 21, 3293–3303 10.1091/mbc.E09-10-0879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter C., West M., Odorizzi G. (2007). Dual mechanisms specify Doa4-mediated deubiquitination at multivesicular bodies. EMBO J. 26, 2454–2464 10.1038/sj.emboj.7601692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. S., Klionsky D. J., Banta L. M., Emr S. D. (1988). Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 8, 4936–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B., Yachdav G., Liu J. (2004). The PredictProtein server. Nucleic Acids Res. 32, Suppl. 2W321–W326 10.1093/nar/gkh377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Row P. E., Liu H., Hayes S., Welchman R., Charalabous P., Hofmann K., Clague M. J., Sanderson C. M., Urbé S. (2007). The MIT domain of UBPY constitutes a CHMP binding and endosomal localization signal required for efficient epidermal growth factor receptor degradation. J. Biol. Chem. 282, 30929–30937 10.1074/jbc.M704009200 [DOI] [PubMed] [Google Scholar]
- Rue S. M., Mattei S., Saksena S., Emr S. D. (2008). Novel Ist1-Did2 complex functions at a late step in multivesicular body sorting. Mol. Biol. Cell 19, 475–484 10.1091/mbc.E07-07-0694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksena S., Wahlman J., Teis D., Johnson A. E., Emr S. D. (2009). Functional reconstitution of ESCRT-III assembly and disassembly. Cell 136, 97–109 10.1016/j.cell.2008.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shestakova A., Hanono A., Drosner S., Curtiss M., Davies B. A., Katzmann D. J., Babst M. (2010). Assembly of the AAA ATPase Vps4 on ESCRT-III. Mol. Biol. Cell 21, 1059–1071 10.1091/mbc.E09-07-0572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim S., Kimpler L. A., Hanson P. I. (2007). Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic 8, 1068–1079 10.1111/j.1600-0854.2007.00584.x [DOI] [PubMed] [Google Scholar]
- Solomons J., Sabin C., Poudevigne E., Usami Y., Hulsik D. L., Macheboeuf P., Hartlieb B., Göttlinger H., Weissenhorn W. (2011). Structural basis for ESCRT-III CHMP3 recruitment of AMSH. Structure 19, 1149–1159 10.1016/j.str.2011.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuchell-Brereton M. D., Skalicky J. J., Kieffer C., Karren M. A., Ghaffarian S., Sundquist W. I. (2007). ESCRT-III recognition by VPS4 ATPases. Nature 449, 740–744 10.1038/nature06172 [DOI] [PubMed] [Google Scholar]
- Tan S. (2001). A modular polycistronic expression system for overexpressing protein complexes in Escherichia coli. Protein Expr. Purif. 21, 224–234 10.1006/prep.2000.1363 [DOI] [PubMed] [Google Scholar]
- Teis D., Saksena S., Emr S. D. (2008). Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev. Cell 15, 578–589 10.1016/j.devcel.2008.08.013 [DOI] [PubMed] [Google Scholar]
- Vida T. A., Emr S. D. (1995). A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128, 779–792 10.1083/jcb.128.5.779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wemmer M., Azmi I., West M., Davies B., Katzmann D., Odorizzi G. (2011). Bro1 binding to Snf7 regulates ESCRT-III membrane scission activity in yeast. J. Cell Biol. 192, 295–306 10.1083/jcb.201007018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T., Hurley J. H. (2010). Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature 464, 864–869 10.1038/nature08849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T., Wunder C., Lippincott-Schwartz J., Hurley J. H. (2009). Membrane scission by the ESCRT-III complex. Nature 458, 172–177 10.1038/nature07836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J., Xia H., Zhou J., Azmi I. F., Davies B. A., Katzmann D. J., Xu Z. (2008). Structural basis of Vta1 function in the multivesicular body sorting pathway. Dev. Cell 14, 37–49 10.1016/j.devcel.2007.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]