Abstract
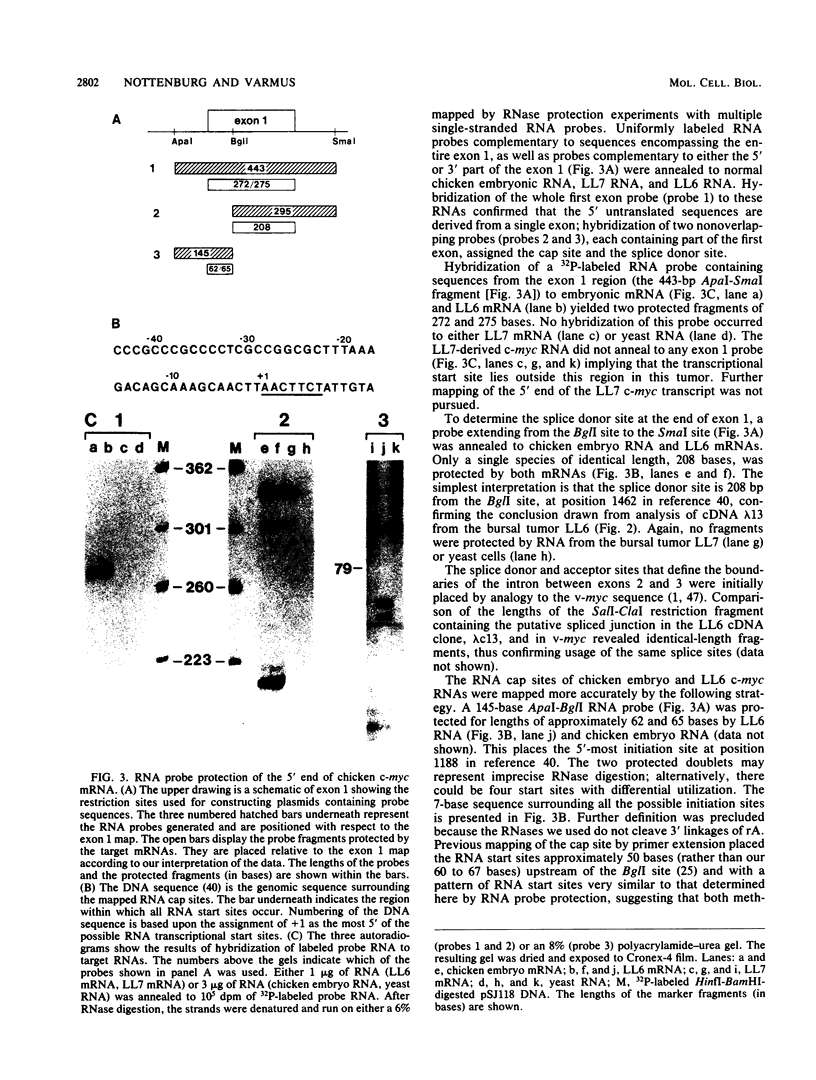
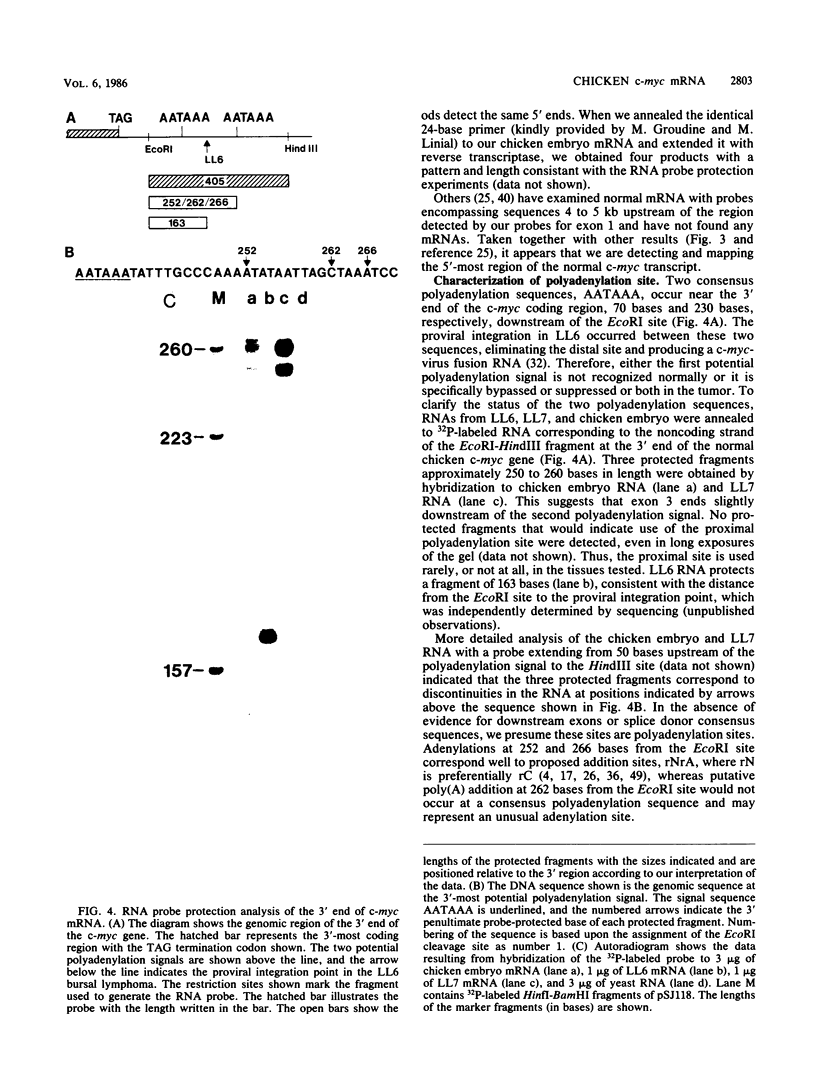
The chicken c-myc gene is the target for proviral insertion mutations in bursal lymphomas and has been transduced to generate several viral oncogenes, but the boundaries of its exons have not been securely established. To define the landmarks of the chicken c-myc gene necessary to produce its mRNA, we used an RNase protection assay and a cDNA clone to analyze the c-myc mRNAs from normal chicken embryos and from two bursal lymphomas: LL6, which contains an avian leukosis virus provirus downstream of the c-myc coding region, and LL7, which contains an avian leukosis virus provirus upstream of the c-myc coding region. Two initiation sites for normal c-myc mRNA are less than 7 bases apart, downstream of a GC-rich region lacking canonical TATA and CAAT sequences. The first exon has two open reading frames for the entire length but no initiator methionine codons. The splice donor and acceptor sites at the boundary of the first intron were assigned by comparing a sequence of an LL6 c-myc cDNA clone with a genomic DNA sequence and confirmed by RNase protection of labeled RNA probes by normal and LL6-derived mRNAs. Two potential polyadenylation signals are located approximately 250 and 400 bases downstream of the c-myc coding region in the third exon, but only the more distal signal is utilized in both normal cells and the LL7 tumor. The proviral integration in the LL6 tumor occurred upstream of the authentic c-myc polyadenylation signal accounting for polyadenylation of this transcript in the proviral long terminal repeat.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alitalo K., Bishop J. M., Smith D. H., Chen E. Y., Colby W. W., Levinson A. D. Nucleotide sequence to the v-myc oncogene of avian retrovirus MC29. Proc Natl Acad Sci U S A. 1983 Jan;80(1):100–104. doi: 10.1073/pnas.80.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alitalo K., Ramsay G., Bishop J. M., Pfeifer S. O., Colby W. W., Levinson A. D. Identification of nuclear proteins encoded by viral and cellular myc oncogenes. Nature. 1983 Nov 17;306(5940):274–277. doi: 10.1038/306274a0. [DOI] [PubMed] [Google Scholar]
- Battey J., Moulding C., Taub R., Murphy W., Stewart T., Potter H., Lenoir G., Leder P. The human c-myc oncogene: structural consequences of translocation into the IgH locus in Burkitt lymphoma. Cell. 1983 Oct;34(3):779–787. doi: 10.1016/0092-8674(83)90534-2. [DOI] [PubMed] [Google Scholar]
- Benoist C., O'Hare K., Breathnach R., Chambon P. The ovalbumin gene-sequence of putative control regions. Nucleic Acids Res. 1980 Jan 11;8(1):127–142. doi: 10.1093/nar/8.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard O., Cory S., Gerondakis S., Webb E., Adams J. M. Sequence of the murine and human cellular myc oncogenes and two modes of myc transcription resulting from chromosome translocation in B lymphoid tumours. EMBO J. 1983;2(12):2375–2383. doi: 10.1002/j.1460-2075.1983.tb01749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Butnick N. Z., Miyamoto C., Chizzonite R., Cullen B. R., Ju G., Skalka A. M. Regulation of the human c-myc gene: 5' noncoding sequences do not affect translation. Mol Cell Biol. 1985 Nov;5(11):3009–3016. doi: 10.1128/mcb.5.11.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J., Gray H. E., Pardee A. B., Dean M., Sonenshein G. E. Cell-cycle control of c-myc but not c-ras expression is lost following chemical transformation. Cell. 1984 Feb;36(2):241–247. doi: 10.1016/0092-8674(84)90217-4. [DOI] [PubMed] [Google Scholar]
- Chamberlin M., Ring J. Characterization of T7-specific ribonucleic acid polymerase. 1. General properties of the enzymatic reaction and the template specificity of the enzyme. J Biol Chem. 1973 Mar 25;248(6):2235–2244. [PubMed] [Google Scholar]
- Corcoran L. M., Adams J. M., Dunn A. R., Cory S. Murine T lymphomas in which the cellular myc oncogene has been activated by retroviral insertion. Cell. 1984 May;37(1):113–122. doi: 10.1016/0092-8674(84)90306-4. [DOI] [PubMed] [Google Scholar]
- Darveau A., Pelletier J., Sonenberg N. Differential efficiencies of in vitro translation of mouse c-myc transcripts differing in the 5' untranslated region. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2315–2319. doi: 10.1073/pnas.82.8.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierks P., van Ooyen A., Cochran M. D., Dobkin C., Reiser J., Weissmann C. Three regions upstream from the cap site are required for efficient and accurate transcription of the rabbit beta-globin gene in mouse 3T6 cells. Cell. 1983 Mar;32(3):695–706. doi: 10.1016/0092-8674(83)90055-7. [DOI] [PubMed] [Google Scholar]
- Donner P., Greiser-Wilke I., Moelling K. Nuclear localization and DNA binding of the transforming gene product of avian myelocytomatosis virus. Nature. 1982 Mar 18;296(5854):262–269. doi: 10.1038/296262a0. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell. 1983 Nov;35(1):79–87. doi: 10.1016/0092-8674(83)90210-6. [DOI] [PubMed] [Google Scholar]
- Eick D., Piechaczyk M., Henglein B., Blanchard J. M., Traub B., Kofler E., Wiest S., Lenoir G. M., Bornkamm G. W. Aberrant c-myc RNAs of Burkitt's lymphoma cells have longer half-lives. EMBO J. 1985 Dec 30;4(13B):3717–3725. doi: 10.1002/j.1460-2075.1985.tb04140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenman R. N., Tachibana C. Y., Abrams H. D., Hann S. R. V-myc- and c-myc-encoded proteins are associated with the nuclear matrix. Mol Cell Biol. 1985 Jan;5(1):114–126. doi: 10.1128/mcb.5.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald M., Shenk T. The sequence 5'-AAUAAA-3'forms parts of the recognition site for polyadenylation of late SV40 mRNAs. Cell. 1981 Apr;24(1):251–260. doi: 10.1016/0092-8674(81)90521-3. [DOI] [PubMed] [Google Scholar]
- Fung Y. K., Crittenden L. B., Kung H. J. Orientation and position of avian leukosis virus DNA relative to the cellular oncogene c-myc in B-lymphoma tumors of highly susceptible 15I5 X 7(2) chickens. J Virol. 1982 Nov;44(2):742–746. doi: 10.1128/jvi.44.2.742-746.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung Y. K., Fadly A. M., Crittenden L. B., Kung H. J. On the mechanism of retrovirus-induced avian lymphoid leukosis: deletion and integration of the proviruses. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3418–3422. doi: 10.1073/pnas.78.6.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerondakis S., Cory S., Adams J. M. Translocation of the myc cellular oncogene to the immunoglobulin heavy chain locus in murine plasmacytomas is an imprecise reciprocal exchange. Cell. 1984 Apr;36(4):973–982. doi: 10.1016/0092-8674(84)90047-3. [DOI] [PubMed] [Google Scholar]
- Hayward W. S., Neel B. G., Astrin S. M. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature. 1981 Apr 9;290(5806):475–480. doi: 10.1038/290475a0. [DOI] [PubMed] [Google Scholar]
- Kelly K., Cochran B. H., Stiles C. D., Leder P. Cell-specific regulation of the c-myc gene by lymphocyte mitogens and platelet-derived growth factor. Cell. 1983 Dec;35(3 Pt 2):603–610. doi: 10.1016/0092-8674(83)90092-2. [DOI] [PubMed] [Google Scholar]
- Levy L. S., Gardner M. B., Casey J. W. Isolation of a feline leukaemia provirus containing the oncogene myc from a feline lymphosarcoma. 1984 Apr 26-May 2Nature. 308(5962):853–856. doi: 10.1038/308853a0. [DOI] [PubMed] [Google Scholar]
- Li Y., Holland C. A., Hartley J. W., Hopkins N. Viral integration near c-myc in 10-20% of mcf 247-induced AKR lymphomas. Proc Natl Acad Sci U S A. 1984 Nov;81(21):6808–6811. doi: 10.1073/pnas.81.21.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linial M., Groudine M. Transcription of three c-myc exons is enhanced in chicken bursal lymphoma cell lines. Proc Natl Acad Sci U S A. 1985 Jan;82(1):53–57. doi: 10.1073/pnas.82.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDevitt M. A., Imperiale M. J., Ali H., Nevins J. R. Requirement of a downstream sequence for generation of a poly(A) addition site. Cell. 1984 Jul;37(3):993–999. doi: 10.1016/0092-8674(84)90433-1. [DOI] [PubMed] [Google Scholar]
- Melton D. W., Konecki D. S., Brennand J., Caskey C. T. Structure, expression, and mutation of the hypoxanthine phosphoribosyltransferase gene. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2147–2151. doi: 10.1073/pnas.81.7.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil J. C., Hughes D., McFarlane R., Wilkie N. M., Onions D. E., Lees G., Jarrett O. Transduction and rearrangement of the myc gene by feline leukaemia virus in naturally occurring T-cell leukaemias. 1984 Apr 26-May 2Nature. 308(5962):814–820. doi: 10.1038/308814a0. [DOI] [PubMed] [Google Scholar]
- Nilsen T. W., Maroney P. A. Translational efficiency of cMyc mRNA in Burkitt lymphoma cells. Mol Cell Biol. 1984 Oct;4(10):2235–2238. doi: 10.1128/mcb.4.10.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne G. S., Bishop J. M., Varmus H. E. Multiple arrangements of viral DNA and an activated host oncogene in bursal lymphomas. Nature. 1982 Jan 21;295(5846):209–214. doi: 10.1038/295209a0. [DOI] [PubMed] [Google Scholar]
- Persson H., Leder P. Nuclear localization and DNA binding properties of a protein expressed by human c-myc oncogene. Science. 1984 Aug 17;225(4663):718–721. doi: 10.1126/science.6463648. [DOI] [PubMed] [Google Scholar]
- Rabbitts P. H., Forster A., Stinson M. A., Rabbitts T. H. Truncation of exon 1 from the c-myc gene results in prolonged c-myc mRNa stability. EMBO J. 1985 Dec 30;4(13B):3727–3733. doi: 10.1002/j.1460-2075.1985.tb04141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds G. A., Basu S. K., Osborne T. F., Chin D. J., Gil G., Brown M. S., Goldstein J. L., Luskey K. L. HMG CoA reductase: a negatively regulated gene with unusual promoter and 5' untranslated regions. Cell. 1984 Aug;38(1):275–285. doi: 10.1016/0092-8674(84)90549-x. [DOI] [PubMed] [Google Scholar]
- Sadofsky M., Alwine J. C. Sequences on the 3' side of hexanucleotide AAUAAA affect efficiency of cleavage at the polyadenylation site. Mol Cell Biol. 1984 Aug;4(8):1460–1468. doi: 10.1128/mcb.4.8.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H., Hayday A. C., Wiman K., Hayward W. S., Tonegawa S. Activation of the c-myc gene by translocation: a model for translational control. Proc Natl Acad Sci U S A. 1983 Dec;80(24):7476–7480. doi: 10.1073/pnas.80.24.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selten G., Cuypers H. T., Zijlstra M., Melief C., Berns A. Involvement of c-myc in MuLV-induced T cell lymphomas in mice: frequency and mechanisms of activation. EMBO J. 1984 Dec 20;3(13):3215–3222. doi: 10.1002/j.1460-2075.1984.tb02281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih C. K., Linial M., Goodenow M. M., Hayward W. S. Nucleotide sequence 5' of the chicken c-myc coding region: localization of a noncoding exon that is absent from myc transcripts in most avian leukosis virus-induced lymphomas. Proc Natl Acad Sci U S A. 1984 Aug;81(15):4697–4701. doi: 10.1073/pnas.81.15.4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton L. W., Fahrlander P. D., Tesser P. M., Marcu K. B. Nucleotide sequence comparison of normal and translocated murine c-myc genes. Nature. 1984 Aug 2;310(5976):423–425. doi: 10.1038/310423a0. [DOI] [PubMed] [Google Scholar]
- Stanton L. W., Watt R., Marcu K. B. Translocation, breakage and truncated transcripts of c-myc oncogene in murine plasmacytomas. Nature. 1983 Jun 2;303(5916):401–406. doi: 10.1038/303401a0. [DOI] [PubMed] [Google Scholar]
- Taub R., Moulding C., Battey J., Murphy W., Vasicek T., Lenoir G. M., Leder P. Activation and somatic mutation of the translocated c-myc gene in burkitt lymphoma cells. Cell. 1984 Feb;36(2):339–348. doi: 10.1016/0092-8674(84)90227-7. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Valerio D., Duyvesteyn M. G., Dekker B. M., Weeda G., Berkvens T. M., van der Voorn L., van Ormondt H., van der Eb A. J. Adenosine deaminase: characterization and expression of a gene with a remarkable promoter. EMBO J. 1985 Feb;4(2):437–443. doi: 10.1002/j.1460-2075.1985.tb03648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmus H. E. The molecular genetics of cellular oncogenes. Annu Rev Genet. 1984;18:553–612. doi: 10.1146/annurev.ge.18.120184.003005. [DOI] [PubMed] [Google Scholar]
- Watson D. K., Reddy E. P., Duesberg P. H., Papas T. S. Nucleotide sequence analysis of the chicken c-myc gene reveals homologous and unique coding regions by comparison with the transforming gene of avian myelocytomatosis virus MC29, delta gag-myc. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2146–2150. doi: 10.1073/pnas.80.8.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt R., Stanton L. W., Marcu K. B., Gallo R. C., Croce C. M., Rovera G. Nucleotide sequence of cloned cDNA of human c-myc oncogene. Nature. 1983 Jun 23;303(5919):725–728. doi: 10.1038/303725a0. [DOI] [PubMed] [Google Scholar]
- Wickens M., Stephenson P. Role of the conserved AAUAAA sequence: four AAUAAA point mutants prevent messenger RNA 3' end formation. Science. 1984 Nov 30;226(4678):1045–1051. doi: 10.1126/science.6208611. [DOI] [PubMed] [Google Scholar]