Abstract
Background
Heterotaxy syndrome is caused by left-right asymmetry disturbances and is associated with abnormal lateralization of the abdominal and thoracic organs. The heart is frequently involved and the severity of the abnormality usually determines the outcome.
Methods
Direct sequence analysis of the coding sequence of genes including Zinc Finger Protein of the Cerebellum 3, Left Right Determine Factor 2, Activin A Receptor Type IIB, and Cryptic was performed in 47 subjects with laterality defects and congenital cardiac disease.
Results
31 (66%) of these subjects had atrioventricular septal defects, 34 (72%) had abnormal systemic venous return, 25 (53%) had transposed or malposed great arteries, and 20 (43%) had pulmonary venous abnormalities. Two novel genetic changes were identified in zinc finger protein of the cerebellum 3 and these variants were not presented in 100 ethnically matched control samples. One previously reported missense mutation in activin A receptor type IIB was identified in 2 unrelated subjects. The genetic changes identified in this study are all located in conserved regions and are predicted to affect protein function in left-right axis formation and cardiovascular development.
Conclusions
Mutations in Zinc Finger Protein of the Cerebellum 3 and Activin A Receptor Type IIB were identified in 4/47 subjects with heterotaxy syndrome for a yield of approximately 8.5%. Our results expand the mutation spectrum of monogenic heterotaxy syndrome with associated cardiac anomalies and suggest that there are other causes of heterotaxy yet to be identified.
Keywords: left-right asymmetry, congenital heart defect, LEFTYA, CFC1
Introduction
The exterior body plan of vertebrates is essentially symmetric along its medial lateral axis while the formation of internal organs displays numerous left-right differences. Left right axis is the third body axis to form, and alteration of the normal pattern of asymmetry can manifest either in mirror-image reversal of all asymmetrical organs or randomized placement of organs.1 The heart is the first organ to develop asymmetrically, and cardiac morphogenesis is sensitive to aberrations in left right positional information.2 Heterotaxy syndrome is often associated with cardiac malformations which cause significant morbidity and mortality. In heterotaxy, the heart is normally patterned and formed in relation to anterior posterior and dorsal ventral axis, indicating that anterior posterior and dorsal ventral specification can act independently from left right specification. Patients often have malposition of the great arteries, systemic or pulmonary venous return abnormalities and abnormal septation such as atrioventricular septal defects.
As a clinically and genetically heterogeneous disorder, heterotaxy syndrome can be associated with chromosome abnormalities such as balanced translocations,3,4 microdeletions or duplications,5 single gene mutations, epigenetic factors, or environment teratogens.6 Monogenetic causes of heterotaxy syndrome segregate as autosomal recessive,7 autosomal dominant 8,9 and X-linked recessive disorders.10 Highly conserved genetic pathways control determination of the left right axis11,12 across species from xenopus,13 zebrafish,14 chick 15 to mouse.16,17 It is suggested that the initial breaking of left right asymmetry occurs in the node, and the nodal flow generated by the monocilia in the node leads to establishment of morphogen gradient including Nodal, Lefty, and Acvr2b in either the left or right side of the body to pattern the internal organs. zinc finger protein of the cerebellum 3 (ZIC3) is a zinc finger transcription factor. Mutations of ZIC3 cause X-linked heterotaxy (HTX1) and isolated congenital heart disease.18–21 Zic3 knockout mice recapitulate human HTX1, and fifty percent of Zic3 null embryos have randomized internal organs and abnormal cardiac looping.22 Mouse homologues of left right determine factor 2 (LEFTYA) (homologue of mouse Lefty-2) and left right determine factor 2 (LEFTYB) (homologue of mouse Lefty-1) mediate Nodal signaling from the node to lateral plate mesoderm. A null mutation of Lefty-2 in mice shows left isomerism.23 Mutations of LEFTYA have been identified in heterotaxy patients as well.24 Activin receptor IIB (Acvr2b) is expressed asymmetrically along left-right axis in mouse and chick. A null mutation of Acvr2b in mice results in situs ambiguous, atrial septal defect, ventricular septal defect and splenic hypoplasia.25 Based on the murine phenotype, 112 sporadic and 14 familial cases of left right axis malformation were analyzed for ACVR2B mutations, and two missense mutations were identified leading to conclusion that ACVR2B mutations are rare causes of human left right axis malformation.26 Cryptic (CFC1) belongs to the EGF (epidermal growth factor)-CFC1 family and encodes extracellular protein that plays a key role in intercellular signaling pathways. CFC1 is expressed around the node and is later found in the intermediate and lateral plate mesoderm during gastrulation in mouse embryo.27 Homozygous Cryptic knockout mice presented laterality defects and complex cardiac malformations. The phenotypes resemble those of mice lacking the type IIB activin receptor or the homeobox-containing factor Pitx2.28 CFC1 mutations and missense variants have been identified in patients with heterotaxy syndrome associated with congenital heart diseases.29–32
To expand the human genetic studies of heterotaxy syndrome with associated congenital heart disease, we screened 47 patients for mutations in ZIC3, LEFTYA, ACVR2B and CFC1 to determine the frequency of mutations, mutation spectrum, and penetrance of the identified mutations, in order to provide data to guide development of future genetic testing for heterotaxy syndrome.
Patient and Methods
Clinical Materials
Subjects included fetuses and children diagnosed with heterotaxy syndrome. Subjects included fetuses and children diagnosed with heterotaxy syndrome defined as segmental discordances of the thoraco-abdominal organs along the left-right axis. All patients seen at the Columbia University Medical Center were offered study entry. The study was approved by the Institutional Review Board at Columbia University Medical Center. Informed consent was obtained from parents or the patients, depending on the age of the patient. Among them, 46 patients had normal karyotypes while one patient had an extra X chromosome (47, XXX). Genomic DNA was extracted from blood, tissue, or amniocytes from each subject using Puregene reagents (Gentra Systems Inc., Minnesota, USA). Clinical data about the type of congenital heart disease and extra-cardiac phenotype was extracted from the medical record. Clinical cardiac information was reviewed by a single cardiologist (SST).
Genotyping Genetic Variations Using PCR Amplification
Primers were designed to amplify all coding regions and at least 20 bp of adjacent intronic sequence to capture the splice junctions for the four genes ZIC3, LEFTYA, ACVR2B and CFC1. Primer sequences are available upon request. In brief, 20ng of genomic DNA from each patient was amplified in a 20 μl volume containing 1xPCR buffer, 40ng of each oligonucleotide primers, 200uM dNTPs and 1.2 U Taq polymerase (Promega). PCR products were purified using ExoSAP-It kit (USB Scientific, OH) and sequenced by Sanger dideoxy sequencing on an ABI 3730xl genetic analyzer according to the manufacturer’s instructions (ABI, CA) using one of the amplification primers. Genetic variants were confirmed on a second PCR reaction with bi-directional sequencing, and all available family members were sequenced. Sequences are analyzed with Sequencher software (Gene Codes, MI). Genetic variants were compared to all reported genetic variants in the literature, in dbSNP, at www.ncbi.nlm.nih.gov/snp and at www.genome.ucsc.edu. All novel genetic variants were independently confirmed using restriction digests. Confirmed novel genetic variants were then genotyped in 100 unrelated normal individuals without congenital heart disease of the same ethnicity.
Results
Clinical Data
The patients consisted of 23 males and 24 females. The ethnicity of the patients was 45% (21/47) Caucasian, 23% (11/47) Hispanic, 20% (9/47) African American, 6% (3/47) Asian and Southeast Asian, and 6% (3/47) other. Seven were fetal cases with prenatal terminations. 44 cases were sporadic and 3 cases were familial. Associated congenital heart defects included atrioventricular septal defect (AVSD) in 31/47 patients, systemic venous anomalies in 34/47 patients, 25/47 patients had transposed or malposed great arteries, outflow tract obstruction were presented in 23/47 patients and pulmonary venous anomalies in 20/47 patients (Supplemental Table 1). Table 1 lists all the cardiac and extracardiac manifestations of the patients in the series.
Table 1.
Cardiac Phenotypes of Heterotaxy Patients
| Cardiac and Extracardiac Manifestatations | Number of Cases |
|---|---|
| Atrioventricular septal defect | 31/47 |
| Anomalous systemic venous connections | |
| Interrupted inferior vena cava | 22/47 |
| Bilateral superior vena cava | 12/47 |
| Malposed/transposed great arteries | 25/47 |
| Outflow tract obstruction | |
| Pulmonary stenosis/atresia | 21/47 |
| Aortic stenosis/atresia | 2/47 |
| Anomalous pulmonary venous connections | |
| Total anomalous pulmonary venous return | 11/47 |
| Ipsilateral or partial pulmonary venous return | 9/47 |
| Visceral organs | |
| Intestinal malrotation | 16/47 |
| Transverse liver | 14/47 |
| Right-sided stomach | 11/47 |
| Asplenia | 10/47 |
| Polysplenia | 5/47 |
Mutation Screening
ZIC3
Sequencing of ZIC3 demonstrated two novel genetic variants. Patient CHD32 was a male with a c.148delG resulting in a frameshift and putative truncated protein p.A50PfsX8 (Figure 1). CHD32 had right ventricle dominant AVSD, double outlet right ventricle, mitral atresia and total anomalous pulmonary venous return (Table 2). The hepatic veins connected directly into the right atrium. Extracardiac manifestations included malrotation, transverse liver and asplenia. The patient was adopted, and no family history or parental samples were available.
Figure 1.
Mutation of ZIC3 exon 1 in CHD32. Sequence of ZIC3 c.148delG in proband was shown. Brackets symbolize adoption.
Table 2.
Patients with ZIC3 and ACVR2B Mutations
| Patients | Gender | Ethnicity | Mutation | Cardiac Phenotype | Visceral Organs |
|---|---|---|---|---|---|
| CHD32 | Male | Caucasian | ZIC3 c.148delG | Right ventricle dominant atrioventricular septal defect, double outlet right ventricle, mitral atresia, total anomalous pulmonary venous return, hepatic veins drain directly into the right atrium | Malrotation, transverse liver and asplenia |
| CHD186 | Female | Caucasian | ZIC3 p.S402P | atrioventricular septal defect, pulmonary atresia, total anomalous pulmonary venous return, hepatic venous drainage to midline of right sided atrium, bilateral superior vena cava | Malrotation |
| CHD218 | Male | Caucasian | ZIC3 p.S402P | Double inlet double outlet single left ventricle, pulmonary stenosis, right sided aortic arch | Situs inversus |
| CHD141 | Female | Hispanic | ACVR2B p.R40H | Common atrium, right ventricular dominant atrioventricular septal defect, pulmonary atresia, ipsilateral pulmonary venous return, inferior vena cava to base of common atrium, bilateral superior vena cava | Not known |
| CHD1067 | Male | African American | ACVR2B p. R40H | atrioventricular septal defect, transposition of the great arteries, interrupted inferior vena cava | Right-sided stomach |
Analysis of ZIC3 in CHD186 identified a heterozygous c.1204T>C transversion which results in substitution of p.S402P (Figure 2). The proband is a female patient with AVSD, pulmonary atresia and total anomalous pulmonary venous return. She had bilateral superior vena cava with hepatic veins connect to the midline of right-sided atrium. She had abdominal malrotation. (Table 2).
Figure 2.
ZIC3 c.1204T>C in CHD186 and the family. Pedigree indicating that the mother (CHD187) is a carrier and the two children (CHD186 and CHD218) are affected. Electropherograms below each family member show that CHD187 (unaffected mother) and CHD186 (proband, female) are heterozygous for c.1204T>C while CHD218 (affected male sibling) carries the hemizygous c.1204T>C transversion resulting in a p.S402P missense mutation. The father (CHD188) and maternal parents (CHD1234 and CHD1235) are normal. The point (CHD187) denotes carrier status.
The brother of CHD 186 (CHD218) also had the c.1204T>C transversion in ZIC3 and presented with complex cardiac disease including double inlet and double outlet single left ventricle, pulmonary stenosis and a right-sided aortic arch. He had abdominal situs inversus and midline bony malformations including hypoplasia of the posterior elements of C1, rotational scoliosis of the upper cervical spine involving C2 and C3, fusion of multiple vertebral bodies including C2-C3 and two lower cervical vertebral bodies.
Analysis of the parents revealed that the mother is heterozygous for the c.1204T>C variant while the father is normal. The mother is clinically unaffected. Because this is X-linked recessive heterotaxy, we performed X chromosome inactivation assay and revealed that the mother had skewed X inactivation while there was random inactivation of X chromosome in the proband (data not shown). Further studies showed that the proband’s maternal grandparents do not carry the c.1204T>C variant which indicates that the amino acid substitution p.S402P in ZIC3 was de novo in proband’s mother. (Figure. 2). The amino acid serine at position 402 lies within the fifth zinc finger domain and is highly conserved among human, mouse and zebrafish (Figure 3).
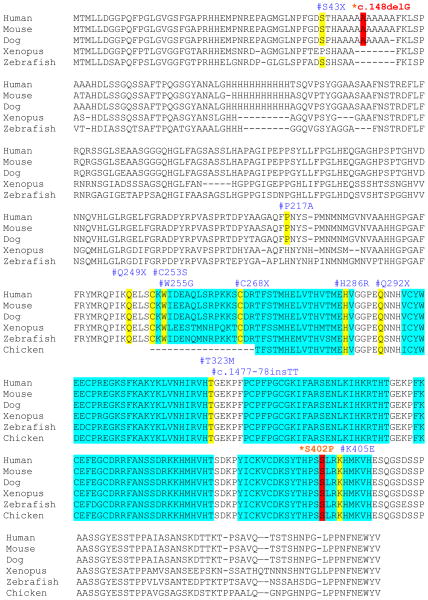
Figure 3.
Cross species comparison of ZIC3 amino acid and locations of putative mutations. Solid rectangles in blue indicate the five C2H2 zinc finger domains. The amino acid positions for each mutation are boxed in red and types of mutations are labeled above each box. The c.148delG mutation causes a frameshift and is predicted to form truncated protein with only 57 amino acids. A missense mutation S402P was detected in the fifth zinc finger domain. Previously reported mutations are boxed in yellow and the amino acid or nucleotide positions are labeled above.
ACVR2B
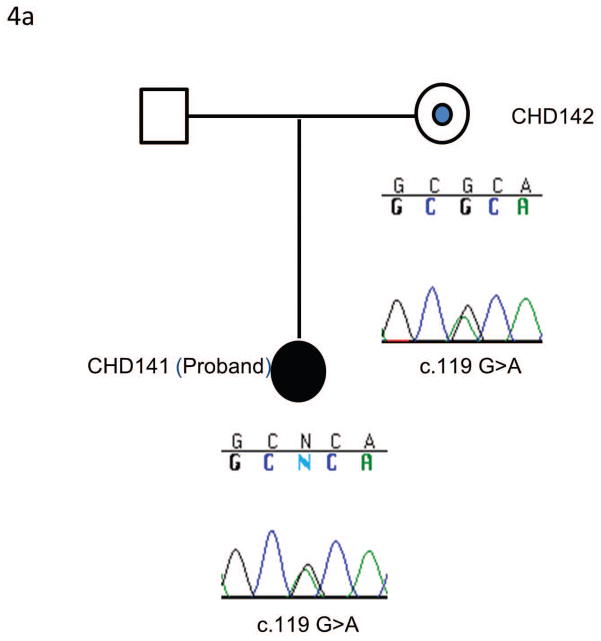
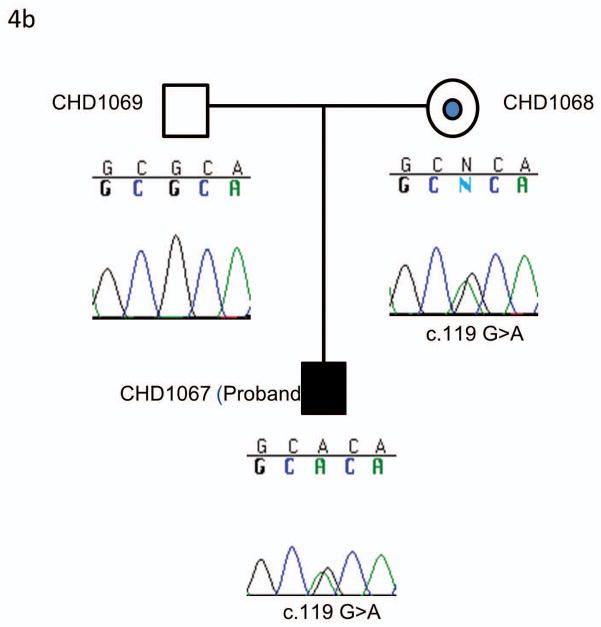
Sequencing of ACVR2B identified two patients (CHD 141 and CHD 1067) with an identical heterozygous c.119G>A variant (p.R40H) (Figure 4). This variant has previously been reported to be a pathogenic mutation (Kosaki R et al., 1999). CHD141 had right ventricle dominant atrioventricular septal defect, pulmonary atresia, ipsilateral pulmonary venous return, inferior vena cava connecting to the base of common atrium, and bilateral superior vena cava (Table 2). CHD1067 was a fetus with atrioventricular septal defect, transposition of great arteries, interrupted inferior vena cava and a right sided stomach (Table 2). The unaffected mothers of both subjects carried the same heterozygous c.119G>A (p.R40H).
Figure 4.
An identical mutation of ACVR2B exon 2 in the two independent families. 4a) Pedigree and sequencing results of CHD 141 and the family. The same mutation was identified in an unrelated family and the pedigree and sequencing results were shown in 4b. In both families the unaffected mothers (CHD142 and CHD1068) are heterozygous for the c.119G>A transversion carried by the affected probands (CHD141 and CHD1067). The c.119G>A transversion results in a p.R40H missense mutation. The dot in the center of the pedigree symbol denotes carrier status.
Controls
The novel variants in ZIC3 were genotyped in 100 ethnically matched normal controls, and were not identified in any of the normal subjects. The mutation R40H in ACVR2B was genotyped in 100 randomly selected individuals in the previous report and has not been identified in normal population (Kosaki R et al., 1999).
Sequencing of CFC1 was previously performed on a portion of these subjects (25 patients) (Selamet Tierney ES et al., 2007) and completed in the remaining 22 subjects in this cohort. No novel mutations were identified. No mutations were identified in LEFTYA.
Discussion
Sequencing of coding regions of ZIC3, LEFTYA, ACVR2B, and CFC1 in 47 heterotaxy patients with associated cardiovascular anomalies identified two novel genetic changes in two patients for ZIC3 and a previously reported mutation in ACVR2B in two unrelated subjects for a total yield of 4/47 positive cases (8.5%). In the family with the S402P variant in ZIC3, S402P segregated with heterotaxy within the family and was associated with skewed X inactivation in the carrier mother. The mother’s mutation was de novo. These data suggested that the S402P variant is pathogenic. All of the novel genetic variants we report are highly conserved across species, are located in functionally important domains of the proteins (Figure 3), and none of the variants were present in 100 randomly selected normal individuals. These data suggested that the two novel variants in ZIC3 and the one previously reported R40H variant in ACVR2B are likely to be pathogenic mutations.
ZIC3 functions in early stages of embryonic development to regulate left right axis formation. ZIC3 p.S402 is located in the α-Helical structure of zinc finger (ZF) 5 for DNA binding. The amino acid is highly conserved among species as well as among Zic/Gli/Glis zinc finger protein superfamily, 33 suggesting that this amino acid maybe important for the ZIC3-DNA complex formation. ZIC3 has been reported to interact with GLI3 through GLI consensus binding site (GLIBS) to regulate multiple aspects of neural and skeletal development.34,35 It is speculated that S402P in ZIC3 may account for the skeletal anomalies in CHD218. CHD186 and CHD218 had different cardiac phenotypes and extracardiac manifestations that could be due to the hemizygous state of the male compared to the heterozygous female with one copy of the wild type allele. CHD32 is a male patient who had a hemizygous c.148delG in ZIC3. This mutation is predicted to produce truncated protein p.A50PfsX8 without formation of Zinc finger domains. CHD32 had multiple complex cardiovascular abnormalities and cardiac transplant was performed at age 10. CHD32 also had extra cardiac manifestations. To date, eleven nonsense, frameshift and missense mutations in ZIC3 gene have been reported among 241 sporadic and familial heterotaxy cases with 85% of mutations being maternally inherited, usually from unaffected mothers.19 The ratio of clinically affected male to female patients with ZIC3 mutations is 2.5:1 and may be explained by skewed X-inactivation.
The frequency of LEFTYA mutations in heterotaxy is low. Only one study identified two missense mutations among 126 patients to date.24 In our current study, no novel mutations were identified in our heterotaxy patients.
One study identified two missense mutations: p.V419I and p.R40H in ACVR2B in 112 sporadic and 14 familial cases.26 Our study identified the R40H mutation in 2/47 cases, both of whom inherited the mutation from an unaffected mother. R40 in ACVR2B is located in the region of extracellular domain within the region of ligand binding and is conserved across human, mouse, dog and elephant. 26 While the patients who carry R40H mutation in ACVR2B had distinct clinical presentations, all exhibited abnormal systemic venous return.
Single genes have been screened in cohorts of patients with heterotaxy, but our study is the most comprehensive genetic study that analyzed ZIC3, LEFTYA, ACVR2B, and CFC1, as a panel in all heterotaxy patients with cardiac manifestations. The total yield for all four genes was 8.5% (4/47). A comparison of the yield for each gene in previous and current studies is provided in Table 3. While this yield from these four genes is modest, it suggests that develop panels of genes in genetic study to evaluate convergent phenotypes in heart that occur in the context of inherited or sporadic diseases such as heterotaxy may be useful for diagnosis and evaluation. Our data also suggests that mutations in any one gene may account for a small percentage of cases and may be incompletely penetrant, making strategies of linkage analysis and association studies less powerful. Our study expands the mutation spectrum in heterotaxy and also suggests that additional genes are likely to be involved in the remaining 90% of cases for whom genetic changes were not identified, such as mutations in NODAL,36–38 NKX2.5 39,40 and CRELD1.41 Our study as well as the results from previous reports 42,43 suggest that genetic changes in more than one gene in the same pathway can converge to produce the same phenotype of heterotaxy with associated congenital heart disease. Patients without identified mutations will provide the substrate for identification of novel genes associated with heterotaxy using high throughput methods of genomic analysis.
Table 3.
Comparison of Frequency of ZIC3, LEFTYA, ACVR2B and CFC1 Mutations across Studies
| Gene | Previous Studies | Frequency of Mutations in Previous Studies | Frequency of Mutations in Current Studies |
|---|---|---|---|
| ZIC3 | 69 heterotaxy with congenital heart disease | 5.8–7.1% | 4.7% |
| LEFTYA | 126 heterotaxy | 1.6% | 0% |
| ACVR2B | 112 sporadic and 14 familial heterotaxy | 2.4% | 4.7% |
| CFC1 | 257 heterotaxy with or without congenital heart disease | 2.2% | 0% |
Supplementary Material
Acknowledgments
We thank the families for their generous contribution.
Sources of Funding
None
References
- 1.Casey B. Two rights make a wrong: human left-right malformations. Hum Molec Genet. 1998;7:1565–1571. doi: 10.1093/hmg/7.10.1565. [DOI] [PubMed] [Google Scholar]
- 2.Kathiriya IS, Srivastava D. Left-right asymmetry and cardiac looping: implications for cardiac development and congenital heart disease. Am J Med Genet. 2000;97:271–279. doi: 10.1002/1096-8628(200024)97:4<271::aid-ajmg1277>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 3.Kato R, Yamada Y, Niikawa N. De novo balanced translocation (6;18)(q21. 3 or q22) in a patient with heterotaxia. Am J Med Genet. 1996;66:184–186. doi: 10.1002/(SICI)1096-8628(19961211)66:2<184::AID-AJMG11>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 4.Fritz B, Kunz J, Knudsen GP, et al. Situs ambiguus in a female fetus with balanced (X;21) translocation – evidence for functional nullisomy of the ZIC3 gene? Eur J Hum Genet. 2005;13:34–40. doi: 10.1038/sj.ejhg.5201213. [DOI] [PubMed] [Google Scholar]
- 5.Schinzel A, Hanson JW, Pagon RA, Hoehn H, Smith DW. Trisomy 3 (p23-pter) resulting from maternal translocation, t (3; 4)(023;q35) Ann Genet. 1978;21:168–171. [PubMed] [Google Scholar]
- 6.Kuehl KS, Loffredo C. Risk factors for heart disease associated with abnormal sidedness. Teratology. 2002;66:242–248. doi: 10.1002/tera.10099. [DOI] [PubMed] [Google Scholar]
- 7.Chen S-C, Monteleone PL. Familial splenic anomaly. J Pediatr. 1977;91:160–161. doi: 10.1016/s0022-3476(77)80476-9. [DOI] [PubMed] [Google Scholar]
- 8.Alonso S, Pierpont ME, Radtke W, et al. Heterotaxia syndrome and autosomal dominant inheritance. Am J Med Genet. 1995;56:12–15. doi: 10.1002/ajmg.1320560105. [DOI] [PubMed] [Google Scholar]
- 9.Casey B, Cuneo BF, Vitali C, et al. Autosomal dominant transmission of familial laterality defects. Am J Med Genet. 1996;61:325–328. doi: 10.1002/(SICI)1096-8628(19960202)61:4<325::AID-AJMG5>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 10.Casey B, Devoto M, Jones KL, Ballabio A. Mapping a gene for familial situs abnormalities to human chromosome Xq24-q27. 1. Nature Genet. 1993;5:403–407. doi: 10.1038/ng1293-403. [DOI] [PubMed] [Google Scholar]
- 11.Fujinaga M. Development of sidedness of asymmetric body structures in vertebrates. Int J Dev Biol. 1997;41:153–186. [PubMed] [Google Scholar]
- 12.Burdine RD, Schier AF. Conserved and divergent mechanism in left-right axis formation. Genes Dev. 2000;14:763–776. [PubMed] [Google Scholar]
- 13.Hyatt BA, Lohr JL, Yost HJ. Initiation of vertebrate left-right axis formation by maternal Vg1. Nature. 1996;384:62–65. doi: 10.1038/384062a0. [DOI] [PubMed] [Google Scholar]
- 14.Essner JJ, Amack JD, Nyholm MK, Harris EB, Yost HJ. Kupffer’s vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left-right development of the brain, heart and gut. Development. 2005;132:1247–1260. doi: 10.1242/dev.01663. [DOI] [PubMed] [Google Scholar]
- 15.Levin M, Johnson RL, Stern CD, Kuehn M, Tabin C. A molecular pathway determining left-right asymmetry in chick embryogenesis. Cell. 1995;82:803–814. doi: 10.1016/0092-8674(95)90477-8. [DOI] [PubMed] [Google Scholar]
- 16.Nonaka S, Tanaka Y, Okada Y, et al. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell. 1998;95:829–837. doi: 10.1016/s0092-8674(00)81705-5. [DOI] [PubMed] [Google Scholar]
- 17.Lowe LA, Supp DM, Sampath K, et al. Conserved left-right asymmetry of nodal expression and alterations in murine situs inversus. Nature. 1996;381:158–161. doi: 10.1038/381158a0. [DOI] [PubMed] [Google Scholar]
- 18.Mathias RS, Lacro RV, Jones KL. X-linked laterality sequence: situs inversus, complex cardiac defects, splenic defects. Am J Med Genet. 1987;28:111–116. doi: 10.1002/ajmg.1320280116. [DOI] [PubMed] [Google Scholar]
- 19.Ware SM, Peng J, Zhu L, et al. Identification and functional analysis of ZIC3 mutations in heterotaxy and related congenital heart defects. Am J Hum Genet. 2004;74:93–105. doi: 10.1086/380998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mégarbané A, Salem N, Stephan E, et al. X-linked transposition of the great arteries and incomplete penetrance among males with a nonsense mutation in ZIC3. Eur J Hum Genet. 2000;8:704–708. doi: 10.1038/sj.ejhg.5200526. [DOI] [PubMed] [Google Scholar]
- 21.Chhin B, Hatayama M, Bozon D, et al. Elucidation of penetrance variability of a ZIC3 mutation in a family with complex heart defects and functional analysis of ZIC3 mutations in the first zinc finger domain. Hum Mutat. 2007;28:563–570. doi: 10.1002/humu.20480. [DOI] [PubMed] [Google Scholar]
- 22.Purandare SM, Ware SM, Kwan KM, et al. A complex syndrome of left-right axis, central nervous system and axial skeletaon defects in Zic3 mutant mice. Development. 2002;129:2293–2302. doi: 10.1242/dev.129.9.2293. [DOI] [PubMed] [Google Scholar]
- 23.Meno C, Takeuchi J, Sakuma R, et al. Diffusion of nodal signaling activity in the absence of the feedback inhibitor lefty2. Dev Cell. 2001;1:127–138. doi: 10.1016/s1534-5807(01)00006-5. [DOI] [PubMed] [Google Scholar]
- 24.Kosaki K, Bassi MT, Kosaki R, et al. Characterization and mutation analysis of human LEFTY A and LEFTY B, homologues of murine genes implicated in left-right axis development. Am J Hum Genet. 1999;64:712–721. doi: 10.1086/302289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oh SP, Li E. The signaling pathway mediated by the type IIB activin receptor controls axial patterning and lateral asymmetry in the mouse. Genes Dev. 1997;11:1812–1826. doi: 10.1101/gad.11.14.1812. [DOI] [PubMed] [Google Scholar]
- 26.Kosaki R, Gebbia M, Kosaki K, et al. Left-right axis malformations associated with mutations in ACVR2B, the gene for human activin receptor type IIB. Am J Med Genet. 1999;82:70–76. doi: 10.1002/(sici)1096-8628(19990101)82:1<70::aid-ajmg14>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 27.Shen MM, Wang H, Leder P. A differential display strategy identifies Cryptic, a novel EGF-related gene expressed in the axial and lateral mesoderm during mouse gastrulation. Development. 1997;124:429–442. doi: 10.1242/dev.124.2.429. [DOI] [PubMed] [Google Scholar]
- 28.Gaio U, Schweickert A, Fischer A, et al. A role of the cryptic gene in the correct establishment of the left-right axis. Curr Biol. 1999;9:1339–1342. doi: 10.1016/s0960-9822(00)80059-7. [DOI] [PubMed] [Google Scholar]
- 29.Bamford RN, Roessler E, Burdine RD, et al. Loss-of-function mutations in the EGF-CFC gene CFC1 are associated with human left-right laterality defects. Nat Genet. 2000;26:365–369. doi: 10.1038/81695. [DOI] [PubMed] [Google Scholar]
- 30.Goldmuntz E, Bamford R, Karkera JD, dela Cruz J, Roessler E, Muenke M. CFC1 mutations in patients with transposition of the great arteries and double-outlet right ventricle. Am J Hum Genet. 2002;70:776–780. doi: 10.1086/339079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ozcelik C, Bit-Avragim N, Panek A, et al. Mutations in the EGF-CFC gene cryptic are an infrequent cause of congenital heart disease. Pediatr Cardiol. 2006;27:695–698. doi: 10.1007/s00246-006-1082-0. [DOI] [PubMed] [Google Scholar]
- 32.Selamet Tierney ES, Marans Z, Rutkin MB, Chung WK. Variants of the CFC1 gene in patients with laterality defects associated with congenital cardiac disease. Cardiol Young. 2007;17:268–274. doi: 10.1017/S1047951107000455. [DOI] [PubMed] [Google Scholar]
- 33.Sakai-Kato K, Ishiguro A, Mikoshiba K, Aruga J, Utsunomiya-Tate N. CD spectra show the relational style between Zic-, Gli-, Glis-zinc finger protein and DNA. Biochim Biophys Acta. 2008;1784:1011–1019. doi: 10.1016/j.bbapap.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 34.Hatayama M, Tomizawa T, Sakai-Kato K, et al. Functional and structural basis of the nuclear localization signal in the ZIC3 zinc finger domain. Hum Mol Genet. 2008;17:3459–3473. doi: 10.1093/hmg/ddn239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu L, Zhou G, Poole S, Belmont JW. Characterization of the interactions of human ZIC3 mutants with GLI3. Human Mutation. 2008;29:99–105. doi: 10.1002/humu.20606. [DOI] [PubMed] [Google Scholar]
- 36.Zlotogora J, Schimmel MS, Glaser Y. Familial situs inversus and congenital heart defects. Am J Med Genet. 1987;26:181–184. doi: 10.1002/ajmg.1320260126. [DOI] [PubMed] [Google Scholar]
- 37.Gebbia M, Ferrero GB, Pilia G, et al. X-linked situs abnormalities result from mutations in ZIC3. Nat Genet. 1997;17:305–308. doi: 10.1038/ng1197-305. [DOI] [PubMed] [Google Scholar]
- 38.Mohapatra B, Casey B, Li H, et al. Identification and functional characterization of NODAL rare variants in heterotaxy and isolated cardiovascular malformations. Hum Mol Genet. 2009;18:861–871. doi: 10.1093/hmg/ddn411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watanabe Y, Benson DW, Yano S, Akagi T, Yoshino M, Murray JC. Two novel frameshift mutations in NKX2. 5 result in novel features including visceral inversus and sinus venosus type ASD. J Med Genet. 2002;39:807–811. doi: 10.1136/jmg.39.11.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dentice M, Cordeddu V, Rosica A, et al. Missense mutation in the transcription factor NKX2–5: a novel molecular event in the pathogenesis of thyroid dysgenesis. J Clin Endocr Metab. 2006;91:1428–1433. doi: 10.1210/jc.2005-1350. [DOI] [PubMed] [Google Scholar]
- 41.Robinson SW, Morris CD, Goldmuntz E, et al. Missense mutations in CRELD1 are associated with cardiac atrioventricular septal defects. Am J Hum Genet. 2003;72:1047–1052. doi: 10.1086/374319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roessler E, Ouspenskaia MV, Karkera JD, et al. Reduced NODAL signaling strength via mutation of several pathway members including FOXH1 is linked to human heart defects and holoprosencephaly. Am J Hum Genet. 2008;83:18–29. doi: 10.1016/j.ajhg.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roessler E, Pei W, Ouspenskaia MV, et al. Cumulative ligand activity of NODAL mutations and modifiers are linked to human heart defects and holoprosencephaly. Mol Genet Metab. 2009;98:225–234. doi: 10.1016/j.ymgme.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.