Abstract
Cell-based therapies for myelin disorders, such as multiple sclerosis and leukodystrophies, require technologies to generate functional oligodendrocyte progenitor cells. Here we describe direct conversion of mouse embryonic and lung fibroblasts to “induced” oligodendrocyte progenitor cells (iOPCs) using sets of either eight or three defined transcription factors. iOPCs exhibit a bipolar morphology and global gene expression profile consistent with bona fide OPCs. They can be expanded in vitro for at least five passages while retaining the ability to differentiate into multiprocessed oligodendrocytes. When transplanted to hypomyelinated mice, iOPCs are capable of ensheathing host axons and generating compact myelin. Lineage conversion of somatic cells to expandable iOPCs provides a strategy to study the molecular control of oligodendrocyte lineage identity and may facilitate neurological disease modeling and autologous remyelinating therapies.
Myelin loss or dysfunction affects millions of people worldwide and causes substantial morbidity and mortality. Diseases of myelin in the central nervous system (CNS) are often severely disabling and include adult disorders such as multiple sclerosis and childhood diseases such as cerebral palsy and congenital leukodystrophies. Oligodendrocyte progenitor cells (OPCs), the predominant source of myelinating oligodendrocytes in the CNS, have shown promise as a cellular therapeutic in animal models of myelin diseases 1–3. However, sources of OPCs have been restricted largely to allogeneic fetal cells with limited expansion capacity 4. Thus, technologies to generate scalable and autologous sources of OPCs are of great interest as they would enable large-scale drug screening and cell-based regenerative medicine. Methods based on pluripotent stem cells and direct lineage reprogramming may meet these requirements. Recently, we showed efficient differentiation of mouse pluripotent stem cells into pure populations of expandable, myelinogenic OPCs using defined developmental signals 5. In the present study, we sought to apply our understanding of oligodendrocyte development to directly convert mouse fibroblasts to expandable OPCs by forced expression of a small number of transcription factors (TFs) (Fig. 1a). Several recent studies have laid the foundation for the use of lineage conversion in regenerative therapies for neurological disorders 6–15. Although these reprogramming technologies have been applied to generate various neuronal fates such as neurons and neural stem cells, production of myelinogenic OPCs has remained elusive. Here we show that defined sets of transcription factors can reprogram mouse fibroblasts into myelinogenic iOPCs. With further optimization, this approach could provide a source of functional OPCs that will complement, and possibly obviate, the use of pluripotent stem cells and fetal cells for cell-based remyelinating therapies.
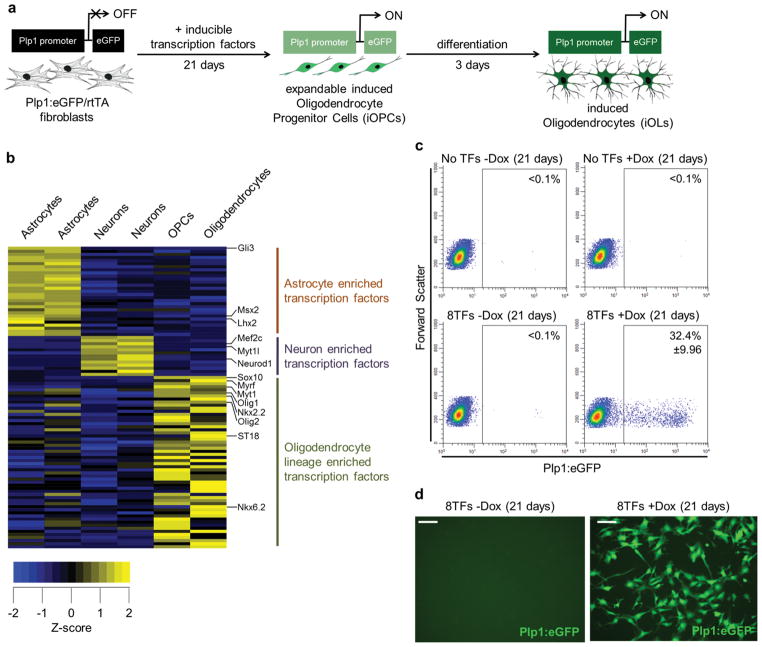
Figure 1. Eight transcription factors can reprogram mouse embryonic fibroblasts to induced oligodendrocyte progenitor cells.
a, Experimental design overview and timeline for transcription factor-mediated reprogramming of Plp1:eGFP/rtTA fibroblasts to induced oligodendrocyte progenitor cells (iOPCs) which are expandable and capable of differentiating to induced oligodendrocytes (iOLs). The modified Plp1:eGFP transgene is expressed in both OPCs and OLs in vivo. b, Gene expression heat map showing transcription factors (TFs) enriched (yellow) in each of the three major CNS lineages (see Supplementary Table 1 for complete list of genes). c, Representative flow cytometry plots and the average percentage of Plp1:eGFP+ cells after 21 days in OPC culture conditions for: non-infected and uninduced cells (No TFs −Dox); non-infected and induced cells (No TFs +Dox); infected with 8TFs but not induced (8TFs −Dox); and infected with 8TFs and induced (8TFs +Dox). Only infection of 8TFs and induction gave rise to Plp:eGFP+ cells at day 21 (8TFs +Dox = 32.4% +/− 9.9%; n = 19 independent biological replicates from 3 independent lots of lentivirus). All other conditions were devoid of Plp1:eGFP+ cells at day 21 (<0.1%). d, Representative live-cell fluorescent images of Plp:eGFP MEF cultures that were infected with inducible lentiviruses containing the 8TF pool and cultured for 21 days in the absence (−Dox) or presence (+Dox) of TF induction. Scale bar, 50μm (d).
RESULTS
Expression of oligodendrocyte lineage TFs in fibroblasts
Using microarray data 5, 16, we identified TFs highly enriched in each of the three major CNS lineages: astrocytes (29 TFs), neurons (13 TFs), and OPCs and oligodendrocytes (52 TFs) (Fig. 1b and Supplementary Table 1). We selected eight TFs from the OPC and oligodendrocyte lists on the basis of their known roles during oligodendrocyte development or their ability to enhance oligodendrogenesis when expressed in neural progenitors 17, 18 and cloned the coding region of each gene individually into a doxycycline-inducible lentiviral vector (Olig1, Olig2, Nkx2.2, Nkx6.2, Sox10, ST18, Gm98 (Myrf), and Myt1; collectively referred to as 8TF) (Supplementary Fig. 1a). The 8TF lentiviral pool was used to infect mouse embryonic fibroblasts (MEFs) isolated from mice constitutively expressing the reverse tetracycline-controlled transactivator (rtTA) and a modified Plp1:eGFP transgene. Plp1 is expressed specifically in both OPCs and oligodendrocytes 19, 20. The Plp1:eGFP/rtTA MEFs were carefully isolated to be free of all neural tissue, as demonstrated by the lack of neural stem cell, neuronal, astrocytic, OPC, and oligodendrocytic markers by immunostaining, qPCR, microarray, and flow cytometry (Fig. 1c, Supplementary Fig. 1b, Supplementary Fig. 2a,b).
In all experiments, we monitored both the percentage of infected cells, by immunostaining of the individual TFs, as well as the transgene induction levels, by qPCR (Supplementary Fig. 1b). Typically, 30–60% of cells were infected with an individual factor. Therefore, when multiple viruses were used, only a small proportion of cells received all TFs. In spite of this, infection and induction (+doxycycline (Dox)) of the MEFs with the 8TF pool consistently resulted in a large percentage of cells expressing the OPC- and oligodendrocyte-specific Plp1:eGFP transgene at day 21 when cells were cultured in defined OPC-promoting culture conditions containing FGF2, PDGF-AA and sonic hedgehog (SHH) supplements (32.4% +/− 9.9%; n = 19 independent biological replicates from 3 independent lots of lentivirus) (Fig. 1c, d). Uninfected (No TFs) and uninduced (−Dox) Plp1:eGFP MEFs cultured under identical conditions for the entire 21 day time course did not express the Plp1:eGFP transgene (Fig. 1c, d).
8TF-induced fibroblasts exhibit properties of OPCs
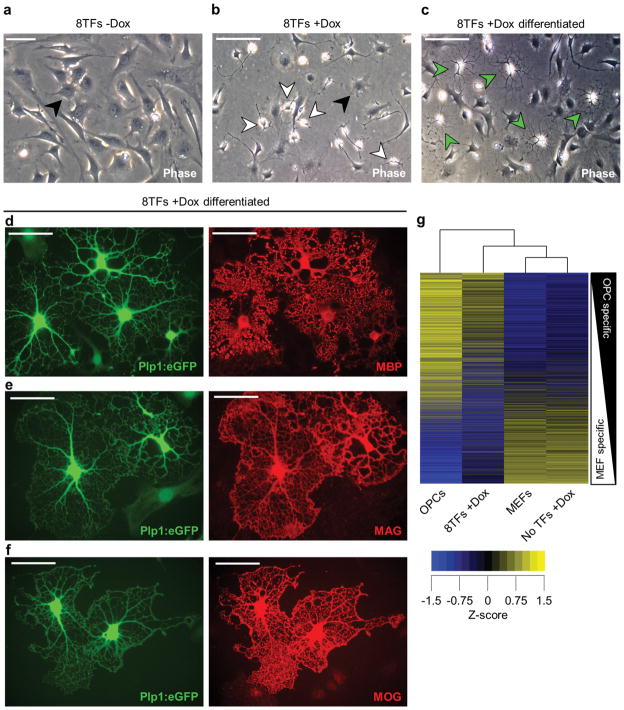
We examined the 8TF-induced MEFs for cellular or molecular features consistent with those of bona fide OPCs. During development, OPCs first emerge from the ventral ventricular zone of the spinal cord, have a bipolar morphology, proliferate in response to PDGF and FGF, express a defined set of oligodendrocyte lineage genes, and are uniquely able to generate myelinating oligodendrocytes required for CNS myelin maintenance and repair 16, 21–27. After induction of the 8TFs, a subpopulation of the cells underwent a marked morphological change within 21 days from large, flat, spindle-shaped cells (fibroblasts) to small, bipolar cells, termed ‘induced’ OPCs (iOPCs) after further characterization (Fig. 2a, b). We assessed whether the 8TF-induced cells could differentiate into oligodendrocytes in response to growth factor removal and the addition of thyroid hormone (T3), a known inducer of oligodendrocyte differentiation, in vitro 28. Notably, within 3 days some of the 8TF-induced cells differentiated into cells with a multiprocessed morphology typical of oligodendrocytes (Fig. 2c), called ‘induced’ oligodendrocytes (iOLs). All iOLs expressed myelin basic protein (MBP), an integral protein component of the myelin sheath, and other defining markers of mature oligodendrocytes, including myelin-associated glycoprotein (MAG) and myelin oligodendrocyte glycoprotein (MOG) (Fig. 2d–f).
Figure 2. Eight transcription factor induced MEFs exhibit properties of bona fide OPCs.
a, b, Phase-contrast images highlighting the dramatic morphological differences between 21 day 8TF-uninduced (a; −Dox) and induced (b; +Dox) MEFs. Non-reprogrammed cells exhibit typical fibroblast morphology (black arrowheads) while a portion of the 8TF-induced cultures show the characteristic bipolar morphology of OPCs (white arrowheads). c, Phase-contrast image of day 21 8TF-induced MEFs passaged into oligodendrocyte differentiation media for 3 days showing the generation of induced oligodendrocytes (iOLs; green arrowheads) that exhibit the distinctive multiprocessed morphology of oligodendrocytes. d–f, Representative immunofluorescent images of iOLs differentiated from 8TF-induced MEFs, containing the Plp1:eGFP reporter and expressing the specific and defining markers of mature oligodendrocytes MBP (d), MAG (e), and MOG (f). g, Clustered heat map of z-scored global gene expression values comparing pluripotent stem cell-derived bona fide OPCs, 8TF-induced MEFs (8TFs +Dox), MEFs, and uninfected MEFs plus doxycycline (No TFs +Dox). Plot is rank ordered with OPC-specific genes at the top and MEF-specific genes at the bottom and includes >13,000 genes for which there was signal above background in at least one of the samples. Scale bars, 50μm (a), 100μm (b, c), 25μm (d–f).
The efficiency of reprogramming fibroblasts to iOPCs and iOLs is difficult to calculate as the cells proliferate during the 21-day induction time course and only 1–2% receive all 8TFs from the initial lentiviral infections. However, we calculated that at day 21 approximately 1 in 900 cells in our bulk 8TF-induced MEF cultures generated multiprocessed MBP+ iOLs after culture in oligodendrocyte differentiation conditions for 3 days (Supplementary Fig. 3a). Generation of iOLs was dependent upon 8TF induction, as uninfected (No TFs +Dox) or uninduced (8TFs −Dox) cells never gave rise to iOLs under identical differentiation conditions (Supplementary Fig. 3a).
8TF-induced fibroblasts globally express OPC genes
Although the bulk 8TF-induced MEF cultures at day 21 contained only ~0.1–1% fully reprogrammed cells, as evidenced by the efficiency of forming MBP+ iOLs, global gene expression analysis of bulk 8TF-induced MEF cultures showed substantial down regulation of the MEF-specific program and large-scale activation of genes specific to the oligodendrocyte lineage (Fig. 2g). As eight of the OPC-specific genes were initially expressed from our inducible lentiviral vectors, we confirmed that the endogenous Olig2 gene was activated using specific qPCR primers (Supplementary Fig. 3b). We functionally annotated the gene expression changes caused by 8TF induction using the Genomic Regions Enrichment of Annotations Tool (GREAT) 29. GREAT analysis of genes up regulated in 8TF-induced MEF cultures showed significant association (all p-values<1×10−5)with ‘Gene Ontology (GO) biological processes’ such as ‘myelination’ and ‘gliogenesis’, with ‘MGI phenotype ontology’ terms associated with ‘oligodendrocyte morphology’ and ‘glial cell morphology’, with ‘MGI expression ontology’ terms such as ‘TS22 spinal cord; lateral wall; ventricular layer’, and with ‘Disease ontology’ terms such as ‘demyelinating disease’ and ‘schizophrenia’ (Supplementary Fig. 3c, see Supplementary Table 2 for the full list of GREAT results). Genes down regulated in 8TF-induced MEF cultures showed significant association with a large number of mesodermal processes, consistent with inactivation of the global fibroblast gene expression program (Supplementary Fig. 3d, see Supplementary Table 2 for the full list of GREAT results).
8TF-induced fibroblasts generate compact myelin
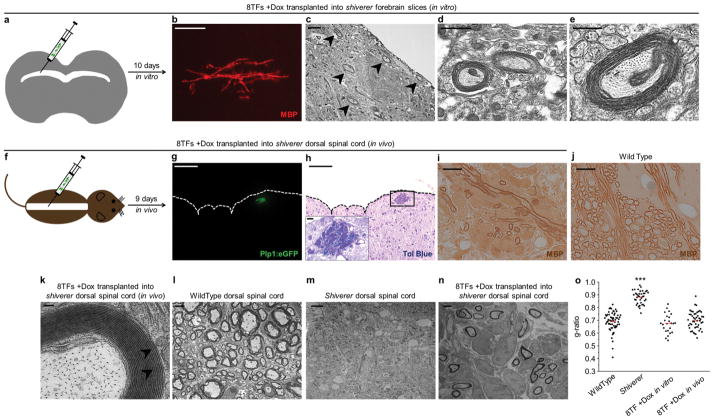
We studied the ability of 8TF-induced MEFs to myelinate axons of hypomyelinated shiverer (MBPshi/shi) mice, which completely lack MBP and compact myelin and serve as a model of congenital dysmyelinating disorders 30. We first transplanted the cells into organotypic slice cultures of early postnatal shiverer forebrain in vitro (Fig. 3a) 5, 31, 32. Transplanted 8TF-induced cells engrafted into forebrain slices, colonized major white matter tracts, including the corpus callosum, and generated characteristically aligned MBP+ myelin sheaths in 10 days (Fig. 3b). Furthermore, ultrastructural analysis by electron microscopy showed that 8TF-induced cells generated multi-layered compact myelin sheaths around hypomyelinated shiverer host axons in slice culture (Fig. 3c–e).
Figure 3. Eight transcription factor induced MEFs function to generate compact myelin.
a, Experimental scheme for testing the ability of 8TF-induced MEFs to myelinate axons in vitro. 8TF-induced MEFs were transplanted into P5 shiverer forebrain slices and cultured for 10 days. b, Representative immunofluorescent image of MBP+ myelin tracts generated from expanded (passage 3, day 32) 8TF-induced MEFs 10 days after transplantation into coronal forebrain slice cultures of shiverer mutant mice (lethally hypomyelinated due to a lack of Mbp). c–e, Electron micrograph images of multi-layered compact myelin (black arrowheads) generated by donor 8TF-induced MEFs 10 days after transplantation into coronal forebrain slice cultures of shiverer mutant mice. f, Experimental scheme for testing the ability of 8TF-induced MEFs to myelinate axons in vivo. 8TF-induced MEFs were transplanted into the dorsal spinal cord of P3-4 shiverer mutant mice. g, h, Representative immunofluorescent image (g) and matched toluidine blue (tol blue) stained section (h) showing localization of 8TF-induced cells (containing Plp1:eGFP transgene) 9 days after transplantation into the dorsal spinal cord of shiverer mutant mice (n=4 mice). The pia mater of the dorsal spinal cord is indicated by the dashed lines. (h, inset) Enlarged view of black box from (h) showing numerous myelinated axons generated from 8TF-induced cells. i–j, Representative immunostaining of MBP showing that the myelin produced in shiverer hosts 9 days after transplantation with 8TF-induced MEFs is of donor origin (i). Identically processed wild type control staining is shown in (j). k, Electron micrograph images of multi-layered compact myelin generated by 8TF-induced MEFs 9 days after transplantation into the dorsal spinal cord of shiverer mutant mice. Major dense lines are evident and indicated by black arrowheads. l–n, Electron micrographs of wild type dorsal column axons (l), untreated shiverer dorsal column axons (m), and shiverer dorsal column axons myelinated by transplanted 8TF-induced MEFs (n). o, g-ratios were calculated for wild type in vivo spinal cord (0.69±0.07), shiverer in vivo spinal cord (0.88±0.05) and 8TF-induced MEF (in vitro slices: 0.68±0.07 and in vivo spinal cord: 0.70±0.07) myelin. Differences between wild type and each of the other groups was compared using a two-tailed Student’s t-test (***p<2.2×10−16 wild type vs. shiverer. All others not significant). Scale bars, 25μm (b), 2μm (c, l–n) 1μm (d), 500nm (e), 100μm (g, h), 100nm (k), and 10μm (i, j)
We next tested whether 8TF-induced cells could function to myelinate shiverer axons in vivo without continued doxycycline induction of the transgenes (−Dox in vivo). 5×104 8TF-induced cells were transplanted into the dorsal region of the spinal cord of early-postnatal (P3-4) shiverer mice (n=4) and analyzed after 9–14 days (Fig. 3f). Transplanted cells colonized the dorsal column white matter of shiverer mice and appeared to generate compact myelin sheaths around dorsal column axons (Fig. 3g, h). As shiverer mice are devoid of MBP and therefore lack compact myelin in the CNS, definitive proof of myelination by transplanted cells requires more detailed analysis. We therefore stained sections of shiverer spinal cord transplanted with 8TF-induced cells and found that the myelin produced was MBP+ (Fig. 3i, j). This shows that the myelin produced was of donor origin and not derived from shiverer host peripheral Schwann cells that may have migrated into the CNS during transplantation. Moreover, electron microscopy analysis of the produced myelin showed clear evidence of ultrastructurally normal myelin and the presence of major dense lines (Fig. 3k). We further analyzed and quantified the myelin produced by 8TF-induced cells by calculating g-ratios—the ratio of axon diameter to total diameter of a myelinated fiber. The g-ratios of myelin produced by 8TF-induced cells was indistinguishable from that of wild-type myelin (wild type 0.69±0.07; shiverer 0.88±0.05; 8TF in vitro 0.68±0.07; 8TF in vivo 0.70±0.07) (Fig. 3l–o).
We noted that the myelin produced from 8TF-induced cells in slice cultures showed clear properties of oligodendrocyte myelin in that individual cells myelinated multiple axons, whereas the same cells transplanted in vivo seemed to myelinate only a single axon, a property consistent with Schwann cell myelination. To explore this issue, we conducted serial block-face scanning electron microscopy on the dorsal column of shiverer mice after transplantation of 8TF-induced cells. Tracing of cell processes and 3-dimensional reconstruction of the resulting images confirmed that individual transplanted cells myelinated only single axons in vivo (Supplementary Movie 1).
Prospective enrichment of expandable iOPCs
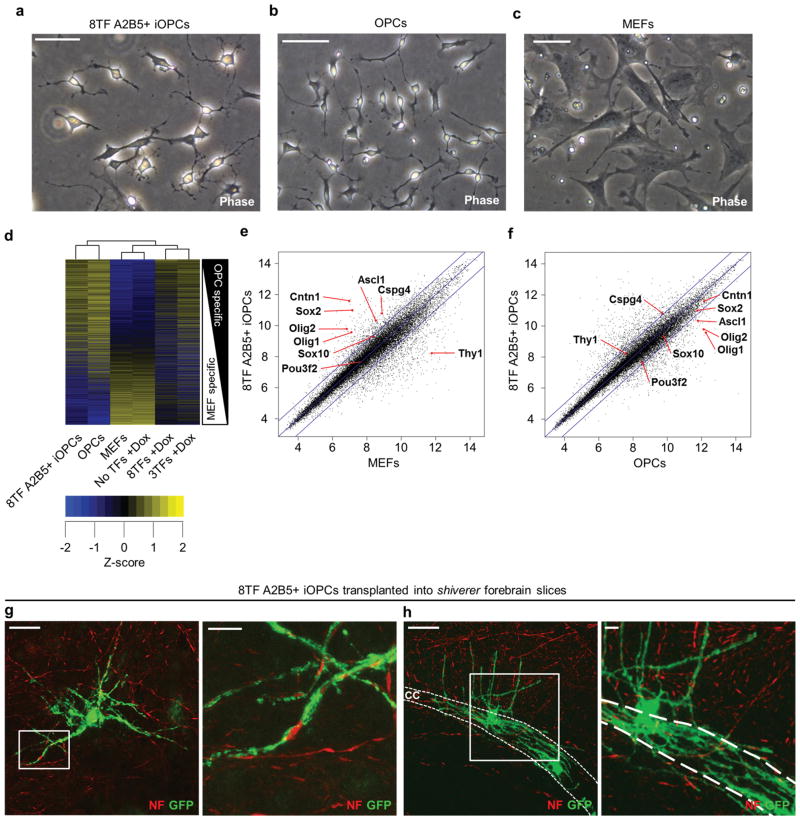
We sought to enrich the fully reprogrammed iOPCs from the bulk 8TF-induced cultures by immunosorting. As OPCs are typically defined by the expression of cell-surface markers, including PDGFRα (CD140a), NG2 (Cspg4) and A2B5, we attempted to prospectively isolate iOPCs using both the Plp1:eGFP transgene and an additional cell-surface marker. Both PDGFRα and NG2 were expressed on uninduced MEFs, but A2B5 was not. We sorted 8TF-induced MEF cultures at day 21 using A2B5 and Plp1:eGFP and found that 2.30±1.62% (8TF: n=4 biological replicates from 2 independent lots of lentivirus) of the cells expressing the Plp1:eGFP reporter were A2B5+. The A2B5+ cells were a near-homogeneous population of bipolar cells with a morphology similar to bona fide OPCs and different from MEFs (Fig. 4a–c). Hierarchical clustering and pair-wise comparisons of global gene expression data showed that 8TF-induced A2B5+ cells correlated tightly with bona fide OPCs (Fig. 4d–f). GREAT analysis of gene expression patterns in 8TF-induced A2B5+ cells showed significant associations with glial, oligodendrocyte- and myelin-related processes, phenotypes and diseases (Supplementary Table 2).
Figure 4. A2B5 immunosorting allows for the prospective enrichment of iOPCs.
a–c, Phase contrast images highlighting the bipolar morphology of 8TF-induced MEFs (sorted at day 21 for both Plp1:eGFP and A2B5; denoted as 8TF A2B5+ iOPCs) (a) compared to bona fide OPCs (b), and MEFs (c). d, Clustered heat map of z-scored global gene expression values comparing 8TF A2B5+ iOPCs to the same three control cell samples in Figure 2g [MEFs, uninfected MEFs plus doxycycline (No TFs +Dox), and pluripotent stem cell-derived bona fide OPCs]. Plot is rank ordered with OPC-specific genes at the top and MEF-specific genes at the bottom and includes >13,000 genes for which there was signal above background in at least one of the samples. e, f, Pairwise comparison of log2-adjusted global gene expression values of 8TF-induced A2B5+ iOPCs with MEFs (e) and bona fide OPCs (f). Blue lines denote a 2-fold difference in gene expression. Characteristic MEF-enriched (Thy1) and OPC-enriched (all others cited) genes are indicated with red arrows. g, h, Confocal images collected 10 days after transplantation of 8TF A2B5+ iOPCs, containing the Plp1:eGFP reporter, at passage 3 into coronal forebrain slice cultures of shiverer mutant mice showing extensive ensheathment of neuron axons by 8TF A2B5+ iOLs. Neurons were visualized with anti-neurofilament (NF) and iOLs with anti-GFP antibodies. The corpus callosum (cc) is indicated with dashed white lines. Zoomed insets are presented to the right of each image. Scale bars, 50μm (a–c, g, h), and 10μm (g, h insets).
The 8TF-induced A2B5+ cells could be stably expanded in culture for at least 5 passages. After 6 passages, 8TF-induced A2B5+ cells began to differentiate, senesce and lose Plp1:eGFP expression; they were not used further. During passages 1–6, 8TF-induced A2B5+ cells could be readily frozen and thawed without any apparent loss of potential. To analyze their myelinogenic potential, we injected 8TF-induced A2B5+ cells into organotypic slice cultures of early postnatal shiverer forebrain. 8TF-induced A2B5+ cells preferentially colonized white matter tracts and differentiated into multiprocessed iOLs displaying extensive ensheathment of NF+ neuron axons within 10 days (Fig. 4g, h).
Reprogramming fibroblasts to iOPCs with three TFs
In an effort to reduce the number of transcription factors required to generate iOPCs, we induced the expression of eight separate 7TF pools, each lacking a single TF from the original 8TF pool (8TFs–1TF; n=3 biological replicates), in the Plp1:eGFP/rtTA MEFs and used the percentage of Plp1:eGFP+ cells at day 21 as a surrogate assay for reprogramming. Only pools lacking either Sox10 or Olig2 had significant decreases in the percentage of Plp1:eGFP+ cells (p<0.05), indicating that these genes may be required for reprogramming (Fig. 5a and Supplementary Fig. 4a). To determine whether these two factors alone were sufficient for reprogramming, we induced Sox10 and Olig2 individually or in combination. Appreciable numbers of Plp1:eGFP+ cells at levels similar to those observed with 8TFs (Fig. 5a and Supplementary Fig. 4b) were not produced, and the Plp1:eGFP+ cells that were produced did not generate any MBP+ iOLs when cultured in differentiation conditions.
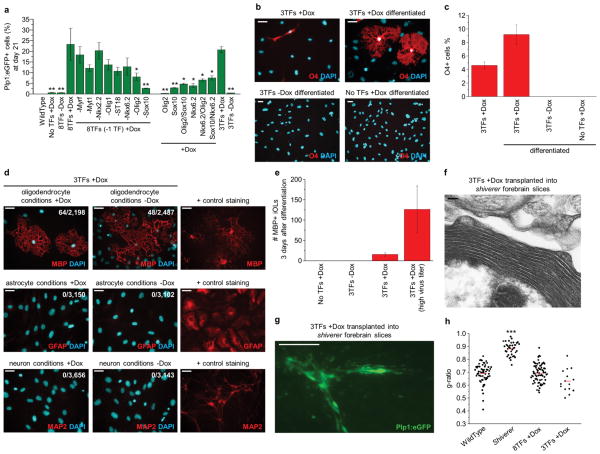
Figure 5. Sox10, Olig2, and Nkx6.2 are sufficient to reprogram fibroblasts to iOPCs.
a, Summary graph quantifying the percentage of Plp1:eGFP+ cells induced by subsets of the original 8TF pool at day 21 (n = 3 independent biological replicates from 1 lot of lentivirus; see Supplementary Figure 4 for representative flow cytometry plots). A 3TF pool of Sox10, Olig2, and Nkx6.2 showed the robust ability to induce Plp1:eGFP+ cells from MEFs (20.8% +/− 1.5%). Differences between groups were compared using a two-tailed Student’s t-test (**p<0.02 and *p<0.05 versus 8TFs +Dox). b, c, Immunofluorescent (b) and quantification (c) data showing the capacity of 3TF-induced MEFs at day 21 (3TFs +Dox) to respond to differentiation signals (3TFs +Dox differentiated) and generate multiprocessed O4+ oligodendrocytes. Note that the undifferentiated cultures (3TFs +Dox) contain a population of O4+ cells which are largely bipolar. 3TF-uninduced MEFs (3TFs −Dox differentiated) and non-infected induced MEFs (No TFs +Dox differentiated) yielded no O4+ cells. O4+ cells were manually scored from triplicate wells and data are presented as mean ± S.E.M. (n = 3 independent biological replicates from 1 lot of lentivirus). d, Representative immunofluorescent images showing the differentiation potential of 3TF-induced MEFs (3TFs +Dox) when exposed to three different lineage inducing conditions. 3TF-induced MEFs differentiated in 3 days into iOLs that expressed MBP when exposed to oligodendrocyte differentiation conditions both in the presence (+Dox) or absence (−Dox) of doxycycline. 3TF-induced MEFs never gave rise to neurons (MAP2) or astrocytes (GFAP) either in the presence (+Dox) or absence (−Dox) of doxycycline when exposed to the respective neuron or astrocyte promoting culture conditions. Positive control cell types that were stained simultaneously to ensure function of each antibody: pluripotent stem cell derived oligodendrocytes (MBP), astrocytes (GFAP), and neurons (MAP2). e, Quantitative efficiency of 3TF-induced MEFs (3TF +Dox) to differentiate into MBP+ oligodendrocytes when exposed to oligodendrocyte differentiation conditions for 3 days. Data are presented as mean ± S.E.M. of MBP+ iOLs per 4×104 cells seeded (n = 10 independent biological replicates from 3 lots of lentivirus). 3TF +Dox cells generated with a high viral titer showed a parallel increase in the efficiency of generating MBP+ iOLs (3TF +Dox high virus titer; n= 8 independent biological replicates from 2 lots of lentivirus). f, Electron micrograph image of multi-layered compact myelin generated from day 21 3TF-induced MEFs 10 days after transplantation into coronal forebrain slice cultures of shiverer mutant mice. g, Immunofluorescent images of engraftment and morphology of Plp1:eGFP+ 3TF-induced MEFs 10 days after transplantation into P5 coronal forebrain slice cultures of shiverer mutant mice. h, g-ratios were calculated from 3TF-induced MEFs transplanted into shiverer forebrain slices (0.63±0.09). Differences between groups and wild type (shown are the same control samples as in Figure 3n: wild type (0.69±0.07), shiverer (0.88±0.05) and 8TF-induced MEFs transplanted into shiverer forebrain slices and dorsal spinal cords (0.69±0.07)) were compared using a two-tailed Student’s t-test (***p<2.2×10−16 wild type vs. shiverer. All others not significant).. Scale bars, 25μm (b, d), 100μm (g), and 100nm (f).
To investigate whether a third factor in combination with Sox10 and Olig2 would be adequate to produce iOPCs from MEFs, we induced the expression of three TF pools. We found that Nkx6.2, when induced with Sox10 and Olig2 (collectively referred to as 3TF), was sufficient to produce Plp1:eGFP+ cells (20.8% +/− 1.5%; n = 3 biological replicates). After culture in oligodendrocyte differentiation conditions for three days, the 3TF-induced cells up regulated the early oligodendrocyte-specific cell-surface marker O4 (9.2% +/− 1.5; n= 3 biological replicates), whereas uninduced and uninfected cells never gave rise to O4+ cells under identical culture conditions (Fig. 5b, c). A subset of the 3TF-induced iOPCs also expressed the mature oligodendrocyte marker MBP under oligodendrocyte differentiation conditions at a rate similar to that of 8TF-induced cells (Fig. 5d–e).
To test whether 3TF-induced iOPCs could differentiate into other CNS cell fates, we cultured the cells in astrocyte- or neuron-promoting conditions in vitro (n=3 replicates). The cells never gave rise to GFAP+ astrocytes or MAP2+ neurons in these conditions, either in the presence or absence of doxycycline (Fig. 5d). As all 3TF-based experiments were matched to the same viral titer as our 8TF-based experiments, we determined whether increasing the viral titer would enhance reprogramming to iOPCs. A 3-fold increase in viral titer resulted in a 5-fold increase in the percentage of MBP+ cells (Fig. 5e and Supplementary Fig. 5a–c).
We evaluated the reprogramming time-course dynamics and the properties of the 3TF-induced cells. The reprogramming process was largely complete in 10–14 days (Supplementary Fig. 5d). Global gene expression analysis showed that the 3TF-induced cells largely expressed the OPC-specific network of genes, including endogenous Olig2, and downregulated the MEF-specific network in a similar manner to 8TF-induced cells (Fig. 4d and Supplementary Fig. 5e). When transplanted into shiverer forebrain slices, 3TF-induced cells colonized the corpus callosum and differentiated into MBP+ iOLs (Fig. 5f and Supplementary Fig. 5f). Notably, the cells generated multilayered compact myelin with proper ultrastructure and g-ratios (0.630±0.09) (Fig. 5g, h).
Finally, as all of our iOPC experiments were first performed on MEFs, we tested the 3TF pool on a different somatic cell type, mouse lung fibroblasts (MLFs) (Supplementary Fig. 6a). 3TF-induced MLFs showed properties consistent with OPCs, were capable of extensive expansion in vitro, and consistently generated MBP+ iOLs at each passage upon growth factor withdrawal and exposure to T3 (Supplementary Fig. 6b, c). Thus, Sox10, Olig2 and Nkx6.2 are sufficient to reprogram two separate somatic cell types in 14–21 days to iOPCs capable of generating MBP+ myelinogenic oligodendrocytes.
DISCUSSION
Here we show that functional iOPCs can be produced by delivering defined sets of transcription factors to mouse fibroblasts. Specifically, expression of three transcription factors, Sox10, Olig2 and Nkx6.2, is sufficient to convert two different sources of mouse fibroblasts to iOPCs that exhibit morphological and molecular features consistent with that of bona fide OPCs. In contrast to the ability of OPCs to form both oligodendrocytes and astrocytes, iOPCs appear lineage restricted to oligodendrocytes as they do not generate neurons or astrocytes when exposed to differentiation conditions in vitro. It remains possible that iOPCs could access neuron or astrocyte fates when exposed to other conditions. The myelination capacity of iOPCs was tested using organotypic slice cultures which provide a complex three-dimensional tissue representative of the CNS. When transplanted to post-natal forebrain slices, iOPCs showed the ability to myelinate multiple host axons and generate compact myelin. In contrast, iOPCs transplanted in vivo into the spinal cord appeared to only myelinate single axons, a property exhibited by Schwann cells in the peripheral nervous system. Although this result was initially surprising, bona fide CNS OPCs are known to have the capacity to produce Schwann-like cells in vivo and myelinate only single axons 33. Collectively, our data show that iOPCs are capable of functioning in vitro and in vivo to generate compact myelin and that different environments may direct them to myelinate single or multiple axons.
As with most current reprogramming strategies, the efficiency of generating iOPCs is low. We demonstrated that increasing the viral titer of the reprogramming factors resulted in an increase in the efficiency of generating functional iOPCs. This suggests that further refinement of the stoichiometry and expression levels of the reprogramming factors will lead to increased efficiency of functional iOPC production. In spite of the low reprogramming efficiencies, we showed that iOPCs could be prospectively isolated from the bulk reprogramming cultures using the monoclonal A2B5 antibody. Sorted A2B5+ iOPCs could be expanded for up to five passages while maintaining the ability to differentiate into oligodendrocytes and myelinate host axons after transplantation. Immunosorting of expandable iOPCs should facilitate the use of iOPCs in molecular and transplantation-based studies that require large numbers of cells.
The potential of cell-based therapies for myelin disorders relies on the ability to generate autologous myelinogenic cells for transplantation. The most promising cell source for such therapies may be OPCs. Mature oligodendrocytes largely fail to remyelinate host axons after transplantation. Although neural stem cells and induced neural stem cells can generate oligodendrocytes, the efficiency of this process is quite low and the cells have a propensity to form neurons and astrocytes. In contrast, iOPCs appear restricted to the oligodendrocyte lineage. We have shown that iOPCs integrate into the CNS and myelinate axons of congenitally dysmyelinated mice in vivo after transplantation. However, for iOPCs to have clinical relevance, future studies will have to extend this reprogramming strategy to human somatic cells and demonstrate extensive CNS myelination and long-term functional benefit to transplant recipients.
METHODS
Isolation of Plp1:GFP/rtTA fibroblasts
Both MEFs and MLFs were isolated at embryonic day 13.5 (E13.5) from embryos generated through timed natural matings between Plp1:eGFP mice19 and rtTA mice (B6.Cg-Gt(ROSA)26Sortm1(rtTA*M2)Jae/J; Jackson Laboratory). For MEFs, the head, spinal cord, and all internal organs were carefully removed to eliminate contamination with any neural precursors. The remainder of the tissue was cut into small pieces and dissociated using 0.125% trypsin-EDTA (Invitrogen). Cells were expanded for one passage and cryopreserved for future use. MLFs were isolated by dissociating pooled lung lobes using 0.125% trypsin-EDTA, expanded for two passages, and cryopreserved for future use. Both MEFs and MLFs were derived in DMEM supplemented with 10% fetal bovine serum (FBS), 2 mM glutamax, 1x nonessential amino acids, and 0.1 mM 2-mercaptoethanol.
Selection of 8TFs
The following publically available GEO datasets were utilized for Figure 1b: GSM241931, GSM241936, GSM241929, GSM241937, GSM241934, and GSM24193316. Putative transcription factors were filtered by selecting genes with both a ‘GO cellular component term’ “nucleus” and a ‘GO molecular function term’ “DNA binding.” Transcription factors that were enriched >3-fold in a particular lineage were selected and cross-referenced with our own microarray data of stem cell-derived OPCs and oligodendrocytes (GEO dataset: GSE31562). Data were then z-scored and plotted in R using the heatmap.2 function of the gplots package.
Production of lentivirus
The mouse coding regions of Myrf, Myt1, Nkx2.2, Olig1, ST18, Nkx6.2, Olig2 and Sox10 were cloned into the pLVX-Tight-Puro vector (Clontech). VSV-G pseudotyped lentivirus was generated according to the manufacturer’s protocol using the Lenti-X HT Packaging Mix and Lenti-Phos or Cal-Phos Mammalian Transfection Kit (all from Clontech). 293T cells (Clontech) cultured on rat tail collagen I coated plastic-ware (BD Biosciences) were seeded between 6.0–8.5×104 cells/cm2 and transfected 16 hours later. Individual supernatants containing virus were harvested and filtered with 0.45μm PVDF membrane (Millipore) 48–72 hours later.
iOPC generation
MEFs or MLFs were seeded at 1.3×104 cells/cm2, allowed to attach overnight, and infected with fresh lentivirus supplemented with polybrene (8μg/ml) four times over a two day period. For 8TF infection, an equal volume of fresh lentiviral supernatant was mixed from each of the 8TFs prior to infection. To facilitate the comparison of data between 3TF and 8TF experiments, 3TF infections were performed by mixing equal volumes of fresh lentiviral supernatant from each of the 3TFs and diluted with 5 equivalent volumes of MEF medium. For ‘high viral titer’ experiments in Figure 5, an equal volume of lentiviral supernatant from the 3TFs was mixed and added directly to cells without further dilution. The end of the virus infection period was termed ‘day 0’. Cells were either uninduced or induced with 2μg/ml doxycycline (Clontech) for 3 days in MEF culture conditions. Cells were then lifted with TrypLE Select (Invitrogen) and either frozen or seeded at 2.0×104 cells/cm2 on Nunclon-Δ plates pre-coated with 0.1mg/ml poly-L-ornithine (Sigma) and 10μg/ml laminin (Sigma; L2020) and cultured in OPC medium which consisted of DMEM/F12 (Invitrogen, 11320) supplemented with 1x N-2 Plus or 1x N-2 Max (R&D Systems), 1x B-27 without vitamin A (Invitrogen), 2mM Glutamax (Invitrogen), 200ng/ml SHH (R&D Systems), 20ng/ml FGF2 (R&D Systems), and 20ng/ml PDGF-AA (R&D Systems). Media was changed every 2 days. Putative iOPCs (Plp1:eGFP+ cells) were typically sorted between day 14–21 using a FACSAria (BD Biosciences) and further expanded in OPC medium with FGF2, PDGF-AA, and SHH in the presence or absence of doxycycline. iOPCs were passaged every 3–5 days with TrypLE Select and could be readily frozen and thawed in DMEM supplemented with 10% FBS and 10% DMSO (Sigma).
iOPC differentiation to iOLs
For differentiation of 8TF-induced or 3TF-induced cells into iOLs, cells were seeded at 2.1×104 cells/cm2 (MEFs) or 1.1×104 cells/cm2 (MLFs) and differentiated with DMEM/F12 supplemented with 1x N-2, 1x B-27 without vitamin A, 2mM Glutamax, 40ng/ml T3 (Sigma), 200ng/ml SHH, 100ng/ml Noggin, 10μM cAMP, 100ng/ml IGF, and 10ng/ml NT3 either in the presence or absence of doxycycline on poly-L-ornithine and laminin treated cultureware. Cultures were fixed after 3 days and stained for MBP. iOLs were quantified by manually counting the number of multiprocessed, MBP+ cells relative to the number of cells seeded. For 3TF lineage differentiation experiments seen in Figure 5d, cells were cultured in either neuronal differentiation medium consisting of Neurobasal (Invitrogen, 21103-049) supplemented with DMEM/F12 (Invitrogen, 11320-033), 1x N-2, 1x B-27 with vitamin A (Invitrogen, 17504004), 2mM Glutamax, and 20ng/ml BDNF (R&D Systems) for 7 days or astrocyte differentiation medium consisting of DMEM/F12 supplemented with 1x N-2, 1x B-27 without vitamin A, 2mM Glutamax, 103units/ml LIF, and 50ng/ml BMP4(R&D Systems) for 3 days.
Immunocytochemistry
Cells were prepared for immunostaining by fixation in 4% paraformaldehyde (Electron Microscopy Sciences) for 15 minutes and subsequent permeabilization for 10 minutes with 0.2% Triton-X in PBS. Cells were then blocked for non-specific binding with filtered 10% normal goat (Abcam) or 10% donkey serum (Abcam) in PBS for 1–2 hours at room temperature. Primary antibodies were diluted in blocking solution and incubated with the samples overnight at 4°C. Samples were rinsed with PBS and incubated with the appropriate fluorescently labeled Alexa-Fluor secondary antibodies (Invitrogen 1:500) for 1 hour at room temperature. For nuclear staining samples were incubated with 1μg/ml DAPI (Sigma) for 5 minutes. Primary antibodies used were: Sox10 (R&D Systems, AF2864; 2μg/ml), Olig2 (Millipore, AB9610; 1:1000), Nkx6.2 (Abcam, ab58708; 1μg/ml), Sox1 (R&D Systems, AF3369; 1μg/ml), Sox2 (R&D Systems, MAB2018; 1μg/ml), Pax6 (Covance, PRB-278P; 0.67μg/ml), Oct3/4 (Santa Cruz, SC-5279; 0.4μg/ml), Nkx2.2 (DSHB, 74.5A5; 4.4μg/ml), GFAP (DAKO, Z0334; 0.58μg/ml), MAP2 (Millipore, AB5622; 2μg/ml), MBP (Covance, SMI-99P; 2μg/ml), MBP (Abcam, ab7349; 1:100), MAG (Millipore, MAB1567;10μg/ml), and MOG (Millipore, MAB5680; 5μg/ml). For O4 staining, cells were incubated live with 10% donkey serum and O4 antibody (Miller Lab; 1:10) for 20 minutes. Cells were then gently rinsed 3 times with cell medium and fixed with 4% PFA in PBS. Staining was then completed as detailed above.
FACS and Flow Cytometry
For Plp1:eGFP expression analysis, cells were analysed on a FACSAria or LSR flow cytometer (BD Biosciences) and plots were generated with WinList 3D 7.0 software. Gates for Plp1:eGFP were set with negative control cells (wild type MEFs without a GFP transgene) at less than 0.1% positive cells. For A2B5 immunosorting, cells were collected from culture and blocked in 10% normal donkey serum for 30 min. Cells were then stained with A2B5 primary antibody (R&D Systems, MAB 1416; 5μg/ml) for 30 min followed by incubation with Alexa Fluor–labeled secondary antibody (Invitrogen; 4 μg/ml) for 20 min. Isotype control antibody was used as a staining control and to set gates (Mouse IgM, Invitrogen; 5μg/ml) with Alexa Fluor secondary antibody (Invitrogen; 4μg/ml). Some experiments were carried out with APC conjugated A2B5 (Miltenyi Biotec, 130-093-582; 1:11) and conjugated isotype control (Miltenyi Biotec, 130-093-176; 1:11). PDGFRα and NG2 flow cytometry was carried out as detailed previously 5.
RNA Isolation and qPCR
Total RNA was isolated as for gene expression analysis. 400ng of RNA was reverse transcribed with SuperScript III Reverse Transcriptase (Invitrogen) and qPCR was performed using 8ng of cDNA with TaqMan Gene Expression Master Mix and TaqMan probes: Sox10 (Mm01300162_m1), Nkx6.2 (Mm00807812_g1), Olig1 (Mm00497537_s1), Nkx2.2 (Mm01275962_m1), Myt1 (Mm00456190_m1), ST18 (Mm01236999_m1), Gm98 (Myrf) (Mm01194959_m1) and Olig2 (AJVI3GC, custom) on the 7300 Real-Time PCR System (Applied Biosystems). Endogenous Olig2 expression was detected using the Olig2 (Mm01210556_m1) TaqMan probe in which one primer sits outside of the CDS and therefore does not detect expression from the Olig2 inducible lentiviral vector. All expression data were normalized to Gapdh (Mm99999915_g1) and samples in which no expression was detected were given an arbitrary Ct value of 40. All analyses were performed with quadruplicate technical replicates for each of a minimum of three independent biological replicates. Relative expression levels were determined by calculating 2−ΔΔCt with corresponding S.E.M.
Global gene expression
Cells were lysed in 1ml TRIzol (Invitrogen) and stored at −80°C until ready for use. Chloroform separation was enhanced with Phase-Lock Gel Tubes (5 Prime). The aqueous phase was collected and the RNA isolation completed with the RNeasy Plus Kit (Qiagen) according to the manufacturer’s protocol. Sample labeling and hybridization to Affymetrix Mouse Gene 1.0 ST arrays (containing probes covering 28,853 mouse genes) were carried out in the Gene Expression and Genotyping Core Facility of the Case Comprehensive Cancer Center. Data were extracted, and RMA normalized using Affymetrix Expression Console software (ver. 1.1). All data are available on the NCBI Gene Expression Omnibus website through GEO Series accession GSEXXXXX. For heat maps in Figures 2g and 4d, data were z-scored and plotted in R using the heatmap.2 function of the gplots package. Plots were rank ordered by the ratio of MEF expression to OPC expression where MEF-specific genes were at the top and OPC-specific genes were at the bottom. Genes were removed from the analysis if signal was not detected above background in any of the six samples analyzed. The plots therefore included values for 13,919 genes. To analyze the global changes during reprogramming, genes increased or decreased 2-fold between 8TF-induced cells or 8TF A2B5+ iOPCs and MEFs were calculated. Files in BED format containing the transcription start sites ±100 base pairs of all the genes within each class (genes up regulated in 8TF-induced cells, genes down regulated in 8TF-induced cells, genes up regulated in A2B5+ iOPCs, and genes down regulated in A2B5+ iOPCs), were assembled and compared to the background set using the Genomic Regions Enrichment of Annotations Tool (GREAT) (http://great.stanford.edu). All significantly associated annotation terms and associated p-values are provided in Supplementary Table 2.
Organotypic slice culture myelination analysis
Experiments were carried out as described previously 5. The forebrains of early postnatal day 5 homozygous shiverer (C3Fe.SWV-Mbpshi/Mbpshi; Jackson Laboratory) were dissected and 300μm slices produced on a Leica Vibratome. Slices were cultured in a DMEM/BME base supplemented with 15% Horse serum, modified N-2 supplement, and PDGF-AA for 3 days 34. 5×104–2×105 cells (8TF-induced, 3TF-induced, A2B5+ 8TF-induced, or uninduced controls) were manually transplanted with a pulled glass pipette into each slice and grown for an additional 10 days in culture. For staining, slices were then fixed in 4% paraformaldehyde, treated with ice-cold 5% acetic acid/95% methanol, and assayed for MBP expression (Covance, SMI99 and/or SMI94; 4μg/ml) with either enzymatic secondary substrate (Jackson Labs, Biotin-anti-mouse IgG; Vector Labs, ABC; Sigma, DAB) or Alexa-Fluor secondary antibody (Invitrogen; 4μg/ml). For confocal imaging of iOPC ensheathment of axons, d10 A2B5+ iOPC-transplanted forebrain slices were fixed in 4% paraformaldehyde, treated with 0.2% Triton-X and labeled with neurofilament cocktail (Covance, SMI-311 and SMI-312; 1:1000) and GFP (Aves Labs, 1020; 10μg/ml) antibodies overnight. Alexa-Fluor secondary antibodies (Invitrogen; 4μg/ml) were used for detection. Slices were mounted using Vectashield (Vector Labs) and images were acquired with a Zeiss LSM 510 META laser scanning confocal microscope (Carl Zeiss MicroImaging, Jenna, Germany) using a 40x C-Apochromat, NA 1.2, water immersion objective. All images presented are maximum intensity projections of a Z series consisting of 1 μm optical slices collected every 0.5 μm. Although Triton-X was necessary for the NF and GFP antibodies, it is known to disrupt membrane proteins (i.e. myelin) and therefore its use was minimized as much as possible when staining forebrain slices. For electron microscopy, slices were fixed in EM fixative (4% paraformaldehyde and 2% glutaraldehyde in 0.1M cacodylate buffer, pH 7.4). Slices were prepared for EM by incubation in 1% osmium tetroxide and staining en bloc in uranyl acetate. Samples were then dehydrated and embedded in Poly/Bed 812 epoxy. Thick (1μm) transverse sections were cut and stained with toluidine blue. Thin (90nm) sections were cut, collected on 300μm nickel grids, stained with uranyl acetate and lead citrate and carbon-coated for imaging on a JEOL JEM-1200-EX electron microscope.
Spinal cord transplantation
Cells were lifted with TrypLE Select, thoroughly rinsed 3 times with cell media, and concentrated at ~1.0×105 cells/μl. Postnatal day 3–4 homozygous shiverer mice were anesthetized with isofluorane and positioned lying prone on a thin roll of gauze such that the animal’s thoracic spine rested higher than the rest of the anterior-posterior body axis. Using the rib cage as a landmark for the lower thoracic spinal level 13, a 30-gauge needle attached to a 10μl Hamilton syringe was inserted between two lamina just below the bone at a depth of approximately 2mm accounting for skin thickness. As slowly as the unassisted hand could manage, 5×104 cells (in a volume not exceeding 1μl) were injected into the dorsal white matter. The needle was left in place for approximately one minute to avoid liquid reflux upon needle withdrawal. After 9–14 days mice were deeply anesthetized with rodent cocktail (ketamine, xylazine and acepromazine), perfused transcardially with 0.9% saline at room temperature and then perfused with ice-cold 4% paraformaldyhyde. Spinal columns were dissected out and post-fixed in ice-cold 4% paraformaldyhyde for 2 hours. Spinal cords were dissected out of the spinal columns and vibratome sectioned into individual 300μm sections that were visualized for the presence of Plp:eGFP+ cells in the dorsal column white matter. Sections containing Plp:eGFP+ cells were imaged and immediately transferred to EM fixative (4% paraformaldehyde and 2% glutaraldehyde in 0.1M cacodylate buffer, pH 7.4) and incubated overnight. Sections were prepared for EM by incubation in 1% osmium tetroxide and staining en bloc in uranyl acetate and then dehydrated and embedded in Poly/Bed 812 epoxy. Thick (1μm) transverse sections were cut and stained with toluidine blue. Thin (90nm) sections were cut, collected on 300μm nickel grids, and carbon-coated for electron microscopy on a JEOL JEM-1200-EX electron microscope. G-ratios were calculated using image analysis software (Adobe Photoshop) by dividing axon diameter with total diameter of the myelinated fiber.
Immunostaining of MBP in epoxy-embedded sections
0.5uM and 1uM thick sections of Poly/Bed 812 embedded wild type and cell-injected shiverer spinal cord were etched using a modification of a process described previously 35. Sections were treated with Target Antigen Retrieval solution (DAKO, pH 9.0) and rinsed with 1x D-PBS (Cellgro). They were then incubated in a 1:200 dilution of anti-Myelin Basic Protein antibody (SMI-94, Covance) for 4 nights at 4°C, rinsed several times with 1x D-PBS (Cellgro), and incubated at ambient temperature in a 1:200 dilution of Horseradish Peroxidase Conjugated Donkey anti-Mouse IgG antibody (Jackson ImmunoResearch) for 1.5 hours. The staining was visualized using Diaminobenzidine (SigmaFast kit). The sample was imaged using transmitted light and oil immersion lens at 100x on a Leica DM 5500B light microscope with a Leica DFC 500 camera.
Serial block face scanning electron microscopy and 3D reconstruction
Sections of cell-injected spinal cord were prepared for 3D EM using a modified version of previously published methods 36, 37. Samples were post-fixed in 2% paraformaldehyde and 2.5% glutaraldehyde with 2mM calcium chloride in 0.1M cacodylate buffer, pH 7.4. Samples were then incubated in 2% osmium tetroxide, subsequently stained en bloc in uranyl acetate and then Walton’s lead aspartate. They were dehydrated and embedded in Durcupan ACM resin (EMS). Thick (0.5 μm) transverse sections were cut and stained with a 1% toluidine blue/1% sodium borate solution, and the region of interest was determined and trimmed. Serial blockface scanning EM was accomplished using a Gatan 3View in-chamber ultramicrotome mounted on a Carl Zeiss Sigma VP scanning electron microscope. Images were obtained in 100 nm increments for a ROI bounded by approximately 50 microns on each side, resulting in a 3D stack of 570 images [actual ROI dimensions: x=64 microns, y=76 microns, z=57 microns]. Images were analyzed using Fiji-Image J (NIH) to align the stack, followed by Amira 5.4.2 for segmentation and 3D reconstruction.
Supplementary Material
Acknowledgments
This research was supported by US National Institutes of Health grants MH087877 (P.J.T. and R.H.M.), NS30800 (R.H.M.), and EY019880 (T.M.), NIH pre-doctoral training grants F31NS083354 (A.M.L.) and T32GM008056 (R.T.K.), Case Western Reserve University School of Medicine (P.J.T.), the New York Stem Cell Foundation (P.J.T.), the Mt. Sinai Health Care Foundation (P.J.T.), and the Cytometry & Imaging Microscopy and the Gene Expression and Genotyping core facilities of the Case Comprehensive Cancer Center (P30CA43703). P.J.T. is a Robertson Investigator of the New York Stem Cell Foundation. We are grateful to Leslie Cooperman, Jonathon Wanta, Midori Hitomi, Janet Krasno, Molly Karl, Sharyl Fyffe-Maricich, Shannon Wu, Maryanne Pendergast, Olivia Corradin, Thomas LaFramboise, and Peter Scacheri for technical assistance, Irene Tsung for artwork, as well as Nanthawan Avishai and Grahame Kidd for 3D SEM imaging and data processing.
Footnotes
AUTHOR CONTRIBUTIONS
F.J.N., A.M.L. and P.J.T. designed the reprogramming strategy and generated all iOPCs; F.J.N., A.M.L., R.T.K. and P.J.T. performed in vitro differentiation experiments; A.V.C., A.Z., F.J.N., A.M.L. and T.M. performed in vivo myelination experiments; A.Z., F.J.N., A.M.L., R.H.M. and P.J.T. performed slice culture myelination experiments; K.W., A.Z. and R.H.M. produced EM images; F.J.N., A.M.L., D.C.F., and P.J.T. generated and analyzed gene expression data; F.J.N., A.M.L., R.H.M., and P.J.T. analyzed all of the data and wrote the paper. All authors edited and approved the final manuscript.
COMPETING FINANCIAL INTERESTS
P.J.T., R.H.M., and F.J.N. have a pending patent application for this technology.
References
- 1.Goldman SA, Nedergaard M, Windrem MS. Glial progenitor cell-based treatment and modeling of neurological disease. Science. 2012;338:491–495. doi: 10.1126/science.1218071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Franklin RJ, Ffrench-Constant C. Remyelination in the CNS: from biology to therapy. Nat Rev Neurosci. 2008;9:839–855. doi: 10.1038/nrn2480. [DOI] [PubMed] [Google Scholar]
- 3.Windrem MS, et al. Neonatal chimerization with human glial progenitor cells can both remyelinate and rescue the otherwise lethally hypomyelinated shiverer mouse. Cell Stem Cell. 2008;2:553–565. doi: 10.1016/j.stem.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sim F, et al. CD140a identifies a population of highly myelinogenic, migration-competent, and efficiently engrafting human oligodendrocyte progenitor cells. Nature Biotechnol. 2011 doi: 10.1038/nbt.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Najm FJ, et al. Rapid and robust generation of functional oligodendrocyte progenitor cells from epiblast stem cells. Nat Methods. 2011;8:957–962. doi: 10.1038/nmeth.1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vierbuchen T, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pang ZP, et al. Induction of human neuronal cells by defined transcription factors. Nature. 2011 doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caiazzo M, et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011 doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- 9.Pfisterer U, et al. Direct conversion of human fibroblasts to dopaminergic neurons. Proc Natl Acad Sci U S A. 2011;108:10343–10348. doi: 10.1073/pnas.1105135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoo AS, et al. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011 doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J, et al. Direct reprogramming of mouse fibroblasts to neural progenitors. Proc Natl Acad Sci U S A. 2011;108:7838–7843. doi: 10.1073/pnas.1103113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Son EY, et al. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell. 2011;9:205–218. doi: 10.1016/j.stem.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han Dong W, et al. Direct Reprogramming of Fibroblasts into Neural Stem Cells by Defined Factors. Cell Stem Cell. 2012 doi: 10.1016/j.stem.2012.02.021. [DOI] [PubMed] [Google Scholar]
- 14.Thier M, et al. Direct Conversion of Fibroblasts into Stably Expandable Neural Stem Cells. Cell Stem Cell. 2012 doi: 10.1016/j.stem.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 15.Lujan E, Chanda S, Ahlenius H, Sudhof TC, Wernig M. Direct conversion of mouse fibroblasts to self-renewing, tripotent neural precursor cells. Proc Natl Acad Sci U S A. 2012;109:2527–2532. doi: 10.1073/pnas.1121003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cahoy JD, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Z, et al. Induction of oligodendrocyte differentiation by Olig2 and Sox10: evidence for reciprocal interactions and dosage-dependent mechanisms. Dev Biol. 2007;302:683–693. doi: 10.1016/j.ydbio.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 18.Zhang X, et al. Induction of oligodendrocytes from adult human olfactory epithelial-derived progenitors by transcription factors. Stem Cells. 2005;23:442–453. doi: 10.1634/stemcells.2004-0274. [DOI] [PubMed] [Google Scholar]
- 19.Mallon BS, Shick HE, Kidd GJ, Macklin WB. Proteolipid promoter activity distinguishes two populations of NG2-positive cells throughout neonatal cortical development. J Neurosci. 2002;22:876–885. doi: 10.1523/JNEUROSCI.22-03-00876.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44:23–28. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
- 21.Bogler O, Wren D, Barnett SC, Land H, Noble M. Cooperation between two growth factors promotes extended self-renewal and inhibits differentiation of oligodendrocyte-type-2 astrocyte (O-2A) progenitor cells. Proc Natl Acad Sci U S A. 1990;87:6368–6372. doi: 10.1073/pnas.87.16.6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noble M, Murray K, Stroobant P, Waterfield MD, Riddle P. Platelet-derived growth factor promotes division and motility and inhibits premature differentiation of the oligodendrocyte/type-2 astrocyte progenitor cell. Nature. 1988;333:560–562. doi: 10.1038/333560a0. [DOI] [PubMed] [Google Scholar]
- 23.Richardson WD, Pringle N, Mosley MJ, Westermark B, Dubois-Dalcq M. A role for platelet-derived growth factor in normal gliogenesis in the central nervous system. Cell. 1988;53:309–319. doi: 10.1016/0092-8674(88)90392-3. [DOI] [PubMed] [Google Scholar]
- 24.Noll E, Miller RH. Oligodendrocyte precursors originate at the ventral ventricular zone dorsal to the ventral midline region in the embryonic rat spinal cord. Development. 1993;118:563–573. doi: 10.1242/dev.118.2.563. [DOI] [PubMed] [Google Scholar]
- 25.Rowitch DH, Kriegstein AR. Developmental genetics of vertebrate glial-cell specification. Nature. 2010;468:214–222. doi: 10.1038/nature09611. [DOI] [PubMed] [Google Scholar]
- 26.Watkins TA, Emery B, Mulinyawe S, Barres BA. Distinct stages of myelination regulated by gamma-secretase and astrocytes in a rapidly myelinating CNS coculture system. Neuron. 2008;60:555–569. doi: 10.1016/j.neuron.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crang AJ, Gilson J, Blakemore WF. The demonstration by transplantation of the very restricted remyelinating potential of post-mitotic oligodendrocytes. J Neurocytol. 1998;27:541–553. doi: 10.1023/a:1006960032023. [DOI] [PubMed] [Google Scholar]
- 28.Barres BA, Lazar MA, Raff MC. A novel role for thyroid hormone, glucocorticoids and retinoic acid in timing oligodendrocyte development. Development. 1994;120:1097–1108. doi: 10.1242/dev.120.5.1097. [DOI] [PubMed] [Google Scholar]
- 29.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chernoff GF. Shiverer: an autosomal recessive mutant mouse with myelin deficiency. J Hered. 1981;72:128. doi: 10.1093/oxfordjournals.jhered.a109442. [DOI] [PubMed] [Google Scholar]
- 31.Gahwiler BH, Capogna M, Debanne D, McKinney RA, Thompson SM. Organotypic slice cultures: a technique has come of age. Trends Neurosci. 1997;20:471–477. doi: 10.1016/s0166-2236(97)01122-3. [DOI] [PubMed] [Google Scholar]
- 32.Bai L, et al. Hepatocyte growth factor mediates mesenchymal stem cell-induced recovery in multiple sclerosis models. Nat Neurosci. 2012;15:862–870. doi: 10.1038/nn.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zawadzka M, et al. CNS-resident glial progenitor/stem cells produce Schwann cells as well as oligodendrocytes during repair of CNS demyelination. Cell Stem Cell. 2010;6:578–590. doi: 10.1016/j.stem.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mi S, et al. Promotion of central nervous system remyelination by induced differentiation of oligodendrocyte precursor cells. Ann Neurol. 2009;65:304–315. doi: 10.1002/ana.21581. [DOI] [PubMed] [Google Scholar]
- 35.Yano S, et al. An antigen retrieval method using an alkaline solution allows immunoelectron microscopic identification of secretory granules in conventional epoxy-embedded tissue sections. J Histochem Cytochem. 2003;51:199–204. doi: 10.1177/002215540305100208. [DOI] [PubMed] [Google Scholar]
- 36.Denk W, Horstmann H. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol. 2004;2:e329. doi: 10.1371/journal.pbio.0020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mikula S, Binding J, Denk W. Staining and embedding the whole mouse brain for electron microscopy. Nat Methods. 2012 doi: 10.1038/nmeth.2213. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.