Abstract
The effective and sustained delivery of DNA locally would increase the applicability of gene therapy in tissue regeneration and therapeutic angiogenesis. One promising approach is to use porous hydrogel scaffolds to encapsulate and deliver nucleotides in the form of nanoparticles to the affected sites. We have designed and characterized micro-porous (µ-pore) hyaluronic acid hydrogels which allow for effective cell seeding in vitro post scaffold fabrication and allow for cell spreading and proliferation without requiring high levels of degradation. These factors, coupled with high loading efficiency of DNA polyplexes using a previously developed caged nanoparticle encapsulation (CnE) technique, then allowed for long-term sustained transfection and transgene expression of incorporated mMSCs. In this study, we examined the effect of pore size on gene transfer efficiency and the kinetics of transgene expression. For all investigated pore sizes (30, 60, and 100 µm), encapsulated DNA polyplexes were released steadily starting by day 4 for up to 10 days. Likewise, transgene expression was sustained over this period, although significant differences between different pore sizes were not observed. Cell viability was also shown to remain high over time, even in the presence of high concentrations of DNA polyplexes. The knowledge acquired through this in vitro model can be utilized to design and better predict scaffold-mediated gene delivery for local gene therapy in an in vivo model where host cells infiltrate the scaffold over time.
Keywords: Porous hydrogel, Controlled release, Non-viral gene delivery, Enzymatically degradable, Cell mediated release
1. Introduction
Vascularization of tissue engineering constructs remains the primary reason for construct failure in vivo [1]. Without the rapid infiltration of blood vessels, diffusion alone is insufficient to sustain migrating endogenous or exogenously implanted cells far from the construct surface. Researchers have recently been focusing on macroscopic biomaterial design to help promote branching from existing blood vessels into the biomaterial. Micro-scale interconnected pores produced through salt-leaching [2, 3], gas foaming [4–6], lyophilization [7–10], and sphere templating [11–14] have shown to be effective in allowing for cellular infiltration and, subsequent, enhanced scaffold vascularization.
In addition to the structural characteristics of the scaffold, the effective local delivery of angiogenic factors, including VEGF and PDGF, are necessary to promote blood vessel formation. For tissue regeneration, localized gene delivery can promote the expression of tissue inductive factors to guide tissue formation. Local gene delivery via hydrogel scaffolds has been studied for nearly a decade primarily through the encapsulation of naked DNA during hydrogel formation [5, 15–19]. Although naked DNA achieves gene expression and guided regeneration in vivo [5, 15], limitations with low gene transfer efficiency and rapid diffusion of the DNA from the hydrogel scaffold motivated the use of DNA nanoparticles instead of naked DNA. DNA condensed either with cationic peptides, lipids, or polymers has previously been introduced into fibrin hydrogels [13, 20–22], enzymatically degradable poly(ethylene glycol) (PEG) hydrogels [23, 24] and PEG-hyaluronic acid hydrogels [25]. Poly(ethylene imine) (PEI) is a widely utilized cationic polymer for non-viral gene delivery; it is able to condense DNA through electrostatic interactions between the positively charged amines on the PEI and the negatively charged phosphates on the DNA, forming nanoparticles (polyplexes) in the range of 50 to 200 nm [26]. PEI has been successfully used in vivo to deliver DNA or siRNA to the brain [27, 28], lungs [29–32], abdomen [33], and tumors [34–36].
Hydrogel properties, such as the type of natural or synthetic polymer used, can likewise be an important factor in the promotion of vascularization. While a synthetic polymer, such as PEG, can be biochemically inert, natural polymers, such as hyaluronic acid (HA), possess intrinsic qualities which can play a role in signaling to surrounding cells. HA, an anionic, non-sulfated glycosaminoglycan and major component of the ECM, is widely distributed in connective, epithelial and neural tissue [37]. HA has recently gained popularity as a biomaterial for tissue engineering and regeneration due to its high biocompatibility and low immunogenicity [38–41]. Moreover, degraded fragments of HA or HA oligomers are known to promote angiogenesis and upregulate MMP expression [42–44]. As a result, several studies have demonstrated that HA-based hydrogels are good candidates for culturing stem cells [45–48]. HA specifically interacts with cell surface receptors, such as CD44, RHAMM (receptor for HA mediated motility) and ICAM-1 (intercellular adhesion molecule 1), and contributes to tissue hydrodynamics, cell proliferation and migration [49, 50]. Semi-synthetic hyaluronic acid (HA) hydrogels which are also degradable by matrix metalloproteinases (MMPs) have previously been developed for culturing mouse mesenchymal stem cells in three-dimensions [51, 52]. MMPs are normally expressed during tissue remodeling and are up-regulated during wound healing, microenvironment remodeling, and in diseased states and can, therefore, serve as triggers for bioactive signal delivery. While peptides and growth factors can be easily incorporated within these hydrogels, rapid degradation by proteases generally limits their effectiveness in long-term cell culture. MMP-degradable hyaluronic acid hydrogels have previously been used to encapsulate DNA/poly(PEI) polyplexes as a means of non-viral gene delivery to stem cells [53]. We found that as the matrix degraded through cell-secreted proteases, the cells were transfected with the polyplexes encountered during their migration. However, direct encapsulation of the polyplexes resulted in aggregation when the concentration exceeded 0.2 µg/µl. Aggregation of polyplexes can result in increased toxicity and inconsistent transfection of encapsulated or infiltrating cells. To overcome this concentration limitation we previously developed a caged nanoparticle encapsulation process (CnE) to incorporate DNA polyplexes inside a variety of hydrogel scaffolds without particle aggregation [54, 55]. This approach utilizes neutral saccharides (sucrose) and polysaccharides (agarose) to protect the polyplexes from inactivation and aggregation during lyophilization and hydrogel formation, respectively.
In this paper, we investigate gene transfer to mouse mesenchymal stem cells (mMSCs) seeded within porous hyaluronic acid hydrogel scaffolds. Porosity in hydrogels has previously been shown to promote cell migration in vitro [9, 56, 57], as well as hydrogel integration and vascularization in vivo [2, 12]. In addition, these porous constructs may serve as non-viral gene carriers by either coating the porous hydrogels or embedding bioactive signals directly within the gel, allowing for cellular uptake [4, 5, 13]. Porous fibrin hydrogels loaded with DNA polyplexes were able to deliver genes to attached fibroblasts [13], however, polyplexes aggregated and elicited cellular toxicity. In the work presented here, the CnE technique was employed to load higher amounts of polyplexes without inducing aggregation. To optimize cell seeding into porous hydrogels in vitro, we propose a two-phase hydrogel system that contains polyplexes in a stiff (3.5%) µ-pore HA-MMP gel and mMSCs in a softer (2.5%) n-pore HA-MMP hydrogel within the µ-sized pores. As the inner gel is degraded quickly in the presence of cell-secreted proteases, it will allow for cell spreading and proliferation, while the polyplexes in the stiff µ-pore backbone will have a more gradual and sustained release upon hydrogel degradation. For in vitro studies, transfection was quantified using a Gaussia luciferase reporter plasmid. Studies were focused on how pore size influenced the efficiency of gene transfer and the kinetics of transgene expression. These factors are important for designing successful scaffolds to mediate gene delivery in vivo. The goal is to transfect either: (i) encapsulated stem cells in a DNA- or siRNA-loaded hydrogel scaffold in vitro or (ii) cells infiltrating an acellular scaffold containing DNA or siRNA in vivo.
2. Materials and Methods
2.1 Materials
Peptides Ac-GCRDGPQGIWGQDRCG-NH2 (HS-MMP-SH) and Ac-GCGYGRGDSPG-NH2 (RGD) were purchased from Genscript (Piscataway, NJ). Sodium hyaluronan (HA) was a gift from Genzyme Corporation (60 kDa, Cambridge, MA). Linear poly(ethylene imine) (PEI, 25kDa) was purchased from Polysciences (Warrington, PA). Vectors expressing mammalian secreted Gaussia Luciferase (pGluc) were obtained from New England Biolabs (Ipswich, MA) and expanded using a Giga Prep kit from Qiagen following the manufacturer’s protocol. All other chemicals were purchased from Fisher Scientific (Pittsburgh, PA) unless otherwise noted.
2.2 Hyaluronic acid modification
Sodium hyaluronan was modified to contain acrylate functionalities as previously described. Briefly, hyaluronic acid (2.0 g, 5.28 mmole, 60 kDa) was reacted with 18.0 g (105.5 mmole) adipic dihydrazide (ADH) at pH 4.75 in the presence of 4.0 g (20 mmole) 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride (EDC) overnight and purified through dialysis (8000 MWCO) in DI water for 2 days. The purified intermediate (HA-ADH) was lyophilized and stored at − 20°C until used. Approximately 56% of the carboxyl groups were modified with ADH, which was determined using 1H-NMR (D2O) by taking the ratio of peaks at δ = 1.6 and 2.3 corresponding to the 8 hydrogens of the methylene groups on the ADH to the singlet peak of the acetyl methyl protons in HA (δ = 1.88). HA-ADH (1.9 g) was reacted with N-Acryloxysuccinimide (NHS-Ac) (1.33 g, 4.4 mmole) in HEPES buffer (10mM HEPES, 150mM NaCl, 10mM EDTA, pH 7.2) overnight and purified through dialysis in DI water for 2 days before lyophilization. The degree of acrylation was determined to be ~10% using 1H-NMR (D2O) by taking the ratio of the multiplet peak at δ = 6.2 corresponding to the cis and trans acrylate hydrogens to the singlet peak of the acetyl methyl protons in HA (δ = 1.88).
2.3 Polyplex lyophilization
For CnE, plasmid DNA (100–250 µg) and L-PEI (91.3–228.3 µg) were mixed in 3.5 mL water in the presence of 35 mg (0.10 mmole) of sucrose (Ultra pure, MP Biomedicals, Santa Ana, CA) and incubated at room temperature for 15 min. Low-melting point agarose (1.0 mg, UltraPureTM Agarose, Tm = 34.5–37.5°C, Invitrogen, Grand Islands, NY) in 1.5 mL water was added before lyophilization. Each aliquot was intended for a 100 µL hydrogel. For smaller hydrogel volumes, the sucrose concentration remained unchanged (relative to DNA levels) while the agarose concentration was proportionally decreased.
2.4 Design template using PMMA microspheres
Microsphere templates for porous hydrogels were prepared as previously described [58]. Briefly, PMMA microspheres (27–33, 53–63, and 90–106 µm, Cospheric, Santa Barbara, CA) were bought dry. Approximately 12–14 mg of microspheres were then added into glass-bottom silicon wells (6 mm × 1 mm, D × H) and covered with a glass slide. The microspheres were then packed by slight tapping for 1–2 min and examined for even packing through phase microscopy. The glass-bottom silicon wells were then placed into an oven and the microspheres were sintered for 22 hr at 150°C.
2.5 Porous (and nano-porous) HA hydrogel formation
Hydrogels were formed by Michael-type addition of acrylate-functionalized HA (HA-Ac) with bis-cysteine containing MMP peptide crosslinkers at pH 7.6–7.8. Prior to reaction, a hydrogel precursor solution was made by mixing HA-Ac with a lyophilized aliquot of cell adhesion peptide, RGD, for 30 min at 37°C. After incubation, HA-RGD was mixed with the remaining HA-Ac and PBS pH 7.4 for a final gel concentration of 3.5 w/v% HA and 500 µM RGD. Finally lyophilized aliquots of the crosslinker (0.91 mg HS-MMP-SH) were diluted in 18.2 µL of PBS buffer pH 7.4 immediately before addition to a mixture of lyophilized (CnE) or fresh (direct encapsulation) DNA/PEI polyplexes and the hydrogel precursor solution. For direct encapsulation, DNA and PEI were mixed to form nanoparticles through vortexing for 15 s and incubating for 15 min at RT before being mixed with the HA solution and crosslinkers to form hydrogels. For porous hydrogels, 20 µl of gel solution was then added directly on top of a PMMA microsphere template, covered with a glass slide, and perfused into the template by centrifugation at 1500 rpm for 6 min at 4°C. The slide was then incubated at 37°C for 30–45 min to induce polymerization. Once complete, the gels were removed from the silicon wells and placed directly into 100% acetone for 48 hr to dissolve the PMMA microsphere template. The acetone solution was replaced 2–3× during this incubation. The gels were then serially hydrated into sterile PBS and left in PBS until ready for use. For nano-porous hydrogels, the gel solution was sandwiched between two Sigmacoted slides using 1mm thick plastic spacers and incubated at 37°C for 30–45 min to induce polymerization. Once complete, the gels were placed directly into sterile PBS and left in PBS until ready for use.
2.6 Subcutaneous implant model
All in vivo studies were conducted in compliance with the NIH Guide for Care and Use of Laboratory Animals and UCLA ARC standards. 6 to 8-week old male Balb/c mice each 20–30 grams were used to study cellular infiltration and blood vessel formation in HA hydrogels since this strain and size has been previously used for wound healing and angiogenesis assays [59, 60]. Nanoporous or single-phase porous hydrogels were made exactly as described above with sucrose and agarose (but without DNA polyplexes) and cut to 6 mm in diameter using a biopsy punch, for final overall dimensions of 6 mm × 1 mm, D × H. All porous hydrogels were made using 100 µm beads. In fabricating the hydrogels, the starting reagents were sterilized through filtering with a 0.22µm filter. After scaffold fabrication, the hydrogels were washed with sterile PBS and kept in PBS with 1% P/S. Immediately prior to surgery, mice were anesthetized with 4–5% isoflurane through a nose cone inhaler. After anesthesia induction, the isoflurane concentration was lowered to 1.5–2.5% for the remainder of the surgery. The back of the mouse was subsequently shaved and washed with Betadine and 70% ethanol. Two incisions appropriate to the size of the implant were made in the skin aside the midline of the animal using scissors. Two subcutaneous pockets were subsequently created by blunt dissection using rounded-end scissors. Within the created pockets, the implants were inserted. After insertion of the hydrogels, each incision was subsequently closed with a single wound clip. All animals were observed daily for signs of inflammation and pain and also administered carprofen injections for the first 48 hrs post survival surgery. After 1, 2, and 3 weeks, mice were sacrificed with CO2 overdose. Two 1 cm2 pieces of tissue were collected from each mouse containing the implant and the surrounding tissue and skin, fixed in 4% PFA overnight at 4°C, dehydrated in 70% EtOH, and finally paraffin embedded. A total of 12 mice were used in this study, with 6 mice per hydrogel condition. Two animals were sacrificed per condition at each time point.
2.7 Immunohistochemisty and Immunofluorescence
Paraffin embedded sections were deparaffinized by incubation in multiple xylene washes followed by serial hydration from 100% ethanol into 100% water. Cell membranes were permeabilized with a 15 min incubation at 37°C in .1 mg/ml proteinase K solution. Sections were then washed with PBS and incubated in blocking buffer (1% goat serum (Jackson Immuno Research Labs, West Grove, PA) + .05% Tween-20 in PBS) for 1 hr at RT before being incubated in primary antibody solution (1:100 dilution in blocking buffer of rat anti-mouse CD31 (BD Pharmingen, San Diego, CA)) overnight at 4°C. Sections were again washed with PBS and incubated in blocking buffer for 10 min at RT before being incubated for 2 hrs at RT in secondary antibody solution (1:200 dilution in blocking buffer of goat anti-rat Alexa 568 (Invitrogen, Grand Islands, NY) which also contained α-smooth muscle actin-FITC (1:500 dilution, Sigma-Aldrich, St. Louis, MO) and DAPI nuclear stain (1:500 dilution, Invitrogen). Sections were then washed twice in PBS, mounted and imaged using an inverted Zeiss fluorescence microscope.
2.8 Gel preparation for SEM imaging
Each porous hydrogel was serially dehydrated in 25% increments of water/EtOH for 10 min at a time. Hydrogels were left in 100% EtOH overnight after which they were placed in a dry holder, air dried and, finally, placed under vacuum until time for imaging. The gels were then coated with a thin layer of gold using a gold sputterer and imaged using a JEOL JSM-6700F FE-SEM in the UCLA MCTP core facility. For SEM images in Figure 1, a Nova 230 Nano SEM in low vacuum mode was used. Hydrogels were slightly dried and imaged directly without further dehydration or metal coating.
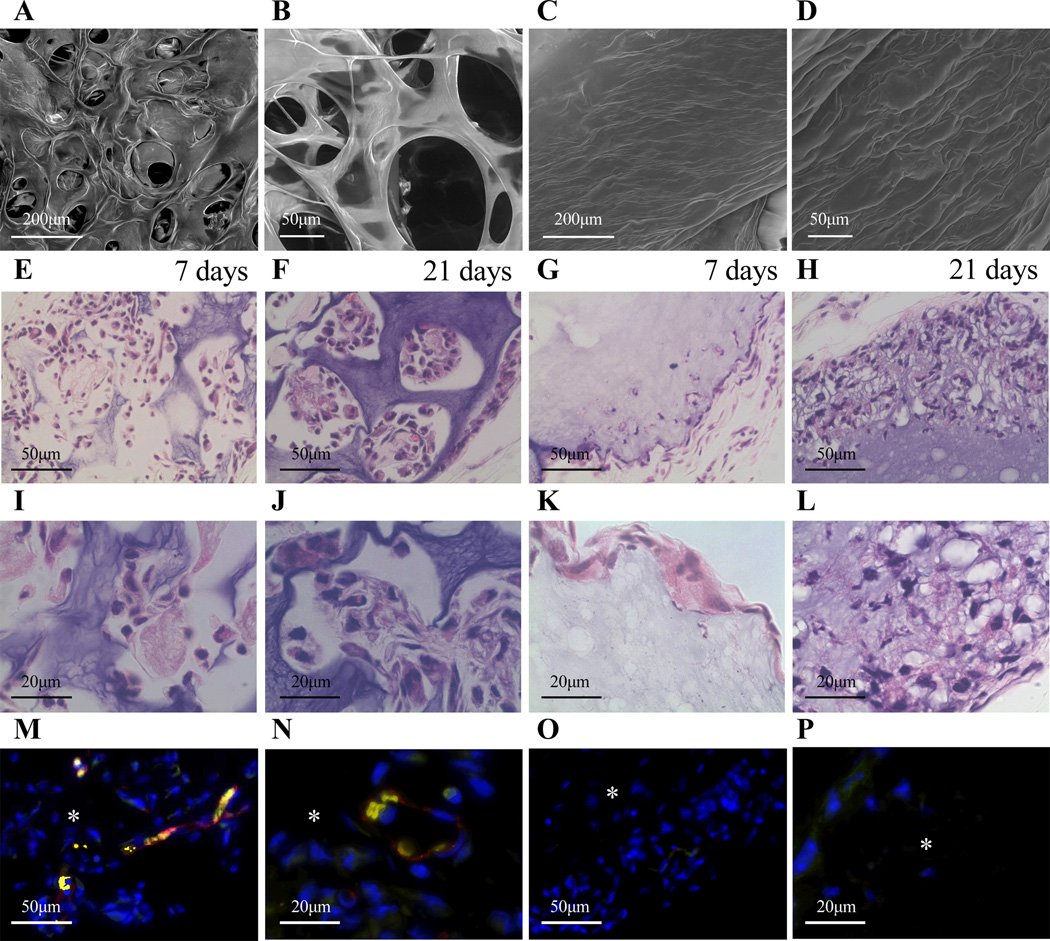
Figure 1.
SEM was used to characterize gel structure of µ-pore hydrogels (A, B) made using the bead template and n-pore hydrogels (C, D) made directly without the template. A, C = 200×, B, D = 750× magnification images. Sections from a mouse subcutaneous implant model show ~100 µm diameter pores (E, F, I, J) allow for enhanced cellular infiltration from the host into the HA hydrogels when compared to n-pore counterparts (G, H, K, L). Staining for endothelial markers at 21 days indicated significant positive staining for µ-pore hydrogel implants (M, N) and not for the n-pore implants (O, P). Red = PECAM positive staining = endothelial cells, yellow appear to be erythrocytes, blue = cell nuclei, asterisks indicate where the gel is present. E-H, M, O = 40×, I-L, N, P = 100× magnification images.
2.9 Preparation and characterization of two-phase gel
Porous hydrogels were prepared exactly as described above, with final overall dimensions of ~8 mm × 1 mm, D × H, after swelling in PBS. After the initial porous gel was formed it was stained with FITC using a 10× dilution in PBS of a 1mg/ml stock in DMSO. The gel was incubated at RT for 30 min in the dark, followed by several washes in PBS until the wash solution was no longer changing color. Prior to formation of the second phase, some HA had been modified with Alexa-350 using NHS-Alexa350 (Invitrogen, Grand Islands, NY). NHS-Alexa350 was added at a 1:5 ratio of HA:NHS. The solution was allowed to react for 2 hrs with mixing in the dark, after which it was dialyzed and lyophilized. To form the second phase, 15 µl per 20 µl porous gel of 2.5% HA-Alexa350 was mixed with MMP crosslinker and immediately added on top of the 3.5% HA porous gel. Due to the fluid nature of the 2.5% HA gel solution prior to gelation, it was able to infiltrate evenly into the pores and form a soft gel after incubation at 37°C for at least 30 min. The two phases were then visualized using fluorescence microscopy.
2.10 Polyplex Visualization
To visualize the polyplex distribution, hydrated gels were stained with ethidium bromide (12 µM) for 1-hr before imaging with a fluorescent (Observer Z1 Zeiss) microscope. To better visualize the distribution throughout the hydrogel, multiple z-stacks 1.9–2.3 µm thick were taken for each image, deconvoluted to minimize background, and presented as maximum intensity projections.
2.11 DNA Loading
In order to determine the extent of release of the encapsulated polyplexes, plasmid DNA was radiolabeled with 3H-dCTP (100 µCi, MP Biomedicals, Santa Ana, CA) using a Nick translation kit (Roche, Indianapolis, IN) as per the manufacturer’s protocol. Briefly, an equimolar mixture of dATP, dGTP, dTTP, and 3H-dCTP was prepared and added to the DNA (5 µg) solution. Once the enzyme solution was added to the mixture, the final solution (200 µl) was gently mixed by pipetting and incubated for 2 hr at 15°C. The reaction was stopped by addition of 10 µl 0.2 M EDTA (pH=8.0) and heating to 65°C for 10 min. The DNA was purified using the mini Prep kit from Qiagen (Valencia, CA) following the manufacturer’s instructions. The final DNA concentration was 0.2 µg/µl. In order to determine the extent of release of the encapsulated polyplexes, gels were formed using the protocols indicated above with 1% radiolabeled DNA. To determine the overall loading, all of the acetone and hydrating washes were collected and analyzed for DNA content. Once completely hydrated, the gels were incubated with 0.25% trypsin/EDTA to completely degrade the gel in order to determine the total amount of DNA still encapsulated. DNA concentrations were measured using a scintillation counter at the UCLA Chemistry core facility. The readout was analyzed using a standard curve.
2.12 Cell culture
Mouse bone marrow derived mesenchymal stem cells (D1, CRL12424) were purchased from ATCC (Manassas, VA) and cultured in DMEM (Invitrogen, Grand Islands, NY) supplemented with 10% bovine growth serum (BGS, Hyclone, Logan, UT) and 1% penicillin/streptomycin (Invitrogen) at 37°C and 5% CO2. The cells were passaged using trypsin following standard cell culture protocols every 2–3 days.
2.13 Preparation of two-phase gels with mMSCs
Hydrogels were prepared exactly as described above with a final HA concentration of 2.5 w/v%, 500 µM RGD, and 5000 cells/µl (final volume). For those two-phase gels that did not contain cells, PBS pH 7.4 was used in place of the cell solution. Immediately after mixing the precursor solution with crosslinker, 15 µl of the gel solution was pipetted directly on top of a previously made 20 µl porous hydrogel (~8 mm × 1 mm, D × H, after swelling in PBS) in a low-attachment 96-well plate for a total of 75,000 cells per gel. Due to the fluid nature of the 2.5% gel solution, it was able to evenly distribute within the pores of porous hydrogel before completely gelling. The gel was allowed to sit at RT for 2–3 min after which it was incubated at 37°C for 30 min to induce polymerization. 100 µl complete DMEM was then added to each well and replaced daily. All two-phase gels containing cells had µ-pore gels with 1 µg/µl DNA.
2.14 Cell viability and spreading
Cell viability was studied with the LIVE/DEAD® viability/cytotoxicity kit (Molecular Probes, Eugene, OR). Briefly, 1 µL of ethidium homodimer-1 and 0.25 µL of calcein AM from the kit were diluted with 500 µL DMEM to make the staining solution. Each gel (2–3 gels per condition per time point) was stained with 150 µL of staining solution for 30 min at 37°C in the dark before imaging. To better analyze cell spreading, gels (2–3 gels per condition per time point) were fixed for 30 min at RT using 4% PFA, rinsed with PBS, treated with .1% triton-X for 10 min and stained for 90 min in the dark with DAPI for cell nuclei (500× dilution from 5 mg/ml stock, Invitrogen, Grand Islands, NY) and rhodamin-phalloidin (5 µl per 200 µl final stain solution, Invitrogen) in 1% bovine serum albumin solution. The samples were then washed with .05% tween-20. For both cell viability and cell spreading, an inverted Observer Z1 Zeiss fluorescence microscope was used to visualize samples. To better visualize the distribution throughout the hydrogel, multiple z-stacks 1.9–2.3 µm thick were taken for each image, deconvoluted to minimize background, and presented as maximum intensity projections.
2.15 Cell proliferation
MTT assay (CellTiter 96R AQueous One Solution Cell Proliferation Assay, Promega, Madison, WI) was used to quantify the cell proliferation rate. 20 µl MTT reagent with 100 µl DMEM was added to each gel in a 96-well plate and incubated at 37°C for 2 hr. The cells were lysed after 2 hr with addition of 10% sodium dodecyl sulfate. The solutions were transferred to a new plate and absorbance was measured at 490 nm using a standard plate reader. Three gels for each condition were analyzed at each time point.
2.16 DNA release in the presence of mMSCs
In order to determine the extent of release of the encapsulated polyplexes, two-phase gels were formed as indicated above with 1% radiolabeled DNA. Gels (n=3) were placed in 150 µl of release solution. At the indicated time points, 150 µl of the solution was removed and an additional 150 µl of fresh release medium was added. Following the final release medium collection, the gels were incubated with 0.25% trypsin/EDTA to result in complete release of the DNA from the gel upon degradation. DNA concentrations were measured using a scintillation counter at the UCLA Chemistry core facility. The readout was analyzed using a standard curve and plotted as percent of total DNA encapsulated.
2.17 Gel degradation in the presence of mMSCs
Gel degradation was determined using a slight modification of a previously established carbazole assay to quantify the uronic acid content in solution [61]. The same two-phase samples used for the DNA release studies were also used to determine degradation. 5 µl of release solution was diluted 20-fold using saturated benzoic acid and added to 600 µl of chilled 25 mM sodium tetraborate in concentrated sulfuric acid. The sample was vortexed, incubated at 100°C for 10 min, and brought back down to 4°C. Next, 20 µl of .125% carbazole in absolute ethanol was added to each sample, vortexed, incubated at 100°C for 15 min, and finally brought back down to 4°C. Sample absorbance was determined at 530 nm with a pure benzoic acid sample used as a blank. Concentration was then obtained in reference to a D-glucuronolactone standard curve and finally plotted as % uronic acid released based on the total amount of HA in both phases. Alternatively, pore size measurements were made manually from phase images of gels with cells growing within the pores over time. At each time point 3 gels were analyzed per condition, with 9 measurements per gel (3 measurements at 3 different z-planes). AxioVision software was used to make measurements.
2.18 Gene transfer from two-phase hydrogels
pGluc/PEI nanoparticle loaded hydrogels with mMSCs were made as described above. Each day the media was collected and frozen immediately at −20°C and fresh media was added to each gel. To quantify secreted Gaussia luciferase levels in the media, the samples were thawed on ice and assayed using a BioLux™ Gaussia Luciferase Assay Kit (New England Biolabs, Ipswich, MA) as per the manufacturer’s protocol. Briefly, 20 µl sample was mixed with 50 µl 1× substrate solution, pipetted for 2–3 sec, and read for luminescence with a 5 sec integration. Background was determined with media from gels that did not contain any DNA and values were expressed as relative light units (RLU).
2.19 Gluc escape from hydrogels
Mouse MSCs were transfected for 24 hrs in tissue culture (TC) plastic and then detached using trypsin/EDTA and either re-plated onto TC plastic or encapsulated within HA-MMP degradable hydrogels at a density of 5000 cells/µl gel. The total number of cells was kept constant. After 48 hrs, conditioned media was collected from all samples and Gaussia luciferase expression was determined for both systems. Total Gaussia luciferase expression in HA hydrogels was normalized to that on TC plastic.
2.20 Statistical Analysis
All statistical analysis was performed using Instat (GraphPad, San Diego, CA). Experiments were statistically analyzed using the Tukey test to compare all pairs of columns using a 95% confidence interval. Outliers in the gene transfer studies were detected using the Grubbs’ outlier test. All errors bars represent the standard error of the mean (SEM).
3. Results and Discussion
Acrylates were conjugated onto the HA backbone through a two-step process, as previously described [53]. Briefly, HA was first modified with ADH by EDC coupling and the resulting hydrazide group was then modified with NHS-acrylate to obtain acrylamide functionalities. Analysis by NMR showed showed that ~10% of the carboxylic acids were modified with acrylates, resulting in approximately 16 acrylates per HA chain. RGD adhesion peptides were incorporated through Michael-type addition of the cysteine side chain in the peptide to the acrylate groups on the HA backbone, followed by addition of an MMP-degradable peptide crosslinker to form the final hydrogels. Porous hydrogels are formed around a PMMA microsphere template which is finally dissolved away using acetone, leaving behind a porous hydrogel structure. Scanning electron microscopy was used to examine the structural differences between gels made with and without the bead template (Fig 1A–D). Hydrogels made with lyophilized agarose and sucrose with the bead template had large, interconnected pores that were uniformly distributed (µ-pore), while those made directly without the template had no visible micro-sized pores (n-pore).
As a preliminary study to determine the potential for porous hydrogels to enhance cellular infiltration in vivo, both µ-pore and n-pore hydrogels were implanted subcutaneously into the back of Balb/c mice. Implants were excised each week for three weeks, paraffin-embedded and sectioned along the height of the gel to include the attached muscle and skin. Hematoxylin and eosin (H&E) stained sections showed micro-scale interconnected pores allowed for enhanced cellular infiltration from the host into the biomaterial increasingly with time (Fig 1E, F, I, J). By one week, cells were able to infiltrate into almost all visible µ-pores without noticeable gel degradation. Conversely, n-pore gels had minimal cellular infiltration along the periphery of the hydrogel even after three weeks (Fig 1G, H, K, L). At sites where cells were present the hydrogel appeared to be degrading and large pores became visible. Staining for PECAM positive endothelial cells showed thin vessel-like structures within µ-pores at three weeks (Fig 1M, N), while no endothelial cells could be observed in any of the n-pore implants at the same time point (Fig 1O, P). Thus, the presence of emerging vasculature could be attributed to the pre-existing interconnected porous structure. These results highlighted the potential for the use of porous hydrogels in vivo. We anticipate that with the addition of pro-angiogenic growth factors through the delivery of non-viral vectors encoding for the factors, blood vessel formation within porous hydrogels could further enhance vessel number, maturity, and rate of formation.
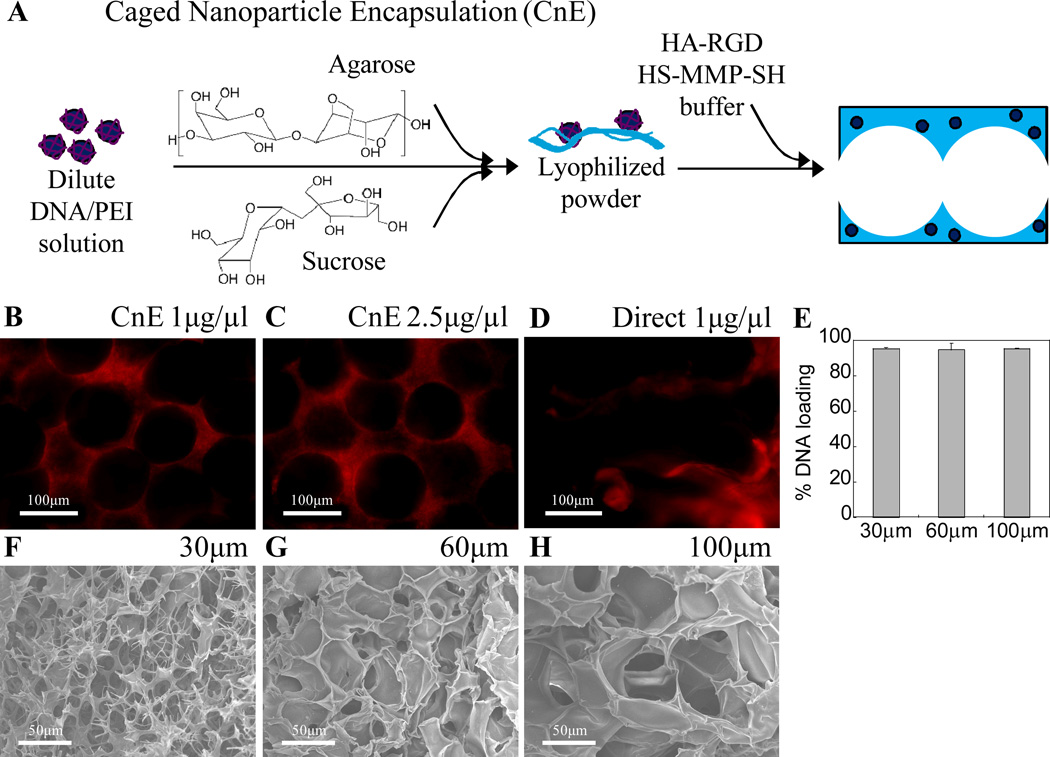
To optimize the design of porous hydrogels with encapsulated non-viral vectors, in vitro studies were initially conducted. Lyophilized polyplexes were incorporated into the hydrogel during hydrogel gelation (Fig 2A) to achieve a final DNA concentration of either 1 or 2.5 µg DNA/µL hydrogel (20 or 50 µg DNA total per 20 µl gel, respectively). The distribution of the polyplexes inside the hydrogel scaffold was determined by staining with ethidium bromide post hydrogel formation. Polyplexes made using the CnE technique were observed mostly as nonaggregated particles and uniformly distributed throughout the gel (Fig 2B, C). In contrast, those made even at 1µg/µl without agarose and sucrose were highly aggregated (Fig 2D). These results confirmed that the CnE technique for incorporating high amounts of DNA into HA hydrogels was essential for nonaggregated distribution. Using radiolabeled DNA, we were also able to verify that there was minimal loss of incorporated DNA during gel formation, acetone washes, and initial swelling in PBS (Fig 2E).
Figure 2.
(A) Schematic of CnE process to incorporate nonaggregated DNA/PEI polyplexes into porous hydrogels. (B–D) Using the CnE technique 1–2.5 µg/µl DNA could be incorporated without significant aggregation, while direct incorporation of 1 µg/µl resulted in highly aggregated particles; DNA was visualized via ethidium bromide staining. (E) 3H-dCTP labeled DNA was used to verify over 95% of loaded DNA remained in the gel after porous gel processing. Porous hydrogels were synthesized by using different sized PMMA template beads. (F–H) SEM was used to characterize gel structure as a function of bead size. F–H = 500× magnification images.
In addition to the incorporation of non-viral DNA nanoparticles, pore size was also assessed as a potential factor in determining gene transfer kinetics. PMMA beads of various sizes (~30, 60, and 100 µm) were used to form the template. To visualize the porous structure and specifically the differences in porous hydrogels made using the various sized microspheres, hydrogels were dehydrated in ethanol and imaged using scanning electron microscopy (Fig. 2F–H). Due to the tight range in bead size, the pore size distribution in each hydrogel was qualitatively uniform. However, exact pore size could not be measured from these SEM images since ethanol dehydration resulted in a significant reduction in overall gel size to ~40% of the original size. Lastly, as a result of sintering the microspheres, interconnectivity was present between almost every neighboring pore.
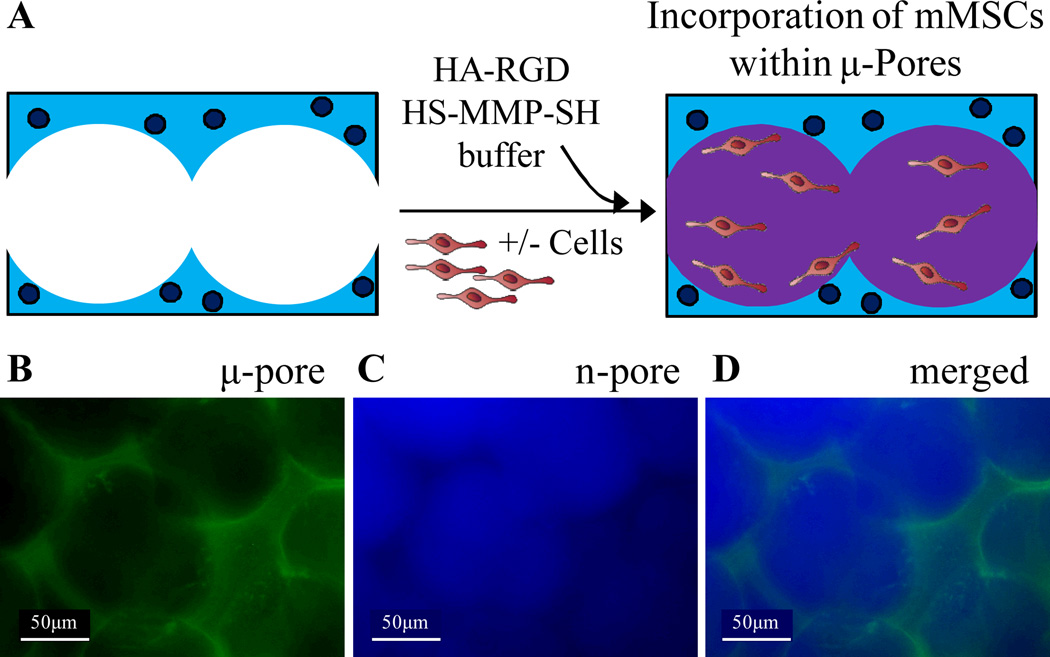
To easily seed cells within pores in vitro, we utilized a two-phase hydrogel technique. Once µ-pore 3.5% HA hydrogels were formed, mouse MSCs were mixed into a thin 2.5% n-pore HA-MMP hydrogel precursor solution and pipetted directly on top of the µ-pore hydrogel (Fig 3A). Due to the fluid nature of the 2.5% hydrogel precursor solution initially at room temperature, the gel solution was able to flow throughout the pores of the porous hydrogel and distribute the cells within the pores. In this situation, the porous hydrogel was used as a template upon which to seed cells in a softer gel phase. To ensure even distribution of the second phase within the pores of the porous hydrogel, two-phase HA hydrogels were prepared in the absence of cells. After the initial porous gel was formed it was stained with FITC. HA modified with Alexa350 was then used to form the inner phase gel. The two phases were then visualized using fluorescence microscopy. Both phases could clearly be distinguished and were present in most parts of the gel system (Fig 3B–D).
Figure 3.
(A) To effectively seed cells and allow for rapid cell spreading without significant gel degradation, cells were seeded within the pores of a 3.5% µ-pore HA gel directly with a soft 2.5% HA gel. To visualize each phase separately the µ-pore phase was stained with FITC (B) and the inner, n-pore phase was stained with Alexa-350 (C). (D) Merged fluorescence image of a two-phase hydrogel made using 100 µm beads.
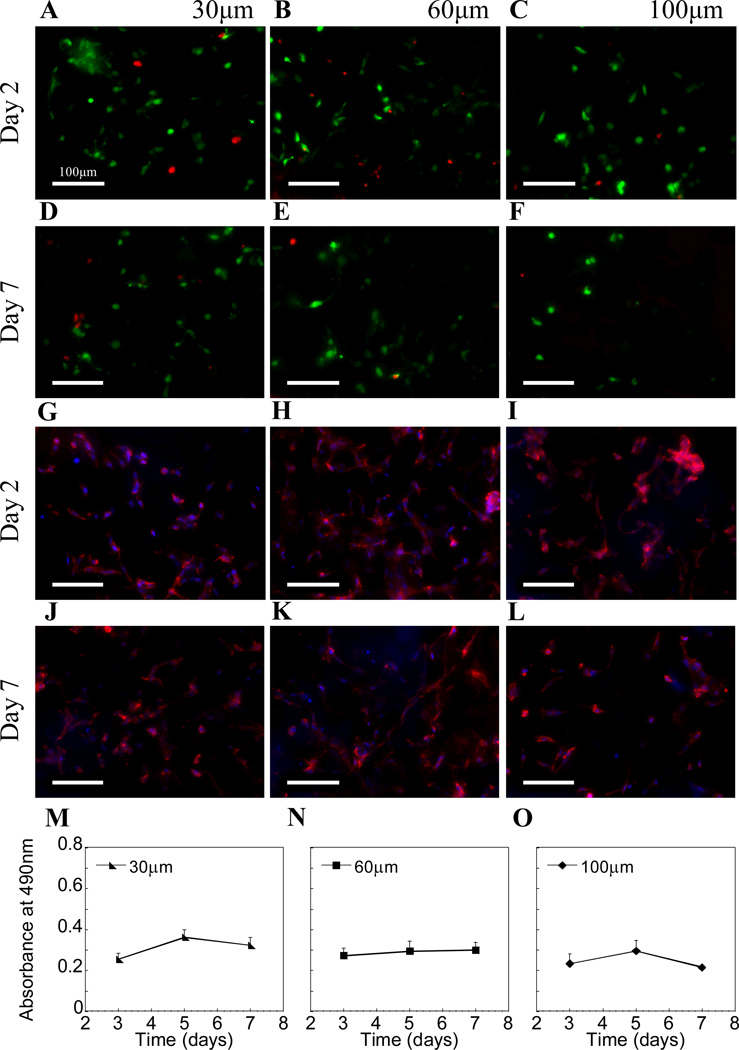
The toxicity of the DNA/PEI polyplexes was assessed using the LIVE/DEAD assay. Since the concentrations used in this study were in general high for in vitro culture systems, some toxicity was expected. However, since most cells were not in direct contact with the polyplexes embedded within the porous phase and were only exposed to the released polyplexes and those near the pore surfaces, toxicity remained low throughout the culture period (Fig 4A–F). With respect to the total number of cells, very few dead (i.e. red) cells could be observed in all pore sizes. However, a number of cells could be seen exiting the hydrogel from the bottom surface and binding to the tissue culture well, especially with increasing pore size. Likewise these cells stained green (S.Fig 1), indicating low levels of toxicity from released DNA polyplexes. Cell spreading as a function of pore size was also investigated. Cell spreading was observed for all conditions even by 2 days as a result of the soft 2.5% HA second phase the cells were seeded within (Fig 4G–L). Last, the metabolic activity of the cells inside the DNA-loaded hydrogels was studied using the MTT assay. No significant changes in metabolic activity were observed within the hydrogel (Fig 4M–O). To measure the metabolic activity of the cells solely within the hydrogels, we placed each gel into a new well immediately prior to measuring. As a result, we cannot directly correlate these findings to a decrease or halt in cell proliferation, but rather maintain that the number of cells within each gel remained relatively constant over time.
Figure 4.
(A–F) Live/dead staining showed most cells were alive (live = green, dead = red) in all pore size hydrogels after 2 and extending through 7 days. (G–L) Cells were also fixed with 4% paraformaldehyde and stained with rhodamine-phalloidin (actin) and DAPI (cell nuclei) to better visualize cell spreading. (M–O) MTT assay indicated similar cellular metabolism between various pore sizes (n=3).
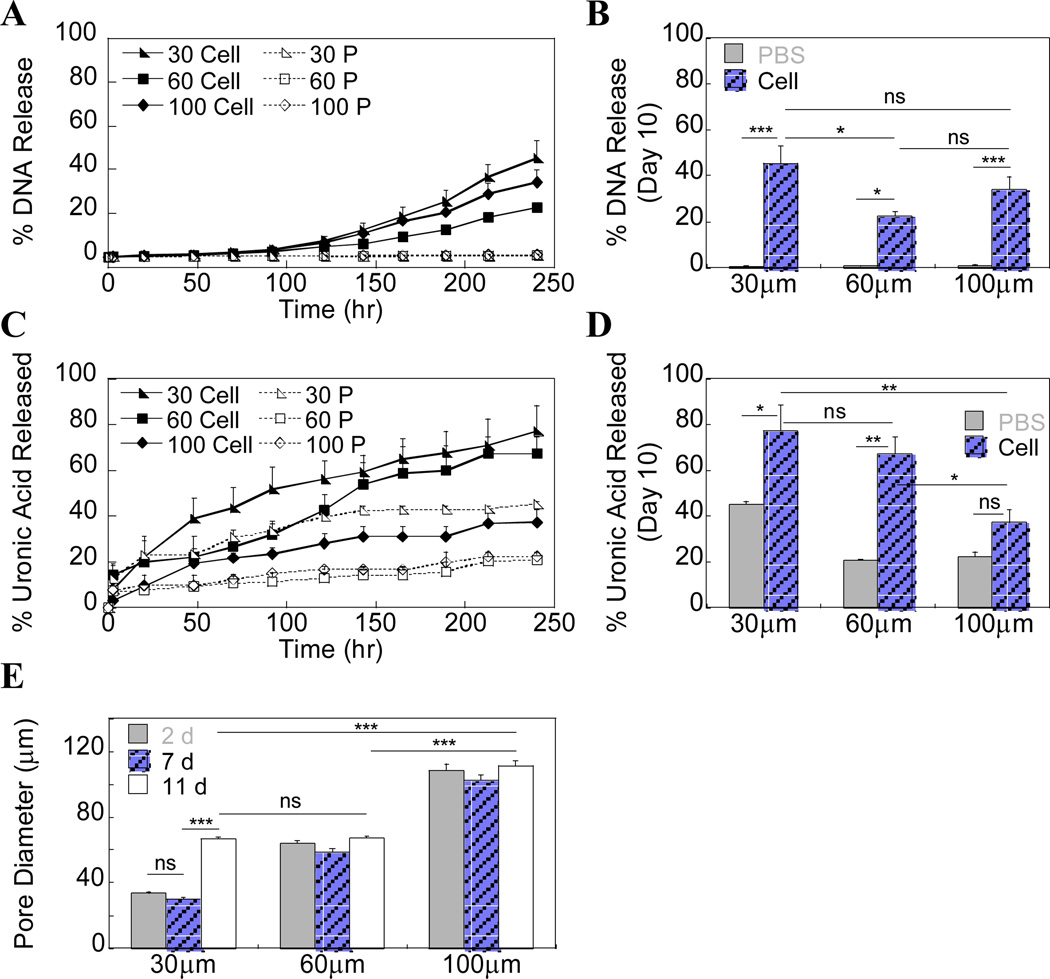
The release of the entrapped polyplexes was assessed using radiolabeled DNA either in the absence (i.e. PBS control) or presence of cells. Release studies indicated a sustained release of polyplexes in the presence of cells over a 10 day period (Fig 5A), while less than 2% of the polyplexes were released in PBS in all pore size hydrogels both due to the absence of enzymes and potentially in part due to the electrostatic interactions of the positively charged polyplexes with the negatively charged HA backbone. By day 10, polyplexes in 30 µm gels had the greatest percentage of release compared to the 60 and 100 µm pore sizes, but because of the increased variability among the 100 µm gels were only significantly greater than the 60 µm gels (Fig 5B, p<.05). The sustained release of DNA polyplexes was a significant improvement over the burst release of naked DNA from porous PEG gels, with over 50% of DNA being released by day 3 [5] in PBS, yet comparable to the release of DNA/PEI polyplexes from porous fibrin hydrogels [13].
Figure 5.
(A) 3H-dCTP radiolabeled DNA was incorporated into the hydrogels to study release kinetics in the presence of mMSCs (Cell). PBS (P) was used as an acellular control. (B) Cumulative release data at Day 10 shows 30 µm pore size gels had the largest percentage of released DNA. (C, D) Two-phase hydrogel degradation was monitored using a carbazole assay to directly quantify uronic acid in solution. Release of uronic acid after 92 hrs is assumed to be primarily from the µ-pore phase. (E) Gel degradation was also assessed by measuring pore size over time (n=27) using phase microscopy.
The same release samples were also used to measure hydrogel degradation by measuring uronic acid content in the release solution using a modified carbazole assay. For all pore sizes, HA gel degradation was greater in the presence of cells compared to the PBS control, although this difference was not significant for the 100 µm pore size (Fig 5C, D). However, for all pore sizes some uronic acid release was observed even in PBS up until about 96 hrs after which no further release was observed (Fig 5C). This directly corresponded to the time at which polyplexes started to be released in the presence of cells. We believe that the majority of the uronic acid released into solution until this point came from the 2.5% inner phase gel, especially since the 2.5% gel is naturally very weak with a storage modulus of only 130.00 ± 4.92 Pa (S.Fig 2). When used as a second-phase gel, it is likely that there was incomplete crosslinking occurring, particularly in the case of smaller pores where you are forcing macromolecular chains into a smaller, more tortuous space and thus the likelihood of the reactive groups encountering one another decreases. After ~96 hrs, it is expected that a larger contribution is made by degradation of the porous, outer phase since degradation is required for polyplex release. Again, we observed that the amount of uronic acid released was the greatest in the 30 µm gels with almost 80% being released. At this point the gels became extremely soft, but were still held together by the remaining crosslinks. We hypothesized that the second-phase was completely gone by this point and those cells still remaining in the gel were then mostly in contact with the porous phase. As a secondary approach to monitoring degradation over time, pore size measurements were made from phase images of two-phase gels with cells at various time points (n=27). The porous, outer structure could still be clearly distinguished even in the presence of the second phase and showed that in only the 30 µm gels did the pore size significantly increase from ~30 to ~60 µm in diameter (Fig 5E, p<.005). Based on the degree of uronic acid release we expected to see an increase in pore size as a result of degradation in the 60 µm gels and a smaller change in the 100 µm gels; however, due to a large degree of error in the carbazole assay so there was no clear discrepancy between the findings of the two separate experiments, as well as those trends found between the DNA release and carbazole assays. Importantly, all the results do indicate that with a 30 µm pore size, significantly more degradation occurs (compared to larger pore size gels.) This difference may be a result of greater confinement of encapsulated cells leading to an increased need for degradation to allow growth and subsequently a greater polyplex release rate.
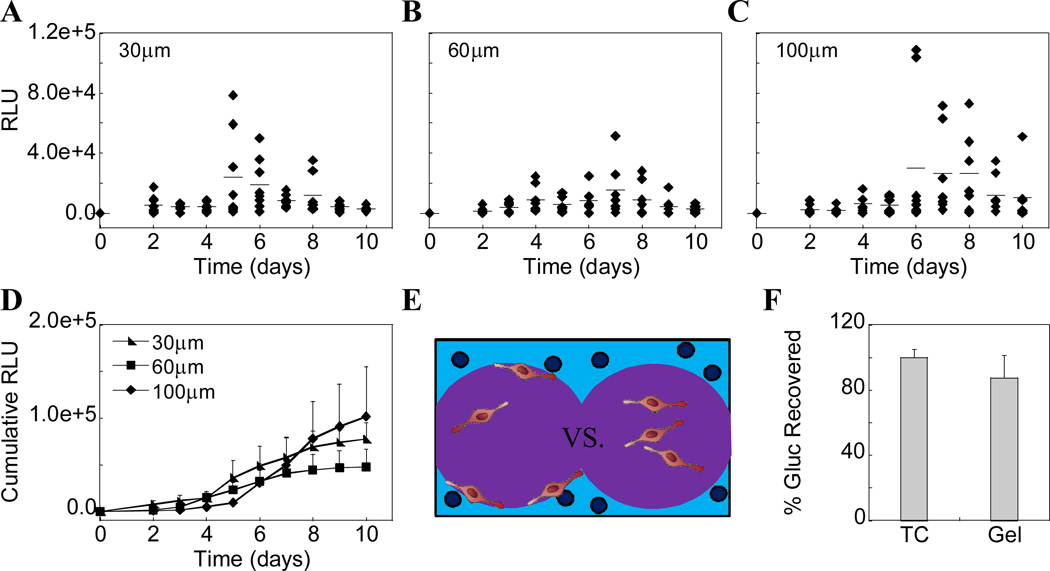
Gene transfer was also assessed as a function of pore size and DNA concentration. Gaussia luciferase (Gluc) reporter plasmid was incorporated into the various pore size hydrogels at a concentration of 1 µg/µl hydrogel solution and transgene expression was quantified using a Gaussia luciferase quantification assay. Histograms show kinetic data of all samples (n=8) over time (Fig 6A–C). Cumulative data shows an increase in Gluc expression with time, although by day 10 the differences between various pore size gels were not statistically significant (Fig 6D). The lack of any trend between pore sizes is most likely a result of the high variability between samples even within a single pore size, as demonstrated in the histograms (Fig 6A–C). We hypothesize that the variability arises from the local distribution of the cells within the pores with respect to the polyplexes in the first gel phase. Cells that are closer to the pore surfaces or are able to infiltrate into the first gel phase are in direct contact with polyplexes and are transfected at higher levels than those far from the pore surfaces (Fig 6E). To alleviate this issue, cells could have been directly seeded onto pore surfaces without the presence of the second gel phase. However, seeding cells in this manner within HA based gels is very difficult because of the lack of immediate adhesion to the pore surfaces even in the presence of 500 µm RGD. Cells then tended to flow through the pores and fall out of the gel more easily, which is specifically why we utilized the two-phase cell seeding technique. Finally, to ensure all expressed protein was being recovered for analysis and not the cause of the transfection variability, cells were first transfected for 24 hrs in tissue culture (TC) plastic and then detached using trypsin/EDTA and either re-plated onto TC plastic or encapsulated within HA-MMP degradable hydrogels. After 48 hrs, Gaussia luciferase expression was determined for both systems and showed similar levels of expression indicating no significant sequestration of Gaussia luciferase within the hydrogel (Fig 6F).
Figure 6.
Gaussia luciferase reporter plasmid was incorporated into the various pore sized hydrogels at 1 µg/µl hydrogel and transgene expression was quantified using a Gaussia luciferase quantification assay. (A–C) Histograms show kinetic data of all samples (n=8) over time for each pore size hydrogel. (D) Cumulative data shows an increase in Gluc expression with time, although by day 10 the differences between gels types are not statistically significant. (E) To verify all protein expressed was being accounted for, previously transfected cells were placed into HA gels and assessed for Gluc expression after 48 hours and compared to those cells on tissue culture (TC) plastic. The results were not significantly different.
4. Conclusions
Porous HA-MMP hydrogels were used to encapsulate DNA/PEI polyplexes and transfect seeded stem cells as they slowly degraded the matrix. Porous hydrogels allowed for effective cell seeding in vitro post scaffold fabrication, and when coupled with high loading efficiency of DNA polyplexes, allowed for long-term sustained transfection and transgene expression of incorporated mMSCs in various pore size µ-pore hydrogels. For all investigated pore sizes transgene expression was sustained for up to 10 days. Cell viability was also shown to remain high over time, even in the presence of high concentrations of DNA polyplexes. We believe the proposed hydrogel system has applications for controlled release of various DNA particles and other gene delivery vectors for in vivo tissue engineering and blood vessel formation. We anticipate that the presence of an open pore structure will increase the rate of vascularization through enhanced cellular infiltration into the gel and that the added delivery of DNA encoding for angiogenic growth factors will result in long lasting angiogenic signals.
Supplementary Material
Acknowledgements
The authors would like to thank James Dorman for his help with SEM imaging, Shiva Gojgini for her assistance in developing the DNA radiolabeling protocol, as well as Jonathan Lam and Maha Rahim for their help modifying hyaluronic acid. The authors acknowledge the NIH (1R21EB007730-01), NSF (CAREER 0747539), and CRCC for funding this work. T. Tokatlian also acknowledges the NIH Biotechnology Training Grant (T32GM067555) for funding.
References
- 1.Novosel EC, Kleinhans C, Kluger PJ. Vascularization is the key challenge in tissue engineering. Advanced Drug Delivery Reviews. 2011;63:300–311. doi: 10.1016/j.addr.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Chiu Y-C, Cheng M-H, Engel H, Kao S-W, Larson JC, Gupta S, et al. The role of pore size on vascularization and tissue remodeling in PEG hydrogels. Biomaterials. 2011;32:6045–6051. doi: 10.1016/j.biomaterials.2011.04.066. [DOI] [PubMed] [Google Scholar]
- 3.Huang X, Zhang Y, Donahue H, Lowe T. Porous thermoresponsive-co-biodegradable hydrogels as tissue-engineering scaffolds for 3-dimensional in vitro culture of chondrocytes. Tissue Engineering. 2007;13:2645–2652. doi: 10.1089/ten.2007.0084. [DOI] [PubMed] [Google Scholar]
- 4.Huang YC, Riddle K, Rice KG, Mooney DJ. Long-term in vivo gene expression via delivery of PEI-DNA condensates from porous polymer scaffolds. Hum Gene Ther. 2005;16:609–617. doi: 10.1089/hum.2005.16.609. [DOI] [PubMed] [Google Scholar]
- 5.Jang JH, Rives CB, Shea LD. Plasmid delivery in vivo from porous tissue-engineering scaffolds: transgene expression and cellular transfection. Mol Ther. 2005;12:475–483. doi: 10.1016/j.ymthe.2005.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nam YS, Yoon JJ, Park TG. A novel fabrication method of macroporous biodegradable polymer scaffolds using gas foaming salt as a porogen additive. Journal of Biomedical Materials Research. 2000;53:1–7. doi: 10.1002/(sici)1097-4636(2000)53:1<1::aid-jbm1>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 7.Iyer P, Walker KJ, Madihally SV. Increased matrix synthesis by fibroblasts with decreased proliferation on synthetic chitosan–gelatin porous structures. Biotechnology and Bioengineering. 2011 doi: 10.1002/bit.24396. n/a-n/a. [DOI] [PubMed] [Google Scholar]
- 8.Kang H-W, Tabata Y, Ikada Y. Fabrication of porous gelatin scaffolds for tissue engineering. Biomaterials. 1999;20:1339–1344. doi: 10.1016/s0142-9612(99)00036-8. [DOI] [PubMed] [Google Scholar]
- 9.Kang JY, Chung CW, Sung J-H, Park B-S, Choi J-Y, Lee SJ, et al. Novel porous matrix of hyaluronic acid for the three-dimensional culture of chondrocytes. International Journal of Pharmaceutics. 2009;369:114–120. doi: 10.1016/j.ijpharm.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 10.Van Vlierberghe S, Cnudde V, Masschaele B, Dubruel P, De Paepe I, Jacobs PJS, et al. Porous gelatin cryogels as cell delivery tool in tissue engineering. Journal of Controlled Release. 2006;116:e95–e98. doi: 10.1016/j.jconrel.2006.09.069. [DOI] [PubMed] [Google Scholar]
- 11.Galperin A, Long TJ, Ratner BD. Degradable, Thermo-Sensitive Poly(N-isopropyl acrylamide)-Based Scaffolds with Controlled Porosity for Tissue Engineering Applications. Biomacromolecules. 2010;11:2583–2592. doi: 10.1021/bm100521x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madden LR, Mortisen DJ, Sussman EM, Dupras SK, Fugate JA, Cuy JL, et al. Proangiogenic scaffolds as functional templates for cardiac tissue engineering. Proceedings of the National Academy of Sciences. 2010;107:15211–15216. doi: 10.1073/pnas.1006442107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saul JM, Linnes MP, Ratner BD, Giachelli CM, Pun SH. Delivery of non-viral gene carriers from sphere-templated fibrin scaffolds for sustained transgene expression. Biomaterials. 2007;28:4705–4716. doi: 10.1016/j.biomaterials.2007.07.026. [DOI] [PubMed] [Google Scholar]
- 14.Stachowiak AN, Bershteyn A, Tzatzaloz E, Irvine DJ. Bioactive Hydrogels with an Ordered Cellular Structure Combine Interconnected Macroporosity and Robust Mechanical Properties. Advanced Materials. 2005;17:5. [Google Scholar]
- 15.Bonadio J, Smiley E, Patil P, Goldstein S. Localized, direct plasmid gene delivery in vivo: prolonged therapy results in reproducible tissue regeneration. Nat Med. 1999;5:753–759. doi: 10.1038/10473. [DOI] [PubMed] [Google Scholar]
- 16.Chun KW, Lee JB, Kim SH, Park TG. Controlled release of plasmid DNA from photo-cross-linked pluronic hydrogels. Biomaterials. 2005;26:3319–3326. doi: 10.1016/j.biomaterials.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 17.Kasper FK, Jerkins E, Tanahashi K, Barry MA, Tabata Y, Mikos AG. Characterization of DNA release from composites of oligo(poly(ethylene glycol) fumarate) and cationized gelatin microspheres in vitro. Journal of Biomedical Materials Research Part A. 2006;78A:823–835. doi: 10.1002/jbm.a.30736. [DOI] [PubMed] [Google Scholar]
- 18.Kong HJ, Kim ES, Huang YC, Mooney DJ. Design of biodegradable hydrogel for the local and sustained delivery of angiogenic plasmid DNA. Pharmaceutical Research. 2008;25:1230–1238. doi: 10.1007/s11095-007-9526-7. [DOI] [PubMed] [Google Scholar]
- 19.Quick DJ, Anseth KS. DNA delivery from photocrosslinked PEG hydrogels: encapsulation efficiency, release profiles, and DNA quality. Journal of Controlled Release. 2004;96:341–351. doi: 10.1016/j.jconrel.2004.01.021. [DOI] [PubMed] [Google Scholar]
- 20.Lei P, Padmashali RM, Andreadis ST. Cell-controlled and spatially arrayed gene delivery from fibrin hydrogels. Biomaterials. 2009;30:3790–3799. doi: 10.1016/j.biomaterials.2009.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trentin D, Hall H, Wechsler S, Hubbell JA. Peptide-matrix-mediated gene transfer of an oxygen-insensitive hypoxia-inducible factor-1 alpha variant for local induction of angiogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:2506–2511. doi: 10.1073/pnas.0505964102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trentin D, Hubbell J, Hall H. Non-viral gene delivery for local and controlled DNA release. Journal of Controlled Release. 2005;102:263–275. doi: 10.1016/j.jconrel.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 23.Lei Y, Ng QKT, Segura T. Two and three-dimensional gene transfer from enzymatically degradable hydrogel scaffolds. Microscopy Research and Technique. 2010;73:910–917. doi: 10.1002/jemt.20840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lei Y, Segura T. DNA delivery from matrix metalloproteinase degradable poly(ethylene glycol) hydrogels to mouse cloned mesenchymal stem cells. Biomaterials. 2009;30:254–265. doi: 10.1016/j.biomaterials.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wieland JA, Houchin-Ray TL, Shea LD. Non-viral vector delivery from PEG-hyaluronic acid hydrogels. Journal of Controlled Release. 2007;120:233–241. doi: 10.1016/j.jconrel.2007.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lungwitz U, Breunig M, Blunk T, Gopferich A. Polyethylenimine-based non-viral gene delivery systems. European Journal of Pharmaceutics and Biopharmaceutics. 2005;60:247–266. doi: 10.1016/j.ejpb.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 27.Abdallah B, Hassan A, Benoist C, Goula D, Behr JP, Demeneix BA. A powerful nonviral vector for in vivo gene transfer into the adult mammalian brain: Polyethylenimine. Human Gene Therapy. 1996;7:1947–1954. doi: 10.1089/hum.1996.7.16-1947. [DOI] [PubMed] [Google Scholar]
- 28.Wang S, Ma N, Gao SJ, Yu H, Leong KW. Transgene expression in the brain stem effected by intramuscular injection of polyethylenimine/DNA complexes. Molecular Therapy. 2001;3:658–664. doi: 10.1006/mthe.2001.0324. [DOI] [PubMed] [Google Scholar]
- 29.Gautam A, Densmore CL, Golunski E, Xu B, Waldrep JC. Transgene expression in mouse airway epithelium by aerosol gene therapy with PEI-DNA complexes. Molecular Therapy. 2001;3:551–556. doi: 10.1006/mthe.2001.0300. [DOI] [PubMed] [Google Scholar]
- 30.Kichler A, Chillon M, Leborgne C, Danos O, Frisch B. Intranasal gene delivery with a polyethylenimine-PEG conjugate. Journal of Controlled Release. 2002;81:379–388. doi: 10.1016/s0168-3659(02)00080-9. [DOI] [PubMed] [Google Scholar]
- 31.Rudolph C, Schillinger U, Plank C, Gessner A, Nicklaus P, Muller RH, et al. Nonviral gene delivery to the lung with copolymer-protected and transferrin-modified polyethylenimine. Biochimica Et Biophysica Acta-General Subjects. 2002;1573:75–83. doi: 10.1016/s0304-4165(02)00334-3. [DOI] [PubMed] [Google Scholar]
- 32.Wiseman JW, Goddard CA, McLelland D, Colledge WH. A comparison of linear and branched polyethylenimine (PEI) with DCChol/DOPE liposomes for gene delivery to epithelial cells in vitro and in vivo. Gene Therapy. 2003;10:1654–1662. doi: 10.1038/sj.gt.3302050. [DOI] [PubMed] [Google Scholar]
- 33.Segura T, Schmokel H, Hubbell JA. RNA Interference Targeting Hypoxia Inducible Factor 1[alpha] Reduces Post-Operative Adhesions in Rats. Journal of Surgical Research. 2007;141:162–170. doi: 10.1016/j.jss.2006.07.045. [DOI] [PubMed] [Google Scholar]
- 34.Aoki K, Furuhata S, Hatanaka K, Maeda M, Remy JS, Behr JP, et al. Polyethylenimine-mediated gene transfer into pancreatic tumor dissemination in the murine peritoneal cavity. Gene Therapy. 2001;8:508–514. doi: 10.1038/sj.gt.3301435. [DOI] [PubMed] [Google Scholar]
- 35.Coll JL, Chollet P, Brambilla E, Desplanques D, Behr JP, Favrot M. In vivo delivery to tumors of DNA complexed with linear polyethylenimine. Human Gene Therapy. 1999;10:1659–1666. doi: 10.1089/10430349950017662. [DOI] [PubMed] [Google Scholar]
- 36.Iwai M, Harada Y, Tanaka S, Muramatsu A, Mori T, Kashima K, et al. Polyethylenimine-mediated suicide gene transfer induces a therapeutic effect for hepatocellular carcinoma in vivo by using an Epstein-Barr virus-based plasmid vector. Biochemical and Biophysical Research Communications. 2002;291:48–54. doi: 10.1006/bbrc.2002.6383. [DOI] [PubMed] [Google Scholar]
- 37.Fraser JR, Laurent TC, Laurent UB. Hyaluronan: its nature, distribution, functions and turnover. J Intern Med. 1997;242:27–33. doi: 10.1046/j.1365-2796.1997.00170.x. [DOI] [PubMed] [Google Scholar]
- 38.Leach JB, Bivens KA, Patrick CW, Schmidt CE. Photocrosslinked hyaluronic acid hydrogels: Natural, biodegradable tissue engineering scaffolds. Biotechnology and Bioengineering. 2003;82:578–589. doi: 10.1002/bit.10605. [DOI] [PubMed] [Google Scholar]
- 39.Park YD, Tirelli N, Hubbell JA. Photopolymerized hyaluronic acid-based hydrogels and interpenetrating networks. Biomaterials. 2003;24:893–900. doi: 10.1016/s0142-9612(02)00420-9. [DOI] [PubMed] [Google Scholar]
- 40.Shu XZ, Liu YC, Palumbo FS, Lu Y, Prestwich GD. In situ crosslinkable hyaluronan hydrogels for tissue engineering. Biomaterials. 2004;25:1339–1348. doi: 10.1016/j.biomaterials.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 41.Yeo Y, Highley CB, Bellas E, Ito T, Marini R, Langer R, et al. In situ cross-linkable hyaluronic acid hydrogels prevent post-operative abdominal adhesions in a rabbit model. Biomaterials. 2006;27:4698–4705. doi: 10.1016/j.biomaterials.2006.04.043. [DOI] [PubMed] [Google Scholar]
- 42.Eldridge L, Moldobaeva A, Wagner EM. Increased hyaluronan fragmentation during pulmonary ischemia. American Journal of Physiology - Lung Cellular and Molecular Physiology. 2011;301:L782–L788. doi: 10.1152/ajplung.00079.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao F, Liu Y, He Y, Yang C, Wang Y, Shi X, et al. Hyaluronan oligosaccharides promote excisional wound healing through enhanced angiogenesis. Matrix Biology. 2010;29:107–116. doi: 10.1016/j.matbio.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 44.Voelcker V, Gebhardt C, Averbeck M, Saalbach A, Wolf V, Weih F, et al. Hyaluronan fragments induce cytokine and metalloprotease upregulation in human melanoma cells in part by signalling via TLR4. Experimental Dermatology. 2008;17:100–107. doi: 10.1111/j.1600-0625.2007.00638.x. [DOI] [PubMed] [Google Scholar]
- 45.Chung C, Burdick JA. Influence of Three-Dimensional Hyaluronic Acid Microenvironments on Mesenchymal Stem Cell Chondrogenesis. Tissue Engineering Part A. 2009;15:243–254. doi: 10.1089/ten.tea.2008.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gerecht S, Burdick JA, Ferreira LS, Townsend SA, Langer R, Vunjak-Novakovic G. Hyaluronic acid hydrogel for controlled self-renewal and differentiation of human embryonic stem cells. Proc Natl Acad Sci U S A. 2007;104:11298–11303. doi: 10.1073/pnas.0703723104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim J, Park Y, Tae G, Lee KB, Hwang CM, Hwang SJ, et al. Characterization of low-molecular-weight hyaluronic acid-based hydrogel and differential stem cell responses in the hydrogel microenvironments. Journal of Biomedical Materials Research Part A. 2009;88A:967–975. doi: 10.1002/jbm.a.31947. [DOI] [PubMed] [Google Scholar]
- 48.Pan LJ, Ren YJ, Cui FZ, Xu QY. Viability and Differentiation of Neural Precursors on Hyaluronic Acid Hydrogel Scaffold. Journal of Neuroscience Research. 2009;87:3207–3220. doi: 10.1002/jnr.22142. [DOI] [PubMed] [Google Scholar]
- 49.Aruffo A, Stamenkovic I, Melnick M, Underhill CB, Seed B. CD44 is the principal cell surface receptor for hyaluronate. Cell. 1990;61:1303–1313. doi: 10.1016/0092-8674(90)90694-a. [DOI] [PubMed] [Google Scholar]
- 50.Sherman L, Sleeman J, Herrlich P, Ponta H. Hyaluronate Receptors - Key Players in Growth, Differentiation, Migration and Tumor Progression. Current Opinion in Cell Biology. 1994;6:726–733. doi: 10.1016/0955-0674(94)90100-7. [DOI] [PubMed] [Google Scholar]
- 51.Lei Y, Gojgini S, Lam J, Segura T. The spreading, migration and proliferation of mouse mesenchymal stem cells cultured inside hyaluronic acid hydrogels. Biomaterials. 2011;32:39–47. doi: 10.1016/j.biomaterials.2010.08.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim J, Park Y, Tae G, Lee KB, Hwang SJ, Kim IS, et al. Synthesis and characterization of matrix metalloprotease sensitive-low molecular weight hyaluronic acid based hydrogels. Journal of Materials Science-Materials in Medicine. 2008;19:3311–3318. doi: 10.1007/s10856-008-3469-3. [DOI] [PubMed] [Google Scholar]
- 53.Gojgini S, Tokatlian T, Segura T. Utilizing Cell-Matrix Interactions To Modulate Gene Transfer to Stem Cells Inside Hyaluronic Acid Hydrogels. Molecular Pharmaceutics. 2011;8:1582–1591. doi: 10.1021/mp200171d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lei Y, Huang S, Sharif-Kashani P, Chen Y, Kavehpour P, Segura T. Incorporation of active DNA/cationic polymer polyplexes into hydrogel scaffolds. Biomaterials. 2010;31:9106–9116. doi: 10.1016/j.biomaterials.2010.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lei Y, Rahim M, Ng Q, Segura T. Hyaluronic acid and fibrin hydrogels with concentrated DNA/PEI polyplexes for local gene delivery. Journal of Controlled Release. 2011;153:255–261. doi: 10.1016/j.jconrel.2011.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peyton SR, Kalcioglu ZI, Cohen JC, Runkle AP, Van Vliet KJ, Lauffenburger DA, et al. Marrow-Derived stem cell motility in 3D synthetic scaffold is governed by geometry along with adhesivity and stiffness. Biotechnology and Bioengineering. 2011;108:1181–1193. doi: 10.1002/bit.23027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stachowiak AN, Irvine DJ. Inverse opal hydrogel-collagen composite scaffolds as a supportive microenvironment for immune cell migration. J Biomed Mater Res A. 2008;85:815–828. doi: 10.1002/jbm.a.31661. [DOI] [PubMed] [Google Scholar]
- 58.Tokatlian T, Shrum CT, Kadoya WM, Segura T. Protease degradable tethers for controlled and cell-mediated release of nanoparticles in 2- and 3-dimensions. Biomaterials. 2010;31:8072–8080. doi: 10.1016/j.biomaterials.2010.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schrementi ME, Ferreira AM, Zender C, DiPietro LA. Site-specific production of TGF-β in oral mucosal and cutaneous wounds. Wound Repair and Regeneration. 2008;16:80–86. doi: 10.1111/j.1524-475X.2007.00320.x. [DOI] [PubMed] [Google Scholar]
- 60.Szpaderska AM, Zuckerman JD, DiPietro LA. Differential Injury Responses in Oral Mucosal and Cutaneous Wounds. Journal of Dental Research. 2003;82:621–626. doi: 10.1177/154405910308200810. [DOI] [PubMed] [Google Scholar]
- 61.Bitter T, Muir HM. A modified uronic acid carbazole reaction. Analytical Biochemistry. 1962;4:330–334. doi: 10.1016/0003-2697(62)90095-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.