Abstract
Objectives
This study sought to investigate into the biologically plausible interaction between the common haptoglobin (Hp) polymorphism rs#72294371 and glycosylated hemoglobin (HbA1c) on risk of coronary heart disease (CHD).
Background
Studies of the association between the Hp polymorphism and CHD report inconsistent results. Individuals with the Hp2-2 genotype produce Hp proteins with an impaired ability to prevent oxidative injury caused by elevated HbA1c.
Methods
HbA1c concentration and Hp genotype were determined for 407 CHD cases matched 1:1 to controls (from the NHS [Nurses' Health Study]) and in a replication cohort of 2,070 individuals who served as the nontreatment group in the ICARE (Israel Cardiovascular Events Reduction With Vitamin E) study, with 29 CHD events during follow-up. Multivariate models were adjusted for lifestyle and CHD risk factors as appropriate. A pooled analysis was conducted of NHS, ICARE, and the 1 previously published analysis (a cardiovascular disease case-control sample from the Strong Heart Study).
Results
In the NHS, Hp2-2 genotype (39% frequency) was strongly related to CHD risk only among individuals with elevated HbA1c (≥6.5%), an association that was similar in the ICARE trial and the Strong Heart Study. In a pooled analysis, participants with both the Hp2-2 genotype and elevated HbA1c had a relative risk of 7.90 (95% confidence interval: 4.43 to 14.10) for CHD compared with participants with both an Hp1 allele and HbA1c <6.5% (p for interaction = 0.004), whereas the Hp2-2 genotype with HbA1c <6.5% was not associated with risk (relative risk: 1.34 [95% confidence interval: 0.73 to 2.46]).
Conclusions
Hp genotype was a significant predictor of CHD among individuals with elevated HbA1c.
Keywords: acute myocardial infarction, coronary disease, epidemiology, genetic association, genotype, glycoproteins
Haptoglobin (Hp) is an abundant plasma protein that protects against oxidative damage mediated by extracorpuscular hemoglobin (1). A common polymorphism (rs#72294371) exists at the Hp locus and consists of 2 alleles, denoted 1 and 2, that are defined by the absence or presence of a 1700 base pair partial intragenic duplication, thus forming 3 possible Hp genotypes (1-1, 2-1, and 2-2) that produce 3 distinctly different proteins that vary in size, shape, and function (2). Interestingly, this common Hp polymorphism is posited to have arisen early in human evolution from a selective advantage of the Hp2 allele against infectious disease, but in modern times, it may confer increased risk of several noninfectious, inflammatory, and chronic disease complications (3). Compared with the Hp1-1 genotype, and to a lesser extent the intermediary Hp2-1 genotype, individuals with the Hp2-2 genotype produce a protein that is dysfunctional in protecting against hemoglobin-driven oxidative damage, leading to increased inflammation and oxidative stress, reduced ability of high-density lipoprotein (HDL) to promote reverse cholesterol efflux, and plaque instability in several in vitro and in vivo systems (4–8). This dysfunction in the Hp2-2 protein is accentuated when hemoglobin is glycosylated (6).
The Hp2-2 genotype has been consistently associated with increased risk of cardiovascular complications, such as myocardial infarction (MI), among individuals with type 2 diabetes (2,9). However, the relationship between Hp genotype and coronary heart disease (CHD) in people without diabetes is unclear and controversial (2,10). The impact of maintaining strict glycemic control on the prevention of cardiovascular complications in individuals with type 2 diabetes is also inconsistent (11,12). A possible explanation for this inconsistency is that strict glycemic control may only be beneficial in reducing CHD risk in a subset of individuals most susceptible to vascular damage from hyperglycemia, such as those with the Hp2-2 genotype.
We explored the potential statistical interaction between the Hp genotype and glycemia, as quantified by glycosylated hemoglobin (HbA1c) concentration, on the risk of CHD within 1 large prospective cohort of apparently healthy women. We then replicated this analysis in a second and more focused cohort of individuals with diabetes to determine if individuals with the Hp2-2 genotype and elevated HbA1c are at increased risk of incident CHD. Finally, we conducted a meta-analysis of results from these 2 cohorts with those from a previously published nested case-control study of Hp genotype and cardiovascular disease (CVD).
Methods
Cohort: the Nurses' Health Study
The NHS (Nurses' Health Study) is a prospective cohort of 121,700 female U.S. registered nurses who were aged 30 to 55 years at baseline in 1976. Information on anthropometric and lifestyle factors is obtained through self-administered questionnaires every 2 years and diet every 4 years. From 1989 to 1990, a blood sample was provided by 32,826 women. Women who had an incident MI (n = 343) or fatal CHD (n = 64) between the date of blood draw and June 2004 were identified and matched 1:1 to controls for age, smoking status, fasting status, and month of blood draw, as described elsewhere (13). The majority (96%) of nurses in this case-control sample are white. Anthropometric and lifestyle variables were derived from the questionnaire administered in 1990, with missing information substituted from previous questionnaires. The validity of the questionnaires and the reproducibility of the measurements have been reported previously (14,15), as has the measurement of the standard biochemical risk variables (13). The institutional review board of the Brigham and Women's Hospital and the Human Subjects Committee Review Board of the Harvard School of Public Health approved the study protocol.
Cohort: the Israel Cardiovascular Events Reduction With Vitamin E Study
The ICARE trial (Israel Cardiovascular Events Reduction With Vitamin E Study; Clinical Trials.gov #NCT00220831) is a large clinical trial of vitamin E in participants with type 2 diabetes; it was conducted within 47 primary healthcare clinics of the Clalit Health Services in Israel, as described previously (16). These participants were white men and women aged 22 to 95 years with type 2 diabetes at baseline who were enrolled in the Clalit Health Plan Diabetes Registry and were followed up prospectively for CHD events. Only participants with the Hp2-2 genotype were randomized to receive vitamin E or placebo; participants with the Hp1-1 and Hp2-1 genotypes were not included in the randomization. The ICARE sample of participants included in the current analysis consists of the 2,070 individuals remaining who did not receive vitamin E (the untreated Hp1-1 and Hp2-1 patients and the Hp2-2 patients receiving placebo). The vitamin E-treated Hp2-2 patients were not included in the current analysis to avoid introducing bias because these patients have been shown to have lower risk of CHD due to the administered vitamin E. The study protocol was approved by the independent ethics committee of the Carmel Medical Center in Clalit Health Services and the Israeli Ministry of Health. All participants provided written informed consent.
Hp typing
Hp genotype, recently given the identifier rs#72294371 (17), was determined in both cohorts by using gel electrophoresis of hemoglobin-enriched serum (18). This procedure produces a signature banding pattern for each Hp type and has been shown to corroborate completely with the Hp genotype determined by polymerase chain reaction (19). An unambiguous Hp type was obtained on >99.5% of all samples. A pilot study to test the Hp genotyping method in the NHS found 99% concordance between duplicate samples of 76 individuals. Genotype frequencies in the NHS were in Hardy-Weinberg equilibrium in the whole sample (p = 0.49) and also within cases (p = 0.84) and controls (p = 0.52) separately. Genotype frequencies in ICARE were not in Hardy-Weinberg equilibrium, but this was driven by the study design that excluded one half of the Hp2-2 participants because they had been given vitamin E treatment in the ICARE clinical trial.
CHD case assessment
As previously described in NHS (20) and ICARE (16), CHD was similarly defined in the 2 datasets as nonfatal MI or fatal CHD. In brief, World Health Organization criteria (21) (symptoms plus either diagnostic electrocardiographic changes or altered levels of cardiac enzymes) were used to diagnose CHD. Nonfatal NHS events were confirmed through review of medical records, with deaths confirmed by using medical records, the National Death Index, and from death certificates. All information was reviewed by NHS investigators who determined the primary cause of death. ICARE cases were ascertained by reviewing all hospitalizations of study participants; the hospitalization discharge summary was used for adjudication of events by a panel of physicians. MI was defined by the typical rise and fall of serum markers of myocardial necrosis with at least 1 of the following: 1) typical ischemic symptoms; 2) development of pathologic Q waves on the electrocardiogram; or 3) electrocardiographic changes diagnostic of ischemia. All death cases were ascertained by using the national death registry. For out-of-hospital deaths, adjudication was based on interviews with the participant's physician and family.
Statistical analysis
Participant characteristics were compared between genotypes by using a general linear model for continuous variables and chi-square tests for categorical variables, unless there was a cell with n < 5, in which case the Fisher exact test was used. For skewed variables, p values from log-transformed analyses, geometric means, and reverse-transformed 95% confidence intervals were reported. All analyses were conducted by using SAS version 9.1 (SAS Institute, Inc., Cary, North Carolina) at a 2-tailed alpha level of 0.05.
Because of the nested case-control study design, relative risks (RRs) of CHD for the NHS were estimated by using incidence rate ratios from logistic regression with adjustment for the matching factors. In the prospective ICARE cohort, Cox proportional hazards models were used, with days in study as the time-dependent variable to estimate hazard ratios as estimates of RR. For the NHS, unconditional logistic regression was used to allow for maximum statistical power in stratified analyses and because 6 participants ended up unmatched due to missing data for their match. Because of limited power to test for interactions across strata of Hp genotype (due to the Hp1-1 genotype frequency [≤15%]), we combined the Hp1-1 genotype with the Hp 2-1 genotype for these analyses.
In addition to matching factors (age, smoking, fasting status, and month of blood draw), NHS analyses were adjusted for body mass index, alcohol intake, diabetes, history of hypercholesterolemia, and history of hypertension. The following variables were considered as potential covariates for the NHS analysis but ultimately were not included in the model because they did not influence the association: marital status, physical activity, cholesterol-lowering medication use, insulin and oral diabetes medication use, menopausal status, hormone therapy, and parental MI at <60 years of age. ICARE analyses were adjusted for sex, age, smoking status, hypertension, MI before enrollment, statin use, and metformin use. The following variables were considered as potential covariates for the ICARE analysis but ultimately were not included in the model: length of time with diabetes, blood pressure medications, and stroke.
For analyses stratified according to HbA1c, we used 6.5% as the cutoff because this has been defined as the value leading to a diagnosis of diabetes, as well as the level at which complications of diabetes arise, as established by the International Expert Committee composed of members of the American Diabetes Association, the European Association for the Study of Diabetes, and the International Diabetes Federation (22).
Pooled analysis
To pool the risk estimates from multiple study cohorts, we used the weighted average of regression estimates in a random-effects meta-analysis, testing for heterogeneity (23). In addition to NHS and ICARE, we searched Medline through March 2012 for published data of Hp genotype and CVD in cohorts with results stratified according to either HbA1c or diabetes status. We also searched the reference lists from the limited number of studies of Hp genotype in the literature. We found 1 suitable publication, and it was from the Strong Heart Study (SHS) (24). The publication contained Hp genotype frequencies for 206 CVD cases and 206 matched controls, and we used these frequencies to calculate the unadjusted odds ratio and 95% confidence intervals (CIs) for the SHS sample. In the SHS sample, the endpoint was total CVD (CHD and incident stroke events), but >85% of events were CHD related. HbA1c concentrations were not available for all participants, but results were published according to diabetes status, which served as a proxy for HbA1c: it has been reported that in the SHS participants specifically, a diabetes diagnosis has similar risk of CVD as the cutoff of HbA1c 6.5% (25). SHS was a population-based prospective longitudinal study of CVD in American Indians. Detailed descriptions of the SHS cohort, survey methods, and CVD case ascertainment and laboratory techniques have been described previously (25).
Results
Nurses' Health Study
Baseline characteristics of the NHS participants are described in Table 1 according to Hp rs#72294371 genotype and case status. The distribution of Hp phenotype frequencies was 15% (Hp1-1), 46% (Hp2-1), and 39% (Hp2-2) (data not shown), and the Hp genotype frequency did not differ between cases and controls. Among cases but not controls, Hp1-1 participants were significantly older than those with the Hp2-1 or Hp2-2 genotype. As expected based on previous reports from this cohort (26), cases and controls differed with respect to classic cardiovascular risk factors (data not shown). When adjusted for covariates and case-control status, serum low-density lipoprotein (LDL) cholesterol was the only biomarker to differ across Hp genotypes (p = 0.05), with lower concentrations (mean ± SE) in Hp1-1 patients (133 ± 3 mg/dl) compared with Hp2-1 patients (140 ± 2 mg/dl) and Hp2-2 individuals (143 ± 2 mg/dl) (Online Table 1).
Table 1. Baseline Characteristics by Hp Genotype and Case Status Among Women Aged 44 to 69 Years at Blood Draw From a Nested Case-Control Study of CHD Events in the NHS, 1990 to 2004.
| Cases | Controls | |||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| Characteristic | Hp1-1 (n = 58) | Hp2-1 (n = 188) | Hp2-2 (n = 161) | P | Hp1-1 (n = 66) | Hp2-1 (n = 189) | Hp2-2 (n = 156) | p |
| Age*(yrs) | 62.1 ± 6.3 | 59.3 ± 6.4 | 59.4 ± 6.6 | 0.01 | 60.8 ± 5.9 | 59.5 ± 6.7 | 59.5 ± 6.5 | 0.33 |
|
| ||||||||
| Smoking status* | ||||||||
| Never | 27 (47) | 63 (34) | 54 (34) | 0.44 | 26 (39) | 66 (35) | 55 (35) | 0.83 |
| Past | 18 (31) | 74 (39) | 62 (38) | 21 (32) | 75 (40) | 62 (40) | ||
| Current | 13 (22) | 51 (27) | 45 (28) | 19 (29) | 48 (25) | 39 (25) | ||
|
| ||||||||
| BMI (kg/m2) | 26.1 (24.9-27.4) | 26.0 (25.3-26.7) | 26.1 (25.3-26.9) | 0.97 | 25.5 (24.5-26.5) | 24.7 (24.1-25.3) | 25.0 (24.4-25.6) | 0.39 |
|
| ||||||||
| Activity (MET h/wk) | 9.2 (6.3-13.5) | 7.4 (6.0-9.17) | 9.8 (7.8-12.3) | 0.20 | 10.7 (7.9-14.6) | 9.6 (8.03-11.6) | 11.9 (9.77-14.6) | 0.30 |
|
| ||||||||
| Alcohol (g/d) | 2.46 (1.58-3.82) | 4.88 (3.97-6.00) | 4.16 (3.31-5.24) | 0.02 | 4.83 (3.42-6.81) | 4.45 (3.59-5.51) | 5.10 (4.09-6.35) | 0.68 |
|
| ||||||||
| Parental MI <60 yrs of age | 9 (16) | 41 (22) | 32 (20) | 0.58 | 7 (11) | 22 (12) | 25 (16) | 0.39 |
|
| ||||||||
| History of hypercholesterolemia | 25 (43) | 96 (51) | 99 (61) | 0.03 | 22 (33) | 79 (42) | 63 (40) | 0.48 |
|
| ||||||||
| History of hypertension | 30 (52) | 94 (50) | 82 (51) | 0.97 | 18 (27) | 47 (25) | 46 (29) | 0.63 |
|
| ||||||||
| History of diabetes | 10 (17) | 24 (13) | 24 (15) | 0.66 | 4 (6) | 13 (7) | 7 (5) | 0.64 |
|
| ||||||||
| Oral diabetic drug use | 2 (3) | 13 (7) | 5 (3) | 0.01 | 0 (0) | 2 (1) | 2 (1) | 0.18 |
|
| ||||||||
| Cholesterol-lowering drug use | 1 (2) | 8 (4) | 13 (8) | 0.12 | 1 (2) | 4 (2) | 5 (3) | 0.70 |
|
| ||||||||
| Postmenopausal hormone use† | 17 (29) | 59 (31) | 50 (31) | 0.43 | 24 (36) | 60 (32) | 59 (38) | 0.83 |
|
| ||||||||
| CRP (mg/L) | 0.30 (0.22-0.41) | 0.24 (0.20-0.28) | 0.26 (0.21-0.31) | 0.36 | 0.19 (0.15-0.25) | 0.19 (0.16-0.22) | 0.16 (0.14-0.19) | 0.49 |
|
| ||||||||
| Triglycerides (mg/dl) | 129.5 (113.7-147.6) | 121.4 (112.9-130.5) | 123.1 (113.8-133.1) | 0.69 | 107.4 (95.7-120.5) | 105.4 (98.5-112.9) | 104.3 (96.7-112.4) | 0.92 |
|
| ||||||||
| HbA1c (%) | 5.96 (5.69-6.24) | 5.76 (5.61-5.91) | 5.76 (5.62-5.94) | 0.43 | 5.46 (5.33-5.59) | 5.53 (5.45-5.61) | 5.43 (5.34-5.51) | 0.23 |
|
| ||||||||
| HbA1c ≥6.5% | 10 (17) | 23 (40) | 25 (43) | 0.001 | 3 (22) | 9 (64) | 2 (14) | 0.01 |
|
| ||||||||
| Total cholesterol (mg/dl) | 228 ± 53 | 232 ± 37 | 237 ± 40 | 0.33 | 222 ± 35 | 227 ± 42 | 229 ± 40 | 0.54 |
|
| ||||||||
| HDL cholesterol (mg/dl) | 53 ± 14 | 52 ± 15 | 52 ± 15 | 0.89 | 60 ± 18 | 59 ± 16 | 60 ± 17 | 0.90 |
|
| ||||||||
| LDL cholesterol (mg/dl) | 139 ± 50 | 143 ± 33 | 147 ± 36 | 0.29 | 130 ± 32 | 136 ± 40 | 137 ± 35 | 0.42 |
|
| ||||||||
| Apolipoprotein B (mg/dl) | 111 ± 36 | 114 ± 34 | 116 ± 33 | 0.68 | 104 ± 33 | 103 ± 30 | 107 ± 30 | 0.45 |
|
| ||||||||
| Adiponectin (ng/ml) | 7,142 (6,325-8,064) | 7,125 (6,661-7,623) | 7,515 (6,986-8,083) | 0.54 | 8,355 (7,553-9,245) | 8,541 (8,045-9,066) | 8,649 (8,099-9,240) | 0.85 |
Values are n (%), mean ± SD, or geometric mean (95% confidence interval [CI]). Participant characteristics were compared between genotypes by using a general linear model for continuous variables and chi-square tests for categorical variables, unless there was a cell with n < 5, in which case the Fisher exact test was used. For skewed variables (physical activity, alcohol, high sensitivity C-reactive protein [CRP], body mass index [BMI], glycosylated hemoglobin [HbA1c], triglycerides, and adiponectin), p values from log-transformed analyses and geometric means with reverse-transformed 95% CIs are displayed.
Case-control matching variable (also matched for fasting status and date at blood draw).
In postmenopausal women only.
CHD = coronary heart disease; HDL = high-density lipoprotein; Hp = haptoglobin; LDL = low-density lipoprotein; NHS = Nurses' Health Study.
Table 2 presents the multivariate RR of CHD for the Hp2-2 genotype compared with the Hp1 allele carriers. Among all Hp2-2 carriers, the RR of CHD was 1.02 (95% CI: 0.76 to 1.36) compared with Hp1 allele carriers (Hp 1-1 and Hp2-1 genotypes). We further stratified the data according to known CHD risk factors. Among participants with HbA1c ≥6.5% (n = 72), the RR for Hp2-2 was 10.12 (95% CI: 1.08 to 94.97), whereas no association for Hp genotype was observed among participants with HbA1c status <6.5% (p for interaction = 0.04). We did not observe a similar interaction for reported diabetes history, but 38% of the women with diabetes had HbA1c <6.5%. Because only 24 participants reported using oral diabetic medication, we could not conduct an analysis further stratified according to medication use.
Table 2.
Multivariate RR* of CHD With 95% CIs for Hp Genotypes, Together and Stratified According to CVD Risk Factors, in a Nested Case-Control Study of CHD in Women Aged 44 to 69 Years at Blood Draw From the NHS, 1990 to 2004
| Hp1-1 + Hp2-1 | Hp2-2 | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Characteristic | Ca/Co | RR | Ca/Co | RR | 95% CI | p for Interaction |
| All participants | ||||||
| Matching factors only | 246/255 | Ref. | 161/156 | 1.07 | 0.81-1.42 | — |
| Multivariate adjusted* | 246/255 | Ref. | 161/156 | 1.02 | 0.76-1.36 | — |
|
| ||||||
| Age at baseline | ||||||
| <60 yrs | 100/112 | Ref. | 77/67 | 1.21 | 0.71-1.78 | 0.40 |
| ≥60 yrs | 146/143 | Ref. | 84/89 | 0.92 | 0.63-1.36 | |
|
| ||||||
| Smoking | ||||||
| Never | 90/92 | Ref. | 54/55 | 0.97 | 0.59-1.60 | 0.87 |
| Past | 92/96 | Ref. | 62/62 | 0.97 | 0.59-1.57 | |
| Current | 64/67 | Ref. | 45/39 | 1.18 | 0.67-2.10 | |
|
| ||||||
| Diabetes† | ||||||
| No | 205/236 | Ref. | 130/146 | 0.97 | 0.71-1.32 | 0.38 |
| Yes | 41/19 | Ref. | 31/10 | 2.05 | 0.73-5.72 | |
|
| ||||||
| HbA1c | ||||||
| <6.5% | 213/243 | Ref. | 136/154 | 0.95 | 0.70-1.29 | 0.04 |
| ≥6.5% | 33/12 | Ref. | 25/2 | 10.12 | 1.08-94.97 | |
|
| ||||||
| History of hypercholesterolemia | ||||||
| No | 125/154 | Ref. | 62/93 | 0.80 | 0.52-1.21 | 0.12 |
| Yes | 121/101 | Ref. | 99/63 | 1.32 | 0.86-2.03 | |
|
| ||||||
| LDL concentrations | ||||||
| Normal (<160 mg/dl) | 173/193 | Ref. | 110/117 | 0.96 | 0.68-1.37 | 0.57 |
| High (≥160 mg/dl) | 73/62 | Ref. | 51/39 | 1.14 | 0.64-2.01 | |
Adjusted model included age, BMI, smoking status, alcohol intake, diabetes, history of hypercholesterolemia, and history of hypertension, unless stratified by 1 of these variables.
For analysis of risk of CHD by diabetes status, cases were updated as having diabetes if they developed diabetes before their CHD event, and controls were updated as having diabetes if diagnosed with diabetes before their matched case had a CHD event.
CI = confidence interval; CVD = cardiovascular disease; Ref. = reference; RR = relative risk; other abbreviations as in Table 1.
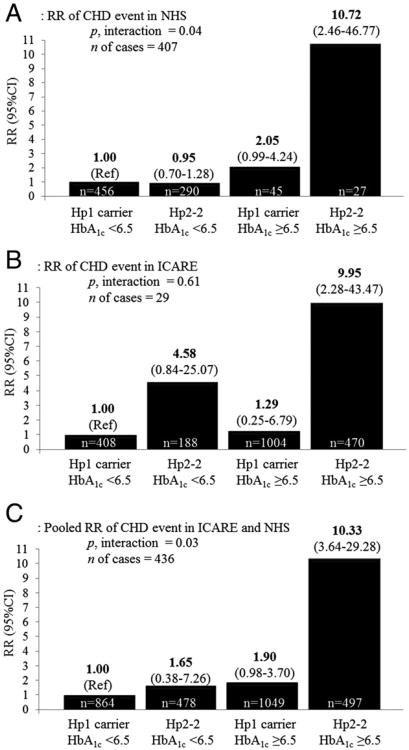
Figure 1A shows the joint effects of HbA1c level and Hp genotype. Among women with Hp2-2 genotype and elevated HbA1c, the RR of a CHD event was 10.72 (95% CI: 2.46 to 46.77) compared with participants with an Hp1 allele and HbA1c <6.5%. Compared with this same reference group, participants with the Hp2-2 genotype who had HbA1c <6.5% were not at increased risk of CHD (RR: 0.95 [95% CI: 0.70 to 1.28]), and participants with an Hp1 allele but with elevated HbA1c had a borderline increased risk (RR: 2.05 [95% CI: 0.99 to 4.24]).
Figure 1. Multivariate RR of CHD Events Figur According to Hp Genotype and HbA1cStatus.
(A) A nested case-control study of coronary heart disease (CHD) in women aged 44 to 69 years from the NHS (Nurses' Health Study) (1990–2004), adjusted for age, body mass index, smoking, alcohol, history of hypercholesterolemia, and history of hypertension. (B) Men and women with diabetes aged 22 to 95 years from the ICARE (Israel Cardiovascular Events Reduction With Vitamin E) study (2005–2006), adjusted for sex, age, smoking status, hypertension, myocardial infarction before enrollment, statin use, and metformin use. (C) ICARE and NHS pooled together. HbA1c = glycosylated hemoglobin; Hp = haptoglobin; RR = relative risk.
Replication in the ICARE Study
The distribution of Hp genotype frequencies was 13% (Hp1-1), 55% (Hp2-1), and 32% (Hp2-2) in the ICARE dataset, and the only baseline characteristics that differed among the genotypes were age and smoking (Table 3). Compared with participants with the Hp1-1 or Hp2-1 genotype, the multivariate RR of CHD for the Hp2-2 genotype was 6.76 (95% CI: 2.88 to 15.87) in this diabetic cohort. Although we had less power, we stratified by similar risk factors as we did for the NHS analyses (Table 4). Among participants <69 years of age (mean age) at baseline, the RR for Hp2-2 was 13.72 (95% CI: 3.06 to 61.54) compared with Hp1 allele carriers, whereas the corresponding RR for the older half of participants was 3.73 (95% CI: 1.27 to 11.01). Among diabetic patients not taking metformin, the RR for CHD was 15.84 (95% CI: 3.54 to 70.83) for the Hp2-2 genotype compared with the Hp1 allele carriers, and those taking metformin had an RR of 3.77 (95% CI: 1.26 to 11.30).
Table 3. Baseline Characteristics According to Hp Genotype Among Men and Women With Diabetes Aged 22 to 95 Years From the ICARE Study, 2005 to 2006*.
| Characteristic | Hp1-1 (n = 270) | Hp2-1 (n = 1,142) | Hp2-2 (n = 658) | p |
|---|---|---|---|---|
| Sex | 125 (46) | 535 (47) | 317 (48) | 0.82 |
| Age (yrs) | 68.9 ± 8.7 | 68.2 ± 8.8 | 69.6 ± 8.1 | 0.007 |
| Smoking (yes) | 16 (6) | 107 (9) | 84 (13) | 0.004 |
| Time since diabetes diagnosis (yrs) | 8.9 (8.1-9.7) | 8.4 (8.0-8.8) | 8.6 (8.1-9.1) | 0.52 |
| Hypertension | 199 (74) | 857 (75) | 496 (75) | 0.86 |
| Statin use | 200 (74) | 882 (77) | 509 (77) | 0.51 |
| Metformin use | 213 (79) | 885 (78) | 511 (78) | 0.88 |
| HbA1c (%) | 7.18 (7.04-7.33) | 7.24 (7.18-7.33) | 7.26 (7.16-7.35) | 0.67 |
| Total cholesterol (mg/dl) | 189 ± 33 | 186 ± 37 | 187 ± 34 | 0.50 |
| LDL cholesterol (mg/dl) | 103 ± 24 | 102 ± 29 | 103 ± 26 | 0.83 |
| HDL cholesterol (mg/dl) | 46 ± 11 | 46 ± 11 | 46 ± 11 | 0.61 |
| MI before study | 31 (11) | 158 (14) | 96 (15) | 0.46 |
| MI during follow-up | 3 (1) | 4 (1) | 17 (3) | <0.0001 |
| CHD death during follow-up | 0 (0) | 0 (0) | 5 (1) | 0.003 |
| Total CHD event† | 3 (1) | 4 (1) | 22 (3) | <0.0001 |
Values are n (%), mean ± SD, or geometric mean (95% CI).
Participant characteristics were compared between genotypes by using a general linear model for continuous variables and chi-square tests for categorical variables, unless there was a cell with n < 5, in which case the Fisher exact test was used. For skewed variables (time since diabetes diagnosis and HbA1c), p values from log-transformed analyses and geometric means with reverse-transformed 95% CIs are displayed.
Myocardial infarction (MI) or coronary heart disease (CHD) death during follow-up.
ICARE = Israel Cardiovascular Events Reduction With Vitamin E; other abbreviations as in Table 1.
Table 4. Multivariate RR* of CHD Event with 95% CIs for Hp Genotypes, Together and Stratified by CVD Risk Factors, in a Prospective Study of Diabetic Men and Women Aged 22 to 95 Years From the ICARE Study, 2005 to 2006.
| Hp1-1 + Hp2-1 | Hp2-2 | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Characteristic | Event/No Event | RR | Event/No Event | RR | 95% CI | p for Interaction |
| All participants | ||||||
| Unadjusted | 7/1405 | Ref. | 22/636 | 6.83 | 2.92-15.98 | — |
| Multivariate adjusted | 7/1405 | Ref. | 22/636 | 6.76 | 2.88-15.87 | — |
|
| ||||||
| Age at baseline | ||||||
| <69 yrs | 2/716 | Ref. | 12/289 | 13.72 | 3.06-61.54 | 0.16 |
| ≥69 yrs | 5/689 | Ref. | 10/347 | 3.73 | 1.27-11.01 | |
|
| ||||||
| Previous MI | ||||||
| No | 5/1218 | Ref. | 15/547 | 6.39 | 2.30-17.74 | 0.89 |
| Yes | 2/187 | Ref. | 7/89 | 8.23 | 1.68-40.46 | |
|
| ||||||
| Smoking | ||||||
| No | 5/1284 | Ref. | 17/557 | 7.64 | 2.81-20.75 | 0.57 |
| Yes | 2/121 | Ref. | 5/79 | 4.50 | 0.80-25.30 | |
|
| ||||||
| HbA1c | ||||||
| <6.5% | 2/406 | Ref. | 4/184 | 4.30 | 0.77-24.01 | 0.61 |
| ≥6.5% | 5/999 | Ref. | 18/452 | 7.55 | 2.79-20.47 | |
|
| ||||||
| Years with diabetes | ||||||
| ≤8 yrs | 5/791 | Ref. | 11/366 | 4.59 | 1.58-13.29 | 0.39 |
| >8 yrs | 2/614 | Ref. | 17/270 | 12.74 | 2.71-58.75 | |
|
| ||||||
| Metformin use | ||||||
| No | 2/312 | Ref. | 13/134 | 15.84 | 3.54-70.83 | 0.16 |
| Yes | 5/1093 | Ref. | 9/502 | 3.77 | 1.26-11.30 | |
To confirm the interaction between Hp genotype and glycemic control, we performed several additional analyses. Among participants with elevated HbA1c (≥6.5%), the RR for CHD was 7.55 (95% CI: 2.79 to 20.47) for Hp2-2 individuals compared with the Hp1 allele carriers, whereas no significant effect of Hp genotype was observed among participants with HbA1c <6.5% (p for interaction = 0.61). When grouped by the combination of Hp genotype and HbA1c status, the RR for CHD was 9.95 (95% CI: 2.28 to 43.47) among those with Hp2-2 genotype and HbA1c ≥6.5%, compared with participants with an Hp1 allele and HbA1c <6.5% (Fig. 1B). Compared with the same reference group, participants with an Hp1 allele and elevated HbA1c were not at significantly increased risk of CHD (RR: 1.29 [95% CI: 0.25 to 6.79]). No sex-based differences in results were present.
Pooled analysis
When the NHS and ICARE cohorts were pooled in a meta-analysis, participants with both the Hp2-2 genotype and high HbA1c had a pooled RR of 10.33 (95% CI: 3.64 to 29.28) compared with participants with both an Hp1 allele and HbA1c <6.5% (pooled p for interaction = 0.03) (Fig. 1C). We did not detect heterogeneity between the studies (p = 0.94).
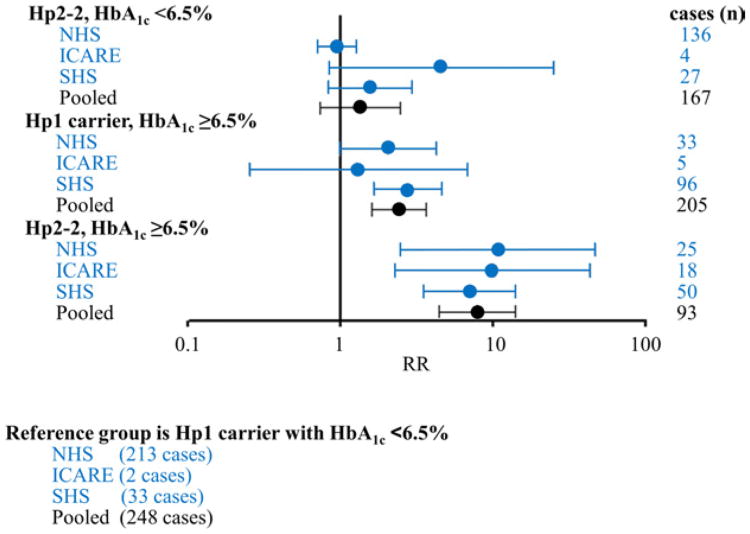
Based on the previously published SHS data (24), we calculated that in the SHS participants, those with both the Hp2-2 genotype and diabetes (as a proxy for elevated high HbA1c because in SHS participants specifically, a diabetes diagnosis has similar risk of CVD as the cutoff of 6.5% for HbA1c) had an RR of 7.01 (95% CI: 3.49 to 14.06) compared with participants with both an Hp1 allele and no diabetes (Fig. 2). When results from the NHS, ICARE, and SHS were combined, this same RR was 7.90 (95% CI: 4.43 to 14.10; heterogeneity p = 0.93, interaction p = 0.004).
Figure 2. RR of CHD Event According to Hp Genotype Figur and HbA1cStatus in Studies to Date.
In a nested case-control study of CHD in women from the NHS, in men and women with diabetes from the ICARE study, in a nested case-control study of cardiovascular disease (CVD) in men and women from the Strong Heart Study (SHS) in which diabetes status serves as a proxy for HbA1c ≥6.5%, and in al 3 studies pooled together (in the pooled analysis, the p for interaction = 0.004). The NHS analysis used logistic regression adjusted for age, body mass index, smoking, alcohol, history of hypercholesterolemia, and history of hypertension; the ICARE analysis used a proportional hazards model adjusted for sex, age, smoking status, hypertension, myocardial infarction before enrollment, statin use, and metformin use. The SHS analysis was unadjusted by using abstracted data (24). Abbreviations as in Figure 1.
Discussion
In 2 independent large prospective cohorts with a broad range of HbA1c concentrations, we found that individuals with the rs#72294371 Hp2-2 genotype and elevated HbA1c (≥6.5%) had a >10-fold increased risk of CHD compared with those with an Hp1 allele and HbA1c <6.5%. Participants with the Hp2-2 genotype and HbA1c <6.5% were not at increased risk of CHD, even in cases of clinically diagnosed diabetes, thus demonstrating that the underlying pathophysiological effects associated with the impaired function of the Hp2-2 protein are greatest among individuals with HbA1c ≥6.5%, regardless of diabetes status. Results from the previously reported SHS data support additional replication. To the best of our knowledge, no other studies have reported results examining Hp genotype, HbA1c, and risk of CHD.
Biological mechanism
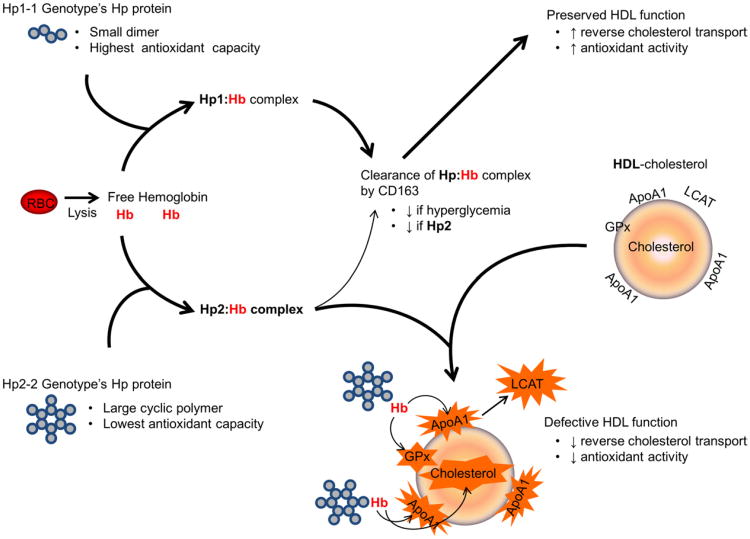
Hp functions to block oxidation by iron associated with hemoglobin (8), and striking differences between the Hp genotypes in their ability to perform this function in the setting of hyperglycemia have been demonstrated both in vitro and in vivo (Fig. 3). In vitro, this functional discrepancy is due to a decreased ability of the Hp2-2 protein to stabilize heme in the heme pocket of HbA1c (6,27). In vivo, both in hyperglycemic animal models and humans, those with the Hp2-2 genotype fail to efficiently clear the Hp2-2–hemoglobin complex via the monocyte/macrophage CD163 Hp–hemoglobin scavenger receptor, resulting in increased plasma redox active iron. Diabetic individuals with the Hp2-2 genotype may be at increased risk of CHD from this increase in plasma redox active Hp–hemoglobin complex if it binds to HDL levels and thus results in the oxidative modification and loss of function of the HDL in promoting reverse cholesterol transport. This hypothesis is supported by evidence demonstrating that the HDL levels of Hp2-2 diabetic individuals are extensively oxidized, and antioxidant therapy among Hp2-2 diabetic individuals and mice prevents HDL oxidative modification and restores HDL function (27). Hp genotype is linked to CHD by additional mechanisms that may be exacerbated by chronically high blood glucose. For example, Hp motivates microangiogenesis (28,29), and because the Hp1-1 genotype has the highest Hp concentrations (10,29), the Hp1 allele may delay onset of MI due to a better developed collateral circulation (18).
Figure 3. Proposed Biological Mechanism to Explain Increased Risk of CHD in Hyperglycemic Individuals With the Hp2-2 Genotype.
Hemoglobin (Hb) released intravascularly from erythrocytes (red blood cells [RBC]) is rapidly bound by haptoglobin (Hp) protein to form an Hp–Hb complex that is cleared by scavenger receptor CD163. However, this clearance by CD163 is impaired in Hp2 as well as under hyperglycemic conditions in vivo, resulting in increased amounts of circulating Hp2:Hb complex in Hp2-2 individuals with hyperglycemia. Moreover, we have shown that glycosylation of Hb impairs the ability of the Hp2-2 protein to act as an antioxidant, thus resulting in increased oxidative activity of the glycosylated Hp2:Hb complex. This pro-oxidant Hp2:Hb complex can bind to high-density lipoprotein (HDL) and produce reactive oxygen species that oxidize cholesterol and its related components such as apolipoprotein A (ApoA1), glutathione peroxidase (GPx), and lecithin-cholesterol acyltransferase (LCAT), thereby decreasing the function of HDL as both an antioxidant and in reverse cholesterol transport. The Hp2-1 protein is a linear polymer, intermediate in size and antioxidant capacity (2). CHD = coronary heart disease. Adapted from Asleh et al. (27).
In the NHS, Hp genotype was associated with LDL cholesterol concentrations. Some previous studies (30), although not all (31), have found a similar association between Hp genotype and cholesterol concentrations. We did not observe an association in the ICARE analysis, but this cohort was limited to diabetic individuals only, the majority of who were receiving cholesterol-lowering medication. If an association between Hp genotype and LDL cholesterol exists, it may be due to linkage disequilibrium because the Hp gene is located in close proximity to the lecithin-cholesterol acyltransferase and cholesteryl ester transfer protein genes (32). However, in that case, we would expect to detect a difference in HDL concentrations among the Hp genotypes, which we did not observe.
Implications
Large randomized controlled trials of intensive glycemic control therapy among diabetic individuals have found significant reduction in cardiovascular outcomes in some (11), but not all (12,33), cohorts. This inconsistency may be due in part to different unknown characteristics among patient subgroups that have not yet been explored, as suggested in a meta-analysis of such trials by Ray et al. (34). The Hp genotype frequencies differ among populations (35) and may potentially explain differences in efficacy of glycemic control reported in the literature. Replication in a randomized trial of glycemic control, such as the ACCORD (Action to Control Cardiovascular Risk in Diabetes) study or the DCCT (Diabetes Control and Complications Trial), is required to confirm this finding. If replicated, Hp genotyping could potentially assist in identifying genetically susceptible individuals who would most benefit from clinical management of HbA1c in the prevention of CHD.
Study strengths and limitations
Strengths of the CURRENT analysis include comprehensive data gathered prospectively with a long duration of follow-up, replication in a second cohort, and a validated Hp genotyping method. The availability of results from a third and previously published study extends the validity of the analysis, although further confirmation in larger cohorts is warranted, especially because the ICARE and SHS cohorts had a limited number of incident CHD cases, and in the pooled analysis in the current study, the majority of cases were derived from the NHS. The RR calculated for the SHS was not multivariate adjusted. However, there is little confounding and attenuation of the multivariate RR in the NHS and ICARE analyses; therefore, the crude RR for the SHS was likely minimally biased by confounding factors. The current study may have been underpowered to detect interactions between Hp genotype and CHD risk factors. Furthermore, for biomarkers assessed in stored plasma, such as HbA1c, we only had a single measurement at baseline. Thus, random error caused by normal fluctuations over time would cause underestimation of true RRs. However, HbA1c reflects glycemic control over an average of the 90-day red blood cell lifespan and is less susceptible to daily changes. We found intraclass correlations of 0.73 for repeated HbA1c samples measured 3 years apart, which suggests good within-person reproducibility.
Conclusions
In 2 independent cohorts, we observed that participants with both the Hp2-2 genotype and an HbA1c ≥6.5% had a 10-fold increased risk of CHD compared with those with at least 1 Hp1 allele and HbA1c <6.5%. This finding, if further replicated in studies such as ACCORD or DCCT, may help explain inconsistencies in the literature for the association of glycemic control with CVD outcomes, as well as inconsistencies in previous studies of Hp genotype and CHD. If replicated, Hp genotyping could potentially assist in targeting cost-efficient clinical management of diabetes among the most genetically sensitive individuals.
Supplementary Material
Acknowledgments
The authors sincerely thank Dr. Mary Townsend for her work running the quality control of the biomarkers, as well as the staff and participants of the 3 cohorts studied in this paper.
This analysis was supported by National Institutes of Health grants CA87969, CA49449, HL34594, DK085226, and TR000101 and by a Canadian Institutes of Health Research (Ottawa, Ontario) Postdoctoral Fellowship to Dr. Cahill. Dr. Levy is the author of a patent owned by his university regarding use of haptoglobin genotypes to predict susceptibility to cardiovascular disease in individuals with diabetes. Dr. Levy also presently serves as a scientific consultant for Haptocure, which has licensed the aforementioned patent from his university. Dr. Blum is a cofounder and CEO of Haptocure.
Abbreviations and Acronyms
- CHD
coronary heart disease
- CI
confidence interval
- CVD
cardiovascular disease
- HbA1c
glycosylated hemoglobin
- HDL
high-density lipoprotein
- Hp
haptoglobin
- LDL
low-density lipoprotein
- MI
myocardial infarction
- RR
relative risk
Footnotes
All other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Appendix: For a supplemental table, please see the online version of this article.
References
- 1.Gutteridge JM. The antioxidant activity of haptoglobin towards haemoglobin-stimulated lipid peroxidation. Biochim Biophys Acta. 1987;917:219–23. doi: 10.1016/0005-2760(87)90125-1. [DOI] [PubMed] [Google Scholar]
- 2.Levy AP, Asleh R, Blum S, et al. Haptoglobin: basic and clinical aspects. Antioxid Redox Signal. 2010;12:293–304. doi: 10.1089/ars.2009.2793. [DOI] [PubMed] [Google Scholar]
- 3.Langlois MR, Delanghe JR. Biological and clinical significance of haptoglobin polymorphism in humans. Clin Chem. 1996;42:1589–600. [PubMed] [Google Scholar]
- 4.Levy AP, Levy JE, Kalet-Litman S, et al. Haptoglobin genotype is a determinant of iron, lipid peroxidation, and macrophage accumulation in the atherosclerotic plaque. Arterioscler Thromb Vasc Biol. 2007;27:134–40. doi: 10.1161/01.ATV.0000251020.24399.a2. [DOI] [PubMed] [Google Scholar]
- 5.Melamed-Frank M, Lache O, Enav BI, et al. Structure-function analysis of the antioxidant properties of haptoglobin. Blood. 2001;98:3693–8. doi: 10.1182/blood.v98.13.3693. [DOI] [PubMed] [Google Scholar]
- 6.Asleh R, Marsh S, Shilkrut M, et al. Genetically determined heterogeneity in hemoglobin scavenging and susceptibility to diabetic cardiovascular disease. Circ Res. 2003;92:1193–200. doi: 10.1161/01.RES.0000076889.23082.F1. [DOI] [PubMed] [Google Scholar]
- 7.Guetta J, Strauss M, Levy NS, Fahoum L, Levy AP. Haptoglobin genotype modulates the balance of Th1/Th2 cytokines produced by macrophages exposed to free hemoglobin. Atherosclerosis. 2007;191:48–53. doi: 10.1016/j.atherosclerosis.2006.04.032. [DOI] [PubMed] [Google Scholar]
- 8.Asleh R, Guetta J, Kalet-Litman S, Miller-Lotan R, Levy AP. Haptoglobin genotype- and diabetes-dependent differences in iron-mediated oxidative stress in vitro and in vivo. Circ Res. 2005;96:435–41. doi: 10.1161/01.RES.0000156653.05853.b9. [DOI] [PubMed] [Google Scholar]
- 9.Levy AP, Larson MG, Corey D, Lotan R, Vita JA, Benjamin EJ. Haptoglobin phenotype and prevalent coronary heart disease in the Framingham offspring cohort. Atherosclerosis. 2004;172:361–5. doi: 10.1016/j.atherosclerosis.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 10.De Bacquer D, De Backer G, Langlois M, Delanghe J, Kesteloot H, Kornitzer M. Haptoglobin polymorphism as a risk factor for coronary heart disease mortality. Atherosclerosis. 2001;157:161–6. doi: 10.1016/s0021-9150(00)00690-0. [DOI] [PubMed] [Google Scholar]
- 11.Patel A, MacMahon S, Chalmers J, et al. Intensive blood glucose control and vascular outcomes in patients with type 2 diabetes. N Engl J Med. 2008;358:2560–72. doi: 10.1056/NEJMoa0802987. [DOI] [PubMed] [Google Scholar]
- 12.Gerstein HC, Miller ME, Genuth S, et al. Long-term effects of intensive glucose lowering on cardiovascular outcomes. N Engl J Med. 2011;364:818–28. doi: 10.1056/NEJMoa1006524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cassidy A, Chiuve SE, Manson JE, Rexrode KM, Girman CJ, Rimm EB. Potential role for plasma placental growth factor in predicting coronary heart disease risk in women. Arterioscler Thromb Vasc Biol. 2009;29:134–9. doi: 10.1161/ATVBAHA.108.171066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colditz GA, Hankinson SE. The Nurses' Health Study: lifestyle and health among women. Nature reviews. 2005;5:388–96. doi: 10.1038/nrc1608. [DOI] [PubMed] [Google Scholar]
- 15.Willett WC, Stampfer MJ, Colditz GA, Rosner BA, Hennekens CH, Speizer FE. Dietary fat and the risk of breast cancer. N Engl J Med. 1987;316:22–8. doi: 10.1056/NEJM198701013160105. [DOI] [PubMed] [Google Scholar]
- 16.Milman U, Blum S, Shapira C, et al. Vitamin E supplementation reduces cardiovascular events in a subgroup of middle-aged individuals with both type 2 diabetes mellitus and the haptoglobin 2-2 genotype: a prospective double-blinded clinical trial. Arterioscler Thromb Vasc Biol. 2008;28:341–7. doi: 10.1161/ATVBAHA.107.153965. [DOI] [PubMed] [Google Scholar]
- 17.Database of Single Nucleotide Polymorphisms (dbSNP) dbSNP accession:{ss1 or ss1 - ss100}, (dbSNP Build ID: {build ID}) Bethesda, MD: National Center for Biotechnology Information, National Library of Medicine; [Accessed May 15, 2012]. Available at: http://www.ncbi.nlm.nih.gov/SNP/ [Google Scholar]
- 18.Hochberg I, Roguin A, Nikolsky E, Chanderashekhar PV, Cohen S, Levy AP. Haptoglobin phenotype and coronary artery collaterals in diabetic patients. Atherosclerosis. 2002;161:441–6. doi: 10.1016/s0021-9150(01)00657-8. [DOI] [PubMed] [Google Scholar]
- 19.Koch W, Latz W, Eichinger M, et al. Genotyping of the common haptoglobin Hp 1/2 polymorphism based on PCR. Clin Chem. 2002;48:1377–82. [PubMed] [Google Scholar]
- 20.Pai JK, Pischon T, Ma J, et al. Inflammatory markers and the risk of coronary heart disease in men and women. N Engl J Med. 2004;351:2599–610. doi: 10.1056/NEJMoa040967. [DOI] [PubMed] [Google Scholar]
- 21.Rose GA, Blackburn H. monograph series no 58. 2nd. Geneva, Switzerland: World Health Organization; 1982. Cardiovascular Survey Methods. [PubMed] [Google Scholar]
- 22.International Expert Committee report on the role of the A1C assay in the diagnosis of diabetes. Diabetes Care. 2009;32:1327–34. doi: 10.2337/dc09-9033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–88. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 24.Levy AP, Hochberg I, Jablonski K, et al. Haptoglobin phenotype is an independent risk factor for cardiovascular disease in individuals with diabetes: the Strong Heart Study. J Am Coll Cardiol. 2002;40:1984–90. doi: 10.1016/s0735-1097(02)02534-2. [DOI] [PubMed] [Google Scholar]
- 25.Wang H, Shara NM, Lee ET, et al. Hemoglobin A1c, fasting glucose, and cardiovascular risk in a population with high prevalence of diabetes: the Strong Heart Study. Diabetes Care. 2011;34:1952–8. doi: 10.2337/dc11-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jensen MK, Mukamal KJ, Overvad K, Rimm EB. Alcohol consumption, TaqIB polymorphism of cholesteryl ester transfer protein, high-density lipoprotein cholesterol, and risk of coronary heart disease in men and women. Eur Heart J. 2008;29:104–12. doi: 10.1093/eurheartj/ehm517. [DOI] [PubMed] [Google Scholar]
- 27.Asleh R, Blum S, Kalet-Litman S, et al. Correction of HDL dysfunction in individuals with diabetes and the haptoglobin 2-2 genotype. Diabetes. 2008;57:2794–800. doi: 10.2337/db08-0450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cid MC, Grant DS, Hoffman GS, Auerbach R, Fauci AS, Kleinman HK. Identification of haptoglobin as an angiogenic factor in sera from patients with systemic vasculitis. J Clin Invest. 1993;91:977–85. doi: 10.1172/JCI116319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chapelle JP, Albert A, Smeets JP, Heusghem C, Kulbertus HE. Effect of the haptoglobin phenotype on the size of a myocardial infarct. N Engl J Med. 1982;307:457–63. doi: 10.1056/NEJM198208193070801. [DOI] [PubMed] [Google Scholar]
- 30.Braeckman L, De Bacquer D, Delanghe J, Claeys L, De Backer G. Associations between haptoglobin polymorphism, lipids, lipoproteins and inflammatory variables. Atherosclerosis. 1999;143:383–8. doi: 10.1016/s0021-9150(98)00330-x. [DOI] [PubMed] [Google Scholar]
- 31.Delanghe J, Langlois M, Duprez D, De Buyzere M, Clement D. Haptoglobin polymorphism and peripheral arterial occlusive disease. Atherosclerosis. 1999;145:287–92. doi: 10.1016/s0021-9150(99)00079-9. [DOI] [PubMed] [Google Scholar]
- 32.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez gene: gene-centered information at NCBI. Nucleic Acids Res. 2011;39:D52–57. doi: 10.1093/nar/gkq1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duckworth W, Abraira C, Moritz T, et al. Glucose control and vascular complications in veterans with type 2 diabetes. N Engl J Med. 2009;360:129–39. doi: 10.1056/NEJMoa0808431. [DOI] [PubMed] [Google Scholar]
- 34.Ray KK, Seshasai SR, Wijesuriya S, et al. Effect of intensive control of glucose on cardiovascular outcomes and death in patients with diabetes mellitus: a meta-analysis of randomised controlled trials. Lancet. 2009;373:1765–72. doi: 10.1016/S0140-6736(09)60697-8. [DOI] [PubMed] [Google Scholar]
- 35.Carter K, Worwood M. Haptoglobin: a review of the major allele frequencies worldwide and their association with diseases. Int J Lab Hematol. 2007;29:92–110. doi: 10.1111/j.1751-553X.2007.00898.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.