Summary
Genome duality in ciliated protozoa offers a unique system to showcase their epigenome as a model of inheritance. In Oxytricha, the somatic genome is responsible for vegetative growth, while the germline contributes DNA to the next sexual generation. Somatic nuclear development removes all transposons and other so-called “junk DNA”, which comprise ~95% of the germline. We demonstrate that Piwi-interacting small RNAs (piRNAs) from the maternal nucleus can specify genomic regions for retention in this process. Oxytricha piRNAs map primarily to the somatic genome, representing the ~5% of the germline that is retained. Furthermore, injection of synthetic piRNAs corresponding to normally-deleted regions leads to their retention in later generations. Our findings highlight small RNAs (sRNAs) as powerful transgenerational carriers of epigenetic information for genome programming.
Introduction
Oxytricha trifallax harbors two types of nuclei within its single cell: a somatic macronucleus and a germline micronucleus. In each sexual conjugation cycle, the old somatic nucleus disintegrates and a new soma develops from the germline. The germline genome contains large quantities of so-called “junk”, including transposons, satellite repeats, intergenic sequences, and internally eliminated sequences [IESs], all of which undergo programmed deletion during somatic nuclear development. This compresses the ~1Gb germline genome to a gene-dense somatic genome of only ~50Mb. Severe chromosome fragmentation necessitates events that fuse and reorder the tens of thousands of gene pieces remaining [macronuclear-destined sequences, MDSs]. Lastly, telomere addition and amplification produce mature macronuclear “nanochromosomes”, typically ~ 3 kb bearing just one gene (Prescott, 2000; Swart et al., in revision).
The precise and reproducible DNA rearrangements in Oxytricha, as well as other ciliates, show a prominent pattern of maternal inheritance (Chalker and Yao, 1996; Duharcourt et al., 1995) mediated by non-coding RNAs (Lepere et al., 2008; Mochizuki et al., 2002; Nowacki et al., 2008; Yao et al., 2003). In the distantly related ciliates Tetrahymena and Paramecium, a class of small RNAs (sRNAs) called scanRNAs (scnRNAs) derive from the germline and scan the macronuclear genome or transcriptome for sequence identity, leaving sRNAs that lack matches in the macronucleus (and thus are micronuclear-limited) to target germline-specific DNA for deletion during nuclear development (Lepere et al., 2008; Mochizuki et al., 2002). We previously demonstrated that long non-coding RNA templates from the maternal somatic nucleus guide whole-genome reorganization in Oxytricha (Nowacki et al., 2008). However, abundant ~27 nucleotide (nt) sRNAs also accumulate during conjugation (Figure 1A, B), suggesting a role for sRNA pathways during Oxytricha genome rearrangement, which we investigate here.
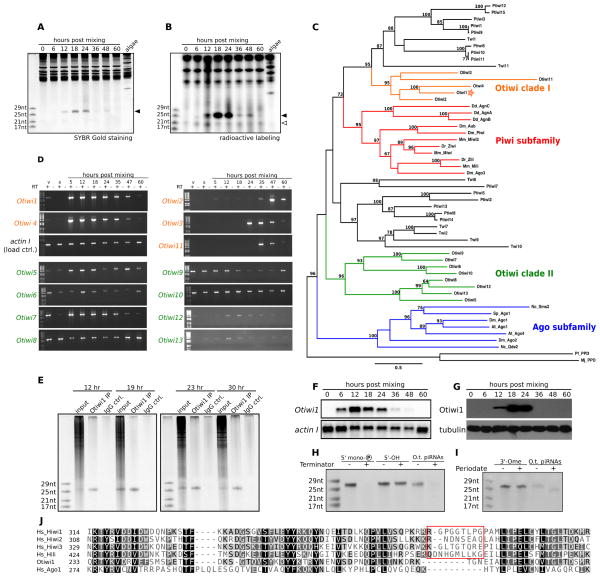
Figure 1. 27 nt sRNAs and Otiwi1 co-express and interact during Oxytrichaconjugation.
(A) 27 nt sRNAs (filled arrowhead) are abundant in early conjugation. Total RNA extracted at different time points during conjugation was separated on a denaturing polyacrylamide gel and stained with SYBR gold. Chlamydomonas (food algae) RNA is also loaded as a control. (B) A class of ~21 nt sRNAs (open arrowhead) is present in weaker abundance in both vegetative and conjugating Oxytricha cells. (C) Phylogenetic analysis of Piwi proteins suggests that Otiwi proteins form two major clades. Archaeal Argonaute proteins were used as outgroups for eukaryotic proteins. Otiwi clade I (Otiwi1–4 and 11) members are labeled orange, and Piwi subfamily members from animals and Dictyostelium are labeled red. Otiwi1, which associates with 27 nt sRNAs in Oxytricha, is indicated by a star. Green: Otiwi clade II (Otiwi5–10, 12, and 13), whose precise phylogeny is not resolved with high confidence (low bootstrap value on relevant nodes); blue: Ago subfamily of Argonaute proteins from plants, fungi and animals. Bootstrap values greater than 60% are shown. Abbreviations: At: Arabidopsis thaliana; Dd: Dictyostelium discodium; Dm: Drosophila melanogaster; Dr: Danio rerio; Mj: Methanocaldococcus jannaschii; Mm: Mus musculus; Nc: Neurospora crassa; Pf: Pyrococcus furiosus; Sp: Schizosaccaromyces pombe. Twi and Ptiwi are Argonaute proteins from Tetrahymena thermophila and Paramecium tetraurelia, respectively. The scale bar represents 0.5 substitution per site (inferred by LG model). (D) Otiwi genes differ in expression profile across Oxytricha’s life cycle. The orange labels indicate Otiwi clade I that clusters with the Piwi subfamily in (C), and these Otiwi genes are strongly induced during conjugation (post mixing). The green labels indicate Otiwi clade II, whose mRNA expression is more uniform. Actin IRT -PCR is used as a loading control. V, vegetative cells; s, starved cells; RT, reverse transcription. (E) Otiwi1 immunoprecipitation enriches for 27 nt sRNAs from total RNA at 12, 19, 23, and 30 hr post mixing of two mating types. (F) Northern analysis of Otiwi1 expression from the conjugation time series shown in (A) and (B). (G) Western analysis of Otiwi1 expression from the conjugation time series shown in (A) and (B). (H) Terminator treatment indicates that Oxytricha piRNAs (O.t. piRNAs) contain a 5′ monophosphate. Control oligonucleotides are 27 nt synthetic RNAs with either a 5′ monophosphate or a 5′-OH. (I) Periodate oxidation followed by β-elimination suggests that Oxytricha piRNAs (O.t. piRNAs) lack 3′ end modification. The control oligonucleotide is a 27 nt synthetic RNA with 2′-O methylation at the 3′ end. (J) An alignment of Otiwi1 with human Piwi and Ago1 proteins suggests that Otiwi1 lacks a Piwi-specific insertion (red box) predicted to help accommodate the 2′-O-methylated 3′ ends of mammalian piRNAs. See also Figure S1 and Table S1.
Small RNAs, in association with an Argonaute/Piwi family protein, can act as sequence-specific guides to regulate gene expression and chromatin structure (Bartel, 2004; Zamore and Haley, 2005). The subclass of Piwi-interacting RNAs (piRNAs) is abundant in the animal germline, and its best-understood function is transposon silencing (Brennecke et al., 2007; Carmell et al., 2007; Grimson et al., 2008; Houwing et al., 2007; Malone and Hannon, 2009; Saito et al., 2006; Vagin et al., 2006). In Drosophila, maternally-deposited piRNAs can prime piRNA production and transposon immunity in daughters, underscoring a role for piRNAs as epigenetic information carriers (Brennecke et al., 2008). Much less is known about piRNAs that do not map to transposon or repetitive DNA, despite the fact that such piRNAs can be overrepresented in mammalian testis, for example (Aravin et al., 2006; Girard et al., 2006; Grivna et al., 2006; Lau et al., 2006; Watanabe et al., 2006). Deep sequencing revealed piRNAs that map to both 3′ UTR and coding regions of unique sets of transcripts, suggesting a role in gene expression regulation (Gan et al., 2011; Robine et al., 2009). However, functional evidence is lacking, except for case studies (Rajasethupathy et al., 2012; Rouget et al., 2010; Saito et al., 2009). A recent study in plant and human cells uncovered a surprising role for sRNAs in double-stranded DNA break repair, relating sRNA function to genome integrity (Wei et al., 2012). Here, by studying Oxytricha, a microbial eukaryote that serves as a paragon for investigations of genome rearrangement, we suggest a genome-wide protective role for piRNAs that map to genic regions. We show that Oxytricha piRNAs derive from the somatic genome, and localization of the associated Piwi protein shifts from the maternal to the developing somatic nucleus during genome rearrangement. Furthermore, injection of sRNAs targeting normally-deleted regions leads to their retention in the sexual offspring across multiple generations, demonstrating a role for these piRNAs in transgenerational epigenetic inheritance.
Results
27 nt sRNAs and Otiwi1 co-express and interact during early conjugation
A time-course examination of total cellular RNA identifies an abundant class of ~27 nt sRNAs exclusively expressed during early conjugation (Figure 1A). A much less abundant class of ~21 nt sRNAs are also detectable by radioactive labeling (Figure 1B), but present in both vegetative and conjugating cells (Figure 1B) and could be siRNAs involved in gene silencing, like the 23–24 nt siRNAs in Tetrahymena (Lee and Collins, 2006) and Paramecium (Lepere et al., 2009). Here, we focus on the development-specific 27 nt sRNAs. To probe the function of these sRNAs, we first analyzed Argonaute proteins in Oxytricha. Using Tetrahymena Twi1p protein as a query, we retrieved thirteen Oxytricha homologs (Otiwi1–13) from the macronuclear genome assembly (Swart et al., in revision). This number of Argonaute proteins is comparable to that in Tetrahymena (Couvillion et al., 2009) and Paramecium (Bouhouche et al., 2011), but higher than most metazoa, reflecting the richness of sRNA-based regulation in single-celled ciliates. The thirteen Oxytricha homologs form two clades (Figure 1C). Clade I (Otiwi1–4 and Otiwi11) groups more closely with Piwi subfamily proteins from metazoa and Dictyostelium, and their mRNA show elevated expression levels during conjugation, based on RT-PCR analysis (Figure 1D). In addition, expression levels of Otiwi1 and Otiwi4 are higher in early conjugation, while Otiwi2, 3, and 11 are more restricted to late conjugation (Figure 1D). Expression of the Clade II Otiwi mRNAs, by contrast, is relatively constitutive across vegetative growth and conjugation (Figure 1D).
Use of an antibody that based on mass spectrometry primarily recognizes Otiwi1 (Figure S1, Table S1) confirmed that the Otiwi1 protein interacts with the ~27 nt sRNAs throughout early conjugation (Figure 1E). Based on this interaction, we denote Otiwi1-interacting sRNAs as a class of piRNAs in Oxytricha. Northern analysis suggests that Otiwi1 mRNA abundance peaks 12 hr after mixing of mating types and then decreases as conjugation proceeds (Figure 1F). In western analysis, Otiwi1 protein levels become detectable at 12 hr, and then peak at 18–24 hr post mixing (Figure 1G), which fits well with the 27 nt piRNA expression pattern (Figure 1A, B).
Oxytricha piRNAs contain 5′ monophosphate but no 3′ end modification
The ends of sRNAs differ among classes. Most microRNAs, siRNAs and piRNAs contain a 5′ monophosphate, but secondary siRNAs in C. elegans (Pak and Fire, 2007; Sijen et al., 2007) and 27 nt sRNAs in Entamoeba histolytica (Zhang et al., 2008)are 5′ polyphosphorylated. At the 3′ end, plant microRNAs (Li et al., 2005), animal siRNAs (Ameres et al., 2010) and piRNAs (Houwing et al., 2007; Kirino and Mourelatos, 2007; Ohara et al., 2007; Saito et al., 2007) can be 2′-O-methylated, but animal microRNAs are not.
We therefore characterized end modifications of the Otiwi1-bound piRNAs in Oxytricha. Terminator exonuclease is specific to RNAs with a 5′ monophosphate. Oxytricha piRNAs are sensitive to Terminator nuclease digestion, suggesting that they contain a 5′ monophosphate (Figure 1H). β-elimination post periodate treatment shortens RNAs without 3′ end modifications by one nucleotide, whereas RNAs with 3′ end modifications are resistant to β-elimination and thus remain the same size. Oxytricha piRNAs are not resistant to periodate oxidation, indicating a lack of 3′ end modifications (Figure 1I). This is surprising because the stabilizing 3′ end methylation is conserved in animal piRNAs (Houwing et al., 2007; Kirino and Mourelatos, 2007; Ohara et al., 2007; Saito et al., 2007) as well as Tetrahymena scnRNAs (Kurth and Mochizuki, 2009). It is possible that Oxytricha piRNAs represent either a more ancestral state prior to the addition of 3′ end modification, or that the end modification was lost in this lineage.
Structural studies of human Piwi PAZ domains suggest that a Piwi-specific insertion, not present in human Ago1, may facilitate formation of a larger binding pocket for the 3′-end of RNA, permitting the Piwi PAZ domain to accommodate the 3′-end methyl group on a piRNA (Tian et al., 2011). An alignment of Otiwi1 with human Piwi and Ago1 proteins suggests that Otiwi1 lacks this insertion (Figure 1J), consistent with the lack of 3′-methylation on Oxytricha 27 nt piRNAs.
Otiwi1 localization shifts from parental to developing macronucleus
If piRNAs are important for Oxytricha genome rearrangement, we would expect to find the Otiwi1-piRNA complex in the developing macronucleus. Immunofluorescence suggests that Otiwi1 is absent from early conjugating pairs, when micronuclei begin meiosis (Figure 2A, see also Figure S2A for details of the Oxytricha sexual cycle), but Otiwi1 protein starts to accumulate in the maternal macronucleus in mid-to-late pairing stage (Figure 2B). Then Otiwi1 transiently localizes in both the cytoplasm and the developing macronucleus after fertilization (Figure 2C). Otiwi1 is abundant in the new somatic nucleus after pair separation (Figure 2D) and then its abundance decreases during late development (Figure 2E). This dynamic localization suggests that the Otiwi1-piRNA complex might transport piRNAs from the maternal nucleus to the developing nucleus where genome rearrangement occurs. In addition, a peptide competition assay using Otiwi1 and Otiwi4 C-terminal peptides further confirms the antibody’s specificity for Otiwi1 (Figure S2B).
Figure 2. Otiwi1 localization shifts from old macronucleus to the developing macronucleus.
Otiwi1 is absent at the initial pairing stage when the micronucleus (MIC) begins meiosis (A), but accumulates in the old macronucleus (MAC) during mid-to-late pair formation (B). Otiwi1 is transiently present in both the cytoplasm and the developing macronucleus (called anlagen, AN) right after its formation (C), and localizes only to the anlagen in early exconjugants (D). (E) Otiwi1 signal decreases dramatically in late anlagen development, and the parental MAC is degrading. Arrowhead in (A) indicates a MIC undergoing meiosis and stained with α-tubulin. Green, Otiwi1; red, 4′, 6-diamidino-2-phenylindole (DAPI); white, α-tubulin. See also Figure S2.
Otiwi1 is essential for the accumulation of 27 nt piRNAs and the viability of sexual progeny
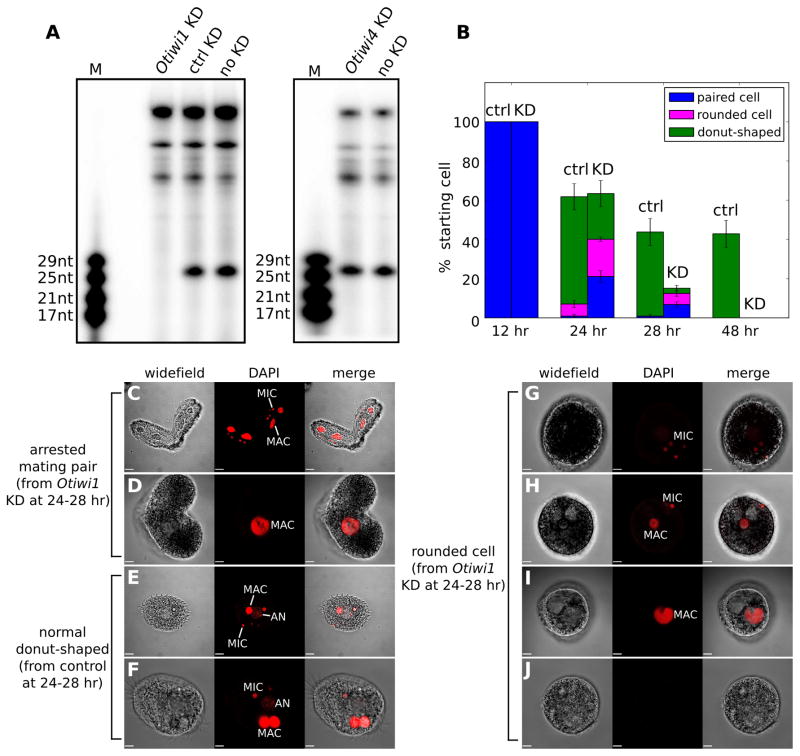
To test if Otiwi1 is essential for new macronuclear development, we injected antisense phosphorothioate-backbone DNA oligonucleotides into cells 2–4 hr post-mixing of mating types. This significantly reduced Otiwi1 mRNA and protein levels compared to control-oligonucleotide injections (Figure S3). The loss of protein signal after Otiwi1 knockdown also confirms the antibody’s specificity. Notably, knockdown of Otiwi1, but not Otiwi4, results in depletion of the 27 nt piRNAs at 18 hr (Figure 3A), suggesting that Otiwi1 is the main interacting partner of this class of Oxytricha sRNAs. It is possible that piRNAs are not stable in the absence of Otiwi1, or alternatively, that the 27 nt piRNAs require Otiwi1 for their biogenesis.
Figure 3. Otiwi1 knockdown (KD) leads to a depletion of 27 nt piRNAs and stalled nuclear development.
(A) 27 nt piRNAs are lost upon Otiwi1 but not Otiwi4 KD. Otiwi1 or Otiwi4 antisense, or control oligonucleotides were injected at 2–4 hr post-mixing, and total RNA was extracted 18 hr post-mixing for radioactive labeling and detection.(B) Cells were arrested at pair stage or shortly after pair separation upon Otiwi1 KD, and died between 24 to 48 hr post-mixing. The quantification of survived cells was based on four independent experiments injecting 20–28 pairs each, for both KD and control. We calculate the percentage of cells based on the starting number of cells, not the population at indicated time points. Error bars indicate standard error of the mean. (C–J) Representative microscopy images of control and KD cells at 24–28 hr post-mixing. Arrested pairs show either nuclear morphology resembling wild-type cells at 12h (C), or degenerated nuclei (D). (E, F) Most control cells show normal donut-shaped morphology, where the round, light-colored “hole” with faint DAPI staining is the developing nucleus, or anlagen (AN). (G–J) Significantly more Otiwi1 KD cells show abnormal spherical structures in which any one or all of the macronucleus (MAC), micronucleus (MIC) and developing macronulceus (AN) are missing. Scale bars are 14 μm in (C) and (E), and 7 μm in (D, F, G-J). See also Figure S3.
Otiwi1 knockdown cells appear normal at 12 hr post-mixing, but at 24 hr, 21% were arrested at pair stage, while only 1.8% were still pairs in control populations (Figure 3B–D). At 24 hr, those that appeared to be morphologically normal exconjugants (donut-shaped) were less active compared to control cells, and died between 24–48 hr post-mixing, with a dramatic decrease in survival between 24–28 hr (Figure 3B). We also observed significantly more (p≤ 0.001) cells with rounded and less transparent morphology (Figure 3G–J). These cells also show degeneration of the developing macronucleus, the old macronucleus and the micronucleus (Figure 3G–J), and eventually disintegrate. It is possible that these spherical structures might be an intermediate stage where cells degrade their nuclei before cell death. We conclude that Otiwi1 is essential for exconjugant survival and new macronuclear development.
Oxytricha piRNAs map primarily to the somatic genome
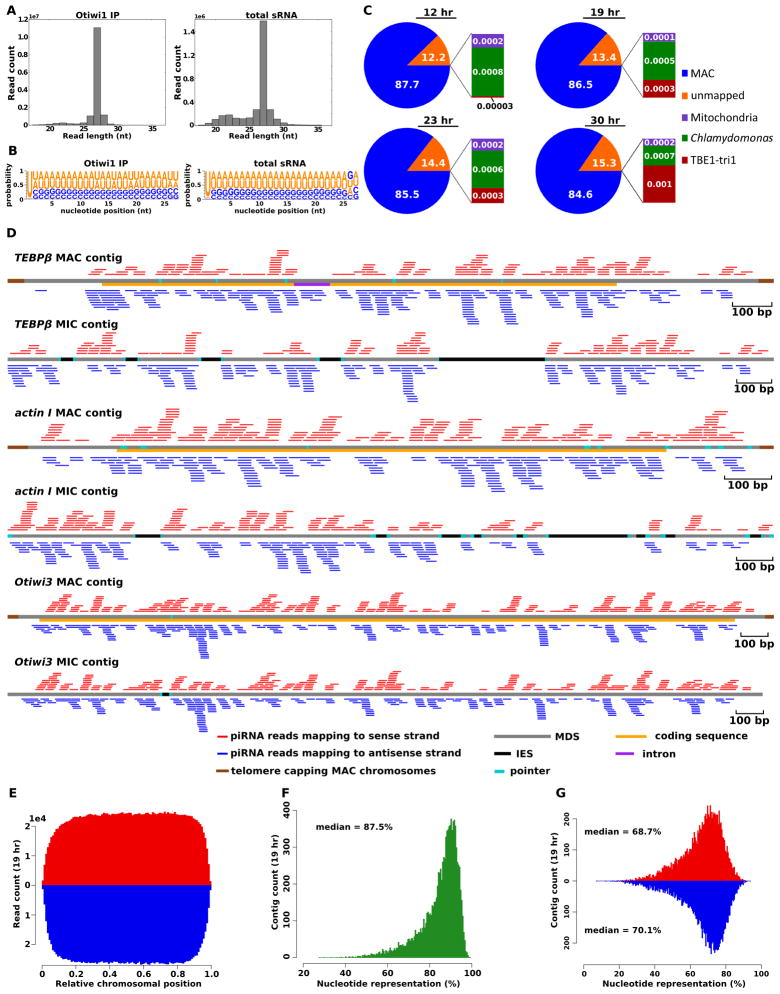
To identify the genomic source of piRNAs in Oxytricha, we deep sequenced sRNAs captured by Otiwi1 co-immunoprecipitation at 12, 19, 23, and 30 hr after mixing of mating types, early conjugation stages when both Otiwi1 and piRNAs are expressed. Otiwi1-bound piRNAs are predominantly 27 nt (Figure 4A) and have a strong 5′-uridine (U) bias (Figure 4B). Sequencing of total sRNA at ~20 hr post-mixing of mating types suggests that the 27 nt 5′-U piRNAs are the major class of sRNAs during conjugation (Figure 4A, B), consistent with the RNA characterization in Figures 1A and B. Total sRNA also displays a minor peak of 21–22 nt (Figure 4A), which reflects the less abundant sRNA class detected by radioactive labeling (Figure 1B). Recently, Zahler et al. (2012) also deep sequenced total sRNA from vegetative and conjugating Oxytricha cells, which provides additional datasets for analysis.
Figure 4. Most Oxytricha piRNAs map to sequences that are retained during genome rearrangement.
(A) The length distribution of Oxytricha piRNAs suggests that they are predominantly 27 nt. Otiwi1 immunoprecipitated sRNA at 19 hr and total sRNA at 20 hr are shown. 19 and 20 hr conjugation populations do not differ significantly regarding staging. (B) Sequence profile of Oxytricha 27 nt sRNA reads from either Otiwi1 immunoprecipitation at 19 hr or total sRNA at 20 hr. (C) Mapping statistics of 27 nt, 5′-U Otiwi1-associated piRNAs at 12, 19, 23, and 30 hr post-mixing. Reads mapping to the Oxytricha mitochondria or Chlamydomonas (food algae) genomes serve as negative controls. (D) Mapping of 27 nt, 5′-U piRNAs to macronuclear and micronuclear loci for TEBPβ, actin I (both 19 hr) and Otiwi3 (12 hr). (E) Metagene analysis (N = 10,497) suggests that piRNAs distribute across the entire macronuclear chromosomes. For each mapping, the 13 nt position of a piRNA read is normalized against the length of the corresponding single-gene macronuclear chromosome, with red indicating the mRNA sense strand for protein-coding genes and blue indicating antisense strand. (F) Genome-wide nucleotide representation by piRNA reads suggests that the whole macronuclear genome is converted into piRNAs. For each macronuclear chromosome surveyed, the nucleotide coverage is calculated as piRNA-covered sequence length divided by the contig length, and the distribution of nucleotide representation is plotted for all 10,497 contigs examined. (G) Genome-wide nucleotide representation by piRNA reads mapping to either sense (red) or antisense (blue) strand of protein-coding genes, as calculated in (F). Only 27 nt 5′-U reads from 19 hr IP were shown for (E), (F) and (G), but the pattern holds for all four time-points. See also Table S2, Figure S4 and Figure S5.
Across all four time points, over 80% of Otiwi1-associated 27 nt 5′-U reads map to the Oxytricha macronuclear genome (Figure 4C), which represents ~5% of the micronuclear genome. Germline-limited TBE1 (Telomere-Bearing Elements) transposons occupy ~1% of the germline genome (Witherspoon et al., 1997), but only constitute less than 0.01% of the reads, making them considerably under-represented. Figure 4D shows representative piRNA mappings to typical macronuclear and micronuclear contigs, illustrating that piRNAs map only to MDS regions (see also Figure S4). Importantly, some piRNAs span MDS-MDS junctions (reads spanning cyan boxes in Figure 4D and Figure S4), suggesting that they do not derive from the micronucleus. To further exclude the possibility that these piRNAs might derive from distinct micronuclear piRNA loci, we mapped these 234,585 MDS-MDS junction-spanning sRNAs to a draft micronuclear genome assembly, sequenced to 50-fold coverage (Chen et al., unpublished data). Only 2 junction spanning piRNAs have a perfect match in the micronuclear genome of just 27 nt, which is not very different from 0.14 expected by chance, based on assumptions of sequence uniformity. This study therefore revealed no evidence of piRNA-specific loci in the micronuclear genome.
We do not observe any position bias in piRNA mapping, and a meta-gene analysis based on 10,497 fully-sequenced single-gene chromosomes (Swart et al., in revision) produces a nearly uniform piRNA distribution across entire chromosomes, except for a small depletion toward the ends (Figure 4E, Figure S5A, D, G, J). At current sequencing depth, the 27 nt 5′-U reads cover 85–91% of these chromosomes at each of the four time points (Figure 4F, Figure S5B, E, H) and the combined reads from all four time points cover 96% (Figure S5K) with a near-equal representation of sense and antisense piRNAs (Figure 4G, Figure S5C, F, I, L). We conclude that the entire macronuclear genome may produce piRNAs. Oxytricha piRNAs either originate from the somatic nucleus or undergo selective amplification based on sequence matching to the macronuclear genome, resulting in their over-representation of sequences derived from the somatic genome.
Although piRNAs seem to map preferentially to coding regions compared to subtelomeric regions (Figure 4D) we do detect many piRNAs mapping to non-coding regions, such as 5′ and 3′ UTR and introns. Furthermore, the piRNA mapping pattern to Otiwi3 at 12 hr post mixing, when there is no Otiwi3 expression (Figure 1D), is indistinguishable from two constitutively expressed genes, TEBPβ and actin I. This argues against a correlation with mRNA expression for these piRNAs.
To further exclude correlation of piRNAs with mRNA levels, we analyzed piRNA mapping to 54 genes that show no expression in Oxytricha at any surveyed time point, based on RNA-seq data from vegetative cells, as well as 0, 10, 20, 40, and 60 hr post mixing of mating types. The mapping of 27 nt 5′-U piRNAs (all four piRNA datasets combined) shows no position bias relative to chromosome location, and the piRNA mapping “coverage” is just slightly lower than that from all 10,497 genes (93% vs. 96%, Figure S5N, O), which again suggests that the 27 nt piRNAs do not primarily derive from mRNA.
Telomere-containing sRNAs were present in the deep-sequenced library (~0.02%, Table S2) but excluded from mapping analysis. A separate analysis of sRNAs containing either “G4T4G4” or “C4A4C4” telomeric repeats suggests that “GT” telomeric reads may be piRNAs, because they are 27 nt with a 5′-U bias (data not shown), but “CA” telomeric reads are not. Given that telomere ends are typically 0.6% of a nanochromosome, the portion of telomeric reads is lower than expected if piRNAs are processed from RNA templates that are telomere-containing transcripts of macronuclear chromosomes (Nowacki et al., 2008). Because “GT” telomeric piRNAs usually start with non-repetitive sequence and end in G4T4 repeats, they more likely derive from macronuclear run-through pre-mRNA or mRNA transcripts, rather than TBE transposons, whose telomeric repeats are internal, or micronuclear telomeres with hundreds of such repeats (Prescott, 2000).
Injection of synthetic RNAs corresponding to normally deleted regions leads to their retention
The data so far argue against a primary role for Oxytricha piRNAs in transposon silencing or DNA elimination. Instead, the predominant mapping of piRNAs to the macronuclear genome hints at an orthogonally-different pathway, in which Oxytricha piRNAs specify somatic regions for retention during DNA rearrangement. To directly test this possibility, we asked whether synthetic piRNAs whose sequences correspond to normally-deleted sequences (IESs) can guide their retention.
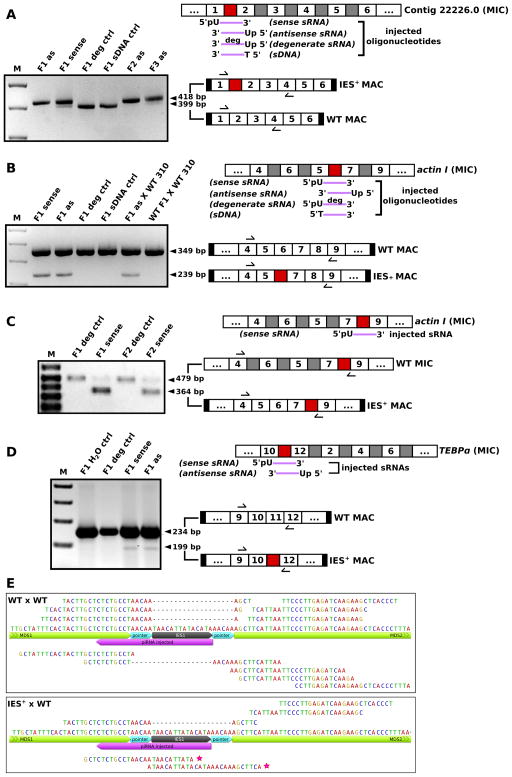
Synthetic piRNA design focused on short IESs that could be covered by a single piRNA. We also required a U at the first position, and chose regions of modestly higher GC content (25–30% GC vs. IES average 16.2%) to facilitate annealing. We injected synthetic 27 nt sRNA targeting an IES into 20–40 mating pairs at 12–18 hr post-mixing, pooled injected pairs and allowed them to finish conjugation and resume vegetative growth for ~1 week (5–8 divisions) before PCR analysis. In injection experiments targeting four different IESs in three genes (Contig22226.0 IES1, actin I IES4 and IES5, and TEBPα IES5, colored red in Figure 5A, B, C, D respectively; see also Figure S6A) the sRNA-injected cells specifically retained the IES whose sequence was present in the injected sRNA, and injection of either sense or antisense RNA protected the corresponding IES against deletion (Figure 5A–D, Figure S6A). Single-cell PCR analysis of Contig22226.0 after injection of a sRNA targeting IES1 further revealed that the majority of macronuclear chromosomes in 2 out of 3 examined cells displayed efficient IES retention (Figure S6B).
Figure 5. Small RNA injection leads to heritable retention of normally-deleted genomic regions.
PCR amplification of total DNA from exconjugant cells after corresponding synthetic sRNA injection reveals programmed IES retention (labeled IES+ MAC). (A) PCR assay for the presence of a larger IES+ MAC PCR product if the targeted conventional IES between MDS1–2 is retained in MAC Contig22226.0. Bands corresponding to wild-type (WT) MAC PCR products are also indicated. (B) PCR assays for a smaller actin I MAC PCR product if the scrambled IES between MDS5–7 is retained instead of being replaced by the longer, scrambled MDS6. (C) Selective PCR assay for the presence of a smaller actin I MAC PCR product if the targeted IES between MDS7–9 is retained; larger bands correspond to the scrambled precursor MIC DNA. (D) PCR assay for the presence of a smaller actin I MAC PCR product if the scrambled IES between MDS10–12 is retained. MIC contig structure, synthetic oligonucleotides injected (purple), and PCR products are shown schematically on the right. MDSs (white boxes with numbers) and IESs (grey and red boxes) are not drawn to scale. Red boxes represent IESs that are targeted by injected RNAs, and black boxes denote short telomeres on nanochromosomes. Arrows schematically indicate primer positions. (E) Exconjugant cells from a backcross between the Contig22226.0 IES1+ strain and a parental wild-type strain (IES+× WT) produce new IES-containing piRNAs, but wild-type mating cells (WT × WT) do not. 25–28 nt 5′-U reads are plotted. Forward and reverse mappings are displayed above and below the relevant portion of the full-length annotated contig, respectively. Stars indicate two new IES-containing piRNAs. Gaps indicate WT reads mapping across the MDS6–7 junction. See also Figure S6 and sequence alignments of all PCR products in a supplemental file.
Injection of a degenerate 27 nt A/U–rich RNA containing all the features of Oxytricha piRNAs showed no IES retention (Figure 5A–D). Furthermore, control injections with a 27 nt single-stranded DNA of the same sequence showed no IES retention in the progeny (Figure 5A,B) confirming that IES retention is a direct effect specific to RNA, and that the protective effect is not simply the consequence of hybridization and subtraction against other functional RNAs that guide DNA rearrangement.
The processing of other IESs in the same gene was unaffected by sRNA injection (Figure 5A–D and supplemental sequence alignment), suggesting that the protective effect requires sequence similarity. On the other hand, sRNAs containing two IES mismatches can have a protective effect, since an injected sRNA corresponding to a single allele also acted on different alleles of the same gene (Figure S6C). Deliberately-introduced substitutions on synthetic sRNAs never transferred to the retained DNA (Figure S6D, S6E), suggesting that the piRNAs are not the templates for RNA-guided DNA repair (Storici, 2008) during genome rearrangement, because substitutions can transfer from the long template RNAs (Nowacki et al., 2008). Instead we propose that the Oxytricha piRNAs prevent excision of a DNA segment.
The IES retention induced by sRNA injection is stable for at least 5 generations of asexual growth in all four injection experiments. We were also able to obtain clonal cell lines with Contig22226.0 IES1 retained, and these were stable for at least 20 doubling times and through an encycstment-excystment cycle (data not shown). Remarkably, when the offspring of injected cells either self-conjugate to produce F2 and F3 cells (Figure 5A, C) or are backcrossed to a wild-type parental strain (Figure 5B), the sexual offspring in the next generation also retain the IES. We therefore conclude that transient availability of Oxytricha piRNAs can effect heritable DNA sequence change in the somatic genome over multiple sexual generations.
To exclude the possibility that the injected RNA was still present in later generations, and to test whether IES-containing piRNAs were produced de novo in the next generation to mediate the IES retention effect, we deep sequenced sRNA at 19 hr post-mixing of the Contig22226.0 IES1+ strain and its wild-type parental strain. Out of 7 million reads, we detected two 5′-U IES-containing piRNAs (26 and 27 nt, respectively; Figure 5E, Figure S6F) neither of which was present in any wild-type sRNA pool, even though those were sequenced to much higher depth (161 million reads, combined). Furthermore, both RNA sequences are different from the original injected synthetic RNA (Figure 5E), which is absent from the sequence pool, suggesting that the transgenerational effect of transient sRNA injection propagates via the production of new, endogenous IES-containing piRNAs. Our discovery that Oxytricha piRNAs can transfer epigenetic information across multiple sexual generations underscores the power of sRNA to influence genome programming and re-programming.
Discussion
piRNA-mediated protection against DNA loss
The most parsimonious model from our experiments is that Oxytricha piRNAs derive from the parental macronucleus, form complexes with Otiwi1 proteins, and inform the developing macronuclear genome which DNA sequences to retain (Figure 6). While this view is conceptually orthogonal to the model where piRNAs target transposon or non-self DNA for silencing or elimination, Oxytricha sRNAs nonetheless provide information to distinguish “self” versus “non-self”, possibly a common theme in piRNA function. Intriguingly, two groups recently proposed that a class of 22 nt 5′-G sRNAs in C. elegans may act as an anti-silencing or “self” signal (Ashe et al., 2012; Lee et al., 2012; Shirayama et al., 2012). In Ascaris, a nematode that undergoes chromatin diminution, sRNAs do not target the eliminated repetitive sequences (Wang et al., 2011). It would be very informative to test if a similar protective role applies to Ascaris and other organisms, either with or without genome rearrangements.
Figure 6. Model: piRNA protects DNA against loss during Oxytricha genome rearrangement.
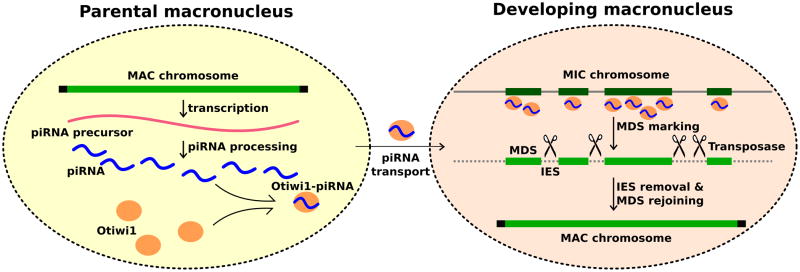
During conjugation, the parental macronuclear genome produces long non-coding RNAs which are subsequently processed into 27 nt piRNAs. Otiwi1 loads 27 nt piRNAs and transport them into the developing macronucleus. The Otiwi1-piRNA complex recognizes and helps mark MDS regions for retention. TBE transposase proteins may introduce DNA breaks in IES regions that are then eliminated, and MDS segments fuse to form new macronuclear chromosomes.
Because piRNAs seem to accumulate relatively early in macronuclear development, and evidence suggests that DNA rearrangements occur later (Mollenbeck et al., 2008) it is possible that the ability to distinguish MDS versus IES information transfers from the piRNAs to chromatin at an early stage of macronuclear development, possibly via chromatin or base modifications. This type of epigenetic information transfer occurs in a wide range of organisms and biological contexts, including sRNA-mediated heteochromatin formation in S. pombe (Verdel et al., 2004; Volpe et al., 2002) and sRNA-guided DNA methylation in plants (Matzke et al., 2009; Wassenegger et al., 1994) and animals (Gu et al., 2012; Watanabe et al., 2011). A mechanism involving histone modification has been suggested downstream of scnRNA-guided DNA elimination in Tetrahymena (Liu et al., 2004; Liu et al., 2007). However, the length of a single MDS or IES in Oxytricha can be shorter than a nucleosome unit, which precludes the ability of a histone-based mechanism to direct precise DNA rearrangements in Oxytricha.
One mechanism we propose for the protective role of piRNAs against DNA deletion in Oxytricha is that piRNAs may “mark” MDSs and prevent cleavage at regions of sequence identity. IfOxytricha piRNAs help introduce DNA modifications, then these might serve, for example, as a signal to prevent TBE transposases that participate in DNA rearrangement (Nowacki et al., 2009) from introducing DNA breaks (Figure 6). This model suggests a new way in which piRNAs may antagonize transposon activity.
Roles for piRNAs and template RNAs in Oxytricha genome rearrangements
We previously reported that long RNA templates can both program MDS order and transfer point substitutions near MDS-MDS junctions from RNA to DNA (Nowacki et al., 2008). Here we propose that piRNAs specify retained regions (MDSs) in early genome rearrangement. Although long template RNA could be processed into sRNA, we propose that the two classes of noncoding RNA act independently for several reasons. First, while we can successfully reprogram the order of DNA segments with long RNA templates, we have been unable to do so with 27 nt sRNAs that span DNA recombination junctions, despite several attempts (data not shown). Second, artificial long RNA templates can transfer substitutions to the reprogrammed DNA sequence near junctions, presumably via RNA-guided DNA repair (Nowacki et al., 2008; Storici, 2008) but substitutions on injected piRNAs did not transfer (Figure S6D and S6E), consistent with a model where Oxytricha piRNAs confer DNA protection from rearrangement. These observations suggest that piRNAs are probably not templates for DNA repair and synthesis. In addition, we also detected fewer than expected telomere-containing piRNAs if telomere-to-telomere transcripts (Nowacki et al., 2008) are the main piRNAs precursors; however, we cannot exclude Illumina sequence bias producing a paucity of telomeric repeats. We suggest that piRNAs and the long template RNAs have distinct features and roles to ensure the integrity of the new somatic genome.
Evolution and comparison to other ciliate piRNAs
Ciliates are over one billion years old, with the genetic distance between Oxytricha and Tetrahymena close to that between humans and fungi (Parfrey et al., 2011). In terms of the Piwi-piRNA system, the conjugation-specific Piwi proteins show a similar expression and localization profile among Oxytricha, Tetrahymena and Paramecium (Bouhouche et al., 2011; Mochizuki et al., 2002), yet the source and function of Oxytricha piRNAs appear orthogonally different from that of Tetrahymena and Paramecium. ScnRNAs in Tetrahymena and Paramecium derive from the germline genome and ultimately target DNA for elimination (Lepere et al., 2008; Lepere et al., 2009; Mochizuki et al., 2002; Mochizuki and Gorovsky, 2004; Schoeberl et al., 2012); whereas Oxytricha piRNAs derive from the somatic genome and target sequences for retention. The opposite piRNA targeting in these species makes economic sense in both lineages, because in each case sRNAs predominantly target the minority class of the germline genome, with roughly 5% of the Oxytricha germline marked for preservation, as opposed to ~ 33% of the Tetrahymena germline that has to be marked for deletion (Tetrahymena Comparative Sequencing Project, Broad Institute of Harvard and MIT (http://www.broadinstitute.org/)). This evolutionary “sign change” suggests that piRNAs as sequence-dependent guides and their related pathways are extremely plastic on a deep evolutionary time scale, permitting the acquisition of new roles in diverse lineages to improve the efficiency of sRNA pathways or to alter the pathways themselves.
In addition to the drastic difference in biogenesis and function, there are two other major differences between Oxytricha piRNAs and Tetrahymena and Paramecium scnRNAs. First, Paramecium 25 nt scnRNAs have a 5′-UNG signature, and the non-5′-U scnRNAs have a “CNA” motif two nucleotides from the 3′ end, indicating that scnRNAs form duplexes with 2 nt 3′ overhangs, processed from dsRNA precursors by Dicer proteins (Lepere et al., 2009). This complementary signature was absent from Oxytricha, and it is unclear whether Oxytricha piRNAs derive from single- or double-stranded RNAs. In addition, experiments in Paramecium that inject sRNA duplexes with a 2 nt 3′ overhang can target IESs for deletion, whereas the Oxytricha experiments shown here demonstrate that single-stranded sRNA can target IES regions for retention, suggesting an opposite mechanism. Secondly, we found that Oxytricha piRNAs lack 3′ modification, whereas Tetrahymena scnRNAs contain 3′ end methylation (Kurth and Mochizuki, 2009). This difference may be part of an evolutionary mechanism leading to two orthogonally different classes of piRNAs in ciliates.
Experimental Procedure
Cell culture and mating
Oxytricha trifallax strains JRB310 and JRB510 were cultured in Pringsheim medium (0.11 mM Na2HPO4, 0.08 mM MgSO4, 0.85 mM Ca(NO3)2, 0.35 mM KCl, pH 7.0) using algae Chlamydomonas reinhardtii as a food source. To induce mating, JRB310 and LRB510 cells were mixed right after food depletion. Cells started to pair between 2–3 hr post mixing, and conjugation efficiency was between 60% and 95%. Post-injection cells were raised in Volvic water.
RNA extraction and small RNA detection
Total RNA was extracted using the mirVana kit (Ambion) following manufacturer’s instructions. To detect small RNA, total RNA was separated on a 15% denaturing polyacrylamide gel in 1× TBE buffer and stained with SYBR Gold (Invitrogen). For radioactive labeling, RNA was treated with calf intestinal alkaline phosphatase (NEB), phenol-chloroform extracted, ethanol precipitated, and labeled with γ-32P-ATP using T4 polynucleotide kinase (NEB). Labeled RNA was separated on a 15% denaturing polyacrylamide gel, which was directly exposed to a storage phosphor screen (GE healthcare) and scanned by Typhoon 9410 (GE healthcare).
Phylogenetic analysis of Oxytricha Argonaute proteins
Oxytricha Argonaute genes (Otiwi1–13) were retrieved by using tBLASTn (Altschul et al., 1997) against the current macronuclear genome assembly (Swart et al., in revision) and Tetrahymena TWI1p (GenBank: BAC02573.1) as the query sequence. RT-PCR followed by clone sequencing was used to generate cDNA and protein sequences. All thirteen Otiwi proteins, selected eukaryotic (accession numbers from (Seto et al., 2007)) and archaeal (accession numbers NP_248321 and NP_578266) Argonaute proteins were aligned by MUSCLE (Edgar, 2004) followed by manual adjustment, which included deletion of multiple gap positions (see the sequence alignment in a separate supplemental file labeled “Argonaute_alignment”). Phylogenetic analysis was performed using maximum likelihood through the PhyML webserver (Guindon et al., 2010) with default settings and the LG substitution matrix. Bootstrap values were produced by 500 re-sampled datasets.
Antibody
The antibody used to detect Otiwi1 in western analysis, immunostaining, and immunoprecipitation is anti-PIWIL1 (Abcam, ab12337), originally raised against the human PIWIL1 protein C-terminal peptide (15aa). We performed knockdown followed by western analysis, peptide competition in immunostaining, and mass spectrometry after immunoprecipitation to verify that the antibody is specific to Otiwi1. Detailed protocols for immunostaining, peptide competition assay, immunoprecipitation, and mass spectrometry analysis are provided in Extended Experimental Procedures. Anti-tubulin antibody is DM1A (Abcam, ab7291).
Biochemical analysis of piRNAs
For detecting a 5′ monophosphate, ~ 40ng Otiwi1 IP-enriched RNA (23 h) or control RNA oligonucleotide was incubated with 0.1U Terminator exonuclease (Epicentre) at 30°C for 1h, and the reaction was quenched by adding EDTA (pH 8.0) to a final concentration of 5 mM. The quenched reaction was directly loaded and separated on a 15% denaturing polyacrylamide gel, and stained with SYBR gold (Invitrogen). To analyze 3′ end modifications, Oxytricha piRNAs were treated with NaIO4, followed by β-elimination as described previously (Vagin et al., 2006).
Antisense knockdown
To increase the efficiency of targeting, mRNA secondary structures were used to guide antisense oligonucleotide design (Patzel et al., 1999). The oligonucleotides, at a final concentration of 30 μg/μl, were injected into cytoplasm right after pair formation (~2–4 hr post mixing). To verify the knockdown effect, total RNA was extracted at 10–11 hr post-mixing for RT-qPCR analysis (Figure S3A). We also collected cells at 10 hr and 18 hr post-mixing for western analysis after Otiwi1 knockdown (Figure S3B). See Extended Experimental Procedures for verification of knockdown by RT-PCR and western analysis.
Survival and developmental characterization of Otiwi1 knockdown
After antisense injection, 20–28 pairs were collected into one droplet (0.2% BSA in Volvic water) on a cover slip, covered with oil. We then observe and counted cells using light microscopy at 12, 24, 28, and 48 hr post mixing. Student’s t-test was used to compare the number of rounded cells at 24 hr based on four independent knockdown injections. In Figures 3C-J, single cells at 24–28 hr post mixing were isolated into droplets, fixed with Methanol:Acetic acid (3:1), stained with DAPI, and imaged with laser scanning confocal microscopy at the Princeton University Microscopy Facility.
sRNA Illumina library construction, high throughput sequencing and analysis
Otiwi1 antibody immunoprecipitated RNA from 12, 19, 23, and 30 hr post mixing, as well as total sRNA isolated at 19 hr after backcrossing the Contig22226.0 IES1+ strain (a clonal line derived from an F1 cell from the population analyzed in Figure 5A lane 1) to wild-type JRB510, were used to construct sRNA libraries, following the Illumina® TruSeq™ Small RNA Sample Preparation protocol, with different barcodes for each sample. The libraries were pooled and sequenced on an Illumina HiSeq 2000 (101 cycles, single-end reads, multiplexed) at the Sequencing Core Facility at Princeton University. The raw reads were barcode-split, adapter trimmed, and collapsed to non-redundant reads before mapping by SHRiMP (David et al., 2011). The total sRNA library at 20 hr post-mixing was prepared following the Illumina® Small RNA v1.5 Sample Preparation guide, and sequenced on an Illumina Genome Analyzer IIX (54 cycles, single-end reads). See Extended Experimental Procedures for details of sequencing data processing and analysis.
Synthetic RNA and DNA oligonucleotide injections for IES retention experiments
RNA and DNA oligonucleotides were synthesized by IDT with standard desalting. Oligonucleotides were dissolved in nuclease-free water (Ambion) to a final concentration of 20 μg/μl, heated to 65 °C for 1 min, and chilled on ice before using. Paired cells between 12 to 18 hr post mixing were isolated in single droplets on cover slips and covered with mineral oil during injection. For each oligonucleotide, the cytoplasm of both cells from 20–40 pairs were injected and all pairs pooled together at the end of injection. Cells were examined for donut shape between 36 to 48 hr post mixing and morphologically vegetative cells were removed. The pooled injected cells gave rise to a starting population of 5–20 cells 3–4 days post-mixing when they started vegetative division. At 7–8 days post-mixing when genomic DNA was extracted, the final population varied between 50 to 300 cells depending on the cell conditions. DNA equivalent of that from 0.5–3 cells was used in each PCR reaction. For single-cell analysis, single cells were hand-isolated in 1 μl medium from the injected population on Day 7, and directly used in PCR.
For F2 analysis, F1 cells were cultured vegetatively for 10 ~ 20 days (or 10 ~ 40 generations) in pools that we described above. We then starved them to induce conjugation. Conjugating pairs were hand-isolated and pooled in a separate well. We allowed the conjugation to proceed and vegetative growth for at least 5 days (5~10 generations) before genomic DNA extraction. When we could not induce F1 cells to mate with each other, we then tried to induce conjugation between F1 cells and the WT JRB310 or JRB510 parental strain, which usually gave higher mating efficiency. Contig 22226.0 IES1+ F2 and F3 derive from a mating between individual F1 and F2 cell lines, respectively.
Supplementary Material
Highlights.
Conjugation in the ciliateOxytricha induces piRNA and Otiwi1 expression.
Localization of Otiwi1 shifts from parental to developing somatic nucleus.
Oxytricha piRNAs map primarily to the somatic genome.
sRNA injections lead to retention of homologous regions over multiple generations.
Acknowledgments
We thank D. Perlman, B. Zee, and B. Garcia for mass spectrometry validation of the Otiwi1 antibody; J. Buckles, W. Wang, D. Storton, and L. Parsons for Illumina sequencing; L. Li and A. D. Goldman for assistance with phylogenetic analysis; X. Chen for searching the micronuclear genome assembly for junction-spanning piRNAs; A. Chen, K. Mochizuki and M. Couvillion for sharing RNA immunoprecipitation protocols; E. Swart, J. Bloom and P. Jiang for advice on computational analysis of deep sequencing results; J. Postberg and J. Goodhouse for immunofluorescence advice; P. Andolfatto, J. Swan, J. Khurana, P. Schedl and L. Beh for discussion or comments on the manuscript; J. Wang for help with Oxytricha cell culture. We also thank three anonymous referees for valuable suggestions. This study was supported by NIH grant GM59708 and NSF grants 0923810 and 0900544 (to L.F.L.) and a DOD pre-doctoral fellowship W81XWH-10-1-0122 (to W.F.).
Footnotes
Accession numbers
Otiwi protein sequences are available with GenBank accession numbers JN604928-JN604940. The raw and processed sRNA sequencing data have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) with the GEO accession numbers GSE35018 and GSE40081.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–1539. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Ashe A, Sapetschnig A, Weick EM, Mitchell J, Bagijn MP, Cording AC, Doebley AL, Goldstein LD, Lehrbach NJ, Le Pen J, et al. piRNAs Can Trigger a Multigenerational Epigenetic Memory in the Germline of C. elegans. Cell. 2012;150:88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bouhouche K, Gout JF, Kapusta A, Betermier M, Meyer E. Functional specialization of Piwi proteins in Paramecium tetraurelia from post-transcriptional gene silencing to genome remodelling. Nucleic Acids Res. 2011;39:4249–4264. doi: 10.1093/nar/gkq1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc’his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Chalker DL, Yao MC. Non-Mendelian, heritable blocks to DNA rearrangement are induced by loading the somatic nucleus of Tetrahymena thermophila with germ line-limited DNA. Mol Cell Biol. 1996;16:365–667. doi: 10.1128/mcb.16.7.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvillion MT, Lee SR, Hogstad B, Malone CD, Tonkin LA, Sachidanandam R, Hannon GJ, Collins K. Sequence, biogenesis, and function of diverse small RNA classes bound to the Piwi family proteins of Tetrahymena thermophila. Genes Dev. 2009;23:2016–2032. doi: 10.1101/gad.1821209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David M, Dzamba M, Lister D, Ilie L, Brudno M. SHRiMP2: sensitive yet practical SHort Read Mapping. Bioinformatics. 2011;27:1011–1012. doi: 10.1093/bioinformatics/btr046. [DOI] [PubMed] [Google Scholar]
- Duharcourt S, Butler A, Meyer E. Epigenetic self-regulation of developmental excision of an internal eliminated sequence on Paramecium tetraurelia. Genes Dev. 1995;9:2065–2077. doi: 10.1101/gad.9.16.2065. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan H, Lin X, Zhang Z, Zhang W, Liao S, Wang L, Han C. piRNA profiling during specific stages of mouse spermatogenesis. RNA. 2011;17:1191–1203. doi: 10.1261/rna.2648411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455:1193–1197. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu SG, Pak J, Guang S, Maniar JM, Kennedy S, Fire A. Amplification of siRNA in Caenorhabditis elegans generates a transgenerational sequence-targeted histone H3 lysine 9 methylation footprint. Nat Genet. 2012;44:157–164. doi: 10.1038/ng.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Kirino Y, Mourelatos Z. Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat Struct Mol Biol. 2007;14:347–348. doi: 10.1038/nsmb1218. [DOI] [PubMed] [Google Scholar]
- Kurth HM, Mochizuki K. 2′-O-methylation stabilizes Piwi-associated small RNAs and ensures DNA elimination in Tetrahymena. RNA. 2009;15:675–685. doi: 10.1261/rna.1455509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- Lee HC, Gu W, Shirayama M, Youngman E, Conte D, Jr, Mello CC. C. elegans piRNAs Mediate the Genome-wide Surveillance of Germline Transcripts. Cell. 2012;150:78–87. doi: 10.1016/j.cell.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SR, Collins K. Two classes of endogenous small RNAs in Tetrahymena thermophila. Genes Dev. 2006;20:28–33. doi: 10.1101/gad.1377006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepere G, Betermier M, Meyer E, Duharcourt S. Maternal noncoding transcripts antagonize the targeting of DNA elimination by scanRNAs in Paramecium tetraurelia. Genes Dev. 2008;22:1501–1512. doi: 10.1101/gad.473008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepere G, Nowacki M, Serrano V, Gout JF, Guglielmi G, Duharcourt S, Meyer E. Silencing-associated and meiosis-specific small RNA pathways in Paramecium tetraurelia. Nucleic Acids Res. 2009;37:903–915. doi: 10.1093/nar/gkn1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Mochizuki K, Gorovsky MA. Histone H3 lysine 9 methylation is required for DNA elimination in developing macronuclei in Tetrahymena. Proc Natl Acad Sci U S A. 2004;101:1679–1684. doi: 10.1073/pnas.0305421101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Taverna SD, Muratore TL, Shabanowitz J, Hunt DF, Allis CD. RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes Dev. 2007;21:1530–1545. doi: 10.1101/gad.1544207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ. RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol. 2009;21:367–376. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell. 2002;110:689–699. doi: 10.1016/s0092-8674(02)00909-1. [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Gorovsky MA. Conjugation-specific small RNAs in Tetrahymena have predicted properties of scan (scn) RNAs involved in genome rearrangement. Genes Dev. 2004;18:2068–2073. doi: 10.1101/gad.1219904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollenbeck M, Zhou Y, Cavalcanti AR, Jonsson F, Higgins BP, Chang WJ, Juranek S, Doak TG, Rozenberg G, Lipps HJ, et al. The pathway to detangle a scrambled gene. PLoS One. 2008;3:e2330. doi: 10.1371/journal.pone.0002330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowacki M, Higgins BP, Maquilan GM, Swart EC, Doak TG, Landweber LF. A functional role for transposases in a large eukaryotic genome. Science. 2009;324:935–938. doi: 10.1126/science.1170023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowacki M, Vijayan V, Zhou Y, Schotanus K, Doak TG, Landweber LF. RNA-mediated epigenetic programming of a genome-rearrangement pathway. Nature. 2008;451:153–158. doi: 10.1038/nature06452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara T, Sakaguchi Y, Suzuki T, Ueda H, Miyauchi K. The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nat Struct Mol Biol. 2007;14:349–350. doi: 10.1038/nsmb1220. [DOI] [PubMed] [Google Scholar]
- Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- Parfrey LW, Lahr DJ, Knoll AH, Katz LA. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc Natl Acad Sci U S A. 2011;108:13624–13629. doi: 10.1073/pnas.1110633108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzel V, Steidl U, Kronenwett R, Haas R, Sczakiel G. A theoretical approach to select effective antisense oligodeoxyribonucleotides at high statistical probability. Nucleic Acids Res. 1999;27:4328–4334. doi: 10.1093/nar/27.22.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott DM. Genome gymnastics: unique modes of DNA evolution and processing in ciliates. Nat Rev Genet. 2000;1:191–198. doi: 10.1038/35042057. [DOI] [PubMed] [Google Scholar]
- Rajasethupathy P, Antonov I, Sheridan R, Frey S, Sander C, Tuschl T, Kandel ER. A Role for Neuronal piRNAs in the Epigenetic Control of Memory-Related Synaptic Plasticity. Cell. 2012;149:693–707. doi: 10.1016/j.cell.2012.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robine N, Lau NC, Balla S, Jin Z, Okamura K, Kuramochi-Miyagawa S, Blower MD, Lai EC. A broadly conserved pathway generates 3′UTR-directed primary piRNAs. Curr Biol. 2009;19:2066–2076. doi: 10.1016/j.cub.2009.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouget C, Papin C, Boureux A, Meunier AC, Franco B, Robine N, Lai EC, Pelisson A, Simonelig M. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature. 2010;467:1128–1132. doi: 10.1038/nature09465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 2009;461:1296–1299. doi: 10.1038/nature08501. [DOI] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Sakaguchi Y, Suzuki T, Suzuki T, Siomi H, Siomi MC. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi- interacting RNAs at their 3′ ends. Genes Dev. 2007;21:1603–1608. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeberl UE, Kurth HM, Noto T, Mochizuki K. Biased transcription and selective degradation of small RNAs shape the pattern of DNA elimination in Tetrahymena. Genes Dev. 2012;26:1729–1742. doi: 10.1101/gad.196493.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto AG, Kingston RE, Lau NC. The coming of age for Piwi proteins. Mol Cell. 2007;26:603–609. doi: 10.1016/j.molcel.2007.05.021. [DOI] [PubMed] [Google Scholar]
- Shirayama M, Seth M, Lee HC, Gu W, Ishidate T, Conte D, Jr, Mello CC. piRNAs Initiate an Epigenetic Memory of Nonself RNA in the C. elegans Germline. Cell. 2012;150:65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Steiner FA, Thijssen KL, Plasterk RH. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- Storici F. RNA-mediated DNA modifications and RNA-templated DNA repair. Curr Opin Mol Ther. 2008;10:224–230. [PubMed] [Google Scholar]
- Tian Y, Simanshu DK, Ma JB, Patel DJ. Structural basis for piRNA 2′-O-methylated 3′-end recognition by Piwi PAZ (Piwi/Argonaute/Zwille) domains. Proc Natl Acad Sci U S A. 2011;108:903–910. doi: 10.1073/pnas.1017762108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Wang J, Czech B, Crunk A, Wallace A, Mitreva M, Hannon GJ, Davis RE. Deep small RNA sequencing from the nematode Ascaris reveals conservation, functional diversification, and novel developmental profiles. Genome Res. 2011;21:1462–1477. doi: 10.1101/gr.121426.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenegger M, Heimes S, Riedel L, Sanger HL. RNA-directed de novo methylation of genomic sequences in plants. Cell. 1994;76:567–576. doi: 10.1016/0092-8674(94)90119-8. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, Minami N, Imai H. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 2006;20:1732–1743. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Tomizawa S, Mitsuya K, Totoki Y, Yamamoto Y, Kuramochi-Miyagawa S, Iida N, Hoki Y, Murphy PJ, Toyoda A, et al. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science. 2011;332:848–852. doi: 10.1126/science.1203919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Ba Z, Gao M, Wu Y, Ma Y, Amiard S, White CI, Danielsen JM, Yang YG, Qi Y. A Role for Small RNAs in DNA Double-Strand Break Repair. Cell. 2012;149:101–112. doi: 10.1016/j.cell.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Witherspoon DJ, Doak TG, Williams KR, Seegmiller A, Seger J, Herrick G. Selection on the protein-coding genes of the TBE1 family of transposable elements in the ciliates Oxytricha fallax and O. trifallax. Mol Biol Evol. 1997;14:696–706. doi: 10.1093/oxfordjournals.molbev.a025809. [DOI] [PubMed] [Google Scholar]
- Yao MC, Fuller P, Xi X. Programmed DNA deletion as an RNA-guided system of genome defense. Science. 2003;300:1581–1584. doi: 10.1126/science.1084737. [DOI] [PubMed] [Google Scholar]
- Zahler AM, Neeb ZT, Lin A, Katzman S. Mating of the Stichotrichous Ciliate Oxytricha trifallax Induces Production of a Class of 27 nt Small RNAs Derived from the Parental Macronucleus. PLoS One. 2012;7:e42371. doi: 10.1371/journal.pone.0042371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- Zhang H, Ehrenkaufer GM, Pompey JM, Hackney JA, Singh U. Small RNAs with 5′-polyphosphate termini associate with a Piwi-related protein and regulate gene expression in the single-celled eukaryote Entamoeba histolytica. PLoS Pathog. 2008;4:e1000219. doi: 10.1371/journal.ppat.1000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.