Abstract
The incidence of esophageal adenocarcinoma (EAC) has risen 600% over the last 30 years. With a five-year survival rate of 15%, identification of new therapeutic targets for EAC is greatly important. We analyze the mutation spectra from whole exome sequencing of 149 EAC tumors/normal pairs, 15 of which have also been subjected to whole genome sequencing. We identify a mutational signature defined by a high prevalence of A to C transversions at AA dinucleotides. Statistical analysis of exome data identified significantly mutated 26 genes. Of these genes, four (TP53, CDKN2A, SMAD4, and PIK3CA) have been previously implicated in EAC. The novel significantly mutated genes include chromatin modifying factors and candidate contributors: SPG20, TLR4, ELMO1, and DOCK2. Functional analyses of EAC-derived mutations in ELMO1 reveal increased cellular invasion. Therefore, we suggest a new hypothesis about the potential activation of the RAC1 pathway to be a contributor to EAC tumorigenesis.
In recent decades, the incidence of esophageal adenocarcinoma (EAC) has increased dramatically in the United States and other Western countries1,2. The increasing frequency and poor prognosis of these cancers is a substantial health concern. EAC does not develop from the naïve esophageal epithelium, but rather originates from intestinal metaplasia of the esophageal epithelium (Barrett’s esophagus) that develops in response to chronic gastroesophageal reflux. While the reason for the dramatic rise in these cancers is unknown, factors influencing the rising rates include gastroesophageal reflux disease (GERD), Barrett’s esophagus, and obesity3. There is great urgency to elucidate the genomic alterations underlying EAC in order to enhance understanding of these tumors, aid in early diagnosis, and identify therapeutic targets.
Knowledge of the somatic mutations in EAC has been limited to studies in small collections of tumors. These studies have identified frequent mutations in TP534 and CDKN2A5. Beyond these two genes, small focused studies have noted sporadic mutations in APC6, BRAF7, CDH18, CTNNB16, EGFR9,10, KRAS7, PIK3CA11, PTEN12, and SMAD412. While comparative whole exome sequencing has been reported for 11 EACs and esophageal squamous cell carcinomas, no clear contributors to EAC were identified at the gene level13.
Here, we describe the landscape and spectrum of genomic alterations in 149 fresh-frozen, surgically-resected cases of EAC including adenocarcinomas arising in the gastric-esophageal junction-GEJ not treated with chemotherapy or radiation prior to surgery. All cases were subjected to whole exome sequencing (WES) with 15 sample pairs also analyzed by whole-genome sequencing (WGS). Examination of the somatic alterations revealed a high frequency of mutations and rearrangements. Additionally, we identify a mutational signature defined by A>C transversions at AA dinucleotide sites (the latter adenine denotes the site of the mutation). Through systematic analysis of the mutated genes, we identify many genes not previously associated with this cancer. These include ELMO1 and DOCK2, upstream modulators of the RAC1 GTPase, and characterize the presence of mutations impacting signal transduction pathways. These results provide a foundation for further study and treatment of these cancers.
Landscape of Esophageal Adenocarcinoma Mutations and Rearrangements
To identify somatic alterations in EAC, we performed WES on tumor-normal pairs from 149 patients and WGS on 16 pairs. Fifteen of the WGS samples have matched WES data, and 14 WGS samples were evaluated on mRNA expression arrays (Supplementary Fig. 1 and Supplementary Table 1). One tumor from which WGS was performed lacked matched WES data due to sequencing failure in this sample. Somatic mutations were identified using the MuTect and Indelocator tools14–16.
For WGS, tumors were sequenced to an average depth of 49X and the matched germline DNA samples were sequenced to 30X coverage with paired 101-basepair reads on Illumina HiSeq instruments (Supplementary Table 2). We identified a median of 26,161 genome-wide mutations per tumor (range 18,881–66,225) corresponding to a median mutation frequency of 9.9/Mb (range 7.1–25.2/Mb) relative to a haploid genome (Supplementary Table 3). The mutation frequency was highest in intergenic regions (17.0/Mb), lower in intronic regions (9.9/Mb) and lowest in coding exons (6.2/Mb) (Supplementary Table 4). This step-wise decrease in mutation frequency was consistently seen in other cancers16. Compared to other cancer types, this overall mutation frequency is high and exceeded only by lung cancer17,18 and melanoma19, diseases that emerge from clear mutagens. By contrast, analogous sequencing of colorectal adenocarcinomas (CRC) identified a mutation frequency of 5.6/Mb20 across the genome. The high mutation frequency in EAC suggests that these tumors may be exposed to significant mutagens, perhaps attributable to the harsh environment created by gastric refluxate and inflammation21.
We also analyzed the WGS data using the dRanger algorithm20 to identify chromosomal rearrangements. A total of 2,952 candidate rearrangements were identified with a median of 172 per tumor (range 77–402) (Supplementary Table 5). Consistent with array data showing a higher degree of structural alterations in EAC compared to CRC22, the number of rearrangements is much greater than observed with a comparable analysis of CRC genomes20. No correlation was observed between the number of mutations and rearrangements (R2=0.0046). Of the rearrangements identified, 20% were interchromosomal translocations. Among the intrachromosomal alterations identified, a majority (55%) involved aberrant fusions of two sequences located within 1 Mb of each other. To identify potential fusion gene products that may contribute to the pathogenesis of EAC, we examined data for predicted in-frame gene fusions. Thirty-eight such events were identified (Supplementary Table 6), but no recurrent gene fusions were detected.
The Mutation Spectrum of EAC Points toward a High Frequency of A to C Transversions at AA Sites across the Entire Genome
Epithelial cancers often display variable mutation spectra pointing toward particular mutagenic stimuli. Therefore, we analyzed the spectrum of mutations in EAC detected by WGS. Earlier exome sequencing of EACs noted A>C transversions to be more common in EAC compared to squamous esophageal carcinoma13. Evaluating the WGS data, we found that A>C base changes comprised an average of 34% of total mutations (Supplementary Table 7). To comprehensively characterize the mutation spectra, we measured the frequencies of base mutations at different trinucleotide contexts and observed a preponderance of C>T transitions (39.2/Mb) as seen in most epithelial cancers. We further investigated the high frequency of A>C transversions (or equivalently T>G transversions on the complementary strand). These events showed preference (20.2/Mb) for the context AA – that is, at adenines flanked by a 5′ adenine and any 3′ nucleotide (Fig. 1a). In total, 84% of A>C mutations are flanked by a 5′ adenine. Expanding upon these findings, A>C transversions at AA dinucltoides were most pronounced when the 3′ base was a guanine (49.3/Mb) and lower when it was an adenine (8.0/Mb), cytosine (16.8/Mb), or thymidine (6.7/Mb) (Supplementary Table 8). To validate these results, genotyping of randomly selected AA>C mutations from intragenic regions showed a concordance rate of 100% (25/25). The high frequency of AA transversions appears to be unique to EAC as equivalent events have not been identified in other cancer types15–19,23,24.
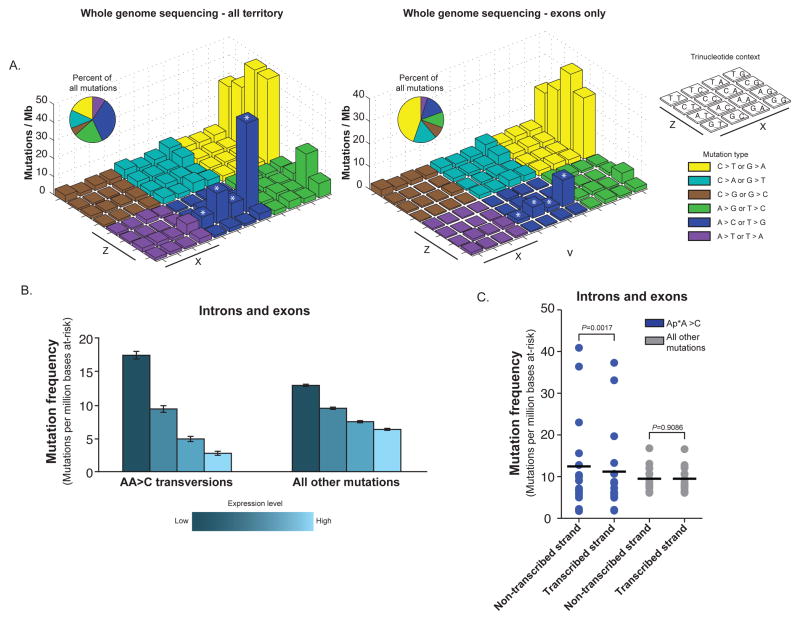
Figure 1. Prominent frequencies of A>C transversions at AA sites identified from whole genome sequencing are observed in less-expressed regions of the genome.
a) “Lego” plots of mutation frequencies across 16 WGS samples for all sequenced territory (left) and exons only (right). Base substitutions are divided into six categories to represent the six possible base changes (each category represented by a different color). Substitutions are further subdivided by the 16 possible flanking nucleotides surrounding the mutated base as listed in “trinucleotide context” legend (X, Z). The inset pie chart indicates the distribution of all mutations for a given middle mutated base across the territory being evaluated. A>C transversions at AA dinucleotides are denoted with an asterisk (*). b) Gene expression was detected from Affymetrix U133 Plus 2 arrays on per sample basis for 14 WGS samples. Mutation frequencies within the introns and exons (y-axis) were calculated as number of mutations detected in per million at-risk bases sequenced for a mutation category. Mutation frequencies as they vary by gene expression (with genes binned into quartiles based on gene expression) are plotted separately for all mutations and for AA transversions. c) The frequency of mutation (number of mutations per million bases at risk for mutation) within intronic and exonic regions are plotted with frequencies separated based upon whether the base at risk is present on the transcribed or non-transcribed strands. For the AA transversions, the mutation frequency is calculated separately for the case of at-risk adenine bases when present on the coding or non-coding strands compared to similar analysis for all other mutations. P-values were calculated by Student’s T-test. All error bars represent S.D.
In total, A to C transversions at AA sites accounted for 29% of the total mutations (Fig. 1a). Within individual tumors, these AA transversions accounted for 5–48% of mutations (Supplementary Table 8), and the event number was correlated with overall mutation frequency (R2=0.92) (Supplementary Fig. 2). When we exclude AA transversions, the mutation frequency is 8.5/Mb and still higher than most tumor types. Thus, AA>C mutations do not fully explain the elevated mutation frequency in EAC relative to other cancers.
We next characterized the distribution of mutations in genomic regions. While A>C transversions remained notable at the AA sites, the percentage of all mutations consisting of these transversions was significantly lower in exons (16%) than seen across the entire genome (AAG, P=0.001; AAT, P=0.0006; AAC, P=0.0007; AAA, P=0.0006; two-tailed Student’s t-test) (Fig. 1a). These results were consistent across the coding regions of all 16 cases evaluated by WGS (Supplementary Tables 9 and 10). In contrast, the attenuation of C>T transitions at CG dinucleotides in coding regions (39.2/Mb vs. 25.4/Mb) was smaller than that seen for AA transversions.
The reduction of AA transversions in coding areas relative to intergenic regions suggested that these mutations may be less likely to occur in transcribed regions or repaired effectively by transcription-coupled repair. To evaluate the potential impact of gene expression on mutation rates, we compared sample-specific frequencies of AA mutations within gene boundaries at varying levels of gene expression in 14 WGS samples from which mRNA was available for microarray expression profiling. Elevated expression was associated with lower global mutation frequency. Additionally, the impact of gene expression upon attenuating mutation rates was three-fold greater at AA sites than for mutations at other nucleotide contexts (Fig. 1b and Supplementary Table 11). This finding demonstrates a strong impact of local gene expression on the development of AA transversions in EAC.
Given the impact of transcription upon these mutations, we analyzed AA transversions for strand bias. The mutation rates for AA transversions in introns and exons were calculated separately depending upon if the adenine base is on the transcribed or non-transcribed strand. The results indicated that AA>C mutations are more common when the AA sites are located on the non-transcribed strand (12.4/Mb vs. 11.2/Mb; P = 0.0016, Student’s T-test, paired). When evaluating all other mutations, a strand bias was not detected (9.5/Mb vs. 9.5/Mb; P = 0.9086, Student’s T-test, paired) (Fig. 1c and Supplementary Table 12). These results suggest that AA transversions may be more effectively recognized and repaired when the mutated adenine is located on the transcribed strand.
Mutations Identified by Whole Exome Sequencing
We next analyzed WES data from 149 tumor/germline pairs (Supplementary Table 13). A mean coverage depth of 83.3x was achieved in neoplastic DNA and 85.9x in the non-cancerous tissue. 89% of exons were covered at 8x or greater depth for normal and at 14x for tumor, a threshold for which MuTect is powered to detect mutation above or equal to an allele fraction of 0.314,16,25. We evaluated mutation calling by comparing candidate coding mutations identified by WES to WGS calls from the same tumor. An 85.1% (2200/2585) concordance was observed for all events and 90% concordance at mutations present at greater than 0.1 allele fraction (Supplementary Table 14).
Four tumors had markedly higher coding mutation frequencies (14.6–50.9/Mb) than other cases. This pattern resembled that of CRC where a subset of tumors were hypermutated, largely attributable to microsatellite instability (MSI). Similarly, MSI-positive tumors have been reported to represent 7% of EAC26. These four cases with the highest mutation rates were found to be MSI-positive with the highest mutation frequency tumor having mutations in two mismatch repair genes MSH6 and MSH3 (Supplementary Table 15). By contrast, none of the 24 EAC samples with the next highest mutation frequency (greater than 5 mutations/Mb) scored positive for MSI. To avoid a potential confounding effect on statistical analysis, we omitted these MSI-positive cases from the final analysis, leaving 145 tumors.
A total of 17,383 mutations, consisting of 16,516 non-silent mutations and 1,954 insertion-deletion/null mutations were detected in the 145-sample cohort, for a median of 104 non-silent coding mutations per tumor (Fig. 2). The overall non-silent median mutation frequency was 3.51/Mb (range 0.97–10.8/Mb). We investigated whether the fraction of AA transversions was associated with clinical variables including age, stage, gender, and tumor location. Interestingly, a trend was seen wherein EACs developing within the tubular esophagus harbored a greater fraction of AA transversions compared to the tumor in the GEJ (P=0.076, two-tailed Student’s t-test); an intriguing result given the possibility of gastric refluxate into the lower esophagus to serve as a mutagenic insult (Fig. 2 and Supplementary Fig. 3). No other significant associations were identified.
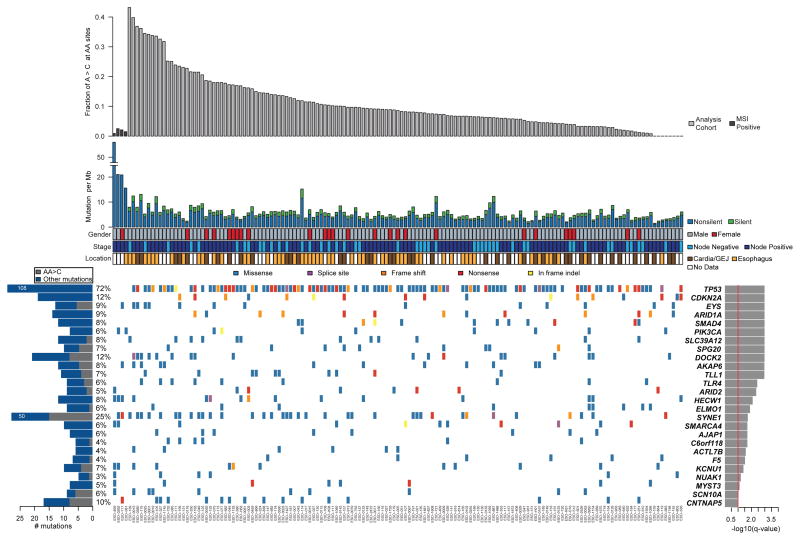
Figure 2. Mutation frequencies and significantly mutated genes in esophageal adenocarcinoma as identified by WES.
a) Mutation frequency of a cohort of 149 primary esophageal adenocarcinomas is sorted by the fraction of mutations consisting of A > C transversions at AA sites. MSI-positive samples labeled in dark gray were not included in mutation significance analysis. b) Key clinical parameters described in Supplementary Table 1. c) Center; mutations in significantly mutated genes, colored by the type of coding mutation. Each column denotes an individual tumor and each row represents a gene. Left; number and percentage of samples with mutations in a given gene. Gray bar represents number of AA transversions in a gene. Right, the negative log of the q values for the significance level of mutated genes is shown for all genes with FDR q < 0.1.
Genes Significantly Mutated in Esophageal Adenocarcinoma
We observed mutations in 8,331 genes, of which 3,639 were mutated in two or more samples (Supplementary Table 16). Among these, 199 genes were mutated in 5% or more of the tumors, including 33 genes mutated in over 10% of cases. To identify genes displaying evidence of positive selection for mutation, we used the mutation significance algorithm, MutSig14–16,25. This tool compares the mutation occurrence in each gene to that which would be expected by chance given a background mutation frequency model that factors in the mutation spectra, presence of silent mutations, mutation frequencies, and regional mutation frequencies along the genome18. We found 26 genes to be significantly mutated (FDR q<0.1) with two known EAC tumor suppressors, TP53 and CDKN2A, being the most significant (Fig. 2). With the exception of ARID1A, PIK3CA, and SMAD4, no other significantly mutated genes have been previously implicated in EAC although several had been implicated in other cancers.
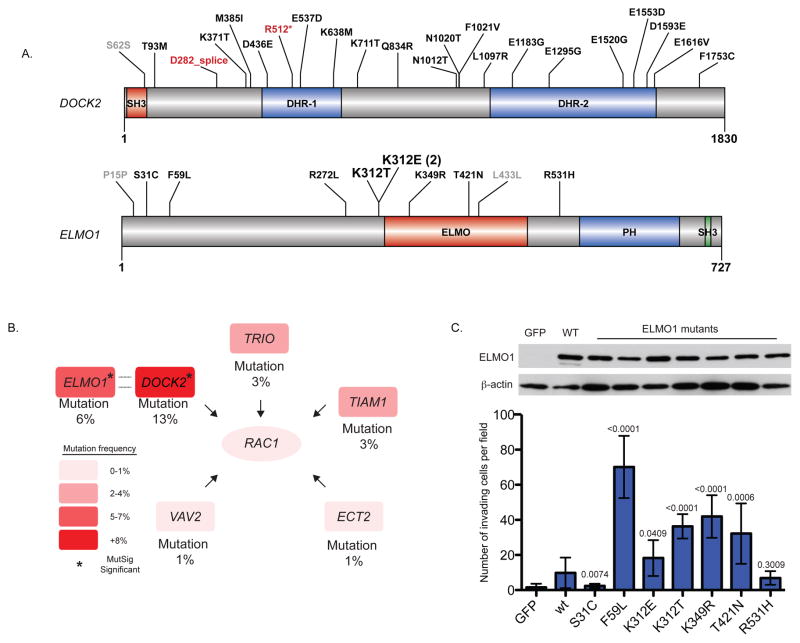
Intriguingly, two significantly mutated genes, ELMO1 and DOCK2, are dimerization partners and intracellular mediators of the Rho family GTPase, RAC127,28. Because aberrant RAC1 activation has been implicated in malignant transformation of other cancer types, mainly by enhancing cellular motility29–33, recurrent mutations in these genes may be functionally important. While no RAC1 or RAC2 mutations were identified, ELMO1 or DOCK2 are mutated in 25 (17%) EAC samples with two samples having mutations in both factors and two samples have two independent mutations in DOCK2 (Fig. 3 and Supplementary Table 16). Notably, a single amino acid, p.K312 of ELMO1, is mutated in three tumors, which suggests a gain of function phenotype. DOCK2 is a guanine nucleotide exchange factor (GEF) that activates RAC1 directly through GTP loading27,34. To fully activate RAC1, DOCK2 and ELMO1 interact to relieve mutual autoinhibition28. In cancer models, ELMO1 and other DOCK family members have been associated with enhanced migration and invasion35,36. Mutations were also present in other RAC1 GEFs (TRIO, TIAM1, VAV2, and ECT2) (Supplementary Fig. 4). Furthermore, we previously observed focal copy-number gain of the 11q13 locus containing the serine/threonine kinase, PAK1, a principal downstream effector of RAC1 that has been shown to be oncogenic in breast cancer22,37. Taken together, the aberrant activation of genes related to RAC1 suggests that the motility pathway may be important to EAC.
Figure 3. Recurrent somatic alterations in ELMO1, DOCK2, and other RAC1 Guanine Nucleotide Exchange Factors (GEFs).
a) Schematic of protein alterations in DOCK2 and ELMO1 detected by WES. Coding alterations in EAC are colored either black (missense) or red (splice site/nonsense); silent mutations are depicted in gray. Conserved domain mapping is from UniProt; SH3, SRC Homology 3; DHR, Dlg homologous region, ELMO, Engulfment and Cell Motility; PH, Pleckstrin homology. b) Sample frequency (left) of candidate ELMO1 and DOCK2 as well as other Rac1-activating guanine nucleotide exchange factors in 145 WES EACs. c) ELMO1 wild-type or mutants (or GFP control) were expressed in NIH/3T3 cells using retroviral transduction with the pBabe vector. Cells were plated in matrigel invasion chambers with full serum containing medium in the lower chamber only, and invading cells from four fields were counted. Invading cells of 3 independent replicates are shown. Error bars represent S.D. P-values compare mutant ELMO1 to wild-type. n.s., not significant. Student’s t-test.
To examine the significance of ELMO1 mutations, wild-type and mutant ELMO1 constructs were generated and introduced into NIH/3T3 cells. Based on studies in glioblastoma demonstrating correlative increase in cellular invasion with overexpression of wild-type ELMO136, we hypothesized that ELMO1 mutations would enhance cell invasion. Compared to GFP control, wild-type ELMO1 increased invasion by 7-fold (P = 0.0040, Student’s T-test, unpaired) (Fig. 3C). ELMO1 mutations (p.F59L, p.K312E, p.K312T, p.K349R, p.T421N) further resulted in a significant increase (2 to 7-fold) in invasion compared to wild-type ELMO1 (P-values in figure). These results suggest that ELMO1 mutations can increase invasiveness and potentially contribute to tumorigenesis in EAC.
Additional significantly mutated genes include members of the SWI/SNF family of chromatin-remodeling factors: ARID1A, SMARCA4, and ARID2. Together, these genes are mutated in 20% of tumors. The enzymatic subunit of the chromatin-remodeling complex, SMARCA4, has been established as a putative tumor suppressor14,38. Likewise, ARID1A and ARID2 have been implicated as tumor suppressors in cancers including gastric cancer39–42. Interestingly, a candidate protein fusion identified by WGS also targets SMARCA4. The predicted fusion between exon 11 of SMARCA4 and exon 14 of DNM2 might point to an alteration that results in a loss-of-function gene-phenotype (Supplementary Table 6). Mutations were also found in other chromatin modifying enzymes: PBRM143 and JARID2. Taken together, 24% (35/145) of EACs harbored mutations in genes encoding chromatin-modifying factors (Supplementary Figs. 4 and 5).
Another intriguing gene is SPG20, mutated in 7% of EACs with five of the mutations generated by AA transversions (Supplementary Fig. 6). Spartin, the gene product of SPG20, was reportedly mutated in Troyer syndrome44, a genetic disorder characterized by progressive muscle stiffness and limb paralysis. The functions of Spartin include endosomal trafficking of growth factor receptors, inhibition of bone morphogenic protein signaling, and ubiquitin targeting45. More recently, SPG20 hypermethylation has been linked to colon cancer progression.46
TLR4 was mutated in 6% of EACs. Germline polymorphisms in TLR4 correlate with risk of in Helicobacter pylori-mediated gastric carcinoma47. In lung models, TLR4 deficiency contributes to enhanced inflammation and tumorigenesis48. TLR4 activates the innate immune response to pathogen exposure through heterodimerization with MD-249. Notably, the mutations in TLR4 fall between amino acids p.D379 and p.F487, a region critical for MD-2 interaction50. One mutation impacts p.E439, a site essential for hydrogen bonding of TLR4 to MD-2 (Supplementary Fig. 6). These mutations suggest disruption of the TLR4/MD-2 complex as a potential driver of tumor progression in EAC.
We also identified other significant candidates, including the protein kinase A anchoring protein, AKAP6 (mutated in 8% of samples), E3 ubiquitin ligase, HECW1 (8%), and AJAP1 (6%), which mediates signaling at adherens junctions and increases invasiveness in cancer cell lines51. NUAK1 (ARK5) is mutated 3% of samples, which is notable given that MYC-overexpressing hepatocellular carcinoma models are dependent on NUAK152. The lysine acetyltransferase MYST3, recurrently targeted for translocation in leukemia53, was also mutated in seven specimens (5%) (Supplementary Fig. 6).
Additional Candidate Genes from Exome Sequencing
Beyond the genes mutated at a statistically significant frequency, we queried the data for mutations of biologic significance given their recurrence in other cancers. We identified mutations in EAC that had been seen two or more times across all cancers in COSMIC database54; we found 22 such genes (Supplementary Table 17). Additionally, ten genes were significantly mutated (FDR q<0.1) in limited analysis of COSMIC gene territory including KRAS, CTNNB1, and ERBB2 (Supplementary Table 18). These results indicate that genes not reaching statistical significance in the cohort may harbor mutations of biologic relevance in individual tumors.
Mutations Targeting Therapeutically Relevant Genes
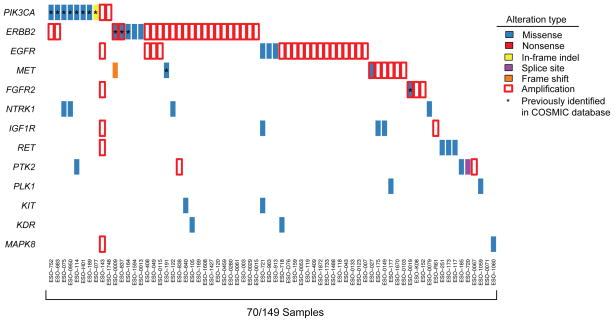
We queried the data for mutations in genes encoding therapeutic targets with inhibitors approved for clinical use or in preclinical development55. Mutations in actionable genes were discovered in 23% of tumors with PIK3CA being the most frequently mutated (Fig. 4). When also evaluating amplification status of genes, 48% of tumors in this cohort have a genomic alteration in a gene with a targeted agent. The high frequency of focally amplified therapeutic targets22 exceeds that of mutation in these same genes in EAC. Therefore, determining how to effectively treat tumors with amplified targets, especially RTKs, should be considered a priority.
Figure 4. Somatic alterations in frequently altered pathways in cancer, putative therapeutic targets, and treatment biomarkers.
a) Potential therapeutic targets or treatment biomarkers are listed by sample. Each column denotes an individual tumor and each row represents a gene. Mutations are colored by the type of mutation event, and genes with amplification of greater than four copies relative to a diploid baseline are marked by red box.
Somatic Alterations in Signal Transduction Pathways
To explore the functional impact of the mutations, we performed unbiased, GO-term enrichment in the overall ranked MutSig list using the 8,356 genes with at least one non-silent mutation56,57. GO processes related to cell adhesion and chemotaxis ranked as enriched near the top of the list (Supplementary Table 19). These findings support the hypothesis that enhanced cellular motility and invasiveness plays an important role in EAC disease progression.
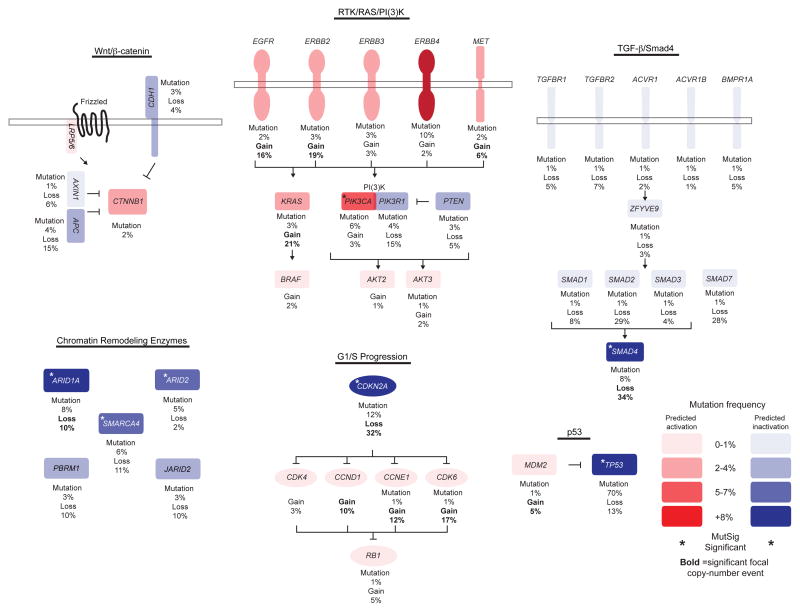
We also studied how cancer-associated pathways were disrupted by mutation in EAC. Cell-cycle control was altered by point mutation in 14% of EACs with most of the mutations occurring in CDKN2A (Fig. 5 and Supplementary Fig. 4). This process was also frequently affected by amplifications at the loci of CCND1, CCNE1, and CDK622. Although activation of β-catenin signaling is ubiquitous in CRC, mutations in this pathway were found in only 9% of EACs with two tumors having APC mutations that co-occur with either CDH1 or AXIN1 (Fig. 5 and Supplementary Fig. 4). Moreover, a potential AXIN1 fusion was identified by WGS in sample ESO-1060 spanning exon 5 of AXIN1 and exon 2 of GALNT7, which might alter normal gene function (Table 1). As in other cancer types, the TGFβ/SMAD signaling pathway was mutated in 18% of EAC tumors. The most recurrently altered gene in this pathway is SMAD4, which mutated in 10 samples and also subject to frequent copy-number loss (Fig. 5 and Supplementary Fig. 4).
Figure 5. Genetic alterations identified by WES across 145 EACs impacting the WNT/CTNNB1 (β-catenin), RTK/RAS/PI(3)K, TGFB1 (TGF-β)/SMAD4, Chromatin Remodeling Enzyme, RB1, and p53 pathways.
Percentages represent number of mutations in a given gene across the cohort. Genes that are predicted to be gain-of-function and loss-of-function are depicted in red and blue, respectively. Frequencies of alteration by mutation or copy-number alteration are shown. Color density of red or blue is based on mutation frequency of a given gene. Genes marked with an asterisk are significant by MutSig analysis. Genes subject to significant focal gain or loss in EAC22 have copy-number frequency marked in bold.
We evaluated the frequency and manner of somatic alterations in mitogen-activated protein kinase (MAPK) and phosphatidylinositol 3-kinase (PI3K), two common pathways required for proliferation and survival of cancer cells. Unlike other epithelial tumor types, where such MAPK-pathway mutations are common, no BRAF mutations were observed, and NF1 and KRAS mutations were seen in only three (2%) and five tumors (3%), respectively. Three of the five KRAS mutations alter p.G12; however one EAC harbored a KRAS c.351A>C (p.K117N) event, a mutation caused by an AA transversion and previously observed in CRC58. The PI3K pathway was the most frequently altered oncogenic pathway altered by mutation (13%). PIK3CA was mutated in seven tumors, followed by PIK3R1 and PTEN in five and four tumors, respectively (Fig. 4b and Supplementary Fig. 4).
We explored mutations in the ErbB family of RTKs, which are important therapeutic targets in many cancer types. Although three samples harbored EGFR mutations, these alterations were not previously annotated in other tumors. Moreover, two of these alterations, p.S447Y and p.S1153I, were predicted by Polyphen-2 score59 to not be deleterious to normal function, and thus of questionable biologic significance. By contrast, ERBB2 mutations were present in five tumors. Three mutations were in the kinase domain including two c.351A>C (p.D769Y) mutations and one c.2327G>T (p.G776V). These alterations have been observed previously in other cancers60–62.
Discussion
Here, through mutation analysis, we provide insight into the somatically-altered genes and signaling pathways as well as confirm the a high rate of A>C transversions in EAC13. We further establish that the rates of these mutations are highest in non-coding areas, and within coding areas are overrepresented in less-expressed genes. Additionally, we demonstrate context specificity showing that A>C transversions are most common when the mutated adenine follows a 5′ flanking adenine (AA) and especially at AAG trinucleotides.
This mutational spectrum appears to be unique to EAC suggesting that these mutations are attributable to gastroesophageal reflux, where the gastric and duodenal contents travel into the lower esophagus creating an environment of inflammation21. Prior studies have linked particular substances such as bile acids, nitrosamines, and reactive oxygen species to the development of metaplasia and carcinomas63, but the precise mutagen(s) remain poorly understood. Experiments in E. coli exploring the mutagenic potential of an oxidatively damaged DNA precursor, 8-hydroxydeoxyguanosine triphosphate, demonstrated that it preferentially induces A>C transversions64. These data suggest that A>C transversions in EAC may arise from oxidative damage induced by GERD; however, experimental evidence is necessary to identify a culprit stimulus. The identification of this mutational signature enables future studies to define specific carcinogen(s) that contribute to EAC and potentially aid in the explanation of the rising incidence.
Statistical analysis also enabled a comprehensive assessment of mutated genes in EAC and identified mutations in cancer-related genes such as TP53, CDKN2A, SMAD4, and PIK3CA. It was notable that most well-annotated cancer genes were not affected by AA transversions. In many cases, it is impossible to generate hotspot mutations such as KRAS p.G12 or PIK3CA p.E545 with an AA transversion. Additionally, given the base composition of stop codons it is difficult to generate nonsense events from AA transversions and impossible to create a stop mutation from such an A>C transversion when it occurs in an AAG trinucleotide context, the most common context for AA mutations in our dataset. Of the 2,570 coding mutations caused by these events, none is a predicted nonsense mutation. Moreover, the data suggest that AA transversions accumulate in lower expressed genes, thus reducing their prevalence in genes contributing to oncogenesis. Despite these caveats, it is likely that mutations caused by AA transversions do impact genes relevant for these tumors. For example, a known transforming mutation in KRAS (c.351A>C; p.K117N) is created by an AA transversion.
Consistent with previous reports38–43, loss-of-function mutations in chromatin-remodeling enzymes are common in EAC. Prior gene studies have also suggested frequent activation of the MAPK, PI3K, and β-catenin pathways. The data presented here verify the presence of frequent mutations in the PI3K cascade, but argue against wide-reaching mutations in these pathways, thus drawing contrasts between EAC and CRC, where β-catenin activation and missense mutations of KRAS and BRAF are highly prevalent65.
For the first time, we detected EAC mutations in regulators of invasion and motility including significantly recurrent mutations in DOCK2 and ELMO1. These mutations may increase tumor fitness through alteration of cytoskeletal structure, increased invasive properties, or mitogenesis. We demonstrate that ELMO1 mutations augment cellular invasiveness; thus, suggesting one mechanism by which these events contribute to tumorigenesis. Given that EAC is a highly-invasive tumor prone to early metastasis, alterations in the RAC1 pathway may contribute to this phenotype.
Although we identified potentially actionable genomic alterations in 48% of samples, the ERBB2 (HER2)-targeted antibody, trastuzumab, is the only targeted agent used in the treatment of EAC/GEJ adenocarcinomas with its use guided by overexpression and genomic amplification of ERBB266. Currently, ERBB2 mutation assessment is not performed for EAC, despite ERBB2 being altered by both co-occurring amplification and mutation in 3% of samples. These data point to a potential role of mutation as an additional biomarker to guide use of ERBB2 (HER2)-targeting agents.
The limited knowledge of the genomic aberrations underlying EAC has hindered the development of new therapies. Numerous candidates, not previously implicated in this disease, have emerged from this analysis. Functional study of these genes will be required to validate and understand their roles in tumorigenesis and to identify the etiology to the unique spectrum of the observed AA transversions. These data provide an enhanced roadmap for the study of EAC and the much-needed development of new therapies for these deadly cancers.
Online Methods
DNA extraction and sample collection
All samples were obtained under institutional IRB approval and with documented informed consent. All samples were fresh frozen primary resections from patients not treated with prior chemotherapy or radiation. Hematoxylin and eosin stained slides were examined by a board-certified pathologist to select cases with estimated carcinoma content >70%. DNA was extracted using salt precipitation or phenol chloroform extraction. DNA was quantified using Picogreen dsDNA Quantitation Reagent (Invitrogen).
Whole exome sequencing
Whole-exome capture libraries were constructed from 100ng of tumor and normal DNA following shearing, end repair, phosphorylation and ligation to barcoded sequencing adapters67. Ligated DNA was size-selected for lengths between 200–350bp and subjected to exonic hybrid capture using SureSelect v2 Exome bait (Agilent). Samples were multiplexed and sequenced on multiple Illumina HiSeq flowcells to average target exome coverage of 83.3x was achieved in neoplastic DNA and 85.9x in the non-cancerous tissue.
Whole genome sequencing
Whole-genome sequencing library construction was done with 500ng of native DNA from primary tumor and germline samples for each patient. The DNA was sheared to a range of 101–700 bp using the Covaris E210 Instrument, and then phosphorylated and adenylated according to the Illumina protocol. Adapter ligated purification was done by preparatory gel electrophoresis (4% agarose, 85 volts, 3 hours), and size was selected by excision of two bands (500–520 bp and 520–540 bp respectively) yielding two libraries per sample with average of 380 bp and 400 bp respectively15,16,20. Qiagen Min-Elute column based clean ups were performed after each step. For a subset of samples, gel electrophoresis and extraction was performed using the automated Pippin Prep system (Sage Science, Beverly MA). Libraries were then sequenced with the Illumina GA-II or Illumina HiSeq sequencer with 101 bp reads, achieving an average of ~30X coverage depth.
Identification of Rearrangements
The dRanger algorithm was used to detect genomic rearrangements by identifying instances where the two read pairs map to distinct regions of the genome or map in a manner that suggests another structural event such as an inversion. Candidate somatic rearrangements were queried in both the matched normal genome and a panel of non-tumor genomes to remove germline events. The final scorings of these somatic reads were then calculated by multiplying the number of supporting read pairs by the estimated quality of the candidate rearrangement (0 to 1). This metric is generated by taking into account the ability to align of the two regions joined by the putative rearrangement and the chance of detecting such a read pair given the library fragment-size distribution. Events with scores ≥4 (observed in at least four read pairs) were included in this analysis.
Validation of selected mutations by mass spectrometry genotyping
A total of 45 intergenic AA>C mutations were selected validation in tumor and germline sample using mass spectrometry genotyping (Sequenom). Mutations were randomly selected across six samples and sites chosen all had estimated mutation allelic fractions exceeding 30% thus enabling mutation detection19,25,68. Of those assays performed, 25 yielded interpretable data with others failing due to lack of PCR amplification or probe hybridization in tumor and/or germline sample.
Sequence data processing and quality control
Exome and whole-genome sequence data processing and analysis were performed using Broad Institute pipelines15,16,25,68. A BAM file aligned to the hg19 human genome build was generated from Illumina sequencing reads for each tumor and normal sample by the “Picard” pipeline. The “Firehose” pipeline was used to manage input and output files and submit analyses for execution in GenePattern69.
Quality control modules in Firehose were used to compare genotypes derived from Affymetrix arrays and sequencing data to ensure concordance. Genotypes from SNP arrays were also used to monitor for low levels of cross-contamination between samples from different individuals in sequencing data using the ContEst algorithm70. One tumor/normal pair (ESO-774) analyzed by WGS was not included in the exome analysis as the exome sequencing from that case failed quality control metrics.
Mutation calling
The MuTect algorithm was used to identify somatic mutations in targeted exons and whole-genome data14,16,25. MuTect identifies candidate somatic mutations by Bayesian statistical analysis of bases and their qualities in the tumor and normal BAMs at a given genomic locus. We required a minimum of 14 reads covering a site in the tumor and 8 in the normal for declaring a site is adequately covered for mutation calling. We determined the lowest allelic fraction at which somatic mutations could be detected on a per-sample basis, using estimates of cross-contamination from the ContEst pipeline70. Small somatic insertions and deletions were detected using the Indelocator algorithm after local realignment of tumor and normal sequences14. All somatic mutations detected by WES were analyzed for potential false-positive calls by performing a comparison to mutation calls from a panel of 2,500 germline DNA samples. Mutations found in 2% of the germline samples or 2% of sequencing reads were removed from analysis. MutSig significant mutations except for all TP53 mutants were reviewed manually in the respective BAM file using the Integrative Genomics Viewer.
Mutation annotation
Somatic point, insertion, and deletion mutations were annotated using information from publicly available databases, including the UCSC Genome Browser’s UCSC Genes track71, miRBase release 1572, dbSNP build 13273, UCSC Genome Browser’s ORegAnno track74, UniProt release 2011_0375, and COSMIC v5154.
Mutation Significance Analysis
For the purpose of discovering recurrently mutated genes, we used the MutSig algorithm, as described in the following studies18. In short, this method builds a background model of mutational processes, which takes into account the genome-wide variability in mutation rates. We achieve this by considering different covariates that have been shown to affect mutation rate: GC content (measured on 100kb windows), local relative replication time76,77, open vs. closed chromatin status as determined by “HiC” – fine-scale mapping of the three-dimensional DNA contacts in the nucleus78, gene expression16, and finally, the local gene density measured in a 1 Mb window. For each gene, we define a set of nearest neighbors according to these covariates, and estimate the background mutation rate from noncoding (flanking and introns) and silent mutations of these neighbors. We then assign a score based on the ratio between the number of nonsilent coding mutation rate of the gene and the noncoding and silent mutation rate of the given gene and its neighbors. Furthermore, in order to increase power in detecting known driver genes, we performed an independent significance analysis that is restricted to events that have been previously reported in the COSMIC database.
Microsatellite Instability Testing
MSI (microsatellite instability) analysis was performed using 10 microsatellite markers (D2S123, D5S346, D17S250, BAT25, BAT26, BAT40, D18S55, D18S56, D18S67 and D18S487) as described previously22.
Copy-number calling from whole exome sequencing
Copy-ratios were calculated as the ratio of tumor read-depth to the average read-depth observed in normal samples for that region using the CapSeg DNA-sequencing based tool (McKenna, et al. in preparation).
Processing of Affymetrix expression arrays
Raw data was processed using the gene chip robust multiarray averaging79 (RMA) approach to provide normalized expression data for each probe set on the arrays.
Cell lines and culture conditions
NIH/3T3 and 293 cells were obtained from American Type Culture Collection (Manassas, VA). All cells were maintained in DMEM plus 10% FBS at 37°C in 5% CO2.
ELMO1 site-directed mutagenesis
The full-length ELMO1 cDNA was obtained from Open Biosystems-Thermo Scientific (Lafayette, CO) and cloned into the EcoRI site of pBabe(puro). Mutants were generated by site-directed mutagenesis using the Quikchange II Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA) according to the manufacturer’s instructions. All mutations were verified by sequencing.
ELMO1 retrovirus production and cell infection
Wild-type ELMO1, ELMO1 mutants, or GFP in the pBabe(puro) vector (1 μg) was co-transfected with 1 μg of pCL-Eco into 293 cells with Fugene HD (Roche, Indianapolis, IN) overnight. The growth media was replaced with new full serum medium after 24 h. After an additional 24 h, the retroviral supernatant was harvested and replaced fresh media. Retroviral supernatant was filtered and incubated with target NIH/3T3 cells in the presence of 5 μg/mL polybrene (hexadimethrine bromide). This procedure was repeated again after 24 h. Stably-infected cells were selected for under puromycin (1μg/mL) pressure for 2 weeks. Positive expression was confirmed by western blotting with an ELMO1 antibody-ab2239 (Abcam, Cambridge, MA).
Matrigel Invasion Assay
Growth Factor Reduced Matrigel-coated Transwell chambers (BD Biosciences, San Jose, CA) were activated in serum-free media at 37°C for 2 h. NIH/3T3 cells (1 × 104) were plated in matrigel invasion chambers with full serum containing medium in the lower chamber only. After 24 h, non-invading cells in the top chamber were removed by cotton swab, and invading cells were fixed and stained using Diff-Quik staining solutions according to the manufacturer’s instructions (VWR International, Radnor, PA). The number of invading cells from each of four fields were counted at 20X magnification.
Supplementary Material
Acknowledgments
We thank Matthew Meyerson for helpful discussions and review of the manuscript and thank members of the Broad Institute Biological Samples Platform, Genetic Analysis Platform, and Genome Sequencing Platform for their assistance. We are also grateful for the physicians and hospital staff whose effort in collecting these samples is essential to this research. This work was supported by the US National Human Genome Research Institute (NHGRI) Large Scale Sequencing Program (U54 HG003067 to the Broad Institute, E.S.L.), National Cancer Institute (K08 CA134931 to A.J.B.), DeGregorio Family Foundation (A.J.B.), Karin Grunebaum Cancer Research Foundation (A.J.B), Target Cancer (A.J.B.), and Connecticut Conquers Cancer (A.J.B.). S.O. and Y.I. are supported by National Cancer Institute (R01 CA151993 to S.O.) and the Dana-Farber/Harvard Cancer Center GI Cancer SPORE (US National Institutes of Health P50 CA127003),. D.G.B is supported by grant CA163059 and CA46592.
Footnotes
Accession codes
Binary sequence alignment/map (BAM) files were deposited in the database of Genotypes and Phenotypes (phs000598.v1.p1). Raw data mRNA expression profiles on 14 EAC samples have been deposited at the Gene Expression Omnibus (GSE42363).
Author Contributions
P.S., S.P., M.S.L., C.F., C.S., S.E.S., A.M., K.C., A.S., S.L.C., G.S., D.V., A.H.R., and R.B. performed computational analyses. E.S., D.A., K.T., C.S., R.C.O., C.G., and S.B.G. processed samples and supervised exome sequencing. A.M.D., S.B., D.Z., L.L., J.L., R.R., A.C., J.D.L., A.P., D.G.B., T.E.G., and A.J.B. coordinated sample acquisition, processing, pathologic review, and analysis. Y.I. and S.O. performed MSI testing. A.M.D., P.S., T.R.G., S.B.G., E.S.L., G.G., and A.J.B. designed the study. A.M.D., P.S., S.P., M.S.L., G.G., and A.J.B. analyzed the data and wrote the manuscript.
URLs
MutSig Algorithm, http://confluence.broadinstitute.org/display/CGATools/MutSig; CCDS, http://www.ncbi.nlm.nih.gov/CCDS/; Broad Institute Picard Sequencing Pipeline, http://picard.sourceforge.net/; Broad Institute Firehose Pipeline, http://www.broadinstitute.org/cancer/cga; Supplementary Table 3: Whole genome sequencing mutation list, http://www.broadinstitute.org/~shouyong/eac/
References
- 1.Holmes RS, Vaughan TL. Epidemiology and pathogenesis of esophageal cancer. Semin Radiat Oncol. 2007;17:2–9. doi: 10.1016/j.semradonc.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 2.Pohl H, Welch HG. The role of overdiagnosis and reclassification in the marked increase of esophageal adenocarcinoma incidence. J Natl Cancer Inst. 2005;97:142–6. doi: 10.1093/jnci/dji024. [DOI] [PubMed] [Google Scholar]
- 3.Wu AH, Wan P, Bernstein L. A multiethnic population-based study of smoking, alcohol and body size and risk of adenocarcinomas of the stomach and esophagus (United States) Cancer Causes Control. 2001;12:721–32. doi: 10.1023/a:1011290704728. [DOI] [PubMed] [Google Scholar]
- 4.Chung SM, Kao J, Hyjek E, Chen YT. p53 in esophageal adenocarcinoma: a critical reassessment of mutation frequency and identification of 72Arg as the dominant allele. Int J Oncol. 2007;31:1351–5. [PubMed] [Google Scholar]
- 5.Hardie LJ, et al. p16 expression in Barrett’s esophagus and esophageal adenocarcinoma: association with genetic and epigenetic alterations. Cancer Lett. 2005;217:221–30. doi: 10.1016/j.canlet.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 6.Choi YW, Heath EI, Heitmiller R, Forastiere AA, Wu TT. Mutations in beta-catenin and APC genes are uncommon in esophageal and esophagogastric junction adenocarcinomas. Mod Pathol. 2000;13:1055–9. doi: 10.1038/modpathol.3880194. [DOI] [PubMed] [Google Scholar]
- 7.Sommerer F, et al. Mutations of BRAF and KRAS2 in the development of Barrett’s adenocarcinoma. Oncogene. 2004;23:554–8. doi: 10.1038/sj.onc.1207189. [DOI] [PubMed] [Google Scholar]
- 8.Wijnhoven BP, de Both NJ, van Dekken H, Tilanus HW, Dinjens WN. E-cadherin gene mutations are rare in adenocarcinomas of the oesophagus. Br J Cancer. 1999;80:1652–7. doi: 10.1038/sj.bjc.6690577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puhringer-Oppermann FA, Stein HJ, Sarbia M. Lack of EGFR gene mutations in exons 19 and 21 in esophageal (Barrett’s) adenocarcinomas. Dis Esophagus. 2007;20:9–11. doi: 10.1111/j.1442-2050.2007.00630.x. [DOI] [PubMed] [Google Scholar]
- 10.Guo M, Liu S, Lu F. Gefitinib-sensitizing mutations in esophageal carcinoma. N Engl J Med. 2006;354:2193–4. doi: 10.1056/NEJMc052698. [DOI] [PubMed] [Google Scholar]
- 11.Phillips WA, et al. Mutation analysis of PIK3CA and PIK3CB in esophageal cancer and Barrett’s esophagus. Int J Cancer. 2006;118:2644–6. doi: 10.1002/ijc.21706. [DOI] [PubMed] [Google Scholar]
- 12.Boonstra JJ, et al. Mapping of homozygous deletions in verified esophageal adenocarcinoma cell lines and xenografts. Genes Chromosomes Cancer. 2012;51:272–82. doi: 10.1002/gcc.20952. [DOI] [PubMed] [Google Scholar]
- 13.Agrawal N, et al. Comparative Genomic Analysis of Esophageal Adenocarcinoma and Squamous Cell Carcinoma. Cancer Discov. 2012;2:899–905. doi: 10.1158/2159-8290.CD-12-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berger MF, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–20. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chapman MA, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–72. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imielinski M, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–20. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammerman PS, et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012 doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berger MF, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–6. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bass AJ, et al. Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat Genet. 2011;43:964–8. doi: 10.1038/ng.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orlando RC. Mucosal Defense in Barrett’s Esophagus. In: Sharma P, editor. Barrett’s Esophagus and Esophageal Adenocarcinoma. Blackwell Publishing, Ltd; Oxford, UK: 2006. pp. 60–72. [Google Scholar]
- 22.Dulak AM, et al. Gastrointestinal adenocarcinomas of the esophagus, stomach and colon exhibit distinct patterns of genome instability and oncogenesis. Cancer Res. 2012 doi: 10.1158/0008-5472.CAN-11-3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banerji S, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486:405–9. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barbieri CE, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–9. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farris AB, 3rd, et al. Clinicopathologic and molecular profiles of microsatellite unstable Barrett Esophagus-associated adenocarcinoma. Am J Surg Pathol. 2011;35:647–55. doi: 10.1097/PAS.0b013e31820f18a2. [DOI] [PubMed] [Google Scholar]
- 27.Sanui T, et al. DOCK2 regulates Rac activation and cytoskeletal reorganization through interaction with ELMO1. Blood. 2003;102:2948–50. doi: 10.1182/blood-2003-01-0173. [DOI] [PubMed] [Google Scholar]
- 28.Hanawa-Suetsugu K, et al. Structural basis for mutual relief of the Rac guanine nucleotide exchange factor DOCK2 and its partner ELMO1 from their autoinhibited forms. Proc Natl Acad Sci U S A. 2012;109:3305–10. doi: 10.1073/pnas.1113512109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gomez del Pulgar T, Benitah SA, Valeron PF, Espina C, Lacal JC. Rho GTPase expression in tumourigenesis: evidence for a significant link. Bioessays. 2005;27:602–13. doi: 10.1002/bies.20238. [DOI] [PubMed] [Google Scholar]
- 30.Hodis E, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kissil JL, et al. Requirement for Rac1 in a K-ras induced lung cancer in the mouse. Cancer Res. 2007;67:8089–94. doi: 10.1158/0008-5472.CAN-07-2300. [DOI] [PubMed] [Google Scholar]
- 32.Pan Y, et al. Expression of seven main Rho family members in gastric carcinoma. Biochem Biophys Res Commun. 2004;315:686–91. doi: 10.1016/j.bbrc.2004.01.108. [DOI] [PubMed] [Google Scholar]
- 33.Sander EE, et al. Matrix-dependent Tiam1/Rac signaling in epithelial cells promotes either cell-cell adhesion or cell migration and is regulated by phosphatidylinositol 3-kinase. J Cell Biol. 1998;143:1385–98. doi: 10.1083/jcb.143.5.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishihara H, et al. Non-adherent cell-specific expression of DOCK2, a member of the human CDM-family proteins. Biochim Biophys Acta. 1999;1452:179–87. doi: 10.1016/s0167-4889(99)00133-0. [DOI] [PubMed] [Google Scholar]
- 35.Sanz-Moreno V, et al. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 2008;135:510–23. doi: 10.1016/j.cell.2008.09.043. [DOI] [PubMed] [Google Scholar]
- 36.Jarzynka MJ, et al. ELMO1 and Dock180, a bipartite Rac1 guanine nucleotide exchange factor, promote human glioma cell invasion. Cancer Res. 2007;67:7203–11. doi: 10.1158/0008-5472.CAN-07-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shrestha Y, et al. PAK1 is a breast cancer oncogene that coordinately activates MAPK and MET signaling. Oncogene. 2012;31:3397–408. doi: 10.1038/onc.2011.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Medina PP, et al. Frequent BRG1/SMARCA4-inactivating mutations in human lung cancer cell lines. Hum Mutat. 2008;29:617–22. doi: 10.1002/humu.20730. [DOI] [PubMed] [Google Scholar]
- 39.Zang ZJ, et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet. 2012;44:570–4. doi: 10.1038/ng.2246. [DOI] [PubMed] [Google Scholar]
- 40.Fujimoto A, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44:760–764. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- 41.Jones S, et al. Somatic mutations in the chromatin remodeling gene ARID1A occur in several tumor types. Hum Mutat. 2012;33:100–3. doi: 10.1002/humu.21633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guan B, Wang TL, Shih Ie M. ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers. Cancer Res. 2011;71:6718–27. doi: 10.1158/0008-5472.CAN-11-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Varela I, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–42. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patel H, et al. SPG20 is mutated in Troyer syndrome, an hereditary spastic paraplegia. Nat Genet. 2002;31:347–8. doi: 10.1038/ng937. [DOI] [PubMed] [Google Scholar]
- 45.Bakowska JC, Jupille H, Fatheddin P, Puertollano R, Blackstone C. Troyer syndrome protein spartin is mono-ubiquitinated and functions in EGF receptor trafficking. Mol Biol Cell. 2007;18:1683–92. doi: 10.1091/mbc.E06-09-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lind GE, et al. SPG20, a novel biomarker for early detection of colorectal cancer, encodes a regulator of cytokinesis. Oncogene. 2011;30:3967–78. doi: 10.1038/onc.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garza-Gonzalez E, et al. Assessment of the toll-like receptor 4 Asp299Gly, Thr399Ile and interleukin-8-251 polymorphisms in the risk for the development of distal gastric cancer. BMC Cancer. 2007;7:70. doi: 10.1186/1471-2407-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bauer AK, et al. Toll-like receptor 4 in butylated hydroxytoluene-induced mouse pulmonary inflammation and tumorigenesis. J Natl Cancer Inst. 2005;97:1778–81. doi: 10.1093/jnci/dji403. [DOI] [PubMed] [Google Scholar]
- 49.Kennedy MN, et al. A complex of soluble MD-2 and lipopolysaccharide serves as an activating ligand for Toll-like receptor 4. J Biol Chem. 2004;279:34698–704. doi: 10.1074/jbc.M405444200. [DOI] [PubMed] [Google Scholar]
- 50.Park BS, et al. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458:1191–5. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 51.Schreiner A, et al. Junction protein shrew-1 influences cell invasion and interacts with invasion-promoting protein CD147. Mol Biol Cell. 2007;18:1272–81. doi: 10.1091/mbc.E06-07-0637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu L, et al. Deregulated MYC expression induces dependence upon AMPK-related kinase 5. Nature. 2012;483:608–12. doi: 10.1038/nature10927. [DOI] [PubMed] [Google Scholar]
- 53.Pelletier N, Champagne N, Stifani S, Yang XJ. MOZ and MORF histone acetyltransferases interact with the Runt-domain transcription factor Runx2. Oncogene. 2002;21:2729–40. doi: 10.1038/sj.onc.1205367. [DOI] [PubMed] [Google Scholar]
- 54.Forbes SA, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–50. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garnett MJ, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–5. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eden E, Lipson D, Yogev S, Yakhini Z. Discovering motifs in ranked lists of DNA sequences. PLoS Comput Biol. 2007;3:e39. doi: 10.1371/journal.pcbi.0030039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith G, et al. Activating K-Ras mutations outwith ‘hotspot’ codons in sporadic colorectal tumours - implications for personalised cancer medicine. Br J Cancer. 2010;102:693–703. doi: 10.1038/sj.bjc.6605534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ikediobi ON, et al. Mutation analysis of 24 known cancer genes in the NCI-60 cell line set. Mol Cancer Ther. 2006;5:2606–12. doi: 10.1158/1535-7163.MCT-06-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee JW, et al. ERBB2 kinase domain mutation in the lung squamous cell carcinoma. Cancer Lett. 2006;237:89–94. doi: 10.1016/j.canlet.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 62.Lee JW, et al. Somatic mutations of ERBB2 kinase domain in gastric, colorectal, and breast carcinomas. Clin Cancer Res. 2006;12:57–61. doi: 10.1158/1078-0432.CCR-05-0976. [DOI] [PubMed] [Google Scholar]
- 63.Badreddine RJ, Wang KK. Barrett esophagus: an update. Nat Rev Gastroenterol Hepatol. 2010;7:369–78. doi: 10.1038/nrgastro.2010.78. [DOI] [PubMed] [Google Scholar]
- 64.Inoue M, et al. Induction of chromosomal gene mutations in Escherichia coli by direct incorporation of oxidatively damaged nucleotides. New evaluation method for mutagenesis by damaged DNA precursors in vivo. J Biol Chem. 1998;273:11069–74. doi: 10.1074/jbc.273.18.11069. [DOI] [PubMed] [Google Scholar]
- 65.MacConaill LE, et al. Profiling critical cancer gene mutations in clinical tumor samples. PLoS One. 2009;4:e7887. doi: 10.1371/journal.pone.0007887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bang YJ, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–97. doi: 10.1016/S0140-6736(10)61121-X. [DOI] [PubMed] [Google Scholar]
- 67.Fisher S, et al. A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol. 2011;12:R1. doi: 10.1186/gb-2011-12-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stransky N, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–60. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reich M, et al. GenePattern 2.0. Nat Genet. 2006;38:500–1. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 70.Cibulskis K, et al. ContEst: estimating cross-contamination of human samples in next-generation sequencing data. Bioinformatics. 2011;27:2601–2. doi: 10.1093/bioinformatics/btr446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fujita PA, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–82. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–7. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Griffith OL, et al. ORegAnno: an open-access community-driven resource for regulatory annotation. Nucleic Acids Res. 2008;36:D107–13. doi: 10.1093/nar/gkm967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 2011;39:D214–9. doi: 10.1093/nar/gkq1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen CL, et al. Impact of replication timing on non-CpG and CpG substitution rates in mammalian genomes. Genome Res. 2010;20:447–57. doi: 10.1101/gr.098947.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stamatoyannopoulos JA, et al. Human mutation rate associated with DNA replication timing. Nat Genet. 2009;41:393–5. doi: 10.1038/ng.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–93. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.