Abstract
Acute myeloid leukemia (AML) remains a therapeutic challenge despite increasing knowledge about the molecular origins of the disease, as the mechanisms of AML cell escape from chemotherapy remain poorly defined. We hypothesized that AML cells are addicted to molecular pathways in the context of chemotherapy and used complementary approaches to identify these addictions. Using novel molecular and computational approaches, we performed genome-wide shRNA screens to identify proteins that mediate AML cell fate after cytarabine exposure, gene expression profiling of AML cells exposed to cytarabine to identify genes with induced expression in this context, and examination of existing gene expression data from primary patient samples. The integration of these independent analyses strongly implicates cell cycle checkpoint proteins, particularly WEE1, as critical mediators of AML cell survival after cytarabine exposure. Knockdown of WEE1 in a secondary screen confirmed its role in AML cell survival. Pharmacologic inhibition of WEE1 in AML cell lines and primary cells is synergistic with cytarabine. Further experiments demonstrate that inhibition of WEE1 prevents S-phase arrest induced by cytarabine, broadening the functions of WEE1 that may be exploited therapeutically. These data highlight the power of integrating functional and descriptive genomics, and identify WEE1 as potential therapeutic target in AML.
Keywords: acute myeloid leukemia, WEE1, genomics, shRNA screen, cell cycle, checkpoint
Introduction
In aggregate, AML continues to have the lowest survival rates of all leukemias (1). In children with AML, long-term survival rates for all patients average around 50–60% and survival rates for older adults are generally worse, and fewer than 10% of elderly patients survive long-term after diagnosis of AML (1, 2). AML is highly molecularly heterogeneous and data suggest at least two cooperating mutations resulting in impaired differentiation (Class II mutation) and enhanced proliferation or survival (Class I mutation) are required for AML genesis (3). MLL rearrangement is an example of Class II mutation, while mutations of several kinases, including Flt3 and c-Kit, are examples of Class I mutations (3).
While the clinical and molecular features of AML can be used to predict response to therapy, with notable exceptions, this information is not used to guide therapy. For over three decades, treatment has remained dependent upon remission induction with cytarabine (ARA-C) and an anthracycline like daunorubicin (4). While attempts to optimize doses of cytotoxic chemotherapeutics have improved outcomes (5), some studies suggest that dose limits may have been reached and that new agents are needed to further improve outcomes (6–8). Cytarabine and daunorubicin are conventional chemotherapeutics with multiple mechanisms of action (9). While some of the mechanisms of cell death in AML cells exposed to these drugs have been determined, the mechanisms by which a minority of cells survives are not as well understood (10).
Traditional approaches to studying cancer have resulted in dramatic improvements in outcomes for certain populations. The quintessential success of the candidate gene approach is that of CML, for which the oncogene BCR-ABL was discovered, its various functions determined, and targeted therapy successfully translated into clinical care (11). However, the majority of cancers, including AML, are much more molecularly heterogeneous than CML, and candidate gene approaches are slow and cumbersome. Thus, novel approaches to more rapidly identify and validate mediators of cell survival have the potential to dramatically alter our understanding of cellular biology and develop novel therapeutic strategies. For example, shRNA libraries have been used to identify positive and negative regulators of cancer cell survival with or without drug treatment (12–18). We have hypothesized that escape pathways become essential to cancer cell survival in the context of specific cancer therapy. Herein we describe the first genome-wide functional genetic screen in AML, which we undertook to identify such escape pathways. Integrating this functional assay with more descriptive analyses of the genome, we have identified that several proteins involved in cell cycle checkpoints are critical mediators of AML cell fate after exposure to cytarabine. One of these molecules in particular, WEE1, can be inhibited pharmacologically, sensitizing AML cells to cytarabine. Contrary to studies in other cancers, inhibition of WEE1 abrogates the S phase arrest induced by cytarabine, broadening the therapeutic potential of WEE1 inhibition. These studies highlight the power of integrating functional and descriptive genomics and implicate WEE1 as a novel therapeutic target in AML.
Materials & Methods
Lentivirus preparation
Virus containing media was prepared as previously described (19). For the genome-wide screen, the HIV based GeneNet Lentiviral Human 50K library was used (pSIH1-H1-Puro, System Biosciences, Mountain View, CA) with minor modifications to the manufacturer’s recommendations. For high-throughput and final validation, TRC HIV based plasmids (pLKO.1) were used.
Cell culture
All cells were cultured at 37° in humidified air supplemented with 5% CO2. 293FT cells were cultured in DMEM with 10% FBS and penicillin/streptomycin. Molm13, MV4-11, and U937 cells were cultured in RPMI with 10% FBS and penicillin/streptomycin. The IC75 of ARA-C for Molm13 and MV4-11 cells was determined to be 20nM and 800nM respectively. Murine bone marrow cells were collected from tibiae and femora of 2 C57Bl/6 mice and cultured in IMDM with 10% FBS, and cytokines as previously described (19). Primary AML samples were collected after informed consent with approval of the Colorado Multiple Institutional Review Board. Ficoll separated mononuclear cells were cultured in StemSpan SFEM (Stem Cell Technologies, Vancouver, BC, Canada) with 20% FBS, initially at 0.6–4×105/ml.
Genome-wide functional genetic screening
Transduction, puromycin selection, and treatment of Molm13 and MV4-11 cells were performed using conditions optimized for each cell line. Three replicates each of untreated and cytarabine treated were incubated for 72 hours. Five-fold more cells were cytarabine treated than left untreated to prevent stochastic loss of representation of shRNA tags. After drug exposure, cells were counted by propidium iodide (Sigma) exclusion and flow cytometry (Guava EasyCyte Plus, Millipore), re-plated in fresh media without drug, and counted and re-plated every 72 hours thereafter, until there were sufficient numbers of cells for harvest of RNA. shRNA tags were isolated and prepared for sequencing and quantification on the Illumina Genome AnalyzerIIx (Illumina, Inc., San Diego, CA; See Supplemental Information for more details).
Targeted high-throughput validation
Pooled, shRNA-expressing plasmids from the TRC1 and TRC1.5 library were provided through the Functional Genomics Core of the University of Colorado Cancer Center. If available, 2 shRNAs per target that were validated by TRC were included in the pool. If validated constructs were not available, 5 shRNAs per target were included. Transduced, puromycin selected Molm13 cells were left untreated or treated with ARA-C, with 5 replicates per condition. After recovery from treatment, shRNA tags were isolated, barcoded per replicate and prepared for sequencing and quantification on the Illumina Genome AnalyzerIIx (Supplemental Information). Genes were considered validated if 1 of 2 shRNAs or if 2 of 3 or more shRNAs were statistically significantly differentially represented in the expected direction. P-values were generated using edgeR (20).
Gene expression analysis
Molm13 cells were treated with cytarabine 20nM for 24 hours, total RNA was isolated, reverse transcribed, analyzed by Affymetrix GeneChip Human Genome U133 Plus 2.0 microarray, and data were analyzed as previously described (21). The top 100 genes with the greatest changes in signal to noise ratios in either direction were considered in subsequent analyses. Oncomine (Oncomine.org) was used to evaluate existing data sets for differential expression of a subset of genes using default settings (22, 23).
Pharmacologic validation
Cytarabine and hydroxyurea were purchased from Sigma-Aldrich (St. Louis, MO); MK1775 from Axon Medchem (Groningen, The Netherlands); and WEE1 inhibitor II (BCHCD) from EMD Chemicals (Philadelphia, PA). AML cells were diluted to 2×105 cells/ml, treated with ARA-C, hydroxyurea (HU), and/or MK1775 or BCHCD at the indicated concentrations, in duplicate or triplicate. DMSO was kept to less than 0.1% final concentration for all experiments. Cells were counted 72 hours later by flow cytometry and PI exclusion. In some experiments, an aliquot of the remaining cells was diluted 1:10, re-plated, incubated for another 72 hours and counted again. The extent of proliferation was calculated as previously described (19). The extent of inhibition relative to DMSO treated cells were input into CalcuSyn (Biosoft, Cambridge, UK) to calculate CI values. CI values less than 1 are considered to represent synergistic inhibition of cellular proliferation (24). Apoptosis was assessed using the GuavaNexin reagent (Millipore) and the Guava EasyCyte Plus.
Cell cycle analyses
MV4-11 cells were treated as indicated for 48 hours in duplicate, after which BrdU was added at 10μM for one hour. Cells were then fixed, stained with FITC linked anti-BrdU antibody and PI, and analyzed by flow cytometry as previously described (25). Data were analyzed with Summit 5.0 (Dako North America, Carpinteria, CA).
Antibodies
Antibodies directed against WEE1, CDK2, and tubulin were purchased from Cell Signaling Technology (Danvers, MA). Antibodies directed against phospho-CDK2 (T14) were purchased from Abcam (Cambridge, MA). Anti-actin antibody was purchased from Millipore. Anti-BrdU antibody was purchased from Dako.
Data analyses
Functional genetic screening data from the deep sequencer was pre-processed using software provided by Illumina. Sequences passing filters for quality and vector specific landmarks were mapped to shRNA tag libraries. EdgeR was used to generate adjusted p-values for each tag (20) and a modified Z score was used to generate p-values and E-scores for each gene (Supplemental Information) for the BINGS ranking of hits. For the RFC ranking, raw data counts were filtered, adjusted, and normalized (Supplemental Information), and genes for which more than one shRNA with greater than 3-fold over- or under-representation after cytarabine exposure were included. The statistical significance of the extent of overlap between hit lists was determined using 10,000 simulations on randomly selected genes. Java Treeview (26) was used to depict hierarchical clustering generated using open source clustering software (27). Ingenuity IPA 9.0 (Ingenuity Systems, www.ingenuity.com) was used to identify networks and functions represented by shRNA tags in the list of hits. Excel and GraphPad Prism 5 (GraphPad Software, San Diego, CA) were used for data sorting, analysis and graphical depiction of data. Students t test was used to compare 2 samples; analysis of variance (ANOVA) and Bonferroni’s post-test was used to compare more than 2 samples. Error bars in each figure depict the standard error of the mean, and may be obscured when narrow.
Results
Genome-wide functional genetic screen identifies mediators of AML cell fate after cytarabine exposure
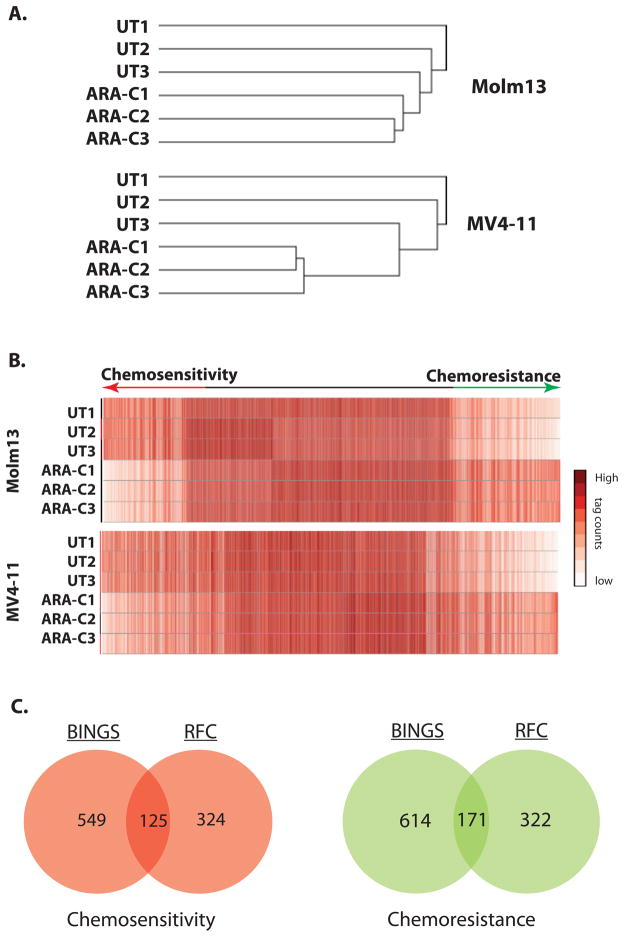
To identify proteins that mediate AML cell survival in the context of treatment with cytarabine, we developed a systematic approach, beginning with a genome-wide, functional genetic screen (Fig. 1A) in 2 AML cell lines (Molm13 and MV4-11), both of which have MLL rearrangement and FLT3 mutation (28, 29). Two independent analysis approaches were applied (Figure 1B). Hierarchical clustering of unfiltered data demonstrates that within cell lines replicates cluster by treatment condition, suggesting that shRNA tag representation is dependent upon the treatment and is not stochastic (Fig. 2A). The relative representation of the shRNA tags was then compared, with the expectation that shRNAs tags under-represented after cytarabine exposure correspond to genes which confer chemosensitivity when inhibited, while those which are over-represented correspond to genes which confer chemoresistance (Figure 2B). To identify “hits” from this screen, we applied a novel statistical and mathematic ranking using a BioInformatics pipeline for Next Generation Sequencing (BINGS; Supplemental Table 1). To complement this, we also developed a hit list based on Redundancy of shRNAs targeting a gene and Fold-Change values over a threshold expected to have biologic relevance (RFC; Supplemental Table 2). These lists were compared and found to have significant overlap (p<0.0001; Fig. 2C). We surmised that the genes in common between the two analyses would be enriched for true positives and concentrated subsequent efforts on this list (Supplemental Table 3).
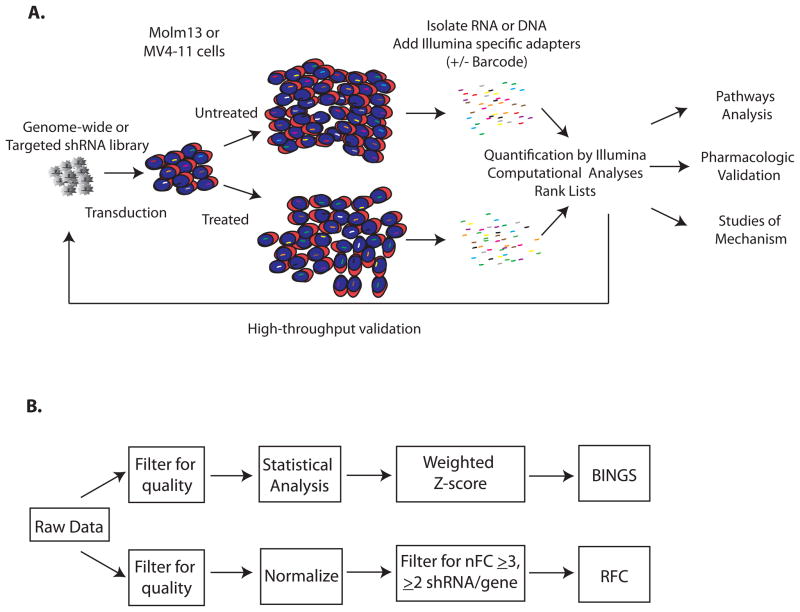
Figure 1. Systematic functional genetic screening.
A. Experimental overview. AML cell lines Molm13 and MV4-11 were transduced with a genome-wide shRNA library and treated with cytarabine at the IC75 dose or left untreated. After recovery in the absence of drug, shRNA tag sequences were isolated and quantified by deep sequencing. Data were analyzed by complementary means generating lists of mediators of chemosensitivity and chemoresistance. A subset of these hits was included in a targeted sub-library for secondary screening. Selected hits were analyzed using Ingenuity Pathways Analysis, validated pharmacologically, and the mechanism of cell fate examined. B. Complementary analyses of differential shRNA tag representation. Data were analyzed by 2 independent processes. The Bioinformatics for Next Generation Sequencing (BINGS) pipeline employed edgeR to identify differentially represented shRNA tags and then applied a weighted z-score to rank genes as providing chemoresistance or chemosensitivity when inhibited (BINGS). A second method takes into consideration the absolute fold-change of normalized shRNA tag counts and redundancy of shRNAs directed against a gene (RFC).
Figure 2. Functional Genetic Screening identifies critical mediators of AML cell fate after cytarabine exposure.
A. shRNA tag representation clusters by treatment condition. Unsupervised hierarchical clustering of unfiltered data demonstrates that within cell lines, shRNA tag representation clusters according to treatment condition, suggesting that differences in shRNA tag representation are due to the treatment condition and are not stochastic. B. Differential shRNA tag representation implicates mediators of AML cell fate after exposure to cytarabine. Heatmaps of data filtered for quality (BINGS method) show differential representation of shRNA tag counts by treatment condition. Mediators of chemosensitivity or chemoresistance are represented at either end. C. Overlap between independent analysis parameters identifies hundreds of mediators of AML cell fate after exposure to cytarabine. The number of genes identified as hits with BINGS and RFC are indicated, as well as the overlap between the two methodologies. The overlapping subset is expected to be enriched for true positive hits.
Validation of hits from functional genetic screening is best performed using complementary and independent methods; however, these experiments can be cumbersome and time consuming when done at large scale. For example, to individually validate 10 chemosensitivity and chemoresistance conferring hits would necessitate the generation of over 100 clones in each cell line, followed by determination of sensitivity to ARA-C and the extent of gene knockdown. This list could be narrowed further based on published data regarding these genes, but to do so would be biased against genes that are understudied. Thus, we developed a strategy of unbiased, high-throughput validation using a sub-library of pooled shRNAs and similar methodology as employed for the original screen. Notably, the shRNAs for high-throughput validation are obtained from a source that uses an entirely different algorithm for shRNA design (30). In this secondary screen, we confirmed that a number of the hits from the genome-wide screen do in fact mediate AML cell survival in the context of cytarabine treatment (Fig. 3A). There were 9 genes confirmed to mediate chemosensitivity when inhibited and 8 confirmed to confer chemoresistance, based on statistical analysis of expected differential representation. Upon closer analyses of these data, though, it is clear that our genome-wide screen analysis was more accurate in identifying sensitivity mediators than resistance mediators. For example, 11 genes expected to confer resistance when knocked down actually conferred sensitivity. Conversely, only three expected sensitivity mediators conferred resistance. Thus, subsequent efforts were concentrated on the list of genes identified as synthetic lethal with cytarabine.
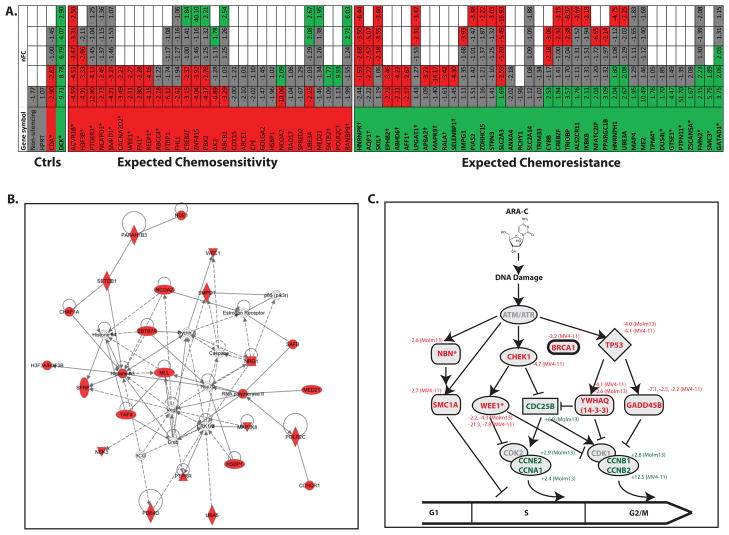
Figure 3. Cell cycle checkpoint proteins are critical mediators of AML cell fate after exposure to cytarabine.
A. High-throughput rescreening confirms the role of several mediators of chemosensitivity. Molm13 cells were transduced with an independent sub-library of shRNAs targeting a subset of 35 hits from the genome-wide screen, as well as negative control shRNAs targeting a non-coding sequence, the housekeeping gene HPRT, and functional controls CDA (which de-activates cytarabine) and DCK (which activates cytarabine). The cells were treated with cytarabine or left untreated and shRNA tags were isolated and quantified as before. Each targeted gene is represented by a column and each shRNA is represented by a row and color-coded for its representation in cytarabine: red (significantly under-represented), green (significantly over-represented) and gray (no change). Empty boxes reflect genes for which fewer than 5 shRNAs were included. The normalized Fold Change (nFC) of each shRNA in cytarabine is indicated within each box. Asterisks indicate genes considered validated. Daggers indicate genes for which shRNA representation was opposite of predicted. B. Pathways analysis of synthetic lethal hits from the genome wide screen identifies a cell cycle network as critical in determination of AML cell fate after exposure to cytarabine. The top scoring network in IPA among those proteins identified as chemosensitizing when inhibited is “Cell Cycle, Cellular Growth and Proliferation, Cancer.” C. Cell cycle checkpoint proteins implicated in the genome-wide screen. Review of data from the genome-wide functional genetic screen revealed several other genes involved in cell cycle checkpoints, for which there were at least one shRNA differentially represented in cytarabine treated cells, but which may not have reached threshold criteria to be included in the original list of hits. A schematic representation of their roles in cell cycle progression is presented with cell cycle inhibitors listed with red letters and cell cycle promoters listed with green letters. The normalized fold change in representation in cytarabine treated cells (and the cell line in which differential representation was noted) are listed next to the gene symbols. Asterisks indicate the genes that met criteria for inclusion in the overlapping list of hits from both analyses.
Pathways analysis of synthetic lethal hits implicates cell cycle regulation as a key determinant of AML cell fate in cytarabine
We then analyzed the list of synthetic lethal hits using Ingenuity Pathways Analysis. The top scoring network from this analysis is “Cell Cycle, Cellular Growth and Proliferation, Cancer” (Figure 3B). The top Molecular and Cellular Function is “Cell Cycle” with p values as low as 7.16×10−6 (Supplemental Table 4). This analysis suggests that cell cycle regulation is a key determinant of AML cell fate after cytarabine exposure. We returned to the original genome-wide screen, to look for additional genes that did not pass criteria to be included as hits and with known function in cell cycle regulation, focusing on those related to DNA damage checkpoints. In so doing, we identified several more genes that may also mediate AML cell survival in the context of cytarabine treatment (Fig. 3C).
Expression of cell cycle control proteins is induced in AML cells exposed to cytarabine
We next asked whether there are changes in gene expression patterns in AML cells exposed to cytarabine, hypothesizing that genes induced in cytarabine and identified as chemosensitizing in the functional screens may be of particular importance in mediating AML cell survival. Molm13 cells were exposed to cytarabine for 24 hours, after which RNA was extracted and analyzed by microarray (Figure 4A and Supplemental Table 5). Surprisingly, there was only one gene (GOLGA2) that was identified as chemosensitizing and was differentially expressed in the presence of cytarabine. GOLGA2 did not appear to mediate AML cell fate in the secondary screen, though, and was not considered for further study. Despite the lack of overlap at the molecular level, differentially regulated genes analyzed by Ingenuity Pathways Analysis identified “Cell Cycle “ as one of the top Molecular and Cellular functions represented by these genes with p-values as low as 5.89×10−4 (Supplemental Figure 1). The functional overlap of genes with altered expression and chemosensitivity when knocked-down highlights the importance of cell cycle control in determining AML cell fate after cytarabine exposure.
Figure 4. Cell cycle regulation proteins are induced in AML cells exposed to cytarabine and over-expressed in patient primary AML samples.
A. Gene expression profiling of AML cells exposed to cytarabine. Molm13 cells were treated with cytarabine or left untreated for 24 hours at the IC75 dose used in the functional genetic screen. Before the initiation of widespread apoptosis, mRNA was harvested and analyzed by microarray. While there was only modest difference in global gene expression, the most differentially expressed genes are enriched for those involved in cell cycle regulation as defined by Ingenuity Pathways Analysis. B. A subset of chemosensitizing hits is over-represented in primary patient samples as compared to normal. Three independent datasets available in Oncomine were queried for the relative expression of chemosensitizing hits. Genes are listed left to right based on the mean relative fold-change as compared to normal from the 3 data sets. The number of asterisks next to the gene name indicates the number of datasets in which differential expression was determined to be statistically significant.
WEE1 is expressed in human primary AML samples
We next considered whether WEE1 and other molecules in our list of chemosensitizing genes are over-expressed in AML cells from primary patient samples, as compared to normal cells, with the hypothesis that differential expression may further implicate mediators of AML cell fate in cytarabine with therapeutic relevance. We used Oncomine to query existing gene expression data from primary patient samples (31). Three datasets are available in this database from which gene expression in AML cells can be compared to “normal” cells (32–34). A subset of genes identified as chemosensitizing, including WEE1, appears to be significantly over-expressed in AML cells as compared to normal cells (Figure 4B).
Inhibition of WEE1 sensitizes AML cell lines to the anti-proliferative effects of cytarabine
The strongest hit from the genome-wide screen among cell cycle control genes was WEE1, which was identified by both analysis techniques, confirmed in the secondary screen, included in the top scoring Network and the top Molecular and Cellular Function as defined by pathways analysis, and is differentially expressed in AML cells as compared to normal. WEE1 is a dual specificity kinase, which in the context of DNA damage, phosphorylates CDK1 at tyrosine 15, inactivating the CDK1/cyclin B complex and preventing mitosis (35). Inhibition of WEE1 is being explored therapeutically in solid tumors, but to our knowledge, has not been examined in AML. To further validate the role of WEE1 in AML cells exposed to cytarabine using non-genetic means, we used a small molecule inhibitor of WEE1, MK1775 (36). Treatment with MK1775 alone has little effect on the proliferation of these cells except at concentrations approaching 200nM (Supplemental Figure 2A). However, in combination with cytarabine, lower doses (20–100 nM) of MK1775 are synergistic in inhibiting AML cellular proliferation after 72 hours of treatment (Fig. 5A; Table 1). Consistent with this finding, when cells are removed from drug treatment after 72 hours, AML cells recover from cytarabine at some doses, but the addition of MK1775 prevents this recovery (Figure 5B). We performed similar experiments in U937 cells which do not have FLT3 mutation or MLL rearrangement (29) and observed similar combinatorial activity of cytarabine with the addition of the WEE1 inhibitor in U937 cells (Figure 5C; Table 1), indicating that susceptibility to this combination of drugs is not specific to FLT3 dependence or MLL rearrangement.
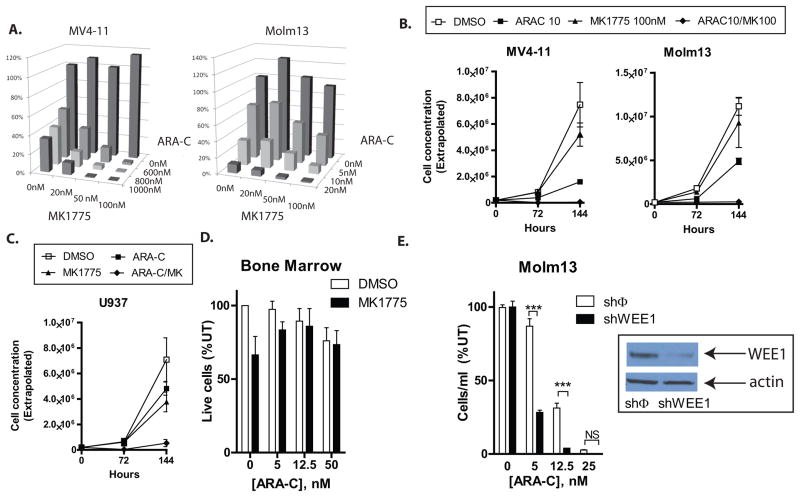
Figure 5. Inhibition of WEE1 sensitizes AML cell lines to cytarabine.
A. MK1775 plus cytarabine results in greater inhibition of AML cell proliferation than cytarabine alone. MV4-11 and Molm13 cells were treated with cytarabine, MK1775, or both at the indicated doses for 72 hours and counted by flow cytometry and propidium iodide exclusion. The concentration of live cells is depicted graphically. MK1775 had little effect at doses up to 100nM, while cytarabine (ARA-C) had a dose dependent inhibitory effect on the number of cells as compared to cells treated with the vehicle (DMSO). However, when the two drugs were combined, inhibition of cellular proliferation was synergistic, as defined by the combination index (Table 1). B. The addition of MK1775 to cytarabine eliminates AML cells to a greater extent than cytarabine alone. MV4-11 and Molm13 cells were treated with cytarabine, MK1775 (100nM), or both for 72 hours, counted and re-seeded at 1/10 dilution without the addition of drugs. 72 hours later they were counted again. At these doses of cytarabine, some cells recover from treatment and begin to proliferate. However, the addition of MK1775 appears to eliminate this population, as there is no evidence of proliferation at the 144 hour time point. P<0.001 for MV4-11; P=0.05 for Molm13). C. Inhibition of WEE1 sensitizes U937 cells to cytarabine. U937 cells were treated with cytarabine and/or MK1775 and counted as in 6C. U937 cells show similar synergistic inhibition of proliferation with both drugs (Table 1; P<0.01)). D. Inhibition of WEE1 does not sensitize normal bone marrow cells to cytarabine. Murine whole BM was cultured in cytokine containing media with cytarabine and/or MK1775 at the indicated concentrations for 72 hours, counted and then allowed to recover for another 72 hours without the addition of more drugs. E. Inhibition of WEE1 by shRNA sensitizes Molm13 cells to cytarabine. Molm13 cells were transduced with a non-silencing shRNA (shφ) or an shRNA directed against WEE1 (shWEE1). Confirmation of knockdown of WEE1 was determined by western blot (inset). Cells were treated with cytarabine at the indicated doses for 72 hours, and then allowed to recover for another 72 hours in the absence of drug. Asterisks indicate statistical significance.
Table 1. Calculated Combination Indexes for MK1775 and cytarabine in AML cell lines.
MK1775 and cytarabine are synergistic in inhibition of AML cellular proliferation.
MV4-11 or Molm13 cells were treated as in Figure 5A and the combination index (CI) was calculated for the various dose combinations. CI values less than one indicate synergistic inhibition of proliferation.
|
| ||||
|---|---|---|---|---|
| MV4-11 | ||||
| ARA-C (nM) | MK1775 (nM) | Fa | CI | |
| 600 | 20 | 0.700 | 0.601 | |
| 600 | 50 | 0.826 | 0.372 | |
| 600 | 100 | 0.975 | 0.089 | |
| 800 | 20 | 0.792 | 0.575 | |
| 800 | 50 | 0.963 | 0.155 | |
| 800 | 100 | 0.986 | 0.079 | |
| 1000 | 20 | 0.857 | 0.529 | |
| 1000 | 50 | 0.991 | 0.075 | |
| 1000 | 100 | 0.994 | 0.055 | |
|
| ||||
| Molm13 | ||||
| ARA-C (nM) | MK1775 (nM) | Fa | CI | |
|
| ||||
| 5 | 20 | 0.300 | 1.672 | |
| 5 | 50 | 0.509 | 1.17 | |
| 5 | 100 | 0.630 | 1.173 | |
| 10 | 20 | 0.625 | 1.32 | |
| 10 | 50 | 0.808 | 0.862 | |
| 10 | 100 | 0.889 | 0.761 | |
| 20 | 20 | 0.902 | 0.838 | |
| 20 | 50 | 0.971 | 0.427 | |
| 20 | 100 | 0.985 | 0.369 | |
|
| ||||
| U937 | ||||
| ARA-C (nM) | MK1775 (nM) | Fa | CI | |
|
| ||||
| 10 | 100 | 0.86 | 0.236 | |
| 10 | 200 | 0.76 | 0.572 | |
| 20 | 100 | 0.95 | 0.173 | |
| 20 | 200 | 0.38 | 1.223 | |
| 50 | 100 | 0.94 | 0.269 | |
| 50 | 200 | 0.71 | 0.845 | |
Treatment of murine hematopoietic progenitor cells ex vivo did not result in combinatorial inhibition of proliferation, suggesting that the combination may not be overly toxic to normal hematopoiesis (Figure 5D). To ensure that the observed combinatorial effect is not an off target effect of MK1775, we treated MV4-11 cells with cytarabine and a second WEE1 inhibitor (BCHCD), and again observed greater inhibition of cellular proliferation with WEE1 inhibition and cytarabine as compared to cytarabine alone (Supplemental Figure 2B). In addition, we used shRNA to knockdown WEE1 in Molm13 cells and observed enhanced sensitivity to cytarabine in the cells in which WEE1 had been knocked down as compared to those expressing a non-silencing shRNA (Figure 5E).
Inhibition of WEE1 abrogates the S-phase replication checkpoint induced by cytarabine
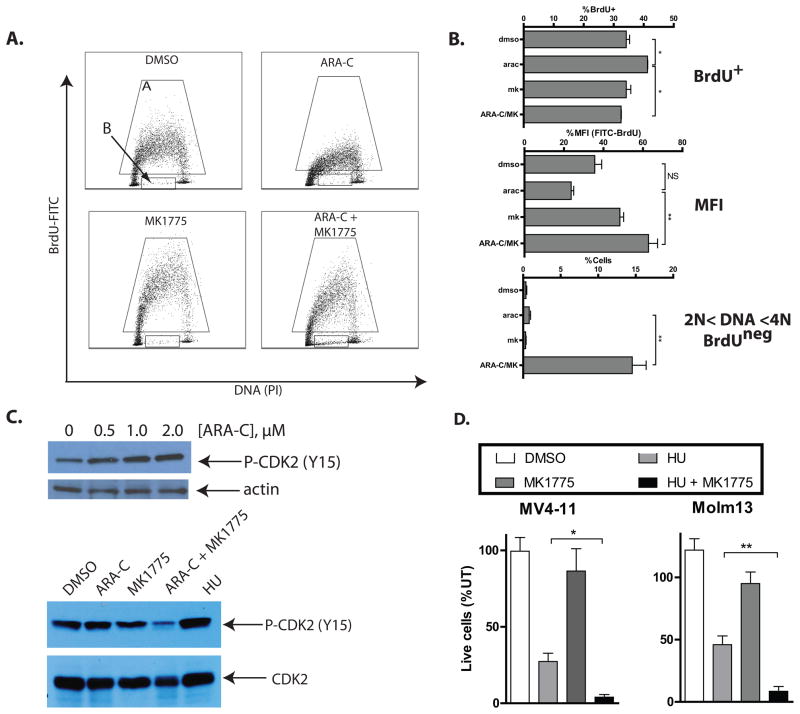
Identification and validation of WEE1 as a chemosensitizing target was somewhat surprising in that inhibition of WEE1 has primarily been examined in the context of abrogation of the G2/M checkpoint (36, 37), whereas cytarabine induces G1 or S-phase arrest (38). To begin to understand the mechanism of the combinatorial effects of cytarabine and WEE1 inhibition, we performed cell cycle analysis of MV4-11 cells by staining with propidium iodide for DNA content. At the doses used in these experiments, there was a considerable increase in the sub-G1 population (Supplemental Figure 3A), but little evidence of cell cycle changes with cytarabine or MK1775 alone or in combination, when only the live population was assessed (Supplemental Figure 3B). To better assess S phase progression, we analyzed BrdU incorporation with or without these drugs (Figure 6A). As expected, in the presence of cytarabine the percentage of cells in S phase was significantly increased (Figure 6B, top), consistent with impaired progression through S phase. In addition, the extent of BrdU incorporation in those cells in S phase was diminished, (Figure 6B, middle), consistent with stalled replication forks and impaired DNA synthesis (39). However, with the addition of MK1775, the impaired progression through S phase induced by cytarabine was abrogated. Moreover, a large population of BrdUneg cells with >2N and <4N DNA content emerged in the context of combination therapy (Figure 6A and B, bottom), suggesting a population of cells that had arrested in S phase for which DNA synthesis had ceased altogether (25). While not identical, we saw similar abrogation of S phase effects induced by cytarabine in the cells in which WEE1 had been knocked down (Supplemental Figure 3C). CDK2 is one of the cell cycle kinases involved in progression through S phase (40). Its activity is inhibited by phosphorylation of threonine 14 (T14) and tyrosine 15 (Y15) residues (41–43). As expected, CDK2 was phosphorylated in AML cells exposed to cytarabine or hydroxyurea, but when cytarabine was combined with MK1775, this phosphorylation was abrogated (Figure 6C). Consistent with activity in the S phase of the cell cycle, WEE1 inhibition with MK1775 markedly sensitized AML cells to hydroxyurea, a DNA synthesis inhibitor which induces S phase arrest (Figure 6D).
Figure 6. Inhibition of WEE1 abrogates S phase arrest induced by cytarabine.
A. Cytarabine and inhibition of WEE1 alter BrdU incorporation in AML cells. MV4-11 cells were treated as in 6A indicated for 48 hours, after which BrdU was added for 1 hour. Cells were then harvested, fixed, stained with a FITC-linked antibody against BrdU and propidium iodide, and analyzed by flow cytometry. Scatter plots are representative of 3 independent experiments. The Brdu+ fraction is indicated by gate A. The Brduneg population with >2N and <4N DNA content is indicated by gate B. B. Cell cycle changes in AML cells treated with cytarabine and/or MK1775. Data from cell cycle analyses as in B are depicted graphically. Cytarabine significantly increases the percentage of BrdU+ cells; the addition of MK1775 abrogates this effect (top). Cytarabine diminishes the mean fluorescence intensity (MFI) of BrdU+ cells as compared to DMSO treated cells; the addition of MK1775 abrogates this effect (middle). The combination of cytarabine and MK1775 significantly increases the percentage of BrdUneg cells which have >2N and <4N DNA content (bottom). C. Inhibition of WEE1 abrogates inhibitory phosphorylation of CDK2 induced by cytarabine. MV4-11 cells were treated with cytarabine at the indicated doses for 24 hours, after which protein lysates were subject to Western blotting with an antibody specific to CDK2 phosphorylated at threonine 14 and actin (top). The cells were also treated with cytarabine (1μM), and/or MK1775 (200nM)for 48 hours and similarly analyzed by Western blot (bottom). Hydroxyurea (50μM) alone was used as a positive control for inhibitory phosphorylation of CDK2. Total CDK2 was used as a loading control. D. Inhibition of WEE1 sensitizes AML cells to hydroxyurea. MV4-11 and Molm13 cells were treated with hydroxyurea (50μM) and/or MK1775 (200nM) for 72 hours and counted by propidium iodide exclusion. The number of live cells relative to untreated is depicted graphically.
Inhibition of WEE1 in combination with cytarabine induces more apoptosis than cytarabine alone in AML cell lines and primary AML cells
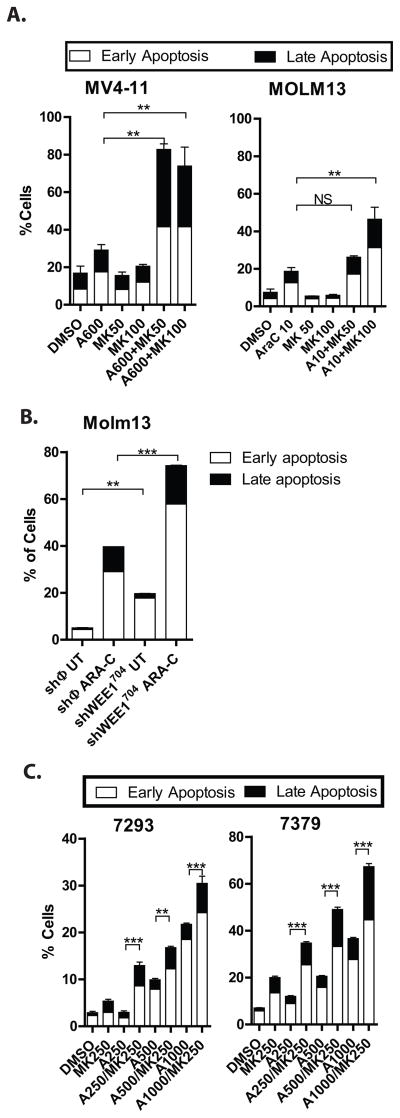
To determine if the alterations in cell cycle control were sufficient to lead to cell death, we used flow cytometry to analyze the proportion of cells undergoing apoptosis after exposure to cytarabine and/or MK1775. Staining for annexin-V and propidium iodide exclusion indicates that the addition of MK1775 to cytarabine leads to more apoptosis than cytarabine alone (Figure 7A). We also measured induction of apoptosis in Molm13 cell lines in which WEE1 had been knocked down, and similarly found a greater percentage of cells undergoing apoptosis in cells in those cells as compared to those expressing a non-silencing control sequence (Figure 7B). To ensure that the observed combinatorial effect is not limited to immortalized AML cell lines, we also exposed primary AML cells to cytarabine and/or MK1775, and found enhanced induction of apoptosis with the combination as compared to either drug alone (Figure 6D and Supplemental Figure 4). We again observed abrogation of inhibitory phosphorylation of CDK2 with WEE1 inhibition, indicating a similar mechanism of action in primary AML cells (Supplemental Figure 4). This effect was not universal, though as 1 of the 4 samples showed no enhancement of apoptosis (Supplemental Figure 4).
Figure 7. Inhibition of WEE1 in combination with cytarabine enhances the apoptotic effects of cytarabine in AML cell lines and primary AML cells.
A. MK1775 plus cytarabine results in more apoptosis than cytarabine alone in AML cell lines. Cells treated as in 6A were stained for annexin V and with 7-AAD and analyzed by flow cytometry. The percentage of early apoptotic (white bar) and late (apoptotic) cells are shown. B. Knockdown of WEE1 sensitizes AML cells to apoptosis induced by cytarabine. Molm13 cells transduced with a non-silencing control vector (shφ) or an shRNA directed against WEE1 (as in Figure 5D) were treated with cytarabine at 25nM for 72 hours. The percentage of apoptotic cells was assessed as in Figure 7A. C. MK1775 plus cytarabine results in more apoptosis than cytarabine alone in primary AML cells. Primary AML cells were cultured with cytarabine and/or MK1775 at the indicated concentrations for 72 hours and the percentage of apoptotic cells was assessed as in Figure 7A. Asterisks indicate statistical significance (ANOVA).
Discussion
The clinical and molecular heterogeneity of AML remain major challenges in improving outcomes for this disease. In order to objectively and rapidly identify proteins which can be modulated to enhance the activity of current standard therapy, we queried the entire genome using complementary functional and descriptive assays. The integration of data from these experiments strongly implicates cell cycle checkpoints as critical determinants of AML cell fate after exposure to cytarabine. WEE1 was among the proteins identified in the genome-wide functional screen, which also appears to be over-expressed in AML cells as compared to normal cells. Genetic and pharmacologic inhibition of WEE1 together with cytarabine inhibited proliferation and induced apoptosis in AML cell lines and primary AML samples, validating the potential of WEE1 as a therapeutic target in AML. Cell cycle analyses and western blotting confirm that the effect is due to the abrogation of the intra-S phase checkpoint, a function of WEE1 that to our knowledge has not been previously considered from a therapeutics perspective.
These data highlight the power of integrating genome-scale experimentation, and functional genomic screening in particular. While gene expression profiling of tumors and analysis of mutations within cancer genomes has provided a wealth of data and may eventually be used prognostically or to guide therapy, the descriptive nature of the data is difficult to interpret and apply clinically. That is, whether the over-expression or mutation of any given gene in a particular tumor may have functional significance or is a viable therapeutic target remains unknown until further experimentation is pursued. In contrast, functional genetic screening identifies molecules based on functional relevance a priori. The approach that we have developed allows for cost-effective, high-throughput validation, rapidly narrowing focus for detailed analyses of true positives. In this particular screen, the false positive rate was higher among shRNAs expected to confer chemoresistance, as compared to chemosensitivity (Figure 3A). The reasons for this are not entirely clear, as others have performed positive selection screening with shRNA libraries with good results (44, 45). Differences in methodology may have contributed to our observations, as continuous or repeated drug exposure may be a more effective method for detecting drug resistance conferring shRNAs (44, 45).
While cell cycle checkpoints have been previously studied in AML, to our knowledge, this is the first report to document the functional importance of WEE1 in AML. Chk1 has similar effects in arresting the cell cycle in the context of chemotherapy, and its inhibition has been explored therapeutically (46). WEE1 may prove to be a better therapeutic target, though, as its effects in cell cycle regulation are direct and specific, while Chk1 integrates from and propagates signals through multiple other molecules, including WEE1. MK1775 is in early phase clinical trials in combination with chemotherapy for solid tumors, and the side effect profile is manageable in preliminary reports (47).
We have demonstrated the importance of WEE1 in inducing the intra-S phase checkpoint in the context of cytarabine, which we propose is critical for AML cell survival during treatment. These data support a model whereby cytarabine treatment results in stalled DNA replication; while some cells will commit to apoptosis, others will recover and successfully complete cell cycle progression. But if inhibition of WEE1 prevents proper recognition of stalled replication, we speculate that replication forks will collapse, leading to S phase arrest and consequent apoptosis. Our results are somewhat surprising, in that the primary function of WEE1 is considered to be the inhibition of cell cycle progression at the G2/M transition of the cell cycle via inhibitory phosphorylation of CDK1 at Y15 (35, 48), while we observed abrogation of CDK2 Y15 phosphorylation with WEE1 inhibition in addition to inhibitory phosphorylation of CDK1 (not shown). Our data are consistent with the conserved function of WEE1 orthologs from Arabidopsis, Xenopus, and human cells in phosphorylation of CDK2 to regulate progression through S-phase (49–51). The S-phase function of WEE1 in AML cells broadens the therapeutic relevance of this molecule, particularly for treatment of AML, which is heavily dependent upon the S phase effects of cytarabine.
Inhibition of WEE1 is primarily being explored in patients with abnormal p53 function, since p53 is primarily responsible for the G1 checkpoint and cells with impaired p53 function are highly dependent on the G2/M checkpoint to maintain genomic integrity (52). Indeed, cells with impaired p53 function can be sensitized to DNA damage by impairing the G2/M checkpoint via inhibition of Chk1 or WEE1 (53). While combining WEE1 inhibition with anthracycline has been shown to be synergistic in solid tumor models (54), whether inhibition of WEE1 in the context of anthracycline is synergistic in AML cells has yet to be demonstrated. While TP53 is not frequently mutated in AML, its function is often impaired (55–57); thus, it is reasonable to hypothesize that WEE1 in combination with anthracyclines would result in synergistic AML cell death. Nonetheless, our data implicating the importance of WEE1 in combination with cytarabine were generated in cell lines that are reported to have functional p53, suggesting that the inhibition of WEE1 can be considered for tumors with functional p53.
We have demonstrated that WEE1 inhibition sensitizes AML cells to cytarabine in vitro; however, we cannot conclude that these findings will be broadly applicable to all patients with AML. Indeed, not all of the primary patient samples we tested provided evidence of combinatorial effects, suggesting underlying molecular susceptibility to this combination. Thus, it will be important to develop appropriate, practical biomarkers with which to predict which patients may benefit the most from such a therapeutic strategy. Animal modeling of AML with assessment of pharmacokinetics will provide further data in evaluating the effectiveness of WEE1 inhibition with cytarabine in eliminating AML cells and prolonging survival.
In summary, using integrated genomic analyses, we have identified and validated WEE1 as a potential therapeutic target in AML. These data highlight the power of integrated genomic analyses which may vastly accelerate the pace of discovery, validation and translation of molecular biology phenomena.
Supplementary Material
Acknowledgments
The authors thank Drs. Chris T Hittinger and Mark Johnston for input in assay design, and Dr. Robert Sclafani for critical review of the manuscript. This work was supported by the Colorado Golfer’s Against Cancer and the AMC Cancer Fund (CCP, ACT, JD), the Leukemia and Lymphoma Society (JD, CCP), and the NCI through The University of Colorado Cancer Center (3P30CA046934-22S4; CCP).
Footnotes
Authorship contributions: CCP, SF, VZ, PP and AVL performed experiments. JK, DG and ACT developed the BINGS pipeline. CCP, JK, ACT, DG, CG, and JD analyzed and interpreted data. CCP, ACT and JD wrote the manuscript.
Disclosure of Conflicts of Interest: The authors have no conflicts of interest to disclose.
Supplementary information is available at Leukemia’s website.
References
- 1.Deschler B, Lubbert M. Acute myeloid leukemia: epidemiology and etiology. Cancer. 2006 Nov 1;107(9):2099–107. doi: 10.1002/cncr.22233. [DOI] [PubMed] [Google Scholar]
- 2.Pui CH, Carroll WL, Meshinchi S, Arceci RJ. Biology, Risk Stratification, and Therapy of Pediatric Acute Leukemias: An Update. J Clin Oncol. 2011 Jan 10; doi: 10.1200/JCO.2010.30.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilliland DG, Tallman MS. Focus on acute leukemias. Cancer Cell. 2002 Jun;1(5):417–20. doi: 10.1016/s1535-6108(02)00081-8. [DOI] [PubMed] [Google Scholar]
- 4.Dohner H, Estey EH, Amadori S, Appelbaum FR, Buchner T, Burnett AK, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010 Jan 21;115(3):453–74. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 5.Fernandez HF, Sun Z, Yao X, Litzow MR, Luger SM, Paietta EM, et al. Anthracycline dose intensification in acute myeloid leukemia. N Engl J Med. 2009 Sep 24;361(13):1249–59. doi: 10.1056/NEJMoa0904544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burnett AK, Hills RK, Milligan DW, Goldstone AH, Prentice AG, McMullin MF, et al. Attempts to optimize induction and consolidation treatment in acute myeloid leukemia: results of the MRC AML12 trial. J Clin Oncol. 2010 Feb 1;28(4):586–95. doi: 10.1200/JCO.2009.22.9088. [DOI] [PubMed] [Google Scholar]
- 7.Lange BJ, Smith FO, Feusner J, Barnard DR, Dinndorf P, Feig S, et al. Outcomes in CCG-2961, a children’s oncology group phase 3 trial for untreated pediatric acute myeloid leukemia: a report from the children’s oncology group. Blood. 2008 Feb 1;111(3):1044–53. doi: 10.1182/blood-2007-04-084293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lowenberg B, Pabst T, Vellenga E, van Putten W, Schouten HC, Graux C, et al. Cytarabine dose for acute myeloid leukemia. N Engl J Med. 2011 Mar 17;364(11):1027–36. doi: 10.1056/NEJMoa1010222. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Carbonero R, Ryan DP, Chabner BA. Cytidine Analogs. In: Chabner BA, Longo DL, editors. Cancer Chemotherapy & Biotherapy, Principles and Practice. 3. Philadelphia, PA, USA: Lippincott Williams and Wilkins; 2001. [Google Scholar]
- 10.Kornblau SM, Qiu YH, Bekele BN, Cade JS, Zhou X, Harris D, et al. Studying the right cell in acute myelogenous leukemia: dynamic changes of apoptosis and signal transduction pathway protein expression in chemotherapy resistant ex-vivo selected “survivor cells”. Cell Cycle. 2006 Dec;5(23):2769–77. doi: 10.4161/cc.5.23.3507. [DOI] [PubMed] [Google Scholar]
- 11.Druker BJ. Translation of the Philadelphia chromosome into therapy for CML. Blood. 2008 Dec 15;112(13):4808–17. doi: 10.1182/blood-2008-07-077958. [DOI] [PubMed] [Google Scholar]
- 12.Gregory MA, Phang TL, Neviani P, Alvarez-Calderon F, Eide CA, O’Hare T, et al. Wnt/Ca2+/NFAT signaling maintains survival of Ph+ leukemia cells upon inhibition of Bcr-Abl. Cancer Cell. 2010 Jul 13;18(1):74–87. doi: 10.1016/j.ccr.2010.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang S, Laoukili J, Epping MT, Koster J, Holzel M, Westerman BA, et al. ZNF423 is critically required for retinoic acid-induced differentiation and is a marker of neuroblastoma outcome. Cancer Cell. 2009 Apr 7;15(4):328–40. doi: 10.1016/j.ccr.2009.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlabach MR, Luo J, Solimini NL, Hu G, Xu Q, Li MZ, et al. Cancer proliferation gene discovery through functional genomics. Science. 2008 Feb 1;319(5863):620–4. doi: 10.1126/science.1149200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009 Nov 5;462(7269):108–12. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scholl C, Frohling S, Dunn IF, Schinzel AC, Barbie DA, Kim SY, et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell. 2009 May 29;137(5):821–34. doi: 10.1016/j.cell.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 17.Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A. 2008 Dec 23;105(51):20380–5. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009 May 29;137(5):835–48. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Porter CC, DeGregori J. Interfering RNA-mediated purine analog resistance for in vitro and in vivo cell selection. Blood. 2008 Dec 1;112(12):4466–74. doi: 10.1182/blood-2008-03-146571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010 Jan 1;26(1):139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He Z, Zhou J. Empirical evaluation of a new method for calculating signal-to-noise ratio for microarray data analysis. Appl Environ Microbiol. 2008 May;74(10):2957–66. doi: 10.1128/AEM.02536-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhodes DR, Anstett M. Data Processing and Normalization. 2010. [Google Scholar]
- 23.Rhodes DR, Anstett M. Computing Fold Change. 2010 Mar, 2010. [Google Scholar]
- 24.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 25.Li FX, Zhu JW, Hogan CJ, DeGregori J. Defective gene expression, S phase progression, and maturation during hematopoiesis in E2F1/E2F2 mutant mice. Mol Cell Biol. 2003 May;23(10):3607–22. doi: 10.1128/MCB.23.10.3607-3622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004 Nov 22;20(17):3246–8. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 27.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004 Jun 12;20(9):1453–4. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 28.Drexler HG, Quentmeier H, MacLeod RA. Malignant hematopoietic cell lines: in vitro models for the study of MLL gene alterations. Leukemia. 2004 Feb;18(2):227–32. doi: 10.1038/sj.leu.2403236. [DOI] [PubMed] [Google Scholar]
- 29.Quentmeier H, Reinhardt J, Zaborski M, Drexler HG. FLT3 mutations in acute myeloid leukemia cell lines. Leukemia. 2003 Jan;17(1):120–4. doi: 10.1038/sj.leu.2402740. [DOI] [PubMed] [Google Scholar]
- 30.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006 Mar 24;124(6):1283–98. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 31.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004 Jan-Feb;6(1):1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andersson A, Ritz C, Lindgren D, Eden P, Lassen C, Heldrup J, et al. Microarray-based classification of a consecutive series of 121 childhood acute leukemias: prediction of leukemic and genetic subtype as well as of minimal residual disease status. Leukemia. 2007 Jun;21(6):1198–203. doi: 10.1038/sj.leu.2404688. [DOI] [PubMed] [Google Scholar]
- 33.Stegmaier K, Ross KN, Colavito SA, O’Malley S, Stockwell BR, Golub TR. Gene expression-based high-throughput screening(GE-HTS) and application to leukemia differentiation. Nat Genet. 2004 Mar;36(3):257–63. doi: 10.1038/ng1305. [DOI] [PubMed] [Google Scholar]
- 34.Valk PJ, Verhaak RG, Beijen MA, Erpelinck CA, Barjesteh van Waalwijk van Doorn-Khosrovani S, Boer JM, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med. 2004 Apr 15;350(16):1617–28. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- 35.Parker LL, Piwnica-Worms H. Inactivation of the p34cdc2-cyclin B complex by thehuman WEE1 tyrosine kinase. Science. 1992 Sep 25;257(5078):1955–7. doi: 10.1126/science.1384126. [DOI] [PubMed] [Google Scholar]
- 36.Hirai H, Iwasawa Y, Okada M, Arai T, Nishibata T, Kobayashi M, et al. Small-molecule inhibition of Wee1 kinase by MK-1775 selectively sensitizes p53-deficient tumor cells to DNA-damaging agents. Mol Cancer Ther. 2009 Nov;8(11):2992–3000. doi: 10.1158/1535-7163.MCT-09-0463. [DOI] [PubMed] [Google Scholar]
- 37.Indovina P, Giordano A. Targeting the checkpoint kinase WEE1: selective sensitization of cancer cells to DNA-damaging drugs. Cancer Biol Ther. 2010 Apr;9(7):523–5. doi: 10.4161/cbt.9.7.11276. [DOI] [PubMed] [Google Scholar]
- 38.Banker DE, Groudine M, Willman CL, Norwood T, Appelbaum FR. Cell cycle perturbations in acute myeloid leukemia samples following in vitro exposures to therapeutic agents. Leuk Res. 1998 Mar;22(3):221–39. doi: 10.1016/s0145-2126(97)00174-4. [DOI] [PubMed] [Google Scholar]
- 39.Vassin VM, Anantha RW, Sokolova E, Kanner S, Borowiec JA. Human RPA phosphorylation by ATR stimulates DNA synthesis and prevents ssDNA accumulation during DNA-replication stress. J Cell Sci. 2009 Nov 15;122(Pt 22):4070–80. doi: 10.1242/jcs.053702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fang F, Newport JW. Evidence that the G1-S and G2-M transitions are controlled by different cdc2 proteins in higher eukaryotes. Cell. 1991 Aug 23;66(4):731–42. doi: 10.1016/0092-8674(91)90117-h. [DOI] [PubMed] [Google Scholar]
- 41.Sebastian B, Kakizuka A, Hunter T. Cdc25M2 activation of cyclin-dependent kinases by dephosphorylation of threonine-14 and tyrosine-15. Proc Natl Acad Sci U S A. 1993 Apr 15;90(8):3521–4. doi: 10.1073/pnas.90.8.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gu Y, Rosenblatt J, Morgan DO. Cell cycle regulation of CDK2 activity by phosphorylation of Thr160 and Tyr15. Embo J. 1992 Nov;11(11):3995–4005. doi: 10.1002/j.1460-2075.1992.tb05493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Booher RN, Holman PS, Fattaey A. Human Myt1 is a cell cycle-regulated kinase that inhibits Cdc2 but not Cdk2 activity. J Biol Chem. 1997 Aug 29;272(35):22300–6. doi: 10.1074/jbc.272.35.22300. [DOI] [PubMed] [Google Scholar]
- 44.Holzel M, Huang S, Koster J, Ora I, Lakeman A, Caron H, et al. NF1 is a tumor suppressor in neuroblastoma that determines retinoic acid response and disease outcome. Cell. 2010 Jul 23;142(2):218–29. doi: 10.1016/j.cell.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hattori H, Zhang X, Jia Y, Subramanian KK, Jo H, Loison F, et al. RNAi screen identifies UBE2D3 as a mediator of all-trans retinoic acid-induced cell growth arrest in human acute promyelocytic NB4 cells. Blood. 2007 Jul 15;110(2):640–50. doi: 10.1182/blood-2006-11-059048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sampath D, Cortes J, Estrov Z, Du M, Shi Z, Andreeff M, et al. Pharmacodynamics of cytarabine alone and in combination with 7-hydroxystaurosporine (UCN-01) in AML blasts in vitro and during a clinical trial. Blood. 2006 Mar 15;107(6):2517–24. doi: 10.1182/blood-2005-08-3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liejen S, Schellens JH, Shapiro G, Pavlick AC, Tibes R, Demuth T, Viscusi J, Cheng Y, Xu Y, Oza AM. A phase I pharmacological and pharmacodynamic study of MK-1775, a Wee1 tyrosine kinase inhibitor, in monotherapy and combination with gemcitabine, cisplatin, or carboplatin in patients with advanced solid tumors. J Clin Oncol. 2010;28(15s) Abstract. [Google Scholar]
- 48.McGowan CH, Russell P. Human Wee1 kinase inhibits cell division by phosphorylating p34cdc2 exclusively on Tyr15. Embo J. 1993 Jan;12(1):75–85. doi: 10.1002/j.1460-2075.1993.tb05633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kiviharju-af Hallstrom TM, Jaamaa S, Monkkonen M, Peltonen K, Andersson LC, Medema RH, et al. Human prostate epithelium lacks Wee1A-mediated DNA damage-induced checkpoint enforcement. Proc Natl Acad Sci U S A. 2007 Apr 24;104(17):7211–6. doi: 10.1073/pnas.0609299104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wroble BN, Finkielstein CV, Sible JC. Wee1 kinase alters cyclin E/Cdk2 and promotes apoptosis during the early embryonic development of Xenopus laevis. BMC Dev Biol. 2007;7:119. doi: 10.1186/1471-213X-7-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cools T, Iantcheva A, Weimer AK, Boens S, Takahashi N, Maes S, et al. The Arabidopsis thaliana checkpoint kinase WEE1 protects against premature vascular differentiation during replication stress. Plant Cell. 2011 Apr;23(4):1435–48. doi: 10.1105/tpc.110.082768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou BB, Bartek J. Targeting the checkpoint kinases: chemosensitization versus chemoprotection. Nat Rev Cancer. 2004 Mar;4(3):216–25. doi: 10.1038/nrc1296. [DOI] [PubMed] [Google Scholar]
- 53.Wang Y, Decker SJ, Sebolt-Leopold J. Knockdown of Chk1, Wee1 and Myt1 by RNA interference abrogates G2 checkpoint and induces apoptosis. Cancer Biol Ther. 2004 Mar;3(3):305–13. doi: 10.4161/cbt.3.3.697. [DOI] [PubMed] [Google Scholar]
- 54.Hirai H, Arai T, Okada M, Nishibata T, Kobayashi M, Sakai N, et al. MK-1775, a small molecule Wee1 inhibitor, enhances anti-tumor efficacy of various DNA-damaging agents, including 5-fluorouracil. Cancer Biol Ther. 2010 Apr;9(7):514–22. doi: 10.4161/cbt.9.7.11115. [DOI] [PubMed] [Google Scholar]
- 55.Seliger B, Papadileris S, Vogel D, Hess G, Brendel C, Storkel S, et al. Analysis of the p53 and MDM-2 gene in acute myeloid leukemia. Eur J Haematol. 1996 Sep;57(3):230–40. doi: 10.1111/j.1600-0609.1996.tb01369.x. [DOI] [PubMed] [Google Scholar]
- 56.Faderl S, Kantarjian HM, Estey E, Manshouri T, Chan CY, Rahman Elsaied A, et al. The prognostic significance of p16(INK4a)/p14(ARF) locus deletion and MDM-2 protein expression in adult acute myelogenous leukemia. Cancer. 2000 Nov 1;89(9):1976–82. doi: 10.1002/1097-0142(20001101)89:9<1976::aid-cncr14>3.3.co;2-e. [DOI] [PubMed] [Google Scholar]
- 57.Bueso-Ramos CE, Yang Y, deLeon E, McCown P, Stass SA, Albitar M. The human MDM-2 oncogene is overexpressed in leukemias. Blood. 1993 Nov 1;82(9):2617–23. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.