Abstract
Background:
Histology sections provide accurate information on atherosclerotic plaque composition, and are used in various applications. To our knowledge, no automated systems for plaque component segmentation in histology sections currently exist.
Materials and Methods:
We perform pixel-wise classification of fibrous, lipid, and necrotic tissue in Elastica Von Gieson-stained histology sections, using features based on color channel intensity and local image texture and structure. We compare an approach where we train on independent data to an approach where we train on one or two sections per specimen in order to segment the remaining sections. We evaluate the results on segmentation accuracy in histology, and we use the obtained histology segmentations to train plaque component classification methods in ex vivo Magnetic resonance imaging (MRI) and in vivo MRI and computed tomography (CT).
Results:
In leave-one-specimen-out experiments on 176 histology slices of 13 plaques, a pixel-wise accuracy of 75.7 ± 6.8% was obtained. This increased to 77.6 ± 6.5% when two manually annotated slices of the specimen to be segmented were used for training. Rank correlations of relative component volumes with manually annotated volumes were high in this situation (P = 0.82-0.98). Using the obtained histology segmentations to train plaque component classification methods in ex vivo MRI and in vivo MRI and CT resulted in similar image segmentations for training on the automated histology segmentations as for training on a fully manual ground truth. The size of the lipid-rich necrotic core was significantly smaller when training on fully automated histology segmentations than when manually annotated histology sections were used. This difference was reduced and not statistically significant when one or two slices per section were manually annotated for histology segmentation.
Conclusions:
Good histology segmentations can be obtained by automated segmentation, which show good correlations with ground truth volumes. In addition, these can be used to develop segmentation methods in other imaging modalities. Accuracy increases when one or two sections of the same specimen are used for training, which requires a limited amount of user interaction in practice.
Keywords: Histology, Segmentation, Classification, Atherosclerosis
BACKGROUND
Stained histological sections contain highly detailed information on tissue composition. Because of a resolution in the order of micrometers and the availability of a variety of specific stains, human observers can accurately indicate which cells or tissue types are present at each location. One of the applications for which histology is used, is the study of atherosclerotic plaques. It has been shown that the composition of such plaques is related to plaque vulnerability, indicating its chance to rupture and possibly causing cerebrovascular events.[1] Histology sections are segmented into different tissue components to improve the understanding of risk factors of stroke,[1] for biomechanical modeling of stress distributions in the plaque,[2] and to develop and evaluate plaque component segmentation methods on ex vivo and in vivo MRI.[3–5]
Manual annotation of histology sections is a time-consuming task. Several studies have automatically segmented histology sections, mainly for oncological applications.[6] To the best of our knowledge, no studies on automated segmentation of atherosclerotic plaque components in histology have been published. Image analysis in histology often consists of segmenting typical structures such as lymphocytes, cell nuclei and glands, and/or indicating a presence or severity of disease.[6] Our approach is different from this in the sense that we aim to divide the complete section into different areas. These segmentations can then be used to determine the stage of atherosclerotic disease, or to train plaque component segmentation methods in registered imaging data.[4,7]
In this paper, we (semi-) automatically segment histology sections of carotid atherosclerotic plaques into fibrous (stable plaque component), necrotic, and lipid (vulnerable components) regions using pixel-wise classification. Although all evaluated sections are stained with the same protocol, variations in appearance occur between sections from different specimens. Therefore, we compare an approach that uses training data from different specimens to training on one or two slices of the specimen to segment, which in practice requires a limited amount of user interaction per specimen. We also evaluate whether these segmentations can reliably be used to train plaque component segmentation methods in ex vivo Magnetic Resonance Imaging (MRI) and in vivo MRI and computed tomography-angiography (CTA), using datasets in which spatial correspondence between these imaging data and histology has been established.
MATERIALS AND METHODS
Data
For 13 patients who were scheduled for carotid endarterectomy, in vivo MRI (pre- and post contrast 3DT1-weighted, pre-contrast T1-weighted, Proton density weighted (PDw), and Time of Flight (TOF)) and CTA scans were acquired. After surgery, the excised specimens were scanned with MRI (3DT1w, PDw, T2w) and μCT, and afterwards decalcified and embedded in paraffin for histology processing. At every 1mm interval, axial slices of 5 μm thickness were taken. These sections were stained with Elastica Von Gieson (EvG, Merck, Germany). This stain colors nuclei brown to black, collagen and elastin pink to red, and muscle and cytoplasm yellow. Lipid regions are characterized by the typically shaped cholesterol esters and the absence of connective tissue. Necrotic tissue contains no cell nuclei and is usually less dense than fibrous tissue. Examples, which also depict the variability between sections, are shown in Figure 1.
Figure 1.
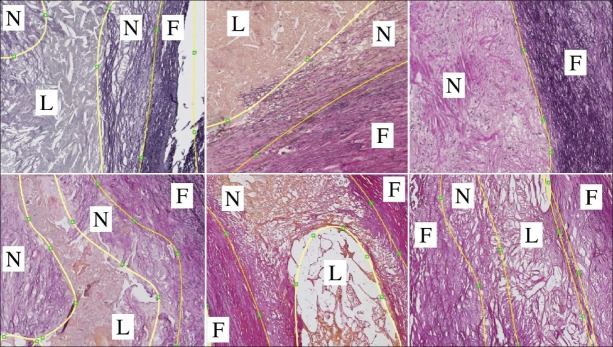
Examples of histology sections and manual segmentations that show the variation in our dataset. F : Fibrous, N : Necrotic, L : Lipid
On the digitized histology slices (resolution 1.8 × 1.8 μm) contours for the lumen, outer vessel wall, necrotic tissue, and lipid regions were manually drawn. Areas within the vessel wall that were not annotated as necrotic or lipid regions were labeled as fibrous tissue. For registration and evaluation purposes lumen and outer wall contours were drawn on the ex vivo MR images and the in vivo aligned MR and CTA images. Sections with low image quality due to histology disruption and sections which contained almost no tissue were excluded. In total, 176 histology sections were used for analysis (13.5 ± 4.9, range 5-21, per specimen).
HISTOLOGY PATTERN CLASSIFICATION
Image Features
Color, texture, and tissue structure are important features to distinguish plaque components [Figure 1], and therefore features of these types were used. Color: We used intensity of the 3 red, green, blue (RGB) color channels before and after Gaussian smoothing (σ = 0.1mm), where in general fibrous tissue is dark purple, necrotic tissue lighter purple and lipid more pink or yellow. Texture: The mean gradient magnitude at σ = 1 pixel and the standard deviation, both of the green channel in square windows of 10 × 10, 20 × 20 and 30 × 30 pixels, were expected to differentiate tissues by tissue heterogeneity and the strength and amount of edges. The structure tensor[8] was calculated (inner scale σ = 1 pixel = 1.8μm, outer scale σ = 72 μm) to differentiate between highly structured tissues (parallel fibrous bands) and less ordered tissues (lipid, and to a lesser extent necrotic tissue). We used the first and second eigenvalue (|λ1| > |λ2|), coherence 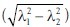 , and the determinant.
, and the determinant.
The original histology slices have large dimensions and are, therefore, time-consuming to process. The color and texture features were calculated from downsampled images that still had a resolution of 0.015 × 0.015 mm. The features of the structure tensor were calculated from the original images, and were subsequently downsampled as well to perform training and classification.
Segmentation
Pixel classification was performed within the manual contours of the vessel wall, with linear discriminant analysis (LDA) for the three components fibrous, lipid, and necrotic tissue. Calcified tissue was not included as the specimens were decalcified and calcifications were not available in the manual annotations. Two approaches were evaluated: 1) Training a classifier on an independent dataset and 2) training a classifier on one or two sections of the specimen to be segmented (details in section Evaluation). Whereas, the second approach requires the annotation of one or two slices for each new specimen, this approach is expected to better account for differences in appearance between specimens. Therefore, of each specimen two histology sections were selected: The first was thought to be most representative for the other sections, and the second to form the most representative combination together with the first selected section. Representative sections contained all three tissue components, and the tissue of these components was similar in appearance to the other sections of that specimen.
Evaluation
Histology Segmentation
Classification accuracy by LDA was compared between: 1) Leave-one-specimen-out cross-validation, where 1% of the pixels from the twelve other specimens were used for training, 2) training on all pixels from the first selected section of the specimen to be segmented, and 3) training on both selected sections. Evaluation was performed on the sections that were never used for training, so on the same sections in every case (number of slices per specimen minus two). The average pixel-wise accuracy for 13 specimens, overall sensitivity for all three components for all specimens together, and correlation of relative component volumes with those obtained from manual segmentations were calculated. A Friedman test was done to test the accuracy for statistical differences.
Ex Vivo MRI
To evaluate the validity of the obtained histology segmentations for training of ex vivo MRI plaque segmentation methods, we use the approach proposed in.[4] This method uses registered three-dimensional (3D) histology to train a voxel classification method. The lipid and necrotic regions from histology were combined to form lipid-rich necrotic cores (LRNC), which is often done in MRI studies.[3–5] Areas for calcification were obtained by thresholding μCT scans, and subtracted from the (automatically determined) fibrous and LRNC segmentations, resulting in three components for classification. Results were compared for training on four different ground truth histology segmentations: 1) The manual histology segmentations, 2) the leave-one-out automated segmentations, 3) the manual histology segmentation for the first selected section per specimen, and the automatic segmentations after training on this section for the other sections, and 4) the manual histology segmentations of both selected sections, and the automatic segmentations after training on these sections for the remaining sections.
Histology, μCT, and ex vivo MRI were registered using a semi-automatic 3D registration approach.[4,9] The same feature set as previously was used,[4] consisting of the available MR images (normalized), first and second order derivatives, and distances to the vessel wall. LDA was evaluated in a leave-one-specimen-out manner, training on 1% of voxels of the other twelve specimens, for each specimen.
For evaluation, voxelwise accuracy and the relative plaque component volumes were determined per specimen (all slices combined) with respect to the manual histology segmentations and thresholded μCT. A Friedman test was done to test the voxelwise accuracy and volume errors for statistical differences.
In Vivo MRI and CTA
Combined MRI and CTA images were segmented as proposed in,[7] using the four different histology segmentations for training, as explained in the previous subsection. Image features were the same as previously[7] and similar to the features used in the ex vivo image segmentation. Registration of all imaging modalities and histology was carried out as described previously.[7,9] As the final registration between histology and in vivo data is less accurate than between histology and ex vivo data, evaluation was only done on Spearman rank correlation of relative volumes and on the error of the relative volume of each component in the result compared to the manual ground truth, per specimen (all slices combined per specimen).
RESULTS
Histology Segmentation
Results are shown in Table 1 and Figure 2. With training on independent data an accuracy of 75.7 ± 6.8% was obtained. Spearman rank correlations were high for fibrous (0.88) and lipid (0.93), and good for necrotic tissue (0.74). Using training data from the same specimen slightly increased both accuracy (77.6 ± 6.3%) and correlation ρ =0.98 for fibrous and lipid and 0.82 for necrotic tissue), although not statistically significant. The main improvement was an increased sensitivity for lipid and necrotic tissue. As for MRI, lipid and necrotic tissues are often combined into one vulnerable component; the combined sensitivity is also given. Specificity for fibrous tissue is then equal to this sensitivity, and vice versa.
Table 1.
Results for histology classification
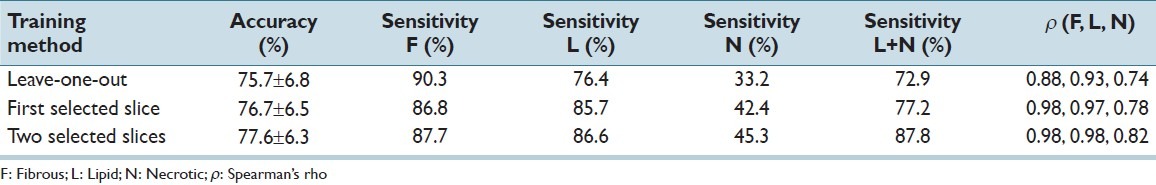
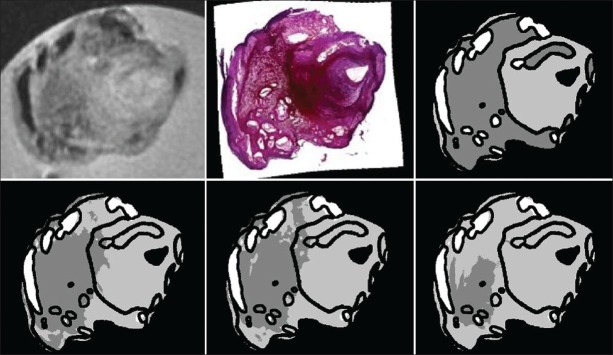
Figure 2.
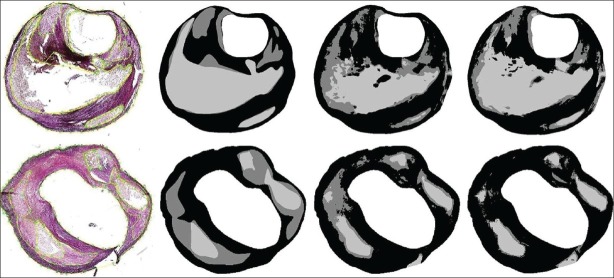
Examples of histology segmentation. From left to right: Histology section, manually annotated ground truth, result by leave-one-out training, and the result for training on two other sections of the same specimen. In the top row, the accuracy is 73.3% with leave-one-out training, and 82.2% when trained on two other sections. All tissue types are well-detected. In the bottom row, the accuracies are 66.7% and 67.8%. The sensitivity for necrotic tissue is lower. Black = fibrous, dark gray = necrotic, and light gray = lipid
Ex Vivo MRI
Results are shown in Table 2 and Figure 3. The results were very similar for training on either manual or automated histology segmentations, and the accuracies and correlations showed no statistically significant differences. The errors in the estimated volumes of the three components were only significantly larger than training on manual histology segmentations when training on leave-one-specimen-out segmented histology, and for calcification. The difference for calcification was so small that it is clinically probably not relevant.
Table 2.
Results on ex vivo MRI
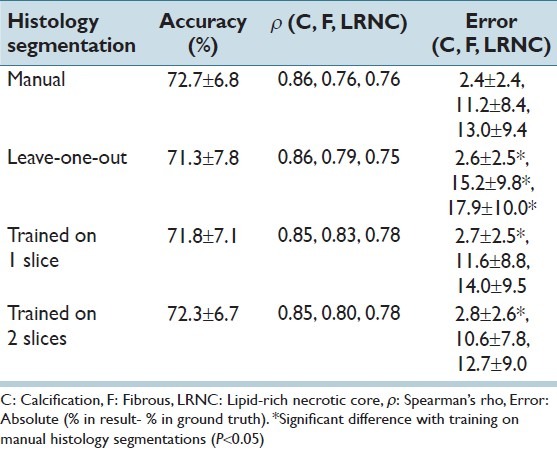
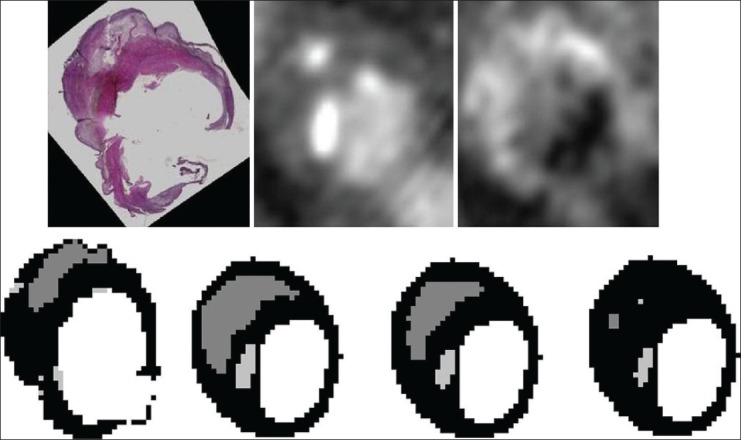
Figure 3.
Example of ex vivo MRI segmentation. Top row from left to right: PDw MRI, histology section and the ground truth. Bottom row: Similar results are obtained by training on manually segmented histology (left figure, accuracy 74.6%) or histology segmentations trained on two slices of that specimen (middle figure, 75.1%). The histology sections segmented using leave-one-out classification result in a reduced accuracy (right figure, 61.4%). Light gray = fibrous, dark gray = LRNC, and white = calcification
In Vivo MRI and CTA
Results are shown in Table 3 and Figure 4. Spearman rank correlation values of relative plaque component volumes for the 13 specimens are similar for training on the automated and manual histology segmentations and even show slight improvements. The absolute error in volume estimation of fibrous and lipid-rich necrotic tissue is somewhat larger when the classifier is trained using the automated histology segmentations. This is not significantly different when the histology is segmented by training on data from one or two slices per specimen.
Table 3.
Results on in vivo MRI
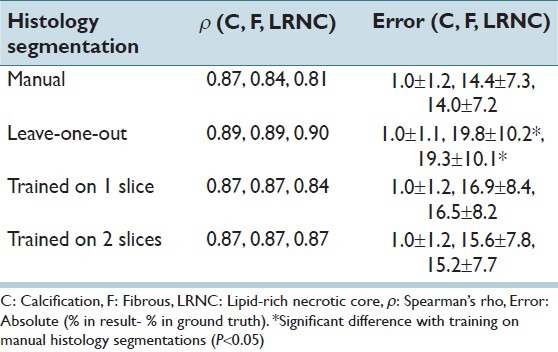
Figure 4.
Example of in vivo MRI/CTA segmentation. Top row from left to right: Histology section, CT-angiography and PDw MRI. Bottom row: The relative size of the LRNC is 15.0% in the ground truth (left Figure), 27.4% using the manual histology segmentations (second Figure), 17.7% for training on two slices of the specimen (third Figure), and 1.0% for leave-one-out histology segmentation (right Figure). Black = fibrous, dark gray = LRNC, and light gray = calcification
CONCLUSIONS
We performed automated segmentation of histology sections of atherosclerotic plaques, and subsequently used these as a ground truth to train segmentation algorithms in ex vivo and in vivo images.
The good accuracy and high Spearman rank correlations of histology segmentation indicate that, although multiple factors are relevant, the automated histology segmentations may be used as a measure in studying plaque vulnerability. The accuracy slightly improved by training on one or two slices of the specimen to be segmented instead of training on data from other specimens, however, differences were not significant.
When these automated histology segmentations were used to train ex vivo and in vivo plaque component segmentation methods, very similar accuracies and correlations were found as when the manual histology segmentations were used for training. This suggests a similar performance for grading of plaque vulnerability. Fibrous tissue was a little more overestimated and LRNC more underestimated when the automated segmentations were used. This is mainly because in the histology sections the difference between fibrous and necrotic tissue is difficult to detect consistently, which results in the classification of necrotic tissue as fibrous. MRI segmentation is slightly better when the automated histology sections for training were obtained by training on one or two annotated sections per specimen. Both better histology segmentations, and the use of the manually annotated sections in the ground truth for MRI classifier training contribute to this. Annotation of two slices still would reduce the number of histology sections that need complete manual segmentation by 85% in the used dataset.
Segmentation of plaques into hard classes may be difficult at specific locations, as gradual changes from one component to another occur. This can be taken into account in future research by including a mixed class or by obtaining posterior probabilities instead of hard segmentations. In addition, empty regions may be present in histology. Lipid tissue is very soft and, therefore, may be disrupted during slicing and observers annotate empty regions usually as lipid. In relation to interobserver reproducibility of histology segmentation our results are in between previously reported values. The same annotations for 92% of all pixels by two observers has been reported,[3] but moderate agreement for observers indicating which components or characteristics are present has also been found[10] (κ: 0.44-0.68 for different components).
This paper uses manual lumen and outer wall contours in all histology sections and is, therefore, not completely automatic. However, as the specimen can clearly be distinguished from the background, we expect automatic segmentation to be relatively easy.
In conclusion, good histology segmentations can be obtained using automated pixel classification. Automatic histology segmentation by using independent training data seems sufficient to indicate plaque vulnerability in clinical studies. These segmentations can also be used as training data to design automatic techniques for segmentation of plaque components in ex vivo MRI and in vivo MRI and CTA data. By using several manually annotated sections for each specimen the segmentations can be improved.
ACKNOWLEDGMENTS
This research was performed within the framework of CTMM, the Center for Translational Molecular Medicine (www.ctmm.nl), project PARISk (grant 01C-202), and supported by the Dutch Heart Foundation. WN, SK, and MdB were financially supported by the Netherlands Organization for Scientific Research (NWO). HG was financially supported by the Interuniversity Cardiology Institute of the Netherlands.
Footnotes
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2013/4/2/3/109844
REFERENCES
- 1.Seeger JM, Barratt E, Lawson GA, Klingman N. The relationship between carotid plaque composition, plaque morphology, and neurologic symptoms. J Surg Res. 1995;58:330–6. doi: 10.1006/jsre.1995.1051. [DOI] [PubMed] [Google Scholar]
- 2.Akyildiz A, Speelman L, van Brummelen H, Gutiérrez MA, Virmani R, van der Lugt A, et al. Effects of intima stiffness and plaque morphology on peak cap stress. Biomed Eng Online. 2011;10:25. doi: 10.1186/1475-925X-10-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clarke SE, Hammond RR, Mitchell JR, Rutt BK. Quantitative assessment of carotid plaque composition using multicontrast MRI and registered histology. Magn Reson Med. 2003;50:1199–208. doi: 10.1002/mrm.10618. [DOI] [PubMed] [Google Scholar]
- 4.van Engelen A, Niessen WJ, Klein S, Groen HC, Verhagen HJ, Wentzel JJ, et al. Multi-feature-based plaque characterization in ex vivo MRI trained by registration to 3D histology. Phys Med Biol. 2012;57:241–56. doi: 10.1088/0031-9155/57/1/241. [DOI] [PubMed] [Google Scholar]
- 5.Liu F, Xu D, Ferguson MS, Chu B, Saam T, Takaya N, et al. Automated in vivo segmentation of carotid plaque MRI with morphology-enhanced probability maps. Magn Reson Med. 2006;55:659–68. doi: 10.1002/mrm.20814. [DOI] [PubMed] [Google Scholar]
- 6.Gurcan MN, Boucheron LE, Can A, Madabhushi A, Rajpoot NM, Yener B. Histopathological image analysis: A review. IEEE Rev Biomed Eng. 2009;2:147–71. doi: 10.1109/RBME.2009.2034865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Engelen A, Niessen WJ, Klein S, Groen HC, Verhagen HJ, Wentzel JJ, van der Lugt A, de Bruijne M. Proceedings of the International Symposium on Biomedical Imaging. Barcelona, Spain: 2012. May 2-5, Supervised in-vivo plaque characterization incorporating class label uncertainty. [Google Scholar]
- 8.Weickert J. Multiscale texture enhancement. In: Hlaváč, Šára, editors. Proceedings of Computer Analysis of Images and Patterns 1995; Lecture Notes in Computer Science. Vol. 970. Berlin/Heidelberg: Springer; 1995. pp. 230–7. [Google Scholar]
- 9.Groen HC, van Walsum T, Rozie S, Klein S, van Gaalen K, Gijsen FJ, et al. Three-dimensional registration of histology of human atherosclerotic carotid plaques to in-vivo imaging. J Biomech. 2010;43:2087–92. doi: 10.1016/j.jbiomech.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 10.Lovett JK, Gallagher PJ, Rothwell PM. Reproducibility of histological assessment of carotid plaque: Implications for studies of carotid imaging. Cerebrovasc Dis. 2004;18:117–23. doi: 10.1159/000079259. [DOI] [PubMed] [Google Scholar]


