Abstract
Introduction:
The notion of local scale was introduced to characterize varying levels of image detail so that localized image processing tasks could be performed while simultaneously yielding a globally optimal result. In this paper, we have presented the methodological framework for a novel locally adaptive scale definition, morphologic scale (MS), which is different from extant local scale definitions in that it attempts to characterize local heterogeneity as opposed to local homogeneity.
Methods:
At every point of interest, the MS is determined as a series of radial paths extending outward in the direction of least resistance, navigating around obstructions. Each pixel can then be directly compared to other points of interest via a rotationally invariant quantitative feature descriptor, determined by the application of Fourier descriptors to the collection of these paths.
Results:
Our goal is to distinguish tumor and stromal tissue classes in the context of four different digitized pathology datasets: prostate tissue microarrays (TMAs) stained with hematoxylin and eosin (HE) (44 images) and TMAs stained with only hematoxylin (H) (44 images), slide mounts of ovarian H (60 images), and HE breast cancer (51 images) histology images. Classification performance over 50 cross-validation runs using a Bayesian classifier produced mean areas under the curve of 0.88 ± 0.01 (prostate HE), 0.87 ± 0.02 (prostate H), 0.88 ± 0.01 (ovarian H), and 0.80 ± 0.01 (breast HE).
Conclusion:
For each dataset listed in Table 3, we randomly selected 100 points per image, and using the procedure described in Experiment 1, we attempted to separate them as belonging to stroma or epithelium.
Keywords: Heterogeneity, local morphological scale, tumor identification
INTRODUCTION
The notion of local scale was introduced to characterize varying levels of image detail so that localized image processing tasks could be performed, yielding an optimal result globally.[1] Pizer, et al., suggested that having a locally adaptive definition of scale was necessary even for moderately complex detailed images. By quantifying these images details, an adaptive local scale image could encode implicit information present in the image intensity values. The underlying concept of localized scale definitions is that homogeneous regions can have computations perform data lower resolution in a similar manner, while more heterogeneous regions can either be examined at higher resolutions using more computationally expensive approaches or broken down into even smaller homogeneous regions.[2] Locally adaptive scale has seen application in a variety of image processing tasks that warrant the identification of locally connected homogeneous regions such as magnetic resonance imaging (MRI) bias field correction,[3] image segmentation,[4] image registration,[5] and image coding.[6]
Previous local scale definitions (e.g., ball, tensor, generalized scale) have either employed prior shape constraints or pre-specified homogeneity criterion to determine locally connected regions. Saha and Udupa introduced the notion of ball-scale (b-scale),[7] which at every spatial location was defined as the value corresponding to the radius of the largest ball encompassing all locations neighboring the location under consideration and satisfying some pre-defined homogeneity criterion. In a previous study,[2] Saha extended the ball-scale idea to a tensor-scale (t-scale), where the t-scale was defined as the largest ellipse at a very spatial location where the pixels within the ellipse satisfied some pre-defined homogeneity criterion. The shape constraints of both (b-scale) and (t-scale) were overcome by Madabhushi and Udupa with the introduction of generalized scale (g-scale).[4] g-scale was defined as the largest connected set associated with every spatial location, such that all spatial locations in this set satisfied a pre-defined homogeneity criterion.
In this work, we have presented a new definition of scale, morphologic scale (MS), which is appropriate for images with high degrees of local complexity, where local scale definitions governed by satisfying homogeneity and pre-defined shape criterion breakdown. Our innovative approach attempts to model the heterogeneity of a local region, allowing for the definition of local, quantitative signatures of heterogeneity. Since MS motivates the definition of local regions as regions which are topographically similar, pixel-level features can be defined from the corresponding MS at that location, features which can then be used for segmentation, registration, or classification. The MS idea also confers several advantages for pixel- and object-level classification compared to a number of other traditional approaches, such as texture features, where the selection of the window size is not only critical but challenging as it is difficult to find a setting which is appropriate for all image regions. In the case of template matching, each homogeneous region is so unique that templates struggle to generalize to higher order classes. MS acts as a novel feature extraction method that overcomes both limitations by being robust across a wide range of window sizes and the ability to generalize well to new, unseen data.
One of the critical challenges in personalized diagnostics is to identify image-based histological markers that are indicative of disease and patient outcome. For instance, lymphocytes contained within tumors (tumor infiltrating, i.e., TILs) have been suggested as a favorable prognostic marker,[8] but morphologically they appear similar to stromal lymphocytes. However, the corresponding topography and, thus, local heterogeneity is actually quite different. In this work, we applied MS to the problem of distinguishing tumor from stromal regions at the pixel level in digitized histopathology images of prostate, breast, and ovarian cancer studies in order to identify these TILs. Although in the context of the problems chosen in this work, we focused on tissue slides stained with standard hematoxylin and eosin (HE), the approach is extensible to tissue samples stained with alternate stains as well.
In Section 2, we have discussed other works in the field of tumor versus stroma identification, followed by Section 3 with the methodology. Section 4 presents the experimental design and associated results. Lastly, Section 5 contains concluding remarks and future work.
Previous Work
In the previous section, we have compared our MS approach to the other pertinent scale approaches. This section reviews previous work specific to the domain of histopathology and our stated application of identifying tumor regions. The desire to separate tumor from stroma regions is not new as attempts at quantifying epithelial volume in tumors by image processing techniques have been reported as far back as the late 1980s.[9,10] Work in automated retrieval for regions of interest from whole-slide imaging has led to the ability to detect cancerous regions via segmentation.[11] Recently, Haralick texture features, local binary patterns, and Gabor filters were employed to build classifiers for the tumor–stroma separation problem.[12] However, most of these approaches have been limited in that the approach has either required specialized staining or the algorithms tend to be computationally prohibitive. Below, we have provided a brief summary of some of these previous approaches and discussed some of their limitations, thus motivating our new approach.
Specialized Staining
Specialized staining techniques have been applied to tissue in order to facilitate tumor-stroma separation. In some cases, it is possible to stain directly for a specific tumor type of interest allowing for a clear separation of regions. In a previous study,[13] a graph-based approach was presented for separating stroma and tumor on fluorescence images, from which the 49,6-diamidino-2-phenylindole (DAPI) channel was extracted. Using topological, morphological, and intensity based features extracted from cell graphs, a support vector machine classifier was constructed to discriminate tumor from stroma. The MS based approach presented in this paper was able to tackle the same problem while operating on solely industry standard hematoxylin (H) or HE stained images, potentially obviating the need for specialized staining for discriminating the stroma and epithelial tissue classes.
Computationally Expensive
An important consideration for automated and computer-based image analysis and quantification procedures developed in the context of digital pathology is that they need to be computationally efficient. For example, N-point correlation functions (N-pcfs) for constructing an appropriate feature space for achieving tissue segmentation in histology stained microscopic images was presented earlier.[14] While the approach showed > 90% segmentation accuracy, the authors acknowledged the need to find more optimal data structures and algorithms to reduce the computational overhead. Another approach, termed C-path,[15] first performed an automated, hierarchical scene segmentation that generated thousands of measurements, including both standard morphometric descriptors of image objects and higher-level contextual, relational, and global image features. Using the concept of superpixels, they measured the intensity, texture, size, and shape of the superpixel and its neighbors and classified them as epithelium or stroma. While the authors report approximately 90% segmentation accuracy for the tumor–stroma separation problem, the approach involves first computing over 6,500 features, suggesting a significant computational overhead to their approach. By contrast, the MS-based approach involves the use of a significantly lower dimensional feature set, resulting in a high-throughput approach, specifically designed to meet clinical needs.
CONCEPTUAL AND METHODOLOGICAL DESCRIPTION OF MS
Brief Overview
Figure 1 presents an overview of the MS creation process as it applies to the problem of tumor versus stromal differentiation. The individual steps involved in the computation of the MS are briefly outlined below and described in greater detail in Sections 3.2-3.3.
Figure 1.
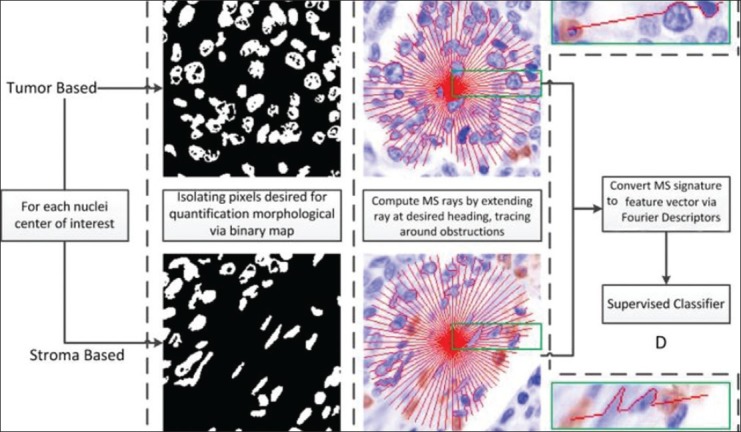
Overview of the MS signature creation process. We can see that the tumor MS signature contains a noticeably increased number of deviations from the straight line trajectory on account of the rays attempting to take the path of least resistance and, hence, overcome obstacles along the way. On the other hand, the MS signature for the non-tumor (stroma) region is much smoother as a result of comprising fewer and smaller objects. Note that a pre-defined stopping criterion is employed such that rays stop when they hit blank spaces
Step 1: Identify both lymphocytes and other cell nuclei centers of interest using a hybrid mean shift and normalized cuts algorithm.[16,17] This is done via direct targeting of the unique color-staining properties. The resulting binarized images are seen in column B. Binarized images indicate which pixels will be incorporated in the calculation of the morphological signature of the point of interest (POI).
Step 2: Calculating MS involves projecting connected paths, radially outward from the POI (nuclear center identified in Step 1). Column C in Figure 1 shows the MS signature (in red) for an illustrative TIL (top) and a non-TIL (bottom). In each image, a green box is used to illustrate the path of a single ray more clearly.
Step 3: The quantification of the average local topology of all these paths (via Fourier descriptors (FD)[18]) yields a characterization of local heterogeneity. These morphological features characterize the local structural topology of the binarized image (Step 1) via the individual rays/paths (Step 2).
Step 4: Use of the MS feature vector created by the FD to train a supervised classifier to identify the POIs, as being located either in tumor or stromal regions.
Radial Path Projection
The paths traversed by an ordered set of rays projected from the POI are used to model the local structural heterogeneity around the POI. Before describing the process of the radial path projection at each POI and subsequent MS computation, we have first introduced some basic definitions and concepts. We started with an image scene defined as C = (C, f), where C is a 2D Cartesian grid of N pixels, c ∈ C, where c = (x, y). We assumed two pixels c, d as adjacent, that is φ(c, d) =1, if c and d differ in exactly one of their components by 1; otherwise φ(c, d) =0. From the image, we can identify a path as an ordered set of m connected pixels, which starts at r and ends at s, or more formally, the following:
pr,s = {<c(1)= r,c(2),….,c(m–1),c(m)=s>:φ(c(i),c(i+1)) = 1, i ∈ {1,…, m}} (1)
We next refined the set of possible paths by limiting which pixels can be included in the path by applying an additional affinity constraint. We considered pr,s to be a μ-path, ṗr,s, if each pair of adjacent pixels conforms to an affinity constraint μ(c(i), c(i+1)) = g(c(i)) + g(c(i+1)) =0, where g(c)∈{0, 1} returns 1 if the pixel is to be considered in the morphological signature. Our affinity constraint simply implies that sequential pixels are both off. The particular ṗr,s that we were interested in is identified by  intuitively making it the shortest path from r to s such that the affinity criteria is met. We can see an example of this in Figure 2a, where the MS path circumvents the obstruction while remaining as close to a straight line as possible given the constraints. This is a single MS ray (Rθ(q)) in direction θ. We could of course perform the same operation in multiple different directions, as shown in Figure 2b.
intuitively making it the shortest path from r to s such that the affinity criteria is met. We can see an example of this in Figure 2a, where the MS path circumvents the obstruction while remaining as close to a straight line as possible given the constraints. This is a single MS ray (Rθ(q)) in direction θ. We could of course perform the same operation in multiple different directions, as shown in Figure 2b.
Figure 2.

An example of an MS ray in (a), where the POI is indicated with X and the target point is indicated with A. We can see that applying an affinity constraint limits the selection of pixels to those which belong to the background, resulting in the shortest path, one which circumvents obstructions
We can see from the examples in Figure 3, the end result of our algorithm is a set of connected pixels (shown in red), which travel from the POI q to δ. The algorithm proceeds by the following:
Figure 3.
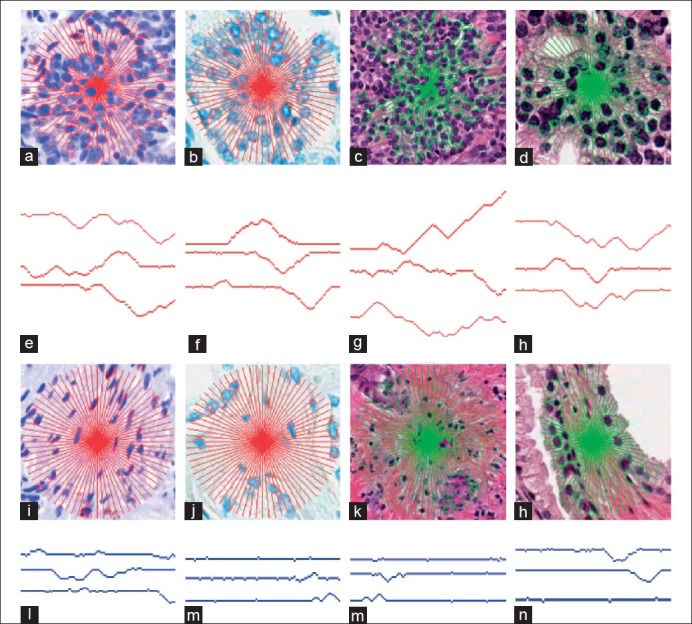
The MS signature overlaid on tumor regions in an (a) ovarian, (b) prostate H, (c) breast HE, and (d) prostate HE image. Corresponding results for benign regions are shown in (i-l), respectively. Three rays from each image (e-h) and (m-p) are extracted and illustrated beneath the corresponding images. We can see that in the presented non-tumor regions (i-l), the MS signature has fewer and smaller objects to obstruct its path, and thus the rays are less tortuous, unlike in the corresponding tumoral regions (a-d)
Step 1: Translating the image such that q is placed at the origin
Step 2: For each of Rθ, θ ∈ {0, ε, 2ε, ..., 2π} (where, ε is used to control resolution of the signature), we determined the location of the end point δ by casting it on a unit circle and multiplying by the window size to get the appropriate magnitude. It is possible that the desired end point δ is not always a background pixel (g(δ) = 1) and thus, we assigned δ to the closest possible pixel in the Euclidian sense, which has the required property (g(δ) = 0).
Step 3: Identify the shortest μ-path, 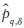 , which is intuitively the path with minimal divergence to circumvent obstacles from q to δ. This is to say when the path hits an object, the resulting affinity function threshold criterion is exceeded and, hence, the path continues along a new direction of lower resistance (satisfying the affinity criterion).
, which is intuitively the path with minimal divergence to circumvent obstacles from q to δ. This is to say when the path hits an object, the resulting affinity function threshold criterion is exceeded and, hence, the path continues along a new direction of lower resistance (satisfying the affinity criterion).
Step 4: We applied these same steps for all desired orientations, as defined by the angle interval parameter ε, and produced R(q), the set of individual rays.
We can see that Rθ(q) models local morphology at point q in direction θ via deviations from a straight line. As μ constrains the MS path to background pixels, we can expect Rθ(q) to become more tortuous as it collides into objects, thereby gaining entropy, resulting in a direct correlation between the pixels selected for Rθ(q) and its associated heterogeneity. For example, if the region contains no obstructions, Rθ(q) is a straight line indicating the most trivial case of homogeneity. On the other hand, if Rθ(q) is computed in an image with a repeating pattern, we would expect to see waves of similar amplitude at equally spaced intervals, encoding the homogeneity of the implicit variables. Lastly, if Rθ(q) is computed in an arbitrarily constructed image, we would expect to see waves of dissimilar amplitudes as the ray must circumvent objects of various sizes. These waves of differing amplitudes would be unevenly spaced as the objects are not uniformly placed, allowing for the modeling of the heterogeneity of these two variables. It is worthwhile to note that since we are not explicitly defining the domain-specific attributes, the ray is subject to varying size, concavities, and morphology.
MS Algorithmic Implementation
The algorithm for computing the MS signature is accompanied with certain computational considerations. When computing 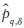 optimally, such that we are guaranteed the shortest past, the application of the approach becomes computationally expensive. This expense is as a result of need to use algorithms such as Dijkstra[19] or a Fast Marching[20] approach to compute the global minimum length path. On the other hand, we can sacrifice some precision and obtain a “short path,” but not the “shortest” path and benefit from orders of magnitude improvement in efficiency. To do so, we have suggested an iterative greedy approach towards solving
optimally, such that we are guaranteed the shortest past, the application of the approach becomes computationally expensive. This expense is as a result of need to use algorithms such as Dijkstra[19] or a Fast Marching[20] approach to compute the global minimum length path. On the other hand, we can sacrifice some precision and obtain a “short path,” but not the “shortest” path and benefit from orders of magnitude improvement in efficiency. To do so, we have suggested an iterative greedy approach towards solving  a sampled approximation of
a sampled approximation of 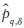 .
.
The approach is outlined by the following three steps:
Step 1: Define the first pixel in the path (c(0)) as the query pixel (q)
Step 2: Iteratively select the next pixel in the path by determining, using Euclidian distance, which of the possible pixels in the κ-neighborhood is closest to its goal (κ = 8).
Step 3: If there are two pixels with the same value, sample from them with equal probability.
Step 4: Return to Step 1 while the target pixel has not been encountered via the ray tracing or the user defined maximum number of iterations has not been exceeded.
A key consideration for the algorithm described above is the valid pixel indication function ƒθ(c), which is specific to the θ being considered. The value of ƒθ(c) is defined intuitively as returning a true value for pixels, which are in the background (g(c) = 0) and on the edge of the objects or if the pixel is on the straight line path, causing the path to be constrained to objected borders or the line from q to δ.
We also noted that ε controls the sampling density of the rays being traced and serves as a trade-off between total computational time and precision of the MS signature. In addition, not only is the approach computationally straight forward (i.e., requiring only the most basic of arithmetic or logic operations), it can be seen that each ray is computed in a deterministic manner and independent of the adjoining rays. These two properties allow for efficient, parallel computation of the individual rays via GPUs.
Fourier Descriptors of MS
FD[18] are a technique for quantifying the morphological structure of a closed curve. FD have the desirable property of being scale, translationally, and rotationally invariant. These properties are important in domains such as biomedical image analysis, where information may often be represented in an orientation-free plane. We used a slightly modified version of the FD, as scale invariance is not criticalto our problem, as all samples are drawn from the same magnification. Unfortunately, at first glance, R(q) is not a closed curve and, thus, these techniques cannot be directly applied. In the following section, we have described a procedure for conversion of the open set of R(q) to a closed curve L(q).
Process for Conversion from R(q) to a Closed Curve
Figure 4a illustrates that the four rays produced by the MS algorithm are not closed curves, as each is a path traveling away from the POI. However, to parameterize the curve in terms of FD, the open curve needs to be converted into a closed contour. To overcome this constraint, for each θ of interest, we adjoined a straight path from δ to q. Observed in Figure 4b as we proceeded from X to A, we followed the MS generated path (in red). Upon reaching A, we proceed back to X by following the green path, a straight line connecting the two points. This now forms a single closed contour as it begins and ends at point X. Since all θ begin and end at X, we have created a single large closed contour from many smaller closed contours.
Figure 4.

Visual example of the conversation from a set of MS rays to a 1D parameterized representation. (a, b) The nuclei stained in deep blue represent obstructions causing local heterogeneity. As the μ-paths are minimized, we can see avoidance of these objects. Later, we formed a closed contour (b) from the MS rays (red) by adding a straight path (green lines) from the end point of each path back to the query point. (c) The 1D representation is displayed. We shrinked the straight lines down to zero, as they provided only redundant information and, thus, (d) retained all heterogeneous information in a compact format
Computation of Fourier Descriptors
The steps followed for the creation of the feature vector from R(q) using FD are presented below and are illustrated via Figure 4.
Step 1: Rotate each Rθ by –θ, resulting in all rays originating at q and having an orientation of 0°
Step 2: Concatenate each Rθ with a straight line from δ to q. We can see the result of this in Figure 4b. We formed L(q), a 1D signal representation of R(q), when these straight lines were reduced to length zero, as shown in Figure 4d.
Step 3: Compute the magnitude of F(L(q)), the Fourier transform of L(q), as F(q). From the proof demonstrated earlier,[18] this leads to a rotationally invariant representation of L(q) in the frequency space, F(q).
Post Processing of Extracted Fourier Descriptors
Typically, the Fourier transforms attempts to quantify the frequency presence in a signal. In the case of using image data, which is discrete, there is often a high degree of saw-tooth wave like properties as shown in the blue curve of Figure 5a. These saw-tooth signals require a high number of FFT coefficients to accurately represent them without adding a large amount of residual noise due to the approximation. To compensate, we smoothened J(q) using a simple moving average filter [Figure 5]. We can see in Figure 5b that the red curve is indeed less susceptible to the discrete nature of the image scene and, thus, could be more easily represented by fewer FFT coefficients.
Figure 5.
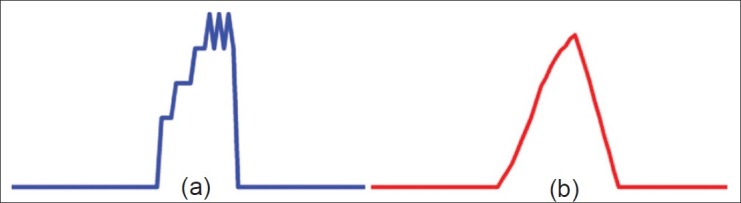
A segment of (a) R (q) shows that it tends to be subject to the discrete nature of the pixel image domain. On the other hand, after applying a smoothing filter, we can see that (b) the function possesses qualities that are better suited for Fourier transform representation
EXPERIMENTAL RESULTS AND DISCUSSION
Application of MS to Classification Problems in Digital Pathology
We applied the MS idea to the problem of detecting tumor infiltrating lymphocytes, which in turn requires separation of epithelial and stromal regions on HE-stained histological images. Ground truth was available in the form of spatial locations of lymphocytes as determined by an expert pathologist. In three experiments involving ovarian, prostate, and breast cancer pathology images, we evaluated whether for any given lymphocyte the MS-based classifier was able to correctly identify it as being within the tumor epithelium (hence a TIL) or within the stroma (and hence a non-TIL).
Training and Testing Methodology
For all our experiments, we sampled a single pixel within the lymphocytic nucleus and attempted to identify the location as being within stroma or epithelium. Consequently, the training and testing sets comprised of individual pixel locations sampled from TILs and non-TILs, respectively. For all the pixel locations so identified, a super-set Q was constructed. From this set Q, approximately 50% of the points were used to construct the training set (Qtr), while the remaining 50% were used as test data (Qte) such that Qte ∪ Qtr = Q and Qte ∩ Qtr = ɸ. Using the training set, a naive Bayesian supervised classifier[21] was built, which involved fitting a multivariate normal density to each class. A pooled estimate of covariance was employed. All pixels in the set Qte were classified as belonging to either the positive (TIL or tumor) or negative class (non-TIL or stroma) and a receiver operating characteristic (ROC) curve was computed. The area under the ROC (AUC) was computed for 50 runs of cross-validation, whereby, in every run, the entire set of samples (Q) was randomly split into training and testing sets. Mean and variance of the AUC was calculated across the different runs.
Experiments in TIL Identification in Ovarian Cancer Slides
Dataset Description
The dataset consisted of 60 whole slide ovarian cancer images of size 1400 × 1050 pixels. Each slide was H-stained, which made the tumor and endothelial cells appear blue, and stained with a CD3-positive T stain, which caused the lymphocytes to appear in red. The images were then scanned using an Aperio slide scanner at ×40 magnification. We have displayed some representative images in Figure 6. In total, 4320 lymphocytes were identified by an expert pathologist, comprising of 1402 TILs and 2918 non-TILs.
Figure 6.
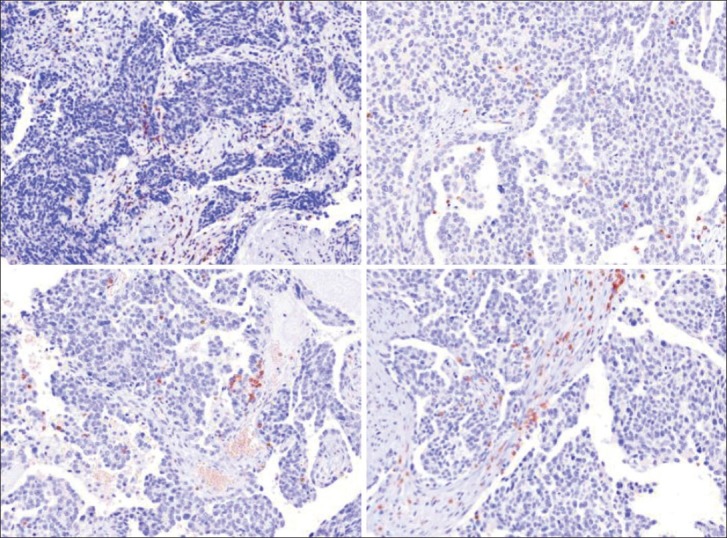
Sample images from the OCa dataset. Each image is 1400 × 1050 pixels, and the blue endothelial and tumor cells are very visible and easily differentiable from the red stained lymphocytes. Note that the lymphocytes occur both within the stromal (non-TIL) and tumor epithelial regions (TIL)
Experiment 1: Distinguishing TILs from Non-TILs Via MS Classifier
We compared the results from the MS-based classifier (Φms) to (a) a classifier employing texture based features (Φt) and (b) a classifier employing the ball scale representation (Φb)[7] for separating TILs from non-TILs. For Φt and Φms (since these employ multi-dimensional attribute vectors), we concatenate the respective pixel (q) level feature vectors, F(q), row-wise to form a matrix M and computed the t rank truncated Singular Value Decomposition such that 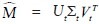 and thus represent each F(q) as its dimensionality reduced Ut(q) counterpart. This allows for the training of classifiers that are simultaneously accurate and computationally efficient.
and thus represent each F(q) as its dimensionality reduced Ut(q) counterpart. This allows for the training of classifiers that are simultaneously accurate and computationally efficient.
Morphological Scale (Φms)
The algorithm proceeds as per the flow chat presented in Figure 1. The only additional pre-processing performed was to use a watershed algorithm[22] to quickly separate large binary regions. To determine the optimal operating parameters, a grid search was conducted for the various variables in the algorithm. The domain space that was searched over is as shown in Table 1. The optimal values for this particular domain and application were found to be ε = 1, t = 5, w = 50, number of coefficients for the Fourier transform = 29, down-sampling = 1, number of points used for smoothing = 10, mask size = 10.
Table 1.
Description of all grid-searched variables used in Φms and their associated attempted values

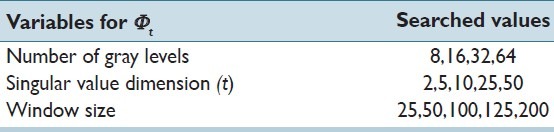
Texture Features (Φt)
Through a grid search of the variables presented in Table 2, we were able to identify the optimal parameters for the texture features as window size set to 50 and the number of gray levels as 16 and the singular value space as 25.
Table 2.
Description of all grid-searched variables used in Φt and their associated attempted values
Ball Scale (Φb)
For this, we used the same implementation as in a previous study,[7] and did not enforce a window size. Briefly, for every pixel location in Q, we computed the corresponding b-scale value that corresponds to the radius of the largest ball at that location, which satisfies a pre-defined homogeneity criterion. The homogeneity criterion employed was the one used in a previous study,[7] involving taking a difference in smoothed image intensities between annular rings.
Results
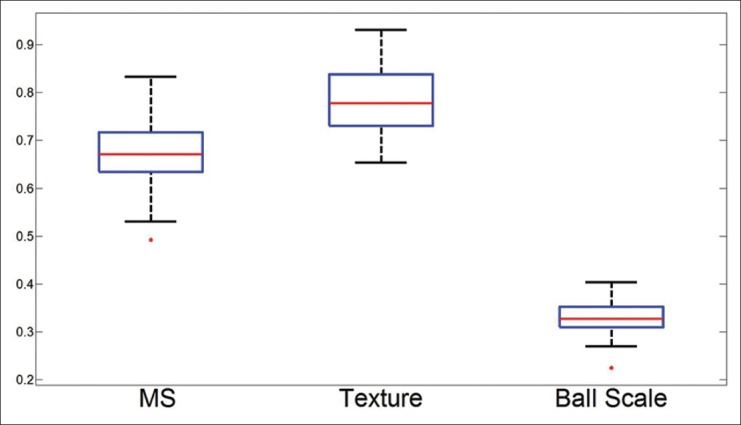
We have presented the box plots for the three approaches in Figure 7, with the associated true positive average in Figure 8 and true negative average value in Figure 9. We can see that with a mean AUC of 0.866, MS yields a slightly better classifier compared to texture features with 0.842. These are comparable to a current state of the art approach,[15] which reported an accuracy of 0.88. Our approach, however, confers the advantage of significantly lower computational cost owing to a significantly smaller-sized feature space in contrast to the nearly 6000 features employed in a previous study.[15] Lastly, we can see that the homogeneity criterion employed in defining b-scale is less than optimal for this task seeing as it performs rather poorly in this classification task.
Figure 7.

Box plots for the AUC across 50 runs from all 3 algorithms (Φms, Φt, Φb). The red line identifies the mean, the blue box encompasses 25th percentile, with the black whiskers extending to the 75th percentile. Red dots are indicative of outliers. We can see that the MS provides a higher mean AUC compared with the texture features, resulting in a significantly a smaller variance. The homogeneity constraint imposed by ball-scale does not appear to provide discrimination between TILs and non-TILs
Figure 8.
Box plots for the true positives across 50 runs from all 3 algorithms. The red line identifies the mean, the blue box encompasses 25th percentile, with the black whiskers extending to the 75th percentile. Red dots are indicative of outliers
Figure 9.
Box plots for the true negatives across 50 runs from all 3 algorithms. The red line identifies the mean, the blue box encompasses 25th percentile, with the black whiskers extending to the 75th percentile. Red dots are indicative of outliers while texture feature-based classifier degrades with increasing scale
Experiment 2: Impact of Window Size
To evaluate the effect of window size on the MS classifier, we performed the identical grid search as in Experiment 1. except that we reported the mean AUC across each tested window size (25, 50, 100, 125, 200) for both MS and texture features.
Results
Figure 10 illustrates the mean AUC across 50 runs using the optimal parameters for each window size. From this experiment, we can clearly see that as the window size varies, the MS approach maintains a consistent AUC.
Figure 10.

Average AUC of Φms using optimal parameters (∈ = I, t = 5) across a set of window sizes w ∈ {25, 50, 100, 150, 200}. The MS maintains a consistent AUC (blue line) even as the window size grows very large. This is in contrast to the texture features graph (green line), which shows a degradation of results along with the expanding window size
Experiment 3: Impact of Interval Size
To evaluate the sensitivity of ε on the classifier results, we performed the identical grid search as in Experiment 1, except that we reported the mean AUC across each ε evaluated (1, 5, 10, 15, 30) for MS.
RESULTS
Figure 11 illustrates (a) the mean AUC across 50 runs using the optimal parameters for each ε interval and the associated time (b). From this experiment, we can see the expected behavior as ε decreases so does the accuracy, but so does the computational time required per sample. However, interestingly, even as ε drops to 1/30th of the original (from 1° to 30° interval), the accuracy only falls about 3%, whereas the computational time required drops by 75%.
Figure 11.
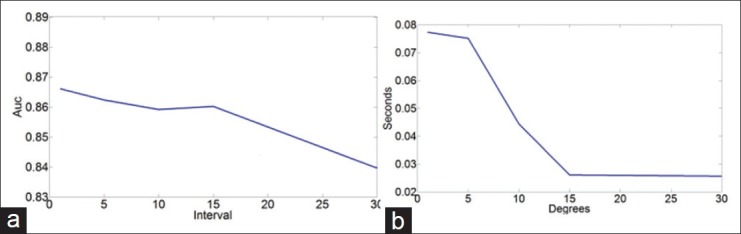
Average AUC of Φms (a) using optimal parameters across a set of 5 varying intervals contrasted with the speed per sample in (b). The total degradation due to smaller sampling is only 3% in exchange for a 75% speed up
Experiment 4: Combined MS and Texture Feature Classifier
Given the results from Experiment 1, we decided to investigate how well a classifier would perform if it used the optimal configurations for MS and texture features in a joint feature space. As such, we concatenate the feature vectors from the two algorithms and used them to train a single classifier (Φms,t) and reported the results across 50 runs.
Results
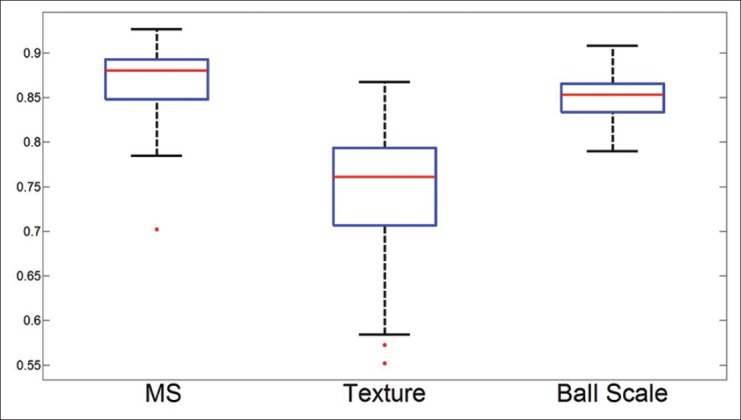
We have presented the AUC, true positive percent and the true negative percent across 50 runs in Figure 12. The true negative and average AUC values approach 0.90, thus conjoining the two classes of features results in a stronger classifier compared to the use of any single feature class. The combination of the sets of features provides potentially orthogonal information to the classifier allowing for a bump in classifier accuracy, with a notable decrease in variance.
Figure 12.

The three box plots associated with the joint classifier Φt, ms. We can see the combination of two of the feature sets produces better results compared to Φt and Φms
Experiments in Discriminating Tumor from Stromal Regions
Dataset Description
For each dataset listed in Table 3 and illustrated in Figure 13, we randomly selected 100 points per image, and using the procedure described in Experiment 1, we attempted to separate them as belonging to stroma or epithelium.
Table 3.
Description of non lymphocyte datasets

Figure 13.
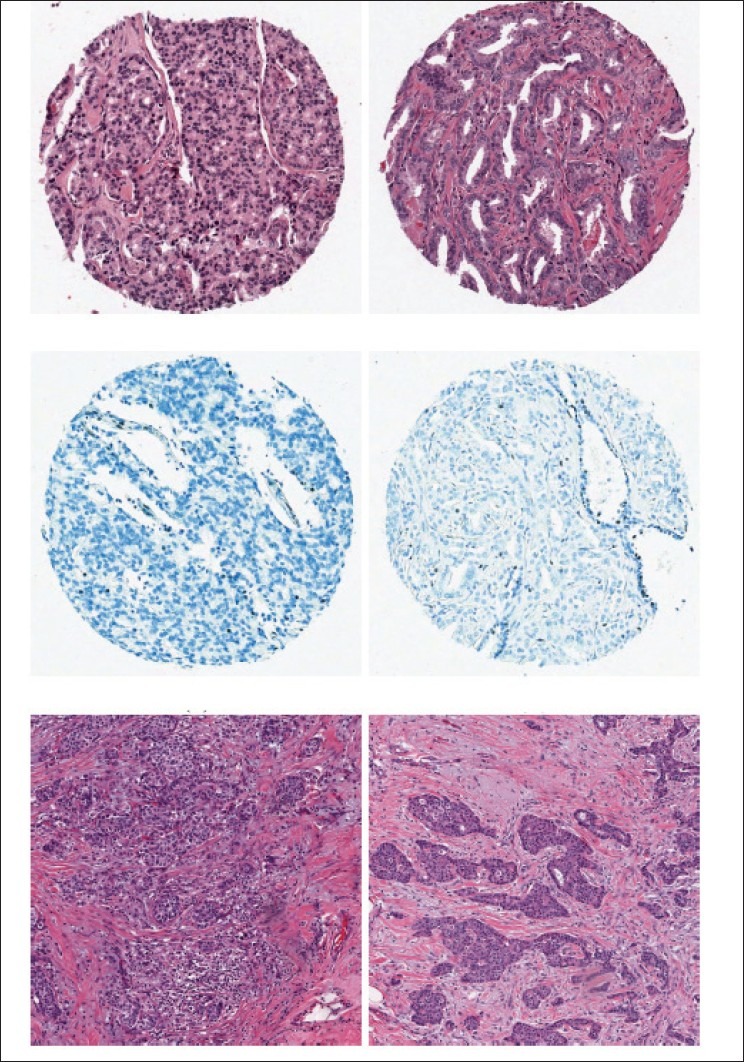
Two sample images from each of the datasets deseribed in Table 3. First row is prostate HE (S1), second row is prostate H (S2) and third row is Breast H (S3)
Experiment 5: Breast and Prostate Pathology Slides
The operating parameters for MS were identical to those employed in Experiment 1.
Results
Although the parameters were not individually tuned for each dataset, we can see from the results in Table 4 that the accuracy values are fairly consistent with those obtained for ovarian cancer (Experiment 1). For datasets S1 and S2, we can see that the mean AUC for Φms is about 0.88, indicating excellent separation between tumoral and stromal regions. It is interesting to note that the HE images did slightly better than the H alone images, probably due to the greater contrast afforded by the counter-staining. Across 51 images of dataset S3, the breast images produced a mean AUC of 0.81 without any parameter tuning.
Table 4.
Bayesian classifier AUC for Φms in distinguishing stromal from tumoral lymphocytes for S1-S3

Qualitative Evaluation
In Figure 3, we have presented MS signatures in red/green overlaid on both tumor and non-tumor regions for representative images from all three datasets. Consistently across tumor-based regions (a-d), the heterogeneity created by the cancer cells is evident by the fluctuations in the MS signature (e-h). Figure 3a reveals that, for a very complex region, the MS paths become increasingly tortuous as they adapt to the local heterogeneity. We can see that, as the complexity of the local region increases, the corresponding MS rays become progressively more convoluted, in turn reflecting the rising entropy. Comparatively, in the stromal regions (i-l), we can see how the homogeneous regions have fewer obstructions as a result of the smaller endothelial cells having less of an impact on the MS paths, with the result that these paths tend to form straighter lines. Figure 3(l) illustrates the unique MS signature for the POI located in a stroma region, but bounded on the left and right sides by the tumor. In this case, the MS signature is able to extend unimpeded in both north and south directions, while being constrained in the west and east directions. Employing a texture-based classifier, where the texture feature is computed using a square neighborhood might not be able to capture this local heterogeneity. Since MS is rotationally invariant, having a few training samples of this type allow easy extension to other similar complex regions.
CONCLUDING REMARKS
In this paper, we have presented a novel MS framework, one that provides a highly generalizable feature extraction model that enables quantification of local morphology through characterization of regional heterogeneity. This represents a departure from previous local scale definitions that have attempted to locally model image homogeneity, as with other local scale definitions. MS is particularly relevant in images with high degrees of local complexity, such as in the context of biological images. In histopathology images, the most relevant or interesting parts of the image usually correspond to those that are characterized by significant local heterogeneity (e.g., cancer nuclei and lymphocytes). Larger homogeneous regions (e.g., benign stroma) may be less interesting or uninformative from a diagnostic or prognostic perspective. As demonstrated across 4 datasets using the same model parameters, the MS signature allows good classifier accuracy for the task of distinguishing nuclei as being tumoral or stromal. For this significantly difficult problem, our classifier achieved an AUC of 0.80-0.89. However, MS is not limited to problems in classification alone. The notion of MS could be easily extended to other applications such as image registration, segmentation, and bias-field correction. Future work will involve leveraging the MS concept for these tasks and extending MS to other problem domains besides digital pathology.
Footnotes
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2013/4/2/8/109865
REFERENCES
- 1.Pizer SM, Eberly D, Morse BS, Fritsch DS. Zoom-invariant vision of figural shape: The mathematics of cores. Comput Vis Image Und. 1998;69:55–71. [Google Scholar]
- 2.Saha PK. Tensor scale: A local morphometric parameter with applications to computer vision and image processing. Comput Vis Image Und. 2005;99:384–413. [Google Scholar]
- 3.Madabhushi A, Udupa JK. New methods of MR image intensity standardization via generalized scale. Med Phys. 2006;33:3426–34. doi: 10.1118/1.2335487. [DOI] [PubMed] [Google Scholar]
- 4.Madabhushi A, Udupa J, Souza A. Generalized scale: Theory, algorithms, and application to image in homogeneity correction. Comp Vis Image Und. 2006;101:100–21. [Google Scholar]
- 5.Nyul LG, Udupa JK, Saha PK. Incorporating a measure of local scale in voxel- based 3d image registration. IEEE Trans Med Imaging. 2003;22:228–37. doi: 10.1109/TMI.2002.808358. [DOI] [PubMed] [Google Scholar]
- 6.Hontsch I, Karam LJ. Locally adaptive perceptual image coding. IEEE Trans Image Process. 2000;9:1472–83. doi: 10.1109/83.862622. [DOI] [PubMed] [Google Scholar]
- 7.Saha PK, Udupa JK, Odhner D. Scale-based fuzzy connected image segmentation: Theory, algorithms, and validation. Comp Vis Image Und. 2000;77:145–74. [Google Scholar]
- 8.Sato E, Olson SH, Ahn J, Bundy B, Nishikawa H, Qian F, et al. Intraepithelial cd8+tumor-infiltrating lymphocytes and a high cd8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc Natl Acad Sci USA. 2005;102:18538–43. doi: 10.1073/pnas.0509182102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schipper NW, Smeulders AW, Baak JP. Quantification of epithelial volume by image processing applied to ovarian tumors. Cytometry. 1987;8:345–52. doi: 10.1002/cyto.990080402. [DOI] [PubMed] [Google Scholar]
- 10.Schipper NW, Smeulders A, Baak JP. Automated estimation of epithelial volume in breast cancer sections. A comparison with the image processing steps applied to gynecologic tumors. Pathol Res Pract. 1990;186:737–44. doi: 10.1016/S0344-0338(11)80264-5. [DOI] [PubMed] [Google Scholar]
- 11.Oger M, Belhomme P, Klossa J, Michels JJ, Elmoataz A. Automated region of interest retrieval and classification using spectral analysis. Diagn Pathol. 2008;(Suppl 1):S17. doi: 10.1186/1746-1596-3-S1-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linder N, Konsti J, Turkki R, Rahtu E, Lundin M, Nordling S, et al. Identification of tumor epithelium and stroma in tissue microarrays using texture analysis. Diagn Pathol. 2012;7:22. doi: 10.1186/1746-1596-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lahrmann B, Halama N, Sinn HP, Schirmacher P, Jaeger D, Grabe N. Automatic tumor-stroma separation in fluorescence TMAs enables the quantitative high-throughput analysis of multiple cancer biomarkers. PLoS One. 2011;6:e28048. doi: 10.1371/journal.pone.0028048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mosaliganti K, Janoos F, Irfanoglu O, Ridgway R, Machiraju R, Huang K, et al. Tensor classification of N-point correlation function features for histology tissue segmentation. Med Image Anal. 2009;13:156–66. doi: 10.1016/j.media.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beck AH, Sangoi AR, Leung S, Marinelli RJ, Nielsen TO, van de Vijver MJ, et al. Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Sci Transl Med. 2011;3:108ra113. doi: 10.1126/scitranslmed.3002564. [DOI] [PubMed] [Google Scholar]
- 16.Janowczyk A, Chandran S, Singh R, Sasroli D, Coukos G, Feldman MD, et al. High-throughput biomarker segmentation on ovarian cancer tissue microarrays via hierarchical normalized cuts. IEEE Trans Biomed Eng. 2012;59:1240–52. doi: 10.1109/TBME.2011.2179546. [DOI] [PubMed] [Google Scholar]
- 17.Xu J, Janowczyk A, Chandran S, Madabhushi A. A high-throughput active contour scheme for segmentation of histopathological imagery. Med Image Anal. 2011;15:851–62. doi: 10.1016/j.media.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Granlund GH. Fourier preprocessing for hand print character recognition. IEEE Trans Comput. 1972;21:195–201. [Google Scholar]
- 19.Dijkstra EW. A note on two problems in connexion with graphs. Numer Math. 1959;1:269–71. [Google Scholar]
- 20.Sethian JA. A fast marching level set method for monotonically advancing fronts. Proc Natl Acad Sci USA. 1996;93:1591–5. doi: 10.1073/pnas.93.4.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seber G. Multivariate observations. New York: Wiley; 1984. [Google Scholar]
- 22.Meyer F. Topographic distance and watershed lines. Signal Processing. 1994;38:113–25. [Google Scholar]


