Abstract
Introduction:
The White Blood Cell (WBC) differential count yields clinically relevant information about health and disease. Currently, pathologists manually annotate the WBCs, which is time consuming and susceptible to error, due to the tedious nature of the process. This study aims at automation of the Differential Blood Count (DBC) process, so as to increase productivity and eliminate human errors.
Materials and Methods:
The proposed system takes the peripheral Leishman blood stain images as the input and generates a count for each of the WBC subtypes. The digitized microscopic images are stain normalized for the segmentation, to be consistent over a diverse set of slide images. Active contours are employed for robust segmentation of the WBC nucleus and cytoplasm. The seed points are generated by processing the images in Hue-Saturation-Value (HSV) color space. An efficient method for computing a new feature, ‘number of lobes,’ for discrimination of WBC subtypes, is introduced in this article. This method is based on the concept of minimization of the compactness of each lobe. The Naive Bayes classifier, with Laplacian correction, provides a fast, efficient, and robust solution to multiclass categorization problems. This classifier is characterized by incremental learning and can also be embedded within the database systems.
Results:
An overall accuracy of 92.45% and 92.72% over the training and testing sets has been obtained, respectively.
Conclusion:
Thus, incremental learning is inducted into the Naive Bayes Classifier, to facilitate fast, robust, and efficient classification, which is evident from the high sensitivity achieved for all the subtypes of WBCs.
Keywords: Incremental learning, naive bayes classifier, number of lobes, white blood cells classification
INTRODUCTION
Over the years, information derived from the White Blood Cell (WBC) differential count has become a cornerstone in Laboratory Hematology and is widely used for screening, case finding, diagnosis, and monitoring of hematological and non-hematological disorders. Human blood consists of five types of white blood cells, namely, Neutrophils (40-60%), Lymphocytes (20-40%), Monocytes (2-8%), Eosinophils (1-4%), and Basophils (0.5-1%).
The WBC differential count is considered to yield clinically relevant information in health and disease.[1] For example, excess of lymphocytes may be caused due to Lymphocytic Leukemia.[2] High Eosinophil and Monocyte count is usually an indicator of bacterial infection in the body.[3] Thus, the WBC count is an important and useful measure, which indicates the health status of the body.
In a typical pathology laboratory, two types of blood counts are performed, namely Complete Blood Count (CBC) and Differential Blood Count (DBC). The CBC is performed using an instrument called the cytometer, based on the principle of ‘flow cytometry’.[4] On the other hand, in DBC, an expert would count 100 WBCs (all categories) on the blood stain slides and compute the percentage occurrence of each type of WBC. The DBC is a much more reliable count than the CBC. However, manual annotation of the WBCs is considered to be an imprecise technique, because of the massive data and tedious nature of the task.[5]
There were a few efforts in the past to automate the solution to this problem. Earlier, Ongun et al.,[6] and Ramesh et al.,[7] had also proposed an automatic DBC system. In the scheme suggested by them, the WBCs were segmented by applying active contours or color-based segmentation. Various geometric and texture features were used by them for classification. Apart from this, several attempts were made in the past for the segmentation of WBCs, such as, the Teager Energy-Based Segmentation by Kumar et al.,[8] and the Watershed Algorithm by Jiang et al.,[9] Rezatofighi et al.,[10] introduced a method based on the orthogonality theory and the Gram-Schmidt process for segmenting the WBC nuclei.
In the opinion of the authors of this article, the previous studies on automation of differential WBC count have not laid emphasis on the development of a fast, parallel, and scalable system. A really fast system is needed, as the number of DBCs a pathology laboratory handles is enormous. This study focuses on building an automatic computer system for performing DBC on digitized peripheral blood stain images. We propose a scheme based on the Bayesian classifier that learns incrementally and that can be embedded inside a database system like MySQL, to create a feasible framework for the development of a scalable learning system.
MATERIALS AND METHODS
Dataset Description
The image dataset used in this process was generated by the digitization of peripheral blood stain slides. The slides used here were Leishman stained.[11] This section describes the process of preparation of the slide. The slides were air dried and thereafter flooded with the Leishman's stain. The stain formulation included methanol, which fixed the cell. The slides were held for two to three minutes before diluting the solution with an equal amount of buffered water at pH 6.8. The water was added slowly with a plastic Pasteur pipette. Such slides were then left to hold for about 12 minutes. The appearance of polychromatic ‘scum’ on the surface of the slides was merely a result of oxidation of the dye components and could be ignored. Following this, the excess stain was washed off with slow running water and the slides were flooded with buffered water at pH 6.8 for another minute. The digitized images of the stained blood smear slide were then captured by using a whole slide scanner at 40X magnification. The images of all the five types of WBCs, as obtained from the scanner, are shown in Figure 1. The dataset is available for download at http://autodbc.wordpress.com/.
Figure 1.
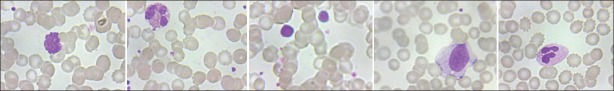
Different types of WBCs in the provided dataset
System Design
The system comprises of the following modules: (1) Stain Normalization, (2) WBC Segmentation, (3) Feature Extraction, and (4) Classification using Naive Bayes Classifier. Each of these has been discussed in the subsequent subsections.
Stain Normalization
For a scalable system, it is necessary for the segmentation to be consistent for a large image dataset. In order to devise a robust segmentation technique, independent of staining in the images, it is essential to stain normalize the images before the segmentation step.
The images were stain normalized with respect to a target image, which was selected on the basis of a larger visible contrast between the WBCs and the background cells as observed in the RGB image. The stain normalization was done as proposed by Reinhard et al.[12]
WBC Segmentation
Image segmentation is pivotal in medical image analysis problems. The segmentation step is decisive, as the subsequent feature extraction and classification steps depend on the accurate segmentation of the white blood cells.
The aim of WBC segmentation was twofold. First was segmentation of the WBCs. Second was extraction of the WBC nucleus and cytoplasm separately. The nucleus and cytoplasm were obtained separately to facilitate computation of the nuclear as well as cytoplasmic features, which were essential for the classification step. Refer to the Feature Extraction section for details of features used for classification. Several attempts for segmentation of WBCs were made in the past, which included scale-space filtering, watershed algorithm as proposed by Jiang, et al.,[9] and Teager energy-based segmentation, as proposed by Kumar et al.,[8] to name a few.
The various steps involved in WBC segmentation are shown in the Figure 2. The nucleus and cytoplasm segmentations have been dealt with individually in the subsequent subsections.
Figure 2.
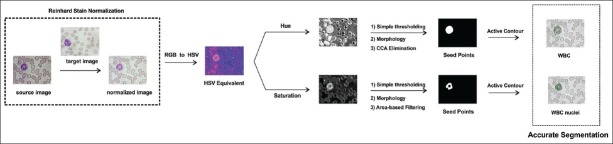
Segmentation scheme
Nuclei Segmentation
Active contours, as proposed by Chan, et al.,[13] were implemented for segmentation of the WBC nuclei. They provided a framework based on the minimization of energy of the contour, which was robust and delineated objects even in the presence of noise. The preliminary task for nuclei segmentation was to obtain seed points for the employment of active contours. In order to acquire seed points, the stain normalized RGB image obtained in the Stain normalization section was converted to its HSV equivalent. The nucleus of the WBCs had higher intensities in the pink hue, therefore, the S channel of the obtained HSV image was used for thresholding.
Morphological opening and area-based filtering, using connected component analysis (CCA), was done, to remove the non-WBCs. The morphological opening was also beneficial in resolving the connected nuclei problem. The structuring element used here was a square matrix of window, size 7 × 7. Thus, the mask containing the seed points was generated.
The active contour model could be implemented because of the difference in the intensity levels of the interior and the exterior of the nucleus. Details of energy function and the minimization of active contour energy can be found in Chan, et al.[13] The active contours enhanced the shape of the nucleus, because some of the features like number of lobes and maximum curvature points depended on the precise shape of the nucleus.
Cytoplasm Segmentation
Cytoplasm segmentation required the extraction of white blood cells. The nucleus was then subtracted from the obtained WBC to get the cytoplasm. The pink hue of the WBCs was separated by simple thresholding of the hue channel of the HSV image. The morphological opening was done to remove protrusions on the obtained cells. Connected component elimination, using the obtained WBC nuclei, was done, to remove the non-WBCs that were obtained in the previous steps. Thus, the seed points were acquired to apply the active contours. The segmentation of the WBC nuclei and the cytoplasm was accurate and no WBC was eliminated at this stage.
Feature Extraction
Choice of features immensely affects the classifier performance. For a robust classification, the features must characterize each WBC subtype and must be independent of each other. A neoteric scheme for determination of the number of lobes in WBCs is presented in this article. Apart from this, other features have also been computed, based on the biological aspects of all kinds of WBCs. The remainder of this section gives information on the features that have been used in the proposed system.
Cellular Features
Size of the White Blood Cells
The size of WBCs is directly proportional to its diameter. The diameter of the lymphocytes lies in the range of 6-10 μm, which is very low, whereas, the monocytes have a very high value of diameter. The basophils, eosinophils, and the neutrophils have intermediate values. The size of each WBC is calculated, to take advantage of the difference in size of all kinds of WBCs.
Compactness of the White Blood Cells
This feature signifies the shape of the WBCs. The monocytes have the highest value of this feature as compared to the other kinds of WBCs.
Nuclear Cytoplasmic Ratio
The Nuclear Cytoplasmic Ratio (NCR) gives the degree of spread of the nucleus with respect to the cytoplasm in a WBC. The NCR is very high for lymphocytes, when compared to the other kinds of WBCs. The NCR for the basophils is close to one.
Nuclear Features
Average Nuclear Roundness
Roundness of any shape refers to how close it is to being a circle. Average Roundness factor for each segment of a nucleus is given by
![]()
where n is the total number of segments of each nucleus. This feature clearly differentiates the WBCs on the basis of the shape of the nuclei. The lymphocytes and basophils have a higher value of this feature, whereas, the eosinophils, monocytes, and neutrophils have a lower value. Among the latter ones, the eosinophils and monocytes (mostly kidney-shaped) have a relatively higher value than the neutrophils. This feature is very vital for the classification of the band neutrophils as they have a very low value of the average roundness factor.
Number of Lobes
The number of lobes in the lymphocytes, basophils, and monocytes has a lower value; the majority of them being single lobed or bi-lobed. On the other hand, eosinophils and neutrophils have a higher number of lobes. Segmented neutrophils have the highest number of lobes. Thus, the number of lobes may be an important distinguishing feature. We have proposed a novel method to estimate the number of lobes in a WBC. The number of lobes have been calculated by splitting the nucleus into Ni regions, where Ni ∈ (2, 3, 4, 5), by using the region splitting algorithm, as proposed by Costas et al.[14] The number of lobes has been computed as follows: If the ratio of the area of the nucleus to that of its bounding box, that is, ‘Extent’ is found to be greater than 0.7, the number of lobes is equal to one, otherwise, the number of lobes is equal to Ni|C(Ni) = Hamonic Mean (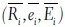 ) has been maximized. Here (
) has been maximized. Here (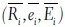 ), are the mean of roundness factors, extents, and eccentricities of the Ni splitted regions, respectively. Figure 3 illustrates the lobe counting method used by us. In this case extent = 0.59, which is less than 0.7.C(2) = 0.72, C(3) = 0.63, C(4) = 0.61, and C(5) = 0.61, therefore, the number of lobes in the given nuclei = 2.
), are the mean of roundness factors, extents, and eccentricities of the Ni splitted regions, respectively. Figure 3 illustrates the lobe counting method used by us. In this case extent = 0.59, which is less than 0.7.C(2) = 0.72, C(3) = 0.63, C(4) = 0.61, and C(5) = 0.61, therefore, the number of lobes in the given nuclei = 2.
Figure 3.

Computation of Lobes
Maximum Curvature Points
This feature gives us a count of the number of sharp bends in the nuclei. The number of maximum curvature points in the lymphocytes and basophils are too low when compared with the eosinophils and monocytes, which have intermediate values of this feature. The segmented neutrophils have the highest value. The curvature is calculated after contour extraction. The points on the boundary of the nuclei, which are above a certain threshold, are counted as the maximum curvature points. The threshold is calculated using the local curvature properties as proposed in.[7] Figure 4 illustrates the maximum curvature points of a nucleus in our dataset.
Figure 4.
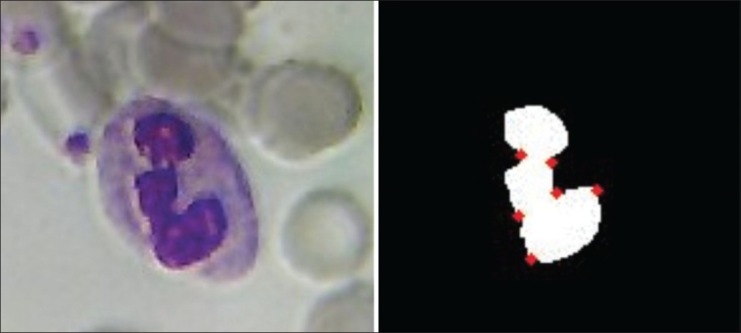
Computation of maximum curvature points
Roughness
Gray-Level Entropy Matrix (GLEM)[15] features were computed from the GLEM matrix. Among the GLEM features, the roughness of the nucleus was calculated. The roughness of the basophil and eosinophil nucleus was higher than the others, because of the nucleus being granular in both the cases.
Cytoplasmic Features
Homogeneity
The degree of homogeneity of the cytoplasm was computed from the Gray-Level Co-occurrence Matrix (GLCM).[16] The basophils and the eosinophils exhibited the lowest values of this feature.
Classification Using Naive Bayes Classifier
The Naive Bayes Classifier is a simple probabilistic induction algorithm that fares well when the classes are easily separable, as in our case. This supervised algorithm comes originally from the study on pattern recognition by Duda and Hart.[17] Fisher's COBWEB algorithm and the AUTOCLASS system outlined by Cheeseman et al.,[18] are also based on the Bayesian ideas.
The categorical data was used for classification. Hence, feature values of each sample were quantized. Quantization was done as low, medium, and high values, based on the characteristics of each of the WBC subtypes, as defined by the medical experts.
The problem of multicategory classification, where sometimes the time or sample size available for training is limited, and where the class apriori probabilities are known or easily estimated, can be typically solved by using the Naive Bayesian classifier with incremental learning.[19]
Incremental learning facilitates the classifier to learn from new training sets, apart from preserving the information acquired from different datasets, thus improving its generalization capabilities on unknown images. Such a learning strategy is beneficial, as it need not store or re-process old instances. It can be applied to situations where the input data comes only in a sequence, and a continuously updating model is crucial for classification in real time.[20,21] In our case (refer to Figure 5), after classification of each WBC in the testing set, it was automatically added to the training set. Thus, incremental learning was accomplished.
Figure 5.
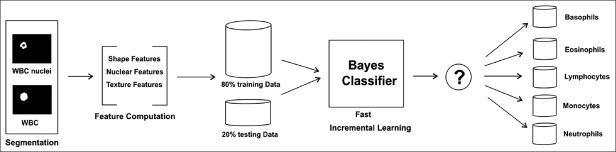
WBC Classification System
The aim of this article was to implement a scalable system to be used in real life situations. Such a system would be implemented on the web and would require the embedding of a classifier within the database system. The Naive Bayes Classifier with Laplacian correction was suitably used.
EVALUATION RESULTS
The total dataset contained 267 WBCs in 237 images. The training was done on 80% of the dataset and 20% was kept for testing purposes. The confusion matrix for training and testing data is shown in Tables 1 and 2, respectively.
Table 1.
Confusion matrix: Training
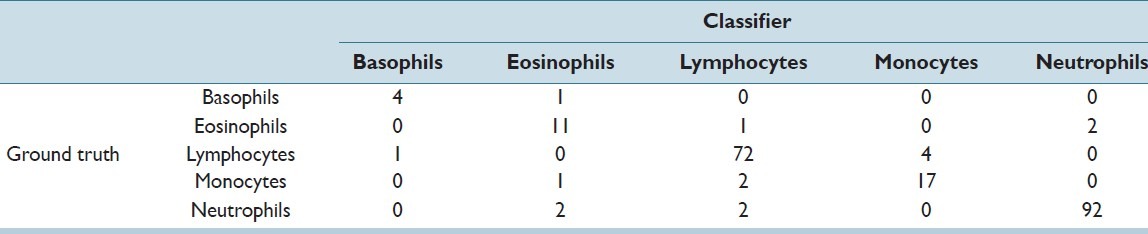
Table 2.
Confusion matrix: Testing
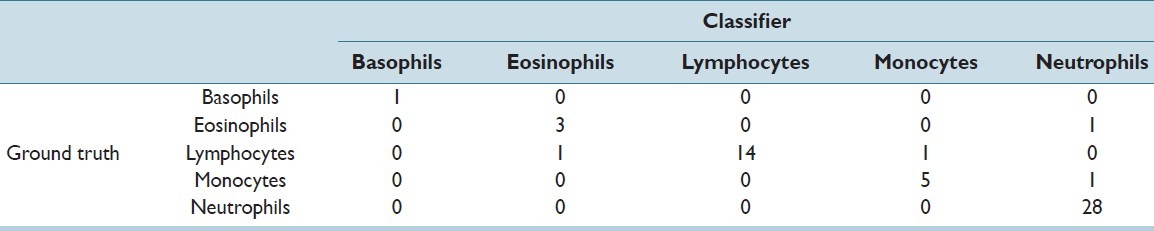
The recall and precision of the classifier for all five types of WBCs in the testing set has been stated in Table 3. An overall accuracy of 92.45% and 92.72% was obtained in the training and testing sets, respectively. The obtained accuracy was found to be far better than the accuracy of 77%, which was achieved by Umpon et al.[22] In multiclass problems such as these, a correct classification of only the majority classes such as neutrophils and lymphocytes would have given a high accuracy, which was not the aim of this study. Hence, the sensitivity (recall) of each class was a more apt measure for classification of WBC subtypes. The individual sensitivities of each WBC subtype, especially minorities like basophils, eosinophils, and monocytes, were found to be better than in many previous studies like that of Ramesh, et al.[7]
Table 3.
Recall and precision of bayes classifier on the testing set
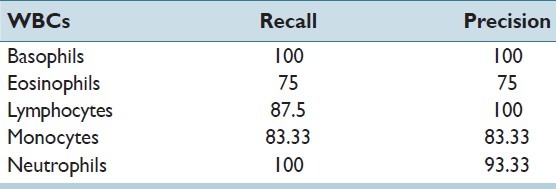
The reasons for the misclassification of the WBCs were studied and it was observed that eosinophils and monocytes were the most misclassified WBC types. The misclassification of eosinophils into neutrophils was due to the similarity in the feature values of eosinophils and segmented neutrophils, except for the homogeneity of the cytoplasm and the roughness of the nucleus. On the other hand, the misclassification of monocytes was basically due to their similarity to band neutrophils. In the opinion of the authors, the misclassification rate could be further reduced by introducing some more texture-based features in the feature set.
Another experiment was performed to test the effectiveness of the novel feature, that is, ‘Number of Lobes’. The Bayesian classifier, trained without this feature, achieved an accuracy of on the testing set, thus, showing the effectiveness and relevance of this novel feature.
CONCLUSION AND FUTURE STUDY
In this study, a scalable, automatic, WBC differential count system is proposed. Segmentation is robust and consistent over a large image dataset. A careful selection of features, based on the domain knowledge of medical experts, has been done. A novel way to estimate the feature, ‘Number of Lobes,’ is proposed. Inclusion of this new feature has shown a drastic improvement in the classification accuracy of the system. Incremental learning is inducted into the Naive Bayes Classifier, to facilitate fast, robust, and efficient classification, which is evident from the high sensitivity achieved for all the subtypes of WBCs.
Currently, studies are being conducted to implement this system as a full-fledged, web-based tool, which can be used in real life clinical practice. This study can also be extended to identify objects in the smear such as ‘microorganisms’. Efforts are also being made for development of implementation of the Bayesian Classifier as a relational database module (MySQL module). A large-scale multisite collaborative study is required to employ such a system, by collaborative filtering, to aid in the development of reliable computerized tools.
Footnotes
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2013/4/2/15/109883
REFERENCES
- 1.Houwen B. The differential cell count. Lab Hematol. 2001;7:89–100. [Google Scholar]
- 2.Hallek M, Cheson BD, Catovsky D, Caligaris-Cappio F, Dighiero G, Döhner H, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: A report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute–Working Group 1996 guidelines. Blood. 2008;111:5446–56. doi: 10.1182/blood-2007-06-093906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oppenheim JJ, Yang D. Alarmins: Chemotactic activators of immune responses. Curr Opin Immunol. 2005;17:359–65. doi: 10.1016/j.coi.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 4.Fulwyler MJ. Electronic separation of biological cells by volume. Science (New York, NY) 1965;150:910. doi: 10.1126/science.150.3698.910. [DOI] [PubMed] [Google Scholar]
- 5.Schaefer M, Rowan RM. The clinical relevance of nucleated red blood cell counts. Sysmex Journal International. 2000 10.2. [Google Scholar]
- 6.Ongun G, Halici U, Leblebicioglu K, Atalay V, Beksac M, Beksac S. An automated differential blood count system. Engineering in Medicine and Biology Society, 2001. Proceedings of the 23rd Annual International Conference of the IEEE, IEEE. 2001 [Google Scholar]
- 7.Ramesh N, Dangott B, Salama ME, Tasdizen T. Isolation and two-step classification of normal white blood cells in peripheral blood smears. J Pathol Inform. 2012;3:13. doi: 10.4103/2153-3539.93895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar BR, Joseph DK, Sreenivas TV. Teager energy based blood cell segmentation. Digital Signal Processing, 2002. DSP 2002. 2002 14th International Conference on, IEEE. [Google Scholar]
- 9.Jiang K, Liao QM, Dai SY. A novel white blood cell segmentation scheme using scale-space filtering and watershed clustering. Machine Learning and Cybernetics, 2003 International Conference on, IEEE [Google Scholar]
- 10.Rezatofighi SH, Soltanian-Zadeh H, Sharifian R, Zoroofi RA. A new approach to white blood cell nucleus segmentation based on gram-schmidt orthogonalization. Digital Image Processing, 2009 International Conference on, IEEE [Google Scholar]
- 11.Leishman W. Note on a simple and rapid method of producing Romanowsky staining in malarial and other blood films. Br Med J. 1901;2:757–8. doi: 10.1136/bmj.2.2125.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reinhard E, Adhikhmin M, Gooch B, Shirley P. Color transfer between images. IEEE Comput Graph Appl. 2001;21:34–41. [Google Scholar]
- 13.Chan TF, Vese LA. Active contours without edges. IEEE Trans Image Process. 2001;10:266–77. doi: 10.1109/83.902291. [DOI] [PubMed] [Google Scholar]
- 14.Panagiotakis C, Ramasso E, Tziritas G. Lymphocyte segmentation using the transferable belief model. Lecture notes in Computer Science (LNCS). Recognizing Patterns in Signals, Speech, Images and Videos. Proceedings of the ICPR 2010 Contests, Springer. 2010:253–62. [Google Scholar]
- 15.Yogesan K, Jørgensen T, Albregtsen F, Tveter KJ, Danielsen HE. Entropy-based texture analysis of chromatin structure in advanced prostate cancer. Cytometry. 1996;24:268–76. doi: 10.1002/(SICI)1097-0320(19960701)24:3<268::AID-CYTO10>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 16.Haralick RM, Shanmugam K, Dinstein H. Textural features for image classification. IEEE Trans Syst Man Cybern B Cybern. 1973;6:610–21. [Google Scholar]
- 17.Duda RO, Hart PE. Pattern recognition and scene analysis. New York: Wiley; 1973. [Google Scholar]
- 18.Stutz J, Cheeseman P, Hanson R, Taylor W. AutoClass: A Bayesian approach to classification. RECON. 1994 (20010122938) [Google Scholar]
- 19.Ratsaby J. Incremental learning with sample queries. IEEE Trans Pattern Anal Mach Intell. 1998;20:883–8. [Google Scholar]
- 20.Bruzzone L, Fernàndez Prieto D. An incremental-learning neural network for the classification of remote-sensing images. Pattern Recognit Lett. 1999;20:1241–8. [Google Scholar]
- 21.Fu L, Hsu HH, Principe JC. Incremental backpropagation learning networks. IEEE Trans Neural Netw. 1996;7:757–61. doi: 10.1109/72.501732. [DOI] [PubMed] [Google Scholar]
- 22.Theera-Umpon N, Dhompongsa S. Morphological granulometric features of nucleus in automatic bone marrow white blood cell classification. IEEE Trans Inf Technol Biomed. 2007;11:353–9. doi: 10.1109/titb.2007.892694. [DOI] [PubMed] [Google Scholar]


