Abstract
The RNA polymerase II (RNAP II) largest subunit contains a C-terminal domain (CTD) with up to 52 Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7 consensus repeats. Serines 2, 5 and 7 are known to be phosphorylated, and these modifications help to orchestrate the interplay between transcription and processing of messenger RNA (mRNA) precursors. Here, we provide evidence that phosphorylation of CTD Thr4 residues is required specifically for histone mRNA 3′ end processing, functioning to facilitate recruitment of 3′ processing factors to histone genes. Like Ser2, Thr4 phosphorylation requires the CTD kinase CDK9 and is evolutionarily conserved from yeast to human. Our data thus illustrate how a CTD modification can play a highly specific role in facilitating efficient gene expression.
The carboxyl-terminal domain (CTD) of the RNA polymerase II (RNAP II) largest subunit (Rpb1) consists of Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7 (YSPTSPS) consensus tandem repeats, conserved from yeast to human. The CTD, through phosphorylation on serine (ser) residues, links transcription to mRNA processing events (1-3). Ser5 is phosphorylated by cyclin-dependent kinase 7, CDK7, a subunit of the general transcription factor TFIIH, and this modification functions to facilitate capping (4-5). During transcriptional elongation, Ser2 is phosphorylated by CDK9/P-TEFb, which helps to coordinate RNA 3′ end processing and transcription termination (6-7). Ser7 phosphorylation has been implicated in transcription and 3′ end processing of genes encoding certain small non-coding RNAs (8, 9). Tyr1 can also be phosphorylated, by the c-Abl kinase (10). While Thr4 has been reported to be phosphorylated in fission yeast (11), there is no evidence that this residue is modified in other species, or what might the function of Thr4 be.
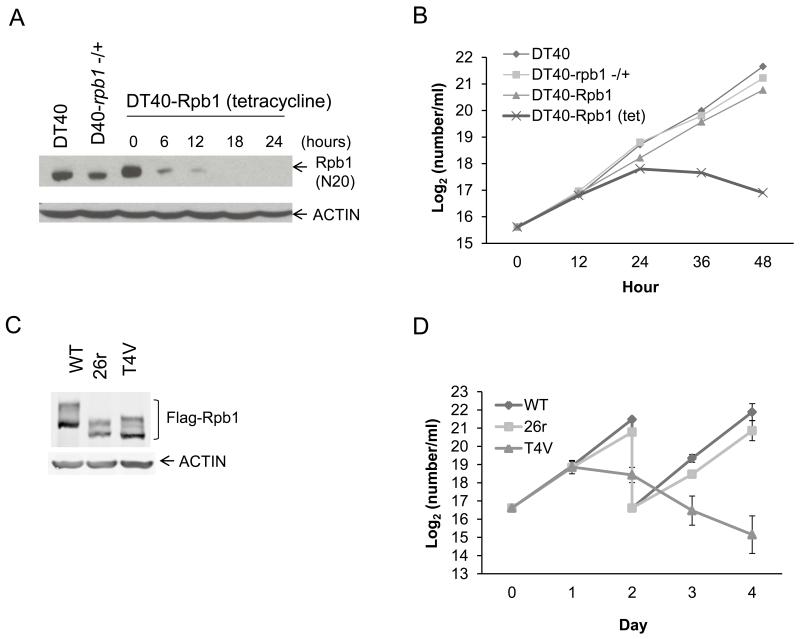
To investigate CTD function in a genetically tractable vertebrate cell system, we created an Rpb1 conditional knock-out chicken cell line, DT40-Rpb1, using methods developed previously to study other conserved proteins (12, 13; Fig. S1). These cells express, as the only source of Rpb1, a tetracycline (tet)-repressible cDNA encoding HA-tagged human Rpb1 (human and chicken Rpb1 are 97% identical, and the CTD is very highly conserved among vertebrates; Fig. S2). After addition of tet, Rpb1 became undetectable between 12 and 18 hours (Figs. 1A and S3), and DT40-Rpb1 cells stopped growing and began to die by 24 hours (Fig. 1B). An important use of DT40 derivatives such as DT40-Rpb1 is for genetic complementation (e.g., 14). Here, a plasmid expressing FLAG-tagged wild-type (wt) Rpb1 was found to restore viability to DT40-Rpb1 cells in the presence of tet. Expression of an Rpb1 derivative (26r) containing 26 all-consensus repeats, plus the ten C-terminal most residues that confer stability (15), also restored viability. However, a comparable derivative containing 30 repeats with all Thr4 residues mutated to Val (T4V) was inviable. All three Rpb1 derivatives were expressed equivalently (Fig. 1C), and wt and 26r-expressing cells grew similarly following addition of tet, while T4V cells stopped growing and began dying by 24 hours (Fig. 1D).
Figure 1. Growth properties of DT40-Rpb1 cells and derivatives.
(A) Western blot of lysates from DT40, DT40 rpb1(+/−) and DT40-Rpb1 cells treated with tet as indicated. Blots were probed with anti-Rpb1 (N20) and anti-actin antibodies. (B) Growth curves of DT40, DT40-rpb1(+/−) and DT40-Rpb1 cells, in the presence or absence of tet. N=2. (C) Western blot of lysates of DT40-Rpb1 cells expressing the indicated Flag-tagged Rpb1 derivative and treated with tet for 24 hours. (D) Growth curves of wt, 26r and T4V cells grown in the presence of tet for 4 days. Cells were split after two days. N=3. Error bars indicate standard deviation.
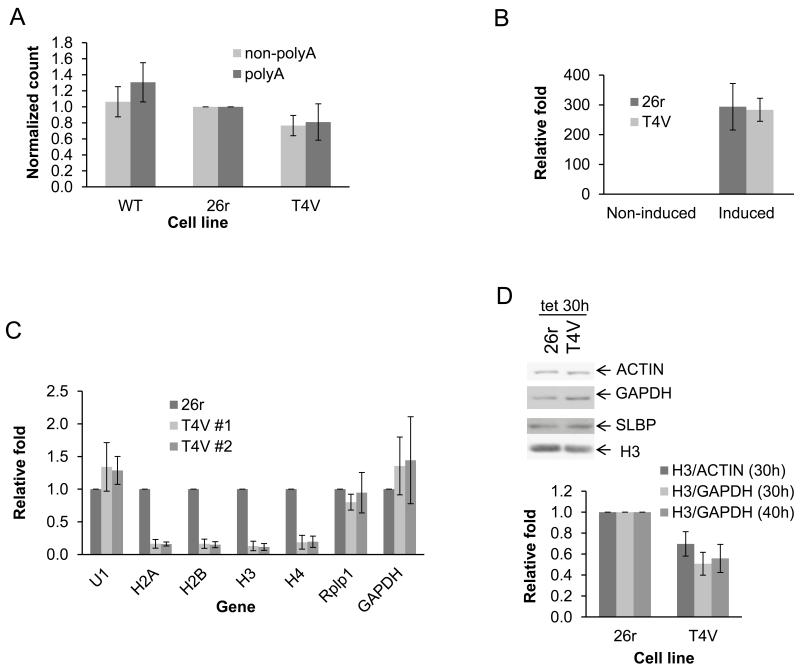
We next investigated the possible basis for T4V cell inviability. Comparison of overall transcription (by 3H pulse labeling) in wt, 26r and TV4 cells treated with tet for 40 hours (Fig. 2A) revealed only a modest decrease (20%) in polyadenylated [poly (A)+] and poly (A)− RNA fractions in T4V compared to 26r cells, indicating that total transcription and synthesis of poly (A)+ mRNA was not significantly affected by the T4V substitutions. A 20-min induction of the inducible Egr1 gene (16) in T4V and 26r cells, grown in tet for 24 hours, resulted in an equivalent ~300-fold mRNA increase in both (Fig. 2B). Additionally, no differences were observed in unspliced/spliced and uncleaved/total (a measure of 3′ processing) Egr1 mRNA (Fig. S4) and 3′ processing of several additional poly (A)+ mRNAs was similar in T4V and 26r cells (Fig. S5). Thus, transcription and processing of poly (A)+ mRNAs was not significantly affected in T4V cells.
Figure 2. T4V cells are defective in histone expression.
(A) Wt, 26r and T4V cells were treated with tet for 40 hours, RNA was labeled for 30 min, purified and poly (A)+ and poly (A)− fractions quantitated by scintillation counting. Counts per minute (CPM) relative to 26r are shown. N=3. (B) T4V and 26r cells were treated with tet for 24 hours before induction with ionomycin and PMA for 20 min. EGR1 mRNA levels were determined by RT-qPCR following normalization to 18S RNA levels. N=3. (C) T4V (two independent lines) and 26r cells were treated with tet for 40 hours. RNA was isolated and indicated RNAs quantitated by RT-qPCR, normalized to 5S RNA levels and displayed relative to 26r. N=3. (D) T4V and 26r cells were treated with tet for 30 or 40 hours. Cell lysates were analyzed by Western blotting (top). Ratios of H3/actin and H3/GAPDH were determined and are displayed relative to values obtained with 26r. N=3. Error bars display standard deviation.
We next measured accumulation of several types of RNAP II products in T4V and 26r cells treated with tet for 40 hours (Fig. 2C). Levels of U1 snRNA were unaffected, as were levels of two poly (A)+ mRNAs, RplPl and GAPDH. However, levels of all four non-poly (A)+ replication-dependent histone mRNAs were significantly reduced in two independent T4V cell lines, to ~15% the levels observed in 26r cells. Consistent with this, histone H3 protein levels were reduced in T4V cells ~50% after 30 or 40 hours tet treatment (Fig. 2D). Levels of other proteins tested were not significantly affected (Figs. 2D and S6).
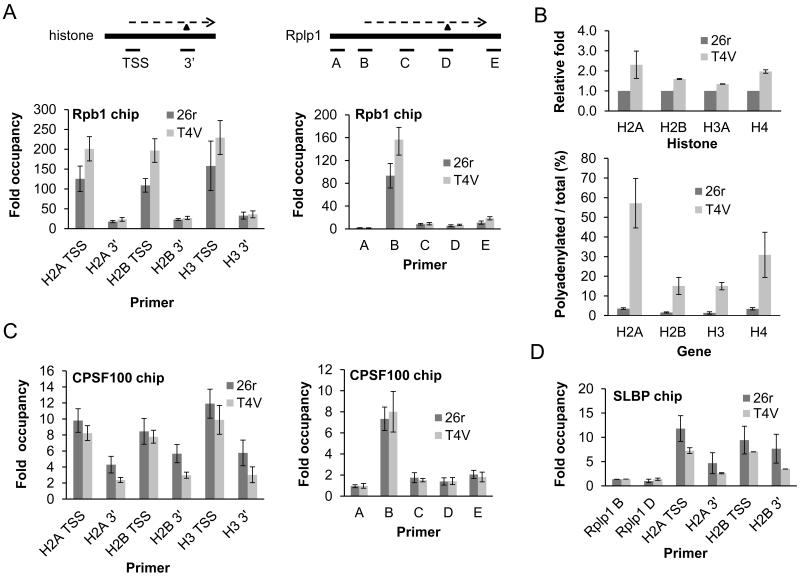
Decreased histone gene transcription could explain the defect in histone mRNA accumulation in T4V cells. However, nuclear run-on analysis showed no differences, or even a slight increase, with three histone genes in 26r and T4V cells after 24 or 40 hours tet treatment (Fig. S7A and B). We also performed chromatin immunoprecipitation (ChIP) with anti-Rpb1 antibodies (N20) in T4V and 26r cells treated with tet for 30 hours. RNAP II levels at the transcription start site (TSS) and 3′ end of H2A, H2B and H3 genes were not reduced, and indeed slightly elevated at the TSS, in T4V cells (Fig. 3A, left). Similar results were observed along the length of the RPLP1 gene (Fig. 3A, right). We conclude that histone gene transcription was unaffected in T4V mutant cells.
Figure 3. T4V cells are defective in histone mRNA 3′ end processing.
(A) Rpb1 CHIP. Diagrams depict genes analyzed. Thick lines represent genes and dashed lines, transcripts. Triangles denote 3′ cleavage sites. Positions of amplicons are indicated. T4V and 26r cells were grown in the presence of tet for 30 hours, then used for ChIP. DNA samples were analyzed by qPCR. Signals were normalized as described in Methods. N=3. (B) T4V and 26r cells were treated with tet for 40 hours, RNA isolated and RT performed using oligo-dT or random hexamer oligos. qPCR was performed, values were normalized to Rplp1 mRNA levels and are shown relative to 26r (top). The ratio of poly (A)+ /total histone transcripts was calculated and plotted (bottom). N=3. (C) CPSF100 CHIP. N=5. (D) SLBP CHIP. N=3. Error bars indicate standard deviation.
The role of the CTD in 3′ end formation suggested that Thr4 might be required specifically in histone mRNA 3′ formation, which uses a distinct mechanism from poly (A)+ mRNAs (17). When histone 3′ processing is defective, cryptic downstream polyadenylation sites can be used, resulting in formation of poly (A)+ histone mRNAs (18). 26r and T4V cells were grown in the presence of tet for 40 hours, and for all four histone genes, ~two fold more poly (A)+ mRNA was recovered from T4V cells than from 26r cells (Fig. 3B, top), and the fraction of histone mRNA in these cells that was poly (A)+ increased 10-50 fold (Fig. 3B, bottom).
Histone pre-mRNA 3′ processing requires a complex set of evolutionarily conserved factors, only some of which are shared with the poly (A)+ mRNA 3′ processing machinery (17). Using ChIP, we examined recruitment of one common (CPSF-100) and one histone-specific (SLBP) factor to several histone genes and to the RPLP1 gene. CPSF-100 levels on the RPLP1 gene peaked at the promoter, consistent with previous observations (e.g., 19), and were equivalent in 26r and T4V cells treated with tet for 30 hours (Fig. 3C, right). On the three histone genes tested, CPSF-100 levels were slightly reduced at the TSS in T4V cells but significantly lower at the 3′ end (Fig. 3C, left), consistent with the defect in 3′ processing. As expected, in 26r cells SLBP was detected at background levels on the RPLP1 gene (Fig. 3D), while significant levels were found associated with the two histone genes tested. However, SLBP levels were significantly reduced both at the TSS and at the 3′ processing site in T4V cells (Fig. 3D). Total levels of SLBP, CPSF-100 and other 3′ processing factors tested were equivalent in the two cell lines (Figs. 2D and S6).
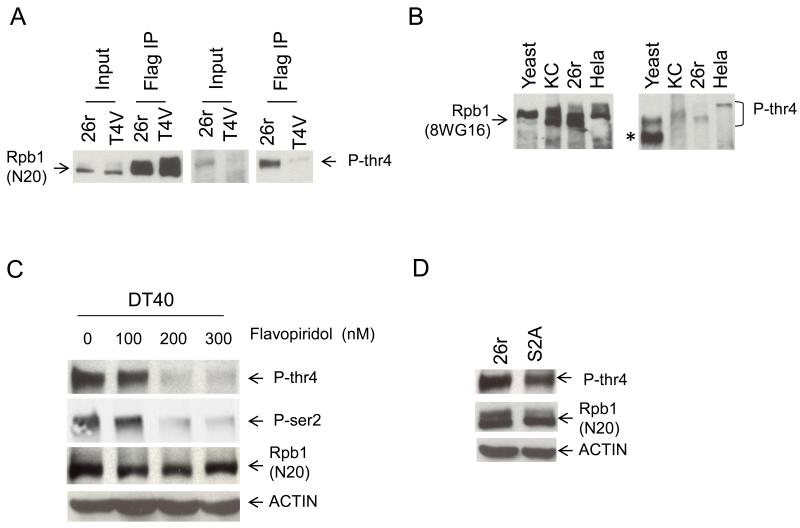
The Thr to Val mutation is a conservative change, and our results thus suggest that modification (e.g., phosphorylation) of Thr4 is important for its function. To investigate whether Thr4 is phosphorylated, we used an antibody raised against and specific for a Thr4-phosphorylated CTD heptad repeat (P-Thr4; fig. S8). IP-Westerns with extracts from 26r and T4V cells, using anti-FLAG for IP and P-Thr4 for Western (Fig. 4A) detected a band corresponding to a protein of the expected size in both input and IP from the 26r cells, but this species was detected only at most at very low levels in the T4V samples, indicating that Thr4 is phosphorylated in the 26r CTD. Thr4 is highly conserved throughout eukaryotes, and Western analysis with P-Thr4 of lysates from yeast, fly (KC), 26r and human (HeLa) cells revealed a band the size expected for Thr4 phosphorylated Rpb1 in all samples (Fig. 4B).
Figure 4. Thr4 is phosphorylated throughout eukaryotes and requires CDK 9.
(A) T4V and 26r cells were treated with tet for 30 hours. Western blotting of input and Flag IP samples was done with P-Thr4 (right) and N20 (left) antibodies. (B) Total cell lysates from yeast, KC, 26r and Hela cells were prepared and used for Western blotting. Total Rpb1 was detected by 8WG16 (left) and Thr4-phosphorylated Rpb1 with P-Thr4 (right). Band indicated with asterisk is a cross-reacting protein. (C) DT40 cells were treated with tet for 24 hours, the indicated concentrations of flavopiridol were added and cells grown for an additional 6 hours. Cell lysates were analyzed by Western blotting. (D) Lysates from 26r and S2A cells treated with tet for 30 hours were analyzed by Western blotting.
To identity the possible kinase responsible for Thr4 phosphorylation, we examined whether Thr4 phosphorylation in DT40 cells was sensitive to the CDK9/P-TEFb inhibitors DRB and flavopiridol (20), and found that it was strongly inhibited by both (Figs. 4C and S9). This did not reflect a requirement for prior Ser2 phosphorylation, as Thr4 phosphorylation was detected in DT40-Rpb1 cells expressing an Rpb1 derivative with all Ser2 residues mutated to Ala (S2A) (Fig. 4D). Linking this to the defect in histone mRNA processing in T4V cells, histone mRNA levels in flavopiridol-treated 26r cells were reduced to levels found in T4V cells (Fig. S10). Consistent with this, knockdown of CDK9 in HEK293 cells was previously shown to impair SLBP recruitment to histone genes and lead to accumulation of poly (A)+ histone mRNA (21).
Our experiments have provided new insight into the intricate mechanisms used by cells to couple transcription by RNAP II to subsequent RNA processing. While our data provide evidence that histone mRNA 3′ end formation specifically requires Thr4 phosphorylation, it is likely that this modification is important for other CTD functions. Although our experiments have not uncovered evidence for this, the fact that Thr4 is phosphorylated in yeast, which produces histone mRNA 3′ ends by the same mechanism as other mRNAs, suggests the existence of additional functions in mRNA synthesis and/or processing.
Supplementary Material
Acknowledgement
We thank T. Kashima for plasmids and help with construction of the DT40-Rpb1 cell line, W. Marzluff for the SLBP antibody, Z. Dominski for the Lsm11 antibody, and Novus Biologicals for the P-Thr4 antibody. This work was supported by NIH grant R01 GM28983.
References and Notes
- 1.Hirose Y, Manley JL. RNA polymerase II and the integration of nuclear events. Genes & Development. 2000;14:1415. [PubMed] [Google Scholar]
- 2.Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 3.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes & Development. 2000;14:2452. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schroeder SC, Schwer B, Shuman S, Bentley DL. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes & Development. 2000;14:2435. doi: 10.1101/gad.836300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahn SH, Kim M, Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Molecular Cell. 2004;13:67. doi: 10.1016/s1097-2765(03)00492-1. [DOI] [PubMed] [Google Scholar]
- 7.Ni Z, Schwartz BE, Werner J, Suarez J-R, Lis JT. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Molecular Cell. 2004;13:55. doi: 10.1016/s1097-2765(03)00526-4. [DOI] [PubMed] [Google Scholar]
- 8.Chapman RD, et al. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science. 2007;318:1780. doi: 10.1126/science.1145977. [DOI] [PubMed] [Google Scholar]
- 9.Egloff S, et al. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science. 2007;318:1777. doi: 10.1126/science.1145989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baskaran R, Dahmus ME, Wang JYJ. Tyrosine phosphorylation of mammalian RNA polymerase II carboxyl-terminal domain. Proc. Natl. Acad. Sci. U.S.A. 1993;90:11167. doi: 10.1073/pnas.90.23.11167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakurai H, Ishihama A. Level of the RNA polymerase II in the fission yeast stays constant but phosphorylation of its carboxyl terminal domain varies depending on the phase and rate of cell growth. Genes to Cells. 2002;7:273. doi: 10.1046/j.1365-2443.2002.00522.x. [DOI] [PubMed] [Google Scholar]
- 12.Wang J, Takagaki Y, Manley JL. Targeted disruption of an essential vertebrate gene: ASF/SF2 is required for cell viability. Genes & Development. 1996;10:2588. doi: 10.1101/gad.10.20.2588. [DOI] [PubMed] [Google Scholar]
- 13.Takagaki Y, Manley JL. Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Molecular Cell. 1998;2:761. doi: 10.1016/s1097-2765(00)80291-9. [DOI] [PubMed] [Google Scholar]
- 14.Wang J, Xiao SH, Manley JL. Genetic analysis of the SR protein ASF/SF2: interchangeability of RS domains and negative control of splicing. Genes & Development. 1998;12:2222. doi: 10.1101/gad.12.14.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chapman RD, Conrad M, Eick D. Role of the mammalian RNA polymerase II C-terminal domain (CTD) nonconsensus repeats in CTD stability and cell proliferation. Molecular and Cellular Biology. 2005;25:7665. doi: 10.1128/MCB.25.17.7665-7674.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen CY, Forman LW, Faller DV. Calcium-dependent immediate-early gene induction in lymphocytes is negatively regulated by p21Ha-ras. Molecular and Cellular Biology. 1996;16:6582. doi: 10.1128/mcb.16.11.6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat. Rev. Genet. 2008;9:843. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sullivan KD, Mullen TE, Marzluff WF, Wagner EJ. Knockdown of SLBP results in nuclear retention of histone mRNA. Rna. 2009;15:459. doi: 10.1261/rna.1205409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rozenblatt-Rosen O, et al. The tumor suppressor Cdc73 functionally associates with CPSF and CstF 3′ mRNA processing factors. Proc. Natl. Acad. Sci. U.S.A. 2009;106:755. doi: 10.1073/pnas.0812023106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chao SH, Price DH. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. The Journal of Biological Chemistry. 2001;276:31793. doi: 10.1074/jbc.M102306200. [DOI] [PubMed] [Google Scholar]
- 21.Pirngruber J, et al. CDK9 directs H2B monoubiquitination and controls replication-dependent histone mRNA 3′-end processing. EMBO reports. 2009;10:894. doi: 10.1038/embor.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.