Supplemental digital content is available in the text.
Key Words: 1p/19q status, Fluorescence in situ hybridization analysis, Glioma, Oligodendroglial tumors, Prognostic factor
Abstract
Evaluation of the molecular status of 1p and 19q is a major relevant diagnostic, prognostic, and predictive tool for oligodendroglial brain tumors. Fluorescence in situ hybridization (FISH) is the most commonly used technique for determining 1p and 19q allelic losses, but it lacks fully standardized criteria for analysis. This lack of standardization has led to interinstitutional disagreement in the interpretation of results, thereby contributing to a “gray prognostic zone” that includes codeleted patients with an unexpectedly unfavorable outcome. To optimize the prognostic potential of 1p/19q status determination, we first compared the actual criteria used for FISH reading (i.e. different ratio cutoff values and the percentage of neoplastic nuclei carrying this chromosomal deletion) in a retrospective series of 143 pure and mixed oligodendroglial tumors. We then created a “weighted” FISH reading based on the merged ratio and percentage of neoplastic cells carrying the deletion that was further differentially modulated for 1p and 19q, respectively. This weighted codeletion setting significantly strengthened the favorable prognostic power of 1p/19q losses by reducing the number of poor outcomes from 42% to 12.5% for patients with codeleted tumors. Thus, by identifying as codeleted only those cases with more than 50% of cells having a combined loss of 1p (using 0.7 ratio cutoff) and 19q (using 0.8 ratio cutoff) arms, we created a molecular report that bears higher clinical impact and strengthens the prognostic potential of 1p/19q allelic loss.
INTRODUCTION
In recent years, progress in molecular analysis has contributed to recognizing important genetic alterations in brain tumorigenesis, some of which are related to patient prognosis (1, 2). The observation of different clinical behavior in gliomas presenting with the same histology and tumor grade has led to the need for a better molecular characterization of these lesions that could identify new useful biomarkers for both tumor classification and patient management (1, 2). Among the molecular markers identified as helpful for neurologic and neuropathologic assessments, 1p/19q status analysis is one of the most important tools with diagnostic, prognostic, and predictive value in oligodendroglial-derived brain tumors (3–21). It is well known that codeletion of chromosome arms 1p and 19q defines a subset of patients with better prognosis most likely because of a higher sensitivity to genotoxic stress (21). Because of the strong association between 1p/19q allelic loss and favorable patient outcome, 1p/19q status is routinely investigated in pure and mixed oligodendroglial tumors.
Fluorescence in situ hybridization (FISH) is the most widely used technique for investigating 1p/19q status because it allows the assessment of 1p/19q allelic loss on paraffin-embedded tissue samples, thereby permitting matching of cell morphology and genetic alterations (15, 22). To date, the interpretation of FISH results for defining 1p/19q status has not been standardized, and laboratories arbitrarily choose their criteria to interpret FISH data based on ratio evaluation (between 1p/1q and 19q/19p), calculation of the percentage of neoplastic nuclei carrying deletion, or according to guidelines defined by the International Society of Pediatric Oncology (ESIOP Neuroblastoma Study Group) (8, 10–12, 15, 19, 22–29). Furthermore, few authors have attempted to integrate the ratio and percentage of deleted nuclei to determine 1p/19q status (25, 30). The lack of a standard procedure for the interpretation of FISH data has led to interinstitutional disagreement on how to characterize 1p/19q molecular status, thereby creating some confusion among clinicians regarding the prognostic and predictive value of codeletion. This confusion most likely accounts for a “gray prognostic zone,” which includes codeleted patients with an unexpected unfavorable outcome that is probably based on false-positive FISH results.
To optimize the prognostic role of 1p/19p status in pure and mixed oligodendroglial tumors, we have tried to establish and compare different criteria for interpreting FISH results to reduce the impact of the gray zone. Specifically, in this retrospective series of 161 oligodendroglial tumors, we assessed 1p/19q status using FISH according to the following parameters: 1) 2 different ratio values; 2) the percentage of neoplastic nuclei carrying chromosomal deletion; and 3) a novel integrated evaluation based on the ratio corrected by the percentage of neoplastic cells carrying codeletion, weighted differently for 1p or 19q. In addition, we evaluated the prognostic impact of an imbalanced 1p/19q deletion, as is observed in the polysomy condition.
MATERIALS AND METHODS
Patients and Follow-up Analysis
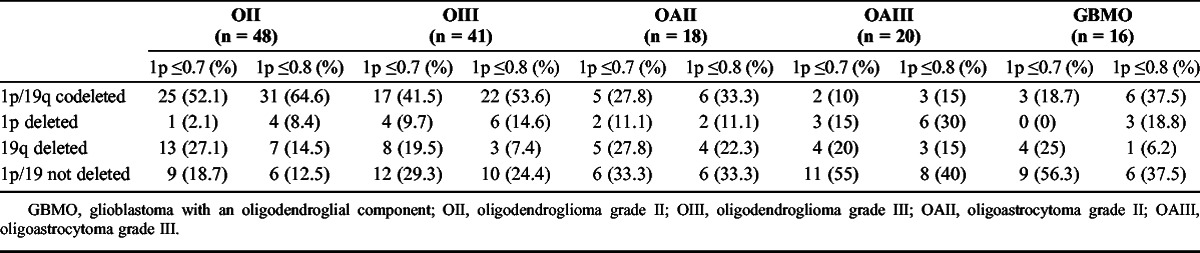
A series of 161 cases of brain tumors with an oligodendroglial component were tested for 1p/19q status by FISH between January 2004 and March 2012 and were retrospectively retrieved from the pathology files of our department. Hematoxylin and eosin–stained slides were independently reviewed by 2 pathologists (Paola Cassoni and Rebecca Senetta) who were blinded to the patient outcome. The histologic diagnosis was made solely on hematoxylin and eosin–stained slides before determining the FISH status. The tumors were classified according to the World Health Organization Classification of Brain Tumors (31). In biopsy specimens, the diagnosis of high-grade gliomas was made when necrosis, microvascular proliferations, and/or mitotic activity (more than 2 mitoses/biopsy) were present. After revision, 18 (11.2%) of 161 cases were excluded from the study because they were recurrences, brain tumors without an undoubtedly oligodendroglial component, or cases with diagnostic disagreement between the 2 observers. The histologic diagnoses of the 143 cases included in the study were as follows: 89 (62.2%) of 143 pure oligodendrogliomas (O), 48 of 89 grade II (OII), and 41 of 89 grade III (OIII); 38 (26.6%) of 143 mixed oligoastrocytomas (OA), 18 of 38 OA grade II (OAII), and 20 of 38 grade III (OAIII); and 16 (11.2%) of 143 glioblastomas with an oligodendroglial component (Table 1).
TABLE 1.
Correlation Between Histologic Diagnosis and 1p/19q Status (Ratio Cutoff for 1p ≤ 0.7 or 0.8 and Ratio Cutoff for 19q ≤ 0.8) in 143 Brain Tumors With an Oligodendroglial Component
None of the patients had been previously treated with chemotherapy or radiotherapy. Follow-up data concerning the overall survival (OS) were available for all 143 cases. Overall survival was defined as months from the date of initial surgery to the date of patient death or last follow-up. Data on disease-free survival (DFS) were obtained in 87 (60.8%) of 143 patients and was calculated from the date of initial surgery to the date of tumor progression/recurrence (defined radiologically or clinically) or last follow-up.
The study was performed according to the standards of the Institutional Ethical Committee and the Helsinki Declaration of 1975, as revised in 1983, and approved by the institutional review board of our institution. All tissue samples were anonymized by staff members who were not involved in the study according to published procedures (32).
Fluorescence In Situ Hybridization
The FISH test was performed using the LSI 1p36/19q13 Dual-Color Probe Sets (Vysis/Abbott, Molecular Europe, Wiesbaden, Germany). The Dual-Color Probe Set consists of 2 separate probe mixtures: 1 probe set contains the LSI 1p36 Spectrum Orange test probe and the LSI 1q25 Spectrum Green control probe; the other probe set contains the LSI 19q13 Spectrum Orange test probe and the LSI 19p13 Spectrum Green control probe. The paraffin sections were deparaffinized, air-dried, incubated in pretreatment solution at 98°C for 15 minutes in Heat Pretreatment Solution (Invitrogen, Carlsbad, CA), and subsequently immersed in purified water. The slides were then treated with Enzyme Reagent (Invitrogen) in a humidified box for 30 to 40 minutes at room temperature and washed in purified water. After air dehydration, 10 μL of probe mixture was applied to each sample. The slides were then coverslipped and sealed with rubber cement. Slides and probes were codenatured at 74°C for 5 minutes and hybridized at 37°C for 16 hours in the dark using the Vysis HYBrite Denaturation/Hibridization Unit (Vysis). A posthybridization wash was performed in 2× SSC–0.3% NP-40 at 73°C for 3 minutes. Finally, the slides were dehydrated and mounted in antifade with DAPI (4′,6-diamidino-2-phenylindole; Abbott Molecular) and stored in the dark before signal evaluation. An automated scanning station, MetaSystems (Carl Zeiss MetaSystems, Thornwood, NY), equipped with an AxioImager-Z1 epifluorescence microscope, was used to determine 1p/19q status. The first step was signal acquisition over 7 to 10 areas that were selected for each slide by a dedicated operator. To choose regions with the most neoplastic cells, areas with a high signal quality and good nuclei preservation were assessed. These areas were automatically scanned, and 13 different consecutive focal planes were made for each FISH signal to form a single bidimensional image. For traditional reading, the automatically acquired images were transferred to Isis software (Zeiss) and stored in dedicated files. The evaluation of hybridization signals was then performed in 200 or more nonoverlapping nuclei, and the nuclei counted displayed at least 2 green signals. The ratio of 1p/1q and 19q/19p was calculated by dividing the number of orange and green signals.
For each case, FISH data were analyzed using criteria as follows: 1) a cutoff ratio of less than or equal to 0.8 was used to define 1p and 19q allelic losses; in addition, for 1p chromosomal arm, we applied a more stringent ratio cutoff equivalent to less than or equal to 0.7, as previously reported (13); and 2) percentage of neoplastic nuclei carrying 1p and 19q deletions using greater than or equal to 50% as the cutoff to define 1p and 19q deletions, as previously reported (24); 3) an integrated evaluation, merging points 1 and 2 with further subcategories, as detailed below. Finally, we investigated the role of codeletion in chromosomal imbalance conditions (polysomy) to determine its possible prognostic value, which is debated in the literature (28).
Establishment of Specific Codeletion Criteria
To identify the most prognostically reliable evaluation of codeletion (intended as a favorable indicator), we set different criteria. As a premise, we specified a different ratio cutoff regarding 1p exclusively, whereas the 19q cutoff for deletion remained unchanged at less than or equal to 0.8. The reason for this choice was based on preliminary evidence that 1p is the most relevant chromosome for favorable prognosis, whereas the 19q deletion can bear unfavorable prognostic value (16, 33).
Therefore, we designated our cutoff criteria for codeletion as follows: 1) codeletion determined as ratio value less than or equal to 0.8 for both 1p and 19q; 2) codeletion determined as ratio value less than or equal to 0.7 for 1p and less than 0.8 for 19q; 3) codeletion determined as more than 50% of neoplastic cells carrying both 1p and 19q deletion irrespective of the ratio value; 4) codeletion determined as ratio value less than or equal to 0.8 for both 1p and 19q + more than 50% of cells carrying the 1p deletion (merging points 1 and 3); and 5) codeletion determined as ratio value less than or equal to 0.7 for 1p and 0.8 for 19q and more than 50% of cells carrying the 1p deletion (merging points 2 and 3).
Statistical Analysis
The data were analyzed with SPSS Version 19 (SPSS, Inc., Chicago, IL). A statistically significant probability value was defined as p < 0.05. Univariate survival analysis was performed using the Mantel-Cox test (34) and depicted by the Kaplan-Meier method (35). Multivariate survival analysis was performed using the Cox model.
RESULTS
Clinical and Pathologic Prognostic Variables: Age, Extent of Surgical Resection, and Histologic Grade
The patients included 81 men and 62 women, aged 22 to 81 years (mean, 51.5 years); 60 (42%) of 143 patients were younger than 50 years and 83 (58%) of 143 were older. Thirty-six (60%) of 60 patients younger than 50 years and 30 (37.5%) of 80 patients older than 50 years had a low-grade glioma. By univariate analysis, age significantly correlated with OS (p < 0.0001), identifying a subgroup of younger patients with a more favorable outcome (Fig. 1A). No correlation was found with DFS (p = 0.27) (Fig. 1B). In addition, young age (<50 years) retained its favorable prognostic value not only in the overall series but also within the distinct not-deleted and codeleted subgroups (p < 0.0001 and p = 0.001, respectively).
FIGURE 1.
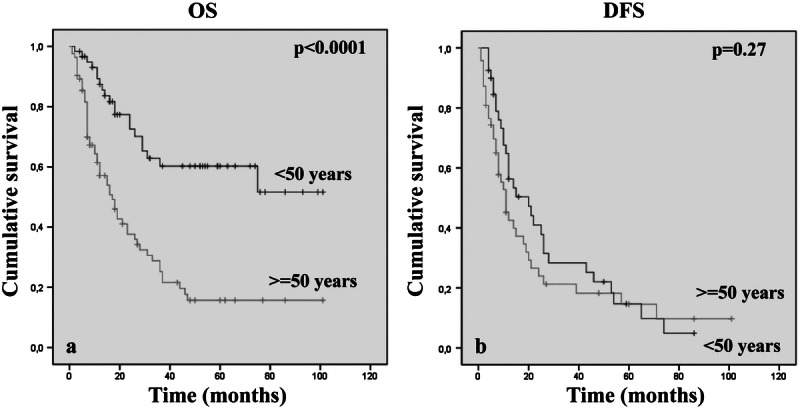
Kaplan-Meier analyses of estimated overall survival OS (A) and disease-free survival (DFS) (B) in relation to patient age in 143 oligodendroglial brain tumors.
Data concerning the extent of surgical resection were available in 99 (69.2%) of 143 cases. More precisely, 19 (19.2%) of 99 patients underwent a biopsy; 59 (59.6%) of 99, a partial resection; 14 (14.2%) of 99, a subtotal resection; and 7 (7%) of 99, a radical resection. There was no significant difference in OS and DFS between these groups (OS, p = 0.07). As expected, a histologic low tumor grade was related to better OS (p < 0.0001) (Fig. 2A). Regarding the DFS, there was a significant difference in tumor recurrence and progression between grades II and IV only (p = 0.008) (Fig. 2B).
FIGURE 2.
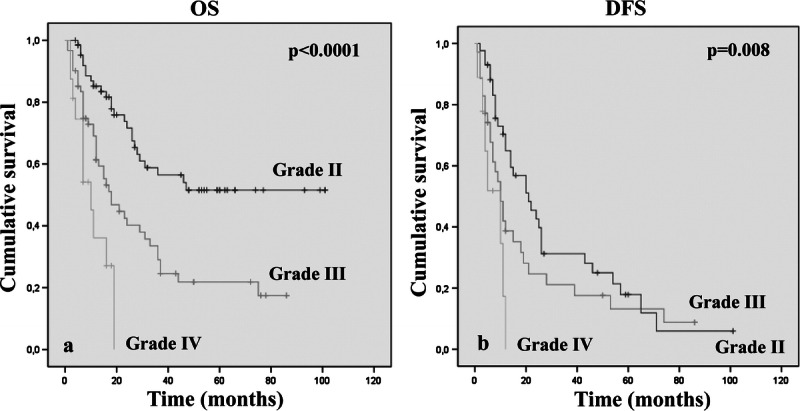
Kaplan-Meier analyses of estimated overall survival (OS) (A) and disease-free survival (DFS) (B) in relation to histologic tumor grade in 143 oligodendroglial brain tumors.
FISH Data Interpretation
Ratio Cutoff Setting 1p ≤ 0.8/19q ≤ 0.8 Versus 1p ≤ 0.7/19q ≤ 0.8
We determined 1p/19q status using 2 different ratio cutoff values to validate whether a more severe (≤0.7) deletion criterion for 1p could reduce the gray zone of unfavorable outcome despite codeletion. According to the 1p ≤ 0.8/19q ≤ 0.8 ratio, there were 68 (47.6%) of 143 cases that had both 1p and 19q allelic losses, 21 (14.7%) of 143 and 18 (12.6%) of 143 cases had isolated loss of 1p or 19q, respectively, and 36 (25.1%) of 143 cases did not have codeletions or single deletions (Table 1).
Using the more stringent ratio cutoff (≤0.7) to define 1p allelic loss, 16 (23.5%) of 68 of the prior codeleted cases were redistributed into different subgroups; thus, codeletion was observed in 52 (36.3%) of 143 cases (Table 1). By univariate analysis independent of the 1p ratio cutoff, the subgroup of cases with 1p/19q codeletion showed longer OS and DFS (1p ≤ 0.8/19q ≤ 0.8, OS, p = 0.002 and DFS, p = 0.03; 1p ≤ 0.7/19q ≤ 0.8, OS, p = 0.002 and DFS, p = 0.005) (Fig. 3). For DFS, the use of the 0.7 cutoff for the 1p deletion strengthened the favorable prognostic value of codeletion. However, despite the use of a different cutoff ratio and patient redistribution, the percentages of codeleted patients who died at the end of follow-up (42%) were equivalent (29 of 68 and 22 of 52 patients in 0.8 and 0.7 cutoff, respectively) (Supplemental Digital Content 1, http://links.lww.com/NEN/A447). These results indicate that restricting the cutoff ratio for 1p is not helpful in reducing the gray zone of codeleted patients with poor outcome. In addition, using the 0.8 ratio cutoff, the 19q allelic loss alone identified a subset of patients with the worst prognosis (Fig. 3A–C). The presence of an isolated 19q loss was not closely associated with any specific tumor histologic type.
FIGURE 3.
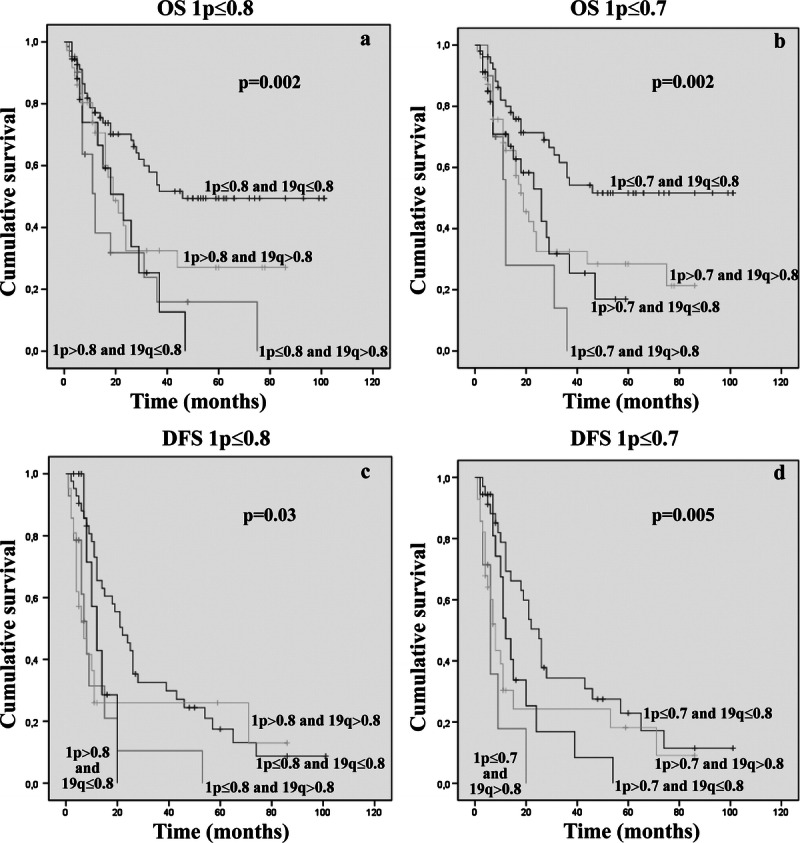
Kaplan-Meier analyses of estimated OS (A, B) and DFS (C, D) in relation to 1p/19q status, as assessed by applying a 1p ≤ 0.8/19q ≤ 0.8 ratio cutoff (A–C) and 1p ≤ 0.7/19q ≤ 0.8 ratio cutoff (B–D).
Ratio and Tumor Histology/Grade
Subdividing all cases according to the histologic type, 1p/19q codeletion (independently of the ratio applied [0.7 or 0.8]) retained prognostic significance only in pure oligodendroglial tumors, identifying oligodendrogliomas with prolonged overall and progression-free survival (1p ≤ 0.8/19q ≤ 0.8, OS, p = 0.005 and DFS, p < 0.0001; 1p ≤ 0.7/19q ≤ 0.8, OS, p = 0.006 and DFS, p < 0.0001) (Table 1). No correlation was found in mixed oligodendroglial tumors of any histologic grade.
Similarly, grouping all cases according to the histologic grade, regardless of tumor histotype, 1p/19q codeletion (with any ratio) identified a subset of patients with a better prognosis within grade II tumors only (1p ≤ 0.8/19q ≤ 0.8, OS, p = 0.03; 1p ≤ 0.7/19q ≤ 0.8, OS, p = 0.01). In addition, we observed a trend for correlation between codeletion and OS in grade IV tumors only when applying the ratio cutoff 1p ≤ 0.7/19q ≤ 0.8 (OS, p = 0.05).
Merging the previously reported data, we analyzed the prognostic impact of 1p/19q status in pure oligodendrogliomas according to histologic grade (grade II vs grade III). Applying the ratio cutoff 1p ≤ 0.8/19q ≤ 0.8, codeletion identified a subset of lesions with better outcome in both OII and OIII (OS, p = 0.04 and p = 0.05, respectively). Conversely, no correlation was found using the more severe cutoff (OS, p = 0.1 and p = 0.2, respectively), although there was a detectable trend toward significance. In terms of DFS, with both ratios, codeletion was found to be significant only in OII (1p ≤ 0.8/19q ≤ 0.8 and 1p ≤ 0.7/19q ≤ 0.8, p < 0.0001).
Percentage of Neoplastic Nuclei Deletion
For the second setting, we defined the 1p/19q molecular status according to the “rough” percentage of neoplastic nuclei carrying the deletion, independent of the ratio (in 11 of 143 cases, the data concerning the percentage of deleted nuclei were not available). The cutoff for codeletion was greater than or equal to 50% of nuclei carrying loss of chromosomal material from both arms (24). Forty-two (31.8%) of 132 cases showed combined 1p and 19q loss; 24 (18.18%) of 132 and 18 (13.6%) of 132 cases showed single 1p or 19q allelic loss, respectively; and 48 (36.3%) of 132 cases displayed 1p/19q maintenance. The subgroup of codeleted lesions showed a better prognosis both in terms of OS and DFS (OS, p = 0.01; DFS, p = 0.02) (Fig. 4). The percentage of codeleted patients who died at the end of follow-up was 31.1% (16 of 42 cases) (Supplemental Digital Content 1, http://links.lww.com/NEN/A447).
FIGURE 4.
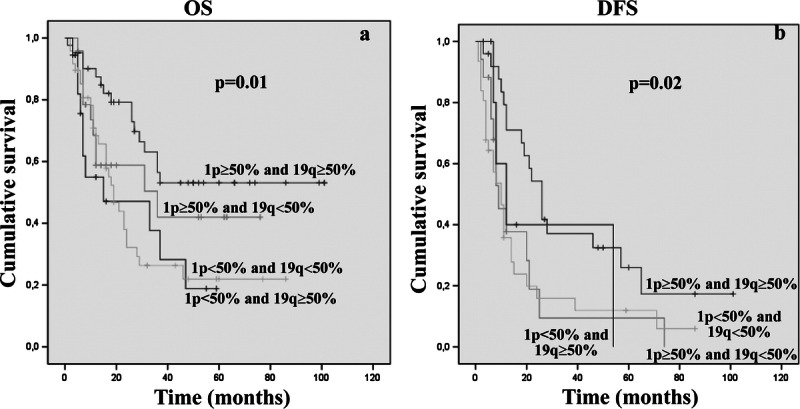
Kaplan-Meier analyses of estimated OS (A) and DFS (B) in relation to 1p/19q molecular status, according to the “rough” percentage of neoplastic nuclei carrying the deletion.
“Weighted” Ratio: Ratio + ≥50% of Neoplastic Nuclei Carrying 1p Deletion in Codeleted Tumors
As previously reported, there was a statistical correlation between codeletion and better patient outcome established by using both the ratio and the rough percentage as independent criteria. Thus, we tried to combine them to optimize the prognostic value of FISH. We identified a subgroup of codeleted tumors according to both previously reported ratio cutoff values and then considered only codeleted cases with greater than or equal to 50% of cells carrying 1p loss. The presence on 1p deletion in greater than or equal to 50% of neoplastic nuclei in codeleted cases identified a subset of patients with better outcome (1p ≤ 0.8/19q ≤ 0.8, OS, p = 0.03 and DFS, p = 0.04; 1p ≤ 0.7/19q ≤ 0.8, OS, p = 0.001) (Fig. 5) (Supplemental Digital Content 1, http://links.lww.com/NEN/A447). Specifically, in the 1p ≤ 0.8/19q ≤ 0.8 codeleted group (63 patients), 9 (56.2%) of 16 patients who had less than 50% of 1p deleted cells died (Fig. 6). Strikingly, in the 1p ≤ 0.7/19q ≤ 0.8 codeleted group (47 patients), all patients (100%) with less than 50% of 1p deleted nuclei died (Fig. 6). In addition, among 8 (12.7%) of 63 and 7 (14.9%) of 47 codeleted patients showing 1p greater than or equal to 50% and a concomitant 19q loss in less than 50% of neoplastic cells, 7 (87.5%, using 1p ≤ 0.8) of 8 and 6 (85.7%, using 1p ≤ 0.7) of 7 respectively, were still living. These data highlight the favorable effect of 1p deletion and the unfavorable effect that 19q loss can have on patient prognosis.
FIGURE 5.
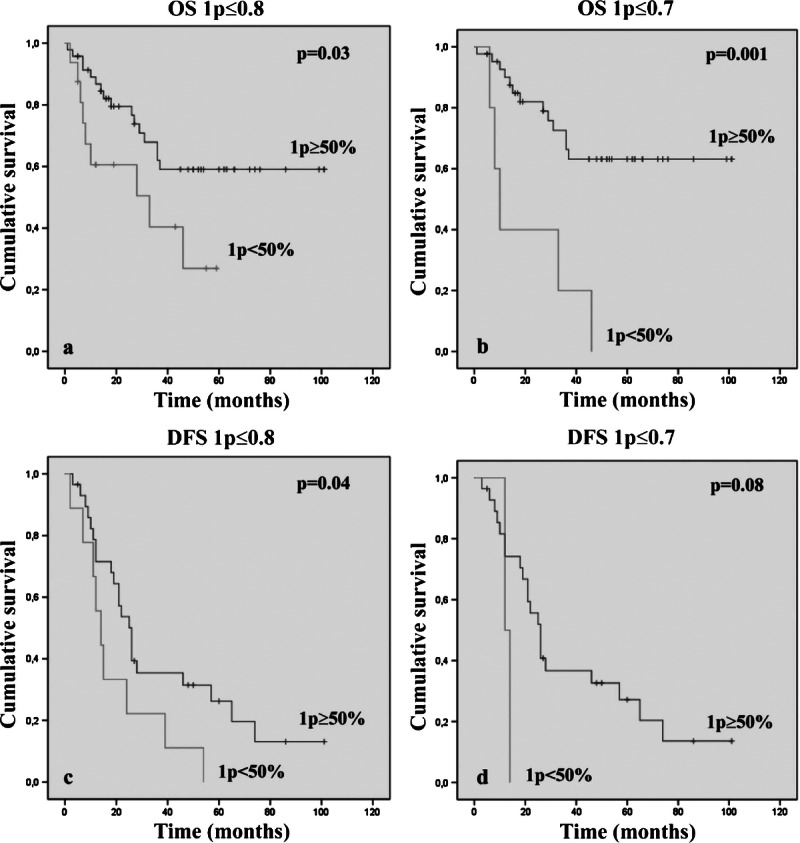
Kaplan-Meier analyses of estimated OS (A, B) and DFS (C, D) in the subgroup of codeleted cases, according to both ratio cutoff values (1p ≤ 0.8, A–C; 1p ≤ 0.7, B–D) in relation to the presence of greater than or equal to 50% of neoplastic nuclei carrying a 1p loss.
FIGURE 6.
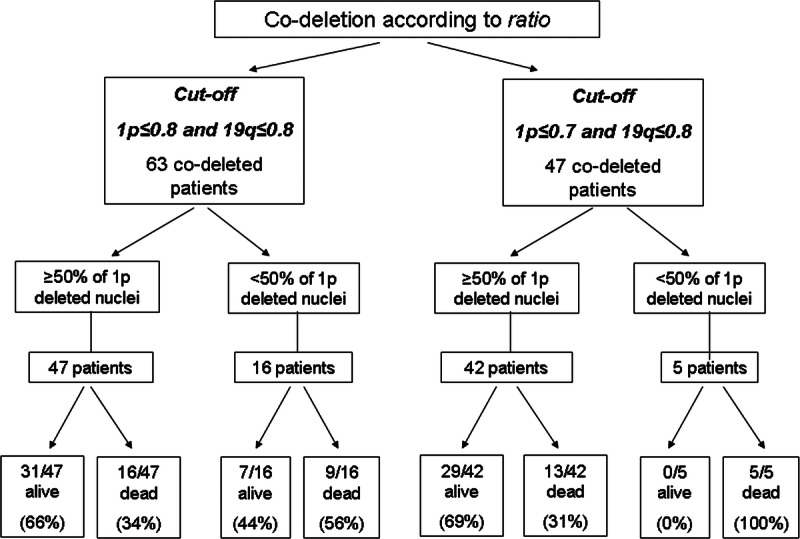
Stratification of follow-up according to ratio cutoff values and percentage of neoplastic nuclei carrying a 1p loss.
Multivariate Analysis
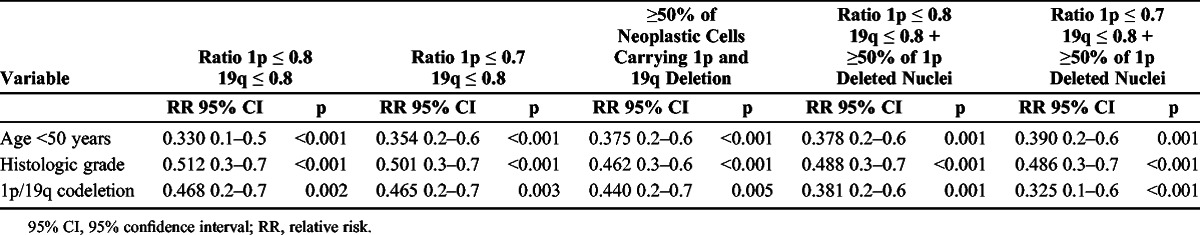
By multivariate Cox regression analysis, among all the clinical, histologic, and molecular variables considered, the age at diagnosis, histologic tumor grade, and 1p/19q deletion status assessed using any reported criteria were independent prognostic factors (Table 2). The combined (weighted) criteria based on ratio plus cell percentage significantly strengthened the favorable prognostic value of FISH, as assessed by codeletion (Table 2).
TABLE 2.
Multivariate Analysis of OS Based on Patient Age, Histologic Tumor Grade, and 1p/19q Codeletion Assessed Using Different Criteria in 143 Brain Tumors With an Oligodendroglial Component
Concurrent 1p/19q Loss and Polysomy: Evaluation of Prognostic Significance
Among the codeleted cases that were identified according to the ratio criteria, 5 lesions showed polysomy (≥30% of neoplastic nuclei carrying 3 or more green signals for both arms) (28). We identified 5 (7.9%) of 63 and 5 (10.6%) of 47 cases using the ratio cutoff of 1p ≤ 0.8/19q ≤ 0.8 or 1p ≤ 0.7/19q ≤ 0.8, respectively. Two (40%) of 5 were grade II and 3 (60%) of 5 were grade III tumors. We did not observe any significant difference in terms of OS or DFS among the subgroups with and without polysomy (OS, p = 0.9 and DFS, p = 0.7 for both ratio values).
DISCUSSION
To date, the 1p/19q codeletion remains the major favorable prognostic/predictive indicator for oligodendroglial tumors (3, 4, 6, 9, 16, 19, 21), but its efficacy may be impaired because of the heterogeneity of methods used for its determination and to the lack of uniform reading criteria within the specific techniques. A good concordance between FISH and polymerase chain reaction–based loss of heterozygosity has been previously reported, and a recent study highlighted that modulation of FISH interpretation criteria and additional determination of 10q status could increase its predictive reliability (25). The need for standardized protocols for FISH is indicated by the relevant percentage of poor outcomes of patients with codeleted tumors, suggesting that interpretation of data could be optimized. To achieve a significant reduction of this gray zone of unexpected unfavorable outcomes in codeleted oligodendroglial tumors, we created a weighted/scored FISH interpretation that proved helpful in increasing the prognostic stratification and that is easy to use in the daily diagnostic assessment of 1p/19q status.
Our search for a gold standard in FISH data analysis was organized through consecutive steps within a series of 143 tumors with an oligodendroglial component, either pure (grade II and III oligodendrogliomas) or mixed (grade II and III oligoastrocytomas and glioblastomas with an oligodendroglial component). First, we compared 2 ratio cutoff values for 1p (0.8 and 0.7, respectively) in the presence of a fixed 0.8 cutoff for 19q and then weighted this analysis by setting a further correction within the codeleted cases, linked to the percentage of neoplastic cells carrying 1p deletion. Specifically, we selected the 0.8 ratio cutoff as the most commonly used and the 0.7 cutoff as the lowest reported in literature (10, 11, 13, 15, 30). This allowed us to identify and compare the variation of relative risk among different subclasses of codeleted tumors. The 2 ratio cutoff values, as well as the cell percentage “restriction criteria,” have been applied within codeleted gliomas to tumors with 1p chromosome loss only because its prognostic weight seems to be greater than 19q (33). Furthermore, there are conflicting results in the literature on the prognostic value of solitary 19q loss: a negative prognostic role in anaplastic gliomas identifying a subgroup of patients at worst prognosis or a favorable role associated with long-term survival in grade IV gliomas (16, 36–38). In our series, 19q solitary deletion identified a subset of patients with a poorer DFS and OS (only using the 0.8 cutoff ratio). Additional information on the role of the 19q could come in the future from the comprehension of the effective clinical relevance of CIC mutations on the retained 19q allele that were recently described to be linked to IDH mutated oligodendogliomas (39).
Our first step was to compare the 0.8 1p/0.8 19q ratio and the more stringent 0.7 1p/0.8 19q ratio. The percentage of unfavorable outcomes within the 2 groups remained identical (42%), indicating that a more severe cutoff ratio was useless for reducing the prognostic gray zone. As a second step, we checked whether codeletion (defined as >50% of cells carrying 1p and 19q deletions) was more effective than ratio cutoff in identifying patients with better prognosis (18, 24, 29). The gray zone of unfavorable outcome was only reduced from 42% to 38% of patients without significant additional decrease of the relative risk. However, when we merged the ratio and cell percentage criteria and restricted the identification of codeleted patients to the cases with an adequate ratio cutoff and more than 50% of cells carrying the 1 p deletion, the favorable impact was increased and the relative risk decreased from 0.468 to 0.325 (Table 2). The most favorable codeletion was defined as more that 50% of tumor cells carrying the 1p deletion in a case showing a ratio cutoff of 0.7 for 1p and 0.8 for 19q; this setting regroups 69% of living patients, irrespective of age, histotype, or tumor grade. Furthermore, codeletion for ratio (according to 0.7 ratio cutoff) combined with less than 50% cells with 1p loss seems to be the most robust indicator of unfavorable prognosis, as 100% of these patients in this series died (5 of 143) (Fig. 6).
Interestingly, further restriction of codeleted patients, including only tumors with a ratio cutoff of 0.8, with more than 50% cells carrying the 1p loss and less that 50% of cells carrying the 19q loss, allowed the identification of 87.5% (7 of 8 cases) (similarly, 85.7% [6 of 7 cases] using 0.7 ratio cutoff) of living patients. These results demonstrate the existence of a balance between the 1p and 19q deletions, where the best prognostic situation is that of an excess of cells with the 1p loss and a minority with the 19q loss, within a codeletion setting.
In conclusion, we suggest that the most prognostically reliable FISH report in oligodendroglial tumors should define as codeleted tumors those with a ratio cutoff less than or equal to 0.7 and more than 50% of cells with the 1p loss, eventually with further a restriction to cases in which a minority of cells have the 19q deletion. A further validation in other dedicated laboratories will help in testing this new weighted FISH reading to assess the consequence of intercenter variability in data interpretation and technical procedures.
Supplementary Material
Footnotes
Send correspondence and reprint requests to: Paola Cassoni, MD, PhD, Department of Medical Sciences, Via Santena 7, University of Turin, Turin, Italy; E-mail: paola.cassoni@unito.it
The authors declare no conflicts of interest.
This work was supported by the following grants: FIRC (Fondazione Italiana per la Ricerca sul Cancro); Compagnia di San Paolo, pratica 4011 SD/cv 2011.0438; Compagnia di San Paolo “Special Project Oncology” and Progetti di Ricerca finanziati dall’Università degli Studi di Torino (ex 60%).
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s Web site (www.jneuropath.com).
REFERENCES
- 1. Ducray F, Idbaih A, Wang XW, et al. Predictive and prognostic factors for gliomas. Expert Rev Anticancer Ther 2011; 11: 781– 89 [DOI] [PubMed] [Google Scholar]
- 2. Sulman EP, Aldape K. The use of global profiling in biomarker development for gliomas. Brain Pathol 2011; 21: 88– 95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aldape K, Burger PC, Perry A. Clinicopathologic aspects of 1p/19q loss and the diagnosis of oligodendroglioma. Arch Pathol Lab Med 2007; 131: 242– 51 [DOI] [PubMed] [Google Scholar]
- 4. Cairncross JG, Ueki K, Zlatescu MC, et al. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J Natl Cancer Inst 1998; 90: 1473– 79 [DOI] [PubMed] [Google Scholar]
- 5. Cairncross G, Berkey B, Shaw E, et al. Phase III trial of chemotherapy plus radiotherapy compared with radiotherapy alone for pure and mixed anaplastic oligodendroglioma: Intergroup Radiation Therapy Oncology Group Trial 9402. J Clin Oncol 2006; 24: 2707– 14 [DOI] [PubMed] [Google Scholar]
- 6. Fallon KB, Palmer CA, Roth KA, et al. Prognostic value of 1p, 19q, 9p, 10q, and EGFR-FISH analyses in recurrent oligodendrogliomas. J Neuropathol Exp Neurol 2004; 63: 314– 22 [DOI] [PubMed] [Google Scholar]
- 7. Felsberg J, Erkwoh A, Sabel MC, et al. Oligodendroglial tumors: Refinement of candidate regions on chromosome arm 1p and correlation of 1p/19q status with survival. Brain Pathol 2004; 14: 121– 30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hirose T, Ishizawa K, Shimada S. Utility of in situ demonstration of 1p loss and p53 overexpression in pathologic diagnosis of oligodendroglial tumors. Neuropathology 2010; 30: 586– 96 [DOI] [PubMed] [Google Scholar]
- 9. Ino Y, Betensky RA, Zlatescu MC, et al. Molecular subtypes of anaplastic oligodendroglioma: Implications for patient management at diagnosis. Clin Cancer Res 2001; 7: 839– 45 [PubMed] [Google Scholar]
- 10. Jeon YK, Park K, Park CK, et al. Chromosome 1p and 19q status and p53 and p16 expression patterns as prognostic indicators of oligodendroglial tumors: A clinicopathological study using fluorescence in situ hybridization. Neuropathology 2007; 27: 10– 20 [DOI] [PubMed] [Google Scholar]
- 11. Kanamori M, Kumabe T, Sonoda Y, et al. Predictive factors for overall and progression-free survival, and dissemination in oligodendroglial tumors. J Neurooncol 2009; 93: 219– 28 [DOI] [PubMed] [Google Scholar]
- 12. Kouwenhoven MC, Kros JM, French PJ, et al. 1p/19q loss within oligodendroglioma is predictive for response to first line temozolomide but not to salvage treatment. Eur J Cancer 2006; 42: 2499– 503 [DOI] [PubMed] [Google Scholar]
- 13. Mahajan D, Prayson RA. Repeated molecular testing in gliomas: A retrospective study of 53 cases. Am J Clin Pathol 2009; 132: 118– 24 [DOI] [PubMed] [Google Scholar]
- 14. Okamoto Y, Di Patre PL, Burkhard C, et al. Population-based study on incidence, survival rates, and genetic alterations of low-grade diffuse astrocytomas and oligodendrogliomas. Acta Neuropathol 2004; 108: 49– 56 [DOI] [PubMed] [Google Scholar]
- 15. Reddy KS. Assessment of 1p/19q deletions by fluorescence in situ hybridization in gliomas. Cancer Genet Cytogenet 2008; 184: 77– 86 [DOI] [PubMed] [Google Scholar]
- 16. Scheie D, Meling TR, Cvancarova M, et al. Prognostic variables in oligodendroglial tumors: A single-institution study of 95 cases. Neuro Oncol 2001; 13: 1225– 33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Senetta R, Trevisan E, Rudà R, et al. Caveolin 1 expression independently predicts shorter survival in oligodendrogliomas. J Neuropathol Exp Neurol 2009; 68: 425– 31 [DOI] [PubMed] [Google Scholar]
- 18. Shukla B, Agarwal S, Suri V, et al. Assessment of 1p/19q status by fluorescence in situ hybridization assay: A comparative study in oligodendroglial, mixed oligoastrocytic and astrocytic tumors. Neurol India 2009; 57: 559– 66 [DOI] [PubMed] [Google Scholar]
- 19. Smith JS, Perry A, Borell TJ, et al. Alterations of chromosome arms 1p and 19q as predictors of survival in oligodendrogliomas, astrocytomas, and mixed oligoastrocytomas. J Clin Oncol 2000; 18: 636– 45 [DOI] [PubMed] [Google Scholar]
- 20. Van den Bent MJ, Looijenga LH, Langenberg K, et al. Chromosomal anomalies in oligodendroglial tumors are correlated with clinical features. Cancer 2003; 97: 1276– 84 [DOI] [PubMed] [Google Scholar]
- 21. Weller M, Berger H, Hartmann C, et al. Combined 1p/19q loss in oligodendroglial tumors: Predictive or prognostic biomarker? Clin Cancer Res 2007; 13: 6933– 37 [DOI] [PubMed] [Google Scholar]
- 22. Gelpi E, Ambros IM, Birner P, et al. Fluorescent in situ hybridization on isolated tumor cell nuclei: A sensitive method for 1p and 19q deletion analysis in paraffin-embedded oligodendroglial tumor specimens. Mod Pathol 2003; 16: 708– 15 [DOI] [PubMed] [Google Scholar]
- 23. Ambros PF, Ambros IMSIOP Europe Neuroblastoma Pathology, Biology, and Bone Marrow Group Pathology and biology guidelines for resectable and unresectable neuroblastic tumors and bone marrow examination guidelines. Med Pediatr Oncol 2001; 37: 492– 504 [DOI] [PubMed] [Google Scholar]
- 24. Hartmann C, Hentschel B, Tatagiba M, et al. Molecular markers in low-grade gliomas: Predictive or prognostic? Clin Cancer Res 2011; 17: 4588– 99 [DOI] [PubMed] [Google Scholar]
- 25. Horbinski C, Nikiforova MN, Hobbs J, et al. The importance of 10q status in an outcomes-based comparison between 1p/19q fluorescence in situ hybridization and polymerase chain reaction-based microsatellite loss of heterozygosity analysis of oligodendrogliomas. J Neuropathol Exp Neurol 2012; 71: 73– 82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Iwadate Y, Matsutani T, Shinozaki N, et al. Anaplastic oligodendroglial tumors harboring 1p/19q deletion can be successfully treated without radiotherapy. Anticancer Res 2011; 31: 4475– 79 [PubMed] [Google Scholar]
- 27. Ren X, Cui X, Lin S, et al. Co-deletion of chromosome 1p/19q and IDH1/2 mutation in glioma subsets of brain tumors in Chinese patients. PLoS One 2012; 7: e32764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Snuderl M, Eichler AF, Ligon KL, et al. Polysomy for chromosomes 1 and 19 predicts earlier recurrence in anaplastic oligodendrogliomas with concurrent 1p/19q loss. Clin Cancer Res 2009; 15: 6430– 37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Woehrer A, Sander P, Haberler C, et al. FISH-based detection of 1p 19q co-deletion in oligodendroglial tumors: Procedures and protocols for neuropathological practice—A publication under the auspices of the Research Committee of the European Confederation of Neuropathological Societies (Euro-CNS). Clin Neuropathol 2011; 30: 47– 55 [DOI] [PubMed] [Google Scholar]
- 30. Molinari C, Iorio P, Medri L, et al. Chromosome 1p and 19q evaluation in low-grade oligodendrogliomas: A descriptive study. Int J Mol Med 2010; 25: 145– 51 [PubMed] [Google Scholar]
- 31.Louis DN, Ohgaki H, Wiestler OD, eds. World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of the Nervous System. Lyon, France: IARC Press, 2007 [Google Scholar]
- 32. Merz JF, Sankar P, Taube SE, et al. Use of human tissues in research: Clarifying clinician and researcher roles and information flows. J Invest Med 1997; 45: 252– 57 [PubMed] [Google Scholar]
- 33. Kujas M, Lejeune J, Benouaich-Amiel A, et al. Chromosome 1p loss: A favorable prognostic factor in low-grade gliomas. Ann Neurol 2005; 58: 322– 26 [DOI] [PubMed] [Google Scholar]
- 34. Cox D. Regressions models and life table. J R Stat Soc 1972; 34: 187– 220 [Google Scholar]
- 35. Kaplan E, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc 1958; 53: 457– 81 [Google Scholar]
- 36. Burton EC, Lamborn KR, Feuerstein BG, et al. Genetic aberrations defined by comparative genomic hybridization distinguish long-term from typical survivors of glioblastoma. Cancer Res 2002; 62: 6205– 10 [PubMed] [Google Scholar]
- 37. Fuller CE, Schmidt RE, Roth KA, et al. Clinical utility of fluorescence in situ hybridization (FISH) in morphologically ambiguous gliomas with hybrid oligodendroglial/astrocytic features. J Neuropathol Exp Neurol 2003; 62: 1118– 28 [DOI] [PubMed] [Google Scholar]
- 38. Perry A, Fuller CE, Banerjee R, et al. Ancillary FISH analysis for 1p and 19q status: Preliminary observations in 287 gliomas and oligodendroglioma mimics. Front Biosci 2003; 8: a1– a9 [DOI] [PubMed] [Google Scholar]
- 39. Yip S, Butterfield YS, Morozova O, et al. Concurrent CIC mutations, IDH mutations, and 1p/19q loss distinguish oligodendrogliomas from other cancers. J Pathol 2012; 226: 7– 16 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


