Abstract
Aim
To compare the levels of Selenomonas sputigena and uncultivated/unrecognized Selenomonas species in subgingival biofilms from generalized aggressive periodontitis (GAgP) and periodontaly healthy (PH) subjects.
Material and Methods
GAgP (n=15) and PH (n=15) subjects were recruited and their clinical periodontal parameters were evaluated. Subgingival plaque samples were collected (9 samples/subject) and analyzed for the levels of 10 bacterial taxa, including cultivated and uncultivated/unrecognized microorganisms using the RNA-oligonucleotide quantification technique (ROQT). Differences in the levels of the test taxa between groups were sought using the Mann-Whitney test.
Results
GAgP subjects showed significantly higher mean counts of Porphyromonas gingivalis, Selenomonas sputigena and Selenomonas oral clone CS002 (Human Oral Microbial Database (HOMD) Oral Taxon 131), while Actinomyces gerencseriae and Streptococcus sanguinis were found in higher mean counts in PH subjects (p<0.01). Selenomonas EW084 (HOMD OT 146) was only detected in the GAgP group. In the GAgP group, levels of P. gingivalis and S. sputigena were higher in sites with probing depth (PD) ≥5mm than in shallow sites (PD ≤3mm) (p<0.01). Furthermore, sites with PD≤3mm in GAgP subjects harbored higher levels of these two species than sites in PH subjects. There were positive correlations between PD and levels of P. gingivalis (r=0.77; p<0.01), S. sputigena (r=0.60; p<0.01) and Selenomonas sp. EW076 (OT 139) (r=042, p<0.05).
Conclusion
S. sputigena, Selenomonas sp. oral CS002 (OT 131) and Selenomonas sp. oral clone EW084 (OT 146) may be associated with the pathogenesis of GAgP, and their role in the onset and progression of this infection should be further investigated.
Keywords: Selenomonas sputigena, molecular biology, 16S rRNA, generalized aggressive periodontitis, not-yet-cultivated species
INTRODUCTION
The role of the oral microbiota in the etiology of periodontal diseases has been well established, and specificity may exist among certain bacterial species or groups and the various forms of periodontal disease (1-5). The complexity and diversity of the periodontal microbiota has been confirmed by numerous studies (6-8). However, only a few species have been recognized as periodontal pathogens, namely Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis and Tannerella forsythia (9).
Aggressive periodontitis is characterized by a rapid rate of tissue destruction and progression, early age of onset, specific patterns of periodontal breakdown and familial aggregation. These features are quite distinct from those observed in chronic periodontitis (10). The microbiota of the localized (LAgP) and generalized forms of aggressive periodontitis (GAgP) present higher proportions of Aggregatibacter actinomycetemcomitans, in comparison with that of chronic periodontitis (ChP); whereas the proportion of red complex pathogens (5) did not differ between these periodontal conditions, despite their clinical differences (11). These observations suggest that further analyses of the microbiota are necessary in order to explain differences in clinical outcomes.
The use of open-ended culture-independent approaches has expanded the breadth of knowledge of the oral microbiota. The oral cavity may harbor more than 700 bacterial species (6,12), of which 35% remains uncultivated (13,14). Even though those taxa have remained undetected in most studies, it is possible that specific uncultivated/unrecognized taxa may play a role in the etiology of oral diseases, or alternatively, they may represent beneficial bacterial species.
Using 16S rRNA sequencing analysis, Faveri et al. (11) observed that Selenomonas species dominated the diseased sites of subjects with GAgP. Selenomonas sputigena was the most frequently detected bacterial species whereas other species of Selenomonas were often present in high levels, including Selenomonas noxia, Selenomonas oral clone EW076, Selenomonas oral clone EW084 and Selenomonas oral clone CS002. High prevalence of Selenomonas species has also been observed in subgingival samples from ChP subjects (6, 16-18). However, none of these studies were able to quantify the levels of multiple uncultivated/unrecognized taxa in large number of individual biofilm samples simultaneously. Information on the absolute numbers and proportions of organisms in samples is important in distinguishing species associated with periodontal health or disease and to evaluate the effects of periodontal therapy (5, 19).
The RNA-oligonucleotide quantification technique (ROQT) allows the quantification of uncultivated/unrecognized and cultivated taxa in multiple subgingival biofilm samples in parallel (20). In addition, the method does not require sample pooling, amplification or dilution, which represent procedures that may introduce biases in the composition of the microbial communities under study (16,19,21). Thus, the purpose of the present study was to evaluate the levels of Selenomonas species, as well as their relationship with P.gingivalis and some host-compatible species (Actinomyces gerensceriae, Streptococcus anginosus/gordonii and Streptococcus sanguinis) in subgingival biofilm samples from GAgP patients and periodontally healthy subjects controls using ROQT.
MATERIAL AND METHODS
Subject population
Fifteen periodontally healthy subjects and 15 individuals presenting GAgP were included in this investigation. All study participants were systemically healthy and were recruited at the Guarulhos University School of Dentistry (Guarulhos, SP, Brazil). The medical and dental histories were obtained and a full-mouth periodontal examination was performed. Based on these data, the periodontal diagnosis was made, and subjects who fulfilled the inclusion/exclusion criteria were invited to participate in the study. The study protocol was explained to each subject, and a signed Informed Consent was obtained. This study protocol was approved by Guarulhos University’s Ethics Committee in Clinical Research.
Clinical examination
One trained and calibrated examiner performed the clinical examination in all subjects. Visible plaque (0/1), gingival bleeding (0/1), bleeding on probing (BOP, 0/1), suppuration (0/1), probing depth (PD, mm) and clinical attachment level (CAL, mm) were measured at six sites per tooth (mesiobuccal, buccal, distobuccal, distolingual, lingual and mesiolingual) in all teeth excluding third molars at the baseline visit. PD and CAL measurements were recorded to the nearest millimeter using a North Carolina periodontal probe (Hu-Friedy, Chicago, IL, USA).
Inclusion criteria
GAgP and PH were diagnosed based on the periodontal classification of the American Academy of Periodontology (18). Subjects had at least 20 teeth and needed to meet the following criteria in order to be included in this study:
Generalized aggressive periodontitis (GAgP): ≤ 35 years of age; minimum of six permanent incisors and/or first molars with at least one site each with PD and CAL ≥ 5 mm; minimum of six teeth other than first molars and incisors with at least one site each with PD and CAL ≥ 5 mm; familial aggregation (at least one other member of the family presenting or with history of periodontal disease) (11).
Periodontal health (PH): ≤ 35 years of age; No sites with PD and CAL measurements > 3 mm and < 10% of sites exhibiting bleeding on probing.
Exclusion criteria
Exclusion criteria were pregnancy, lactation, smoking, subgingival periodontal therapy in the past 12 months, any systemic condition that could affect the progression of periodontal disease (e.g. diabetes and immunological disorders), long-term administration of anti-inflammatory medication, and antibiotic therapy in the previous 6 months.
Microbiological examination
Sample collection
Individual subgingival plaque samples were collected from nine non-contiguous interproximal sites per subject. For the GAgP group, three sites with PD ≤ 3mm and six sites with PD ≥ 5mm were selected. Nine sites with PD ≤ 3mm were collected from the PH group. The selected sites were randomized in different quadrants. After the clinical parameters had been recorded, the supragingival plaque was removed and the samples were taken with individual sterile Gracey curettes and immediately placed in separate polypropylene tubes containing 100 μl RNAse-free TE (10mM Tris-HCl, 1 mM EDTA, pH 7.6) and kept at -80 °C until extraction of total nucleic acids (TNA).
RNA-oligonucleotide quantification technique
Counts of the 10 bacterial species/phylotypes (Table 1) were determined in each sample, using the RNA-oligonucleotide quantification technique (ROQT) (20). The microbiological analysis was performed at the Laboratory of Microbiology of Guarulhos University.
Table 1.
Sequences for oligonucleotide probes and for the standards for quantification of the species/phylotypes evaluated.
| Species/phylotypes | Sequence (5’-3’) | Control (3’-5’) |
|---|---|---|
| Selenomonas sputigena | CCGTCACCCAAACTCAAT | GGCAGTGGGTTTGAGTTA |
| Selenomonas noxia | CTATTCGCATTAGGCACG | GATAAGCGTAATCCGTGC |
| Selenomonas sp. EW084 (OT 146) | GGACTCATCTCTGAGTCT | CCTGAGTAGAGACTCAGA |
| Selenomonas sp. EW076 (OT 139) | CTCTGCATGCTTCAGTCA | GAGACGTACGAAGTCAGT |
| Selenomonas sp. CS002 (OT 131) | GGCGCAACATTCGGTATT | CCGCGTTGTAAGCCATAA |
| A.actinomycetemcomitans | TTAAAGGTCCGCCTACGT | AATTTCCAGGCGGATGCA |
| Porphyromonas gingivalis | GTGGAAGCTTGACGGTAT | CACCTTCGAACTGCCATA |
| Actinomyces gerensceriae | ACCCCAGAAGCCCGTT | TGGGGTCTTCGGGCAA |
| S. anginosus/gordonii | CAACTCACAGTCTATGGTGTAG | GTTGAGTGTCAGATACCACATC |
| Streptococcus sanguinis | CAATAATCAATTTTATGCGGT | GTTATTAGTTAAAATACGCCA |
| 16S rRNA Universal | CTGCTGCCTCCCGTAGG | GACGACGGAGGCATCC |
A subset of the probes employed were ‘combination probes’, in that they could not distinguish species/phylotypes. This was the case for S. anginosus/gordonii.
OT : oral taxon ; oral taxon designations for uncultivated/unrecognized taxa are provided in accord with the Human Oral Microbiome Database, (www.homd.org), when available; GenBank assession numbers can also be found in the HOMD website.
Extraction of total nucleic acids
Extraction of TNA from all samples was performed using a Masterpure RNA & DNA purification kit (Epicentre, Madison, WI, USA). Cells were pelleted by centrifugation and resuspended in 25 μl TE. One microliter of proteinase K (50 μg μl-1) and 300 μl of tissue and cell lysis buffer were added followed by incubation at 65 °C for 15 min. After 5 min in ice, 175 μl MCP protein precipitation reagent (Epicentre) was added to each sample. After centrifugation, 500 μl isopropanol added to the supernatant and TNA was precipitated by centrifugation. The pellets were rinsed twice with 70% ethanol, air dried and ressupended. TNA samples were kept at -80 °C until analysis.
Probe and standards preparation
Oligonucleotide probes targeting the 16S ribosomal DNA (rRNA) gene of 7 cultivated and 3 uncultivated/unrecognized bacterial taxa were synthesized (Table 1). The probe panel also included a universal (eubacterial) probe, based on a conserved region of the bacterial 16S rRNA gene. The universal probe is only a positive control in the test and the levels were not considered. Probes sequences were 18-22 nucleotides in length and had minimal secondary structure. The sequences employed in ROQT were those routinely used in the Human Oral Microbial Identification Microarray (HOMIM) and are published elsewhere (21). One hundred picomolar (pM) of each oligonucleotide probe were labeled using a digoxigenin 3’end labeling kit (Roche, Indianapolis, IN, USA). The standards for detection of the oligonucleotide probes were a mixture of sequences complementary to each oligonucleotide probe (Table 1). The final mixtures of standards had 0.004 and 0.04 pM of each sequence, corresponding to 105 and 106 bacterial cells, respectively. The oligonucleotide probes and their ‘complementary’ sequences were synthesized by Invitrogen (São Paulo, SP, Brazil).
Hybridization
Ninety microliters of 2% glutaraldehyde (Ted Pella, Redding, CA, USA) and 910 μl 6 × SSC (1× SSC = 150mM NaCl, 15 mM sodium citrate, pH 7.0) were added to each TNA sample and standards. The final solutions were deposited in individual lanes of a Minislot (Immunetics, Cambridge, MA) concentrated onto a nylon membrane (Boehringer Mannheim, Indianapolis, IN, USA) and fixed to the membrane. The membranes were pre-hybridized into a plastic hybridization bag at 42°C for at least 90 min in 35ml of a pre-hybridization solution containing 50% formamide, 5 × SSC, 1% casein (Vetec, Rio de Janeiro, RJ, Brazil), 5x Denhardt’s reagent (Sigma, St. Louis, MO, USA), 25mM sodium phosphate (pH6.5) and 0.5mg ml-1 yeast RNA (Boehringer Mannheim). Then, the membranes were placed in a Miniblotter 45 (Immunetics), with the “lanes” of TNA at 90° to the channels of the device. The digoxigenin-labeled oligonucleotide probes were diluted in hybridization buffer (Ultrahyb Oligo buffer; Ambion, Austin, TX, USA), and added to individual lanes of the Miniblotter 45 at final concentrations ranging from 2 to 40 pM. The membranes were hybridized at 42°C for 80 min and then washed with 2 × SSC, 0.5% sodium dodecyl sulfate at 37°C for 60 min. After blocking with maleic acid buffer containing 10% casein, hybrids were detected by incubation with 1:2,500 dilution of anti-digoxigenin antibody conjugated with alkaline phosphate (Roche) for 30 min. The membranes were washed with maleic acid buffer thrice for 15 min and then with ‘buffer 3’ [0.04% MgCl2 and 2.1 diethanolamine (pH 9.5), equal volumes] for 5 min. Finally, 1 ml of a chemiluminescent substrate (CDP Star; Tropix, Bedford, MA, USA) diluted in ‘buffer 3’ was deposited onto the membrane surface, which was then exposed to an X-ray film (IBF-Medix, AGFA, Rio de Janeiro, RJ, Brazil). The films were scanned and signal intensities of the samples and the standards were measured using the TotalLab software (NonLinear USA, Durham, NC). Signals were converted to absolute “counts” by comparison with standards on the membrane. Absence of signal detection was recorded as zero.
Statistical analysis
The mean age and the mean percentage of sites with visible plaque, gingival bleeding, BOP and suppuration, as well as mean PD and CAL were computed for each subject and then averaged across subjects in each group separately. Mean estimated ‘counts’ (× 105) of individual bacterial species were computed for each site individually, averaged within each subject and then across subjects in each group separately. Prevalence of the test taxa was computed by determining the percentage of sites per subject colonized by ≥105 cells of each species. The mean percentage of “positive” sites was calculated for each subject and then averaged across subjects in each group. The significance of differences between the two groups for age and the clinical and microbiological parameters was sought using the Mann–Whitney U-test. The Wilcoxon test was used to detect statistically significant differences within PD categories in GAgP group. Chi-square test was employed to compare the differences in the frequency of gender and prevalence of subjects colonized by bacterial species. Spearman correlation was used to assess possible associations between PD and mean ‘counts’ of bacterial species.
RESULTS
Clinical findings
The demographic characteristics and clinical parameters of the studied population are presented in Table 2. No statistically significant differences were observed between groups for age and gender. The GAgP group displayed significantly higher mean PD, mean CAL, % BOP as well as levels of plaque and number of moderate and deep periodontal pockets (p< 0.05) in comparison with the periodontally healthy group.
Table 2.
Demographic characteristics and mean (± SD) full-mouth clinical parameters in both groups.
| Clinical variables | PH (n=15) | GAgP (n=15) |
|---|---|---|
| AgeNS | 26.6 ± 3.6 | 27.4 ± 4.3 |
| Gender (M/F)NS | 9/6 | 8/7 |
| Probing depth (mm)* | 1.96 ± 0.6 | 4.62 ± 0.81 |
| Clinical Attachment level (mm)* | 1.04 ± 0.5 | 4.53 ± 1.23 |
| Percentage of sites with | ||
| Plaque accumulation* | 34.5 ± 9.9 | 74.1 ± 10.4 |
| Gingival bleeding* | 4.5 ± 2.9 | 24.2 ± 10.9 |
| Bleeding on probing* | 6.1 ± 3.2 | 74.1 ± 15.7 |
| Suppuration* | 0.0 ± 0.0 | 4.5 ± 6.2 |
| PD≤3 mm* | 100 | 37.3 ± 20.1 |
| PD 4-6 mm* | 0 | 39.9 ± 28.2 |
| PD≥7 mm* | 0 | 22.8 ± 15.7 |
PH, periodontal health; GAgP, generalized aggressive periodontitis;
p<0.05;
Mann-Whitney test; NS, not significant (p>0.05); PD, probing depth; SD, standard deviation.
Microbiological findings
The mean estimated ‘counts’ (×105) of the 10 species evaluated in the subgingival plaque samples from the PH and GAgP groups are shown in Table 3. Subjects with GAgP showed significantly higher mean ‘counts’ of P. gingivalis (p<0.001), S. sputigena (p<0.001) and Selenomonas sp. CS002 (OT 131) (p<0.01). Selenomonas sp. EW084 (OT 146) was detected only in the GAgP group (n= 4 subjects). Throughout this manuscript, OT or oral taxon designations for uncultivated/unrecognized taxa are provided in accord with the Human Oral Microbiome Database (13, HOMD; www.homd.org) when available. GenBank assession numbers can also be found in the HOMD website.
Table 3.
Mean estimated ‘counts’ (×105 ± SD) of 10 bacterial taxa in subgingival biofilm samples obtained from subjects with generalized aggressive periodontitis and subjects with periodontal health.
| Species/phylotypes | Experimental groups
|
*p-value | |
|---|---|---|---|
| PH (n=15) | GAgP (n=15) | ||
| S. sputigena | 0.3 ± 0.5 | 4.5 ± 2.9 | 0.001 |
| S. noxia | 0.2 ± 0.4 | 1.2 ± 2.0 | NS |
| Selenomonas sp. EW084 | 0 | 0.1 ± 0.4 | NS |
| Selenomonas sp. EW076 | 0.05 ± 0.9 | 0.5 ± 1,0 | NS |
| Selenomonas sp. CS002 | 0.01 ±0.02 | 0.12 ± 0.09 | 0.01 |
| A. actinomycetemcomitans | 0.06 ± 0.01 | 0.2 ± 0.4 | NS |
| P. gingivalis | 0.1 ± 0.3 | 7.8 ± 3.9 | 0.001 |
| A. gerencseriae | 1.7 ± 0.6 | 0.3 ± 0.4 | 0.001 |
| S. anginosus/gordonii | 1.3 ± 1,0 | 1.4 ± 1.8 | NS |
| S. sanguinis | 1.3 ± 1.5 | 0.3 ± 0.01 | 0.01 |
p<0.05;
Mann-Whitney test; PH, periodontal health; GAgP, generalized aggressive periodontitis; NS, not significant; PD, probing depth; SD, standard deviation.
Levels of A. actinomycetemcomitans were more elevated in GAgP subjects than in PH, however the difference between the groups did not reach statistical significance (p>0.05). Mean ‘counts’ of Actinomyces gerencseriae and Streptococcus sanguinis were significantly higher in subjects with PH (p<0.01).
The mean estimated ‘counts’ of the bacterial species detected in shallow and deep periodontal pockets (PD ≤3mm and ≥5mm) in subjects with GAgP are shown in Table 4. P. gingivalis and S. sputigena were detected in significantly higher mean ‘counts’ in deep pockets than in shallow sites. In addition, both shallow and deep sites in subjects with GAgP harbored higher mean ‘counts’ of these two pathogens, compared with the shallow sites of PH subjects (Figure 1).
Table 4.
Mean estimated ‘counts’ (×105 ± SD) of 10 bacterial taxa in subgingival biofilm samples obtained baseline PD ≤3mm and PD ≥5mm at subjects with generalized aggressive periodontitis.
| Species/phylotypes | GAgP
|
*p-value | |
|---|---|---|---|
| PD ≤3mm | PD ≥5mm | ||
| S. sputigena | 1.7 ± 1.7 | 6.7 ± 5.5 | 0.027 |
| S. noxia | 0.6 ± 0.9 | 1.3 ± 1.1 | NS |
| S. oral clone EW084 | 0.01 ± 0.01 | 0.3 ± 0.8 | NS |
| S. oral clone EW076 | 0.04 ± 0.09 | 1.2 ± 2.9 | NS |
| S.oral clone CS002 | 0.08 ± 0.01 | 0.02 ± 0.05 | NS |
| A. actinomycetemcomitans | 0.01 ± 0.04 | 0.3 ± 0.8 | NS |
| P. gingivalis | 2.4 ± 2.7 | 12.2 ± 6.1 | 0.001 |
| A. gerencseriae | 0.2 ± 0.5 | 0.5 ± 0.8 | NS |
| S. anginosus/gordonii | 1.4 ± 2.8 | 1.4 ± 1.8 | NS |
| S. sanguinis | 0.5 ± 0.09 | 0.1±0.02 | NS |
p<0.05;
Wilcoxon test; GAgP, generalized aggressive periodontitis; NS, not significant; PD, probing depth; SD, standard deviation.
Figure 1.
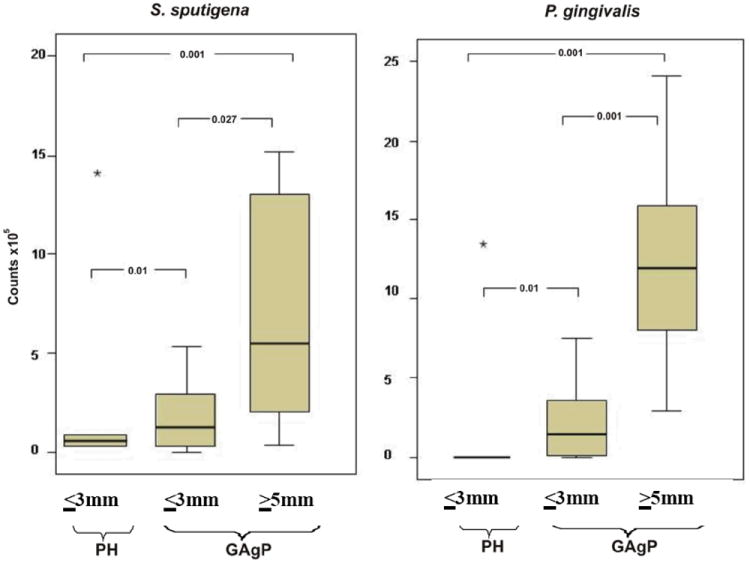
Mean estimated ‘counts’ (×105) of S. sputigena and P.gingivalis in the PH subjects and in sites with PD ≤3mm and PD ≥4mm in the GAgP group. Boxplots show the median, minimum, and maximum values. The significance of differeces between groups was assessed using Mann-Whitney and Wilcoxon test. The star (*) represent extreme outliers. PH, periodontal health; GAgP, generalized aggressive periodontitis;
The percentage of sites colonized by the 10 species/phylotypes evaluated in subgingival plaque samples taken from subjects with GAgP and PH are presented in Table 5. Subjects with GAgP showed higher mean percentage of sites colonized by S. sputigena (p<0.005), Selenomonas sp. CS002 (OT 131) (p<0.041) and P. gingivalis (p<0.001) in comparison with those with PH. No differences in the prevalence of subjects colonized by all the species analyzed were observed between groups, except for P. gingivalis, which was more prevalent in subjects with GAgP (data not show, p<0.05). Spearman correlations revealed positive correlations between PD and mean ‘counts’ of P. gingivalis (r=0.77, p=0.0001), S. sputigena (r=0.60, p< 0.009) and Selenomonas sp. EW076 (OT 139) (r= 0.42, p=0.031).
Table 5.
Percentage of sites colonized by 10 bacterial taxa at levels ≥ 105 in subgingival plaque samples taken from subjects with generalized aggressive periodontitis and subjects with periodontal health.
| Species/phylotypes | Experimental groups
|
*p-value | |
|---|---|---|---|
| PH (n=15) | GAgP (n=15) | ||
| S. sputigena | 15.4 ± 18.9 | 51.4± 32.7 | 0.005 |
| S. noxia | 12.4± 19.4 | 27.8 ± 28.4 | NS |
| Selenomonas sp. EW084 | 0 | 7.3 ± 10.7 | NS |
| Selenomonas sp. EW076 | 2.9 ± 8.0 | 15.4 ± 13.7 | NS |
| Selenomonas sp. CS002 | 6.1 ± 5.7 | 22.1 ± 11.9 | 0.041 |
| A. actinomycetemcomitans | 3.6 ± 6.7 | 13.9 ± 16.8 | NS |
| P. gingivalis | 4.4 ± 8.1 | 65.5 ± 30.9 | 0.001 |
| A. gerencseriae | 38.2 ± 29.6 | 22.9 ± 15.0 | NS |
| S. anginosus/gordonii | 41.8 ± 30.4 | 27.8 ± 28.4 | NS |
| S. sanguinis | 27.9 ± 29.2 | 24.9 ± 26.1 | NS |
p<0.05;
Mann-Whitney test; PH, periodontal health; GAgP, generalized aggressive periodontitis; NS, not significant; PD, probing depth; SD, standard deviation.
DISCUSSION
Previous investigations using the 16S rRNA cloning and sequencing strategy had shown that species of Selenomonas are present in greater proportions in the subgingival biofilm samples of subjects with chronic (6, 16, 18, 22, 23) and aggressive periodontitis (Faveri et al, 2008). Recent studies have reported contrasting findings regarding the potential role of Selenomonas species in periodontitis. Whilst a high levels of Selenomonas noxia, in association with other bacterial species, was observed in a subset of young periodontitis patients (23), the use of a specific oligonucleotide probe targeting the majority of all oral Selenomonas spp. indicated that members of this genus were ubiquitous both in periodontitis resistant and diseased patients (24). Therefore, the association between species of Selenomonas and periodontal disease remains unclear. Thus, the main goal of the present study was to determine the levels of selected Selenomonas species, including uncultivated/unrecognized phylotypes in subgingival biofilm samples from subjects with GAgP and PH individuals.
The present study used the RNA-oligonucleotide quantification technique (ROQT) (20) to determine the levels of the test taxa in subgingival biofilm samples from GAgP and PH subjects. The use of ROQT can overcome some of the limitations presented by other molecular biology techniques, including checkerboard DNA-DNA hybridization, real time PCR and 16S rRNA cloning analysis. None of these techniques has the ability to quantify the levels of multiple uncultivated species in large numbers of individual biofilm samples simultaneously. ROQT is a high-throughput method for bacterial enumeration in clinical biofilm samples. In addition, it does not require sample pooling, amplification or dilution, because an entire individual sample is laid onto the membrane. Therefore, introduction of biases in the microbial profiles of the ecosystem under study due to PCR-associated procedures can be eliminated (16, 25, 26).
In agreement with previous studies that used other microbiological techniques such as culture techniques (4, 27, 28, 29), PCR (30,31), checkerboard DNA-DNA hybridization (5, 7, 11, 32) the data from the present study by ROQT support the notion that P. gingivalis plays an important role in the etiology of GAgP, as well as, A. gerensceriae and S. sanguinis were elevated in the healthy in comparison with periodontally diseased subjects.
The results of the present study reinforce the association of S. sputigena with the etiology of GAgP. This species was found in significantly higher levels (Table 3) and prevalence (% of sites colonized) in subjects with GAgP than PH group (Table 5; p<0.01), as well as in higher levels in deep sites compared with shallow sites in GAgP subjects (Table 4). S. sputigena is a Gram-negative, multiflagellated, motile, anaerobic rod (33). An early study showed that periodontitis patients showed high antibodies titers against S. sputigena (34). Later, investigations demonstrated that this species was also associated with necrotizing ulcerative periodontitis (35), aggressive periodontitis (27), active periodontitis lesions (36, 37) and GAgP (15). Studies, using cloning and sequencing, as well as culture, also identified this species as a highly prevalent (38) and dominant member of the periodontal pocket microbiota of ChP subjects (28, 29, 39, 40).
Despite its possible association with disease, the components and products of S. sputigena involved in the colonization of the oral cavity and the mechanisms of induce tissue destruction are unknown. This species presented a bone resorption activity in experimentally colonized rats (41). It is noteworthy that Lipopolysaccharide/lipid A of S. sputigena differs from that generally found in other Gram-negative bacteria, which consisted mainly of fatty acids such as undecanoic, trideanoic, tridecenoic, 3-hydroxytridecanoic and 3-hydroxytetradecanoic acid. In addition, Lipid A from S. sputigena is able to induce IL-1α and IL-6 in murine macrophages (42), suggesting its role in inducing inflammation. However, there is no evidence that Lipid A from S. sputigena leads to a different inflammatory phenotype compared with other Gram-negative bacterial species.
Members of the genus Selenomonas were frequently detected as part of the not-yet-cultivated species of the oral cavity (6, 17, 22, 43). The present analysis of uncultivated/unrecognized Selenomonas phylotypes revealed that these taxa are frequently part of the subgingival microbiota of GAgP patients. Selenomonas sp. EW084 (OT 146) was only detected in GAgP subjects, Selenomonas sp. CS002 (OT 131) was more prevalent and present in higher levels in GAgP than in periodontal healthy subjects. This species has also been associated with other anaerobic sites of the oral cavity such as deep dentin caries (44). In addition, the levels of Selenomonas sp. EW076 were positively correlated with the increase of PD, which is in accord with the findings reported by Kumar et al (17). Drescher et al. (24) analyzed the topography of the subgingival biofilm by fluorescence in situ hybridization (FISH) and electron microscopy in subjects with GAgP, ChP and periodontitis-resistant subject and revealed that Selenomonas spp. appeared in large numbers in all parts of the collected biofilms and seemed to make a relevant contribution to their structural organization. The authors reported that it is hard to imagine that a group of organisms constituting an important part of the biomass does not contribute to the pathogenetic process of periodontal disease.
As a whole, the data indicated that the evaluated Selenomonas species/phylotypes are heterogeneous in their ability to colonize oral sites, and cannot be seen as a single taxonomic unit. Although their role in inducing periodontal destruction cannot be drawn unless longitudinal studies are performed and their phenotypic traits are evaluated, the data indicated that S. sputigena and Selenomonas sp. CS002 (OT 131) are part of the subgingival microbiota associated with aggressive periodontitis and may play a role in the disease onset and progression.
CONCLUSION
S. sputigena and Selenomonas sp. CS002 (OT 131) may be associated with the pathogenesis of GAgP, and therefore, their role in the onset and progression of this infection merits further investigation.
Acknowledgments
This study was supported by Research Grants 2005/59443-2 and 2009/12358-1 from Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP, Brazil) and in part by NIH/NIDCR grant R03-DE-021742 (F.T.) and the Eleanor and Miles Shore Fellowship Program for Scholars in Medicine (The Forsyth Institute/Harvard Medical School) (F.T.). We want to thank Dr. Bruce Paster (The Forsyth Institute/Harvard Medical School) for his assistance with the oligonucleotide sequences employed in the present study.
Footnotes
We declare that there are no conflicts of interests for any author in the present paper.
References
- 1.Albandar JM, Brown LJ, Löe H. Putative periodontal pathogens in subgingival plaque of young adults with and without early-onset periodontitis. J Periodontol. 1997;68:973–981. doi: 10.1902/jop.1997.68.10.973. [DOI] [PubMed] [Google Scholar]
- 2.Haraszthy VI, Hariharan G, Tinoco EM, Lally ET, Davis E, Zambon JJ. Evidence for the role of highly leukotoxic Actinobacillus actinomycetemcomitans in the pathogenesis of localized juvenile and other forms of early-onset periodontitis. J Periodontol. 2000;71:912–922. doi: 10.1902/jop.2000.71.6.912. [DOI] [PubMed] [Google Scholar]
- 3.Kamma JJ, Nakou M, Baehni PC. Clinical and microbiological characteristics of smokers with early onset periodontitis. J Periodontal Res. 1999;34:25–33. doi: 10.1111/j.1600-0765.1999.tb02218.x. [DOI] [PubMed] [Google Scholar]
- 4.Moore WE, Holdeman LV, Cato EP, Smibert RM, Burmeister JA, Palcanis KG, Ranney RR. Comparative bacteriology of juvenile periodontitis. Infect Immun. 1985;48:507–519. doi: 10.1128/iai.48.2.507-519.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 6.Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, Sahasrabudhe A, Dewhirst FE. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001;183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Socransky SS, Haffajee AD. Periodontal microbial ecology. Periodontol 2000. 2005;38:135–187. doi: 10.1111/j.1600-0757.2005.00107.x. [DOI] [PubMed] [Google Scholar]
- 8.Ledder RG, Gilbert P, Huws SA, Aarons L, Ashley MP, Hull PS, McBain AJ. Molecular analysis of the subgingival microbiota in health and disease. Appl Environ Microbiol. 2007;73:516–523. doi: 10.1128/AEM.01419-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.American Academy of Periodontology. Consensus report. Periodontal diseases: pathogenesis and microbial factors. Ann Periodontol. 1996;1:926–932. doi: 10.1902/annals.1996.1.1.926. [DOI] [PubMed] [Google Scholar]
- 10.Armitage GC. Development of a classification system for periodontal diseases and conditions. Ann Periodontol. 1999;4:1–6. doi: 10.1902/annals.1999.4.1.1. [DOI] [PubMed] [Google Scholar]
- 11.Faveri M, Figueiredo LC, Duarte PM, Mestnik MJ, Mayer MP, Feres M. Microbiological profile of untreated subjects with localized aggressive periodontitis. J Clin Periodontol. 2009;36:739–749. doi: 10.1111/j.1600-051X.2009.01449.x. [DOI] [PubMed] [Google Scholar]
- 12.Paster BJ, Dewhirst FE. Molecular microbial diagnosis. Periodontol 2000. 2009;51:38–344. doi: 10.1111/j.1600-0757.2009.00316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen T, Yu WH, Izard J, Baranova OV, Lakshmanan A, Dewhirst FE. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford) 2010;6:baq013. doi: 10.1093/database/baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. The human oral microbiome. J Bacteriol. 2010;192:5002. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faveri M, Mayer MP, Feres M, de Figueiredo LC, Dewhirst FE, Paster BJ. Microbiological diversity of generalized aggressive periodontitis by 16S rRNA clonal analysis. Oral Microbiol Immunol. 2008;23:112–118. doi: 10.1111/j.1399-302X.2007.00397.x. [DOI] [PubMed] [Google Scholar]
- 16.de Lillo A, Ashley FP, Palmer RM, Munson MA, Kyriacou L, Weightman AJ, Wade WG. Novel subgingival bacterial phylotypes detected using multiple universal polymerase chain reaction primer sets. Oral Microbiol Immunol. 2006;21:61–68. doi: 10.1111/j.1399-302X.2005.00255.x. [DOI] [PubMed] [Google Scholar]
- 17.Kumar PS, Griffen AL, Moeschberger ML, Leys EJ. Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J Clin Microbiol. 2005;43:3944–3955. doi: 10.1128/JCM.43.8.3944-3955.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olson JC, Cuff CF, Lukomski S, Lukomska E, Canizales Y, Wu B, Crout RJ, Thomas JG, McNeil DW, Weyant RJ, Marazita ML, Paster BJ, Elliott T. Use of 16S ribosomal RNA gene analyses to characterize the bacterial signature associated with poor oral health in West Virginia. BMC Oral Health. 2011;1:11–17. doi: 10.1186/1472-6831-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haffajee AD, Socransky SS. Microbiology of periodontal diseases: introduction. Periodontol 2000. 2005;38:9–12. doi: 10.1111/j.1600-0757.2005.00112.x. [DOI] [PubMed] [Google Scholar]
- 20.Teles FR, Teles RP, Siegelin Y, Paster B, Haffajee AD, Socransky SS. RNA-oligonucleotide quantification technique (ROQT) for the enumeration of uncultivated bacterial species in subgingival biofilms. Mol Oral Microbiol. 2011;26:127–139. doi: 10.1111/j.2041-1014.2010.00603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colombo AP, Boches SK, Cotton SL, Goodson JM, Kent R, Haffajee AD, Socransky SS, Hasturk H, Van Dyke TE, Dewhirst F, Paster BJ. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J Periodontol. 2009;80:1421–1432. doi: 10.1902/jop.2009.090185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar PS, Brooker MR, Dowd SE, Camerlengo T. Target region selection is a critical determinant of community fingerprints generated by 16S pyrosequencing. PLoS One. 2011;6:e20956. doi: 10.1371/journal.pone.0020956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.López R, Dahlén G, Baelum V. Subgingival microbial consortia and the clinical features of periodontitis in adolescents. Eur J Oral Sci. 2011;19:455–462. doi: 10.1111/j.1600-0722.2011.00875.x. [DOI] [PubMed] [Google Scholar]
- 24.Drescher J, Schlafer S, Schaudinn C, Riep B, Neumann K, Friedmann A, Petrich A, Göbel UB, Moter A. Molecular epidemiology and spatial distribution of Selenomonas spp. in subgingival biofilms. Eur J Oral Sci. 2010;118:466–474. doi: 10.1111/j.1600-0722.2010.00765.x. [DOI] [PubMed] [Google Scholar]
- 25.Polz MF, Cavanaugh CM. Bias in template-to-product ratios in multitemplate PCR. Appl Environ Microbiol. 1998;64:3724–3730. doi: 10.1128/aem.64.10.3724-3730.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suzuki MT, Giovannoni SJ. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamma JJ, Nakou M, Manti FA. Predominant microflora of severe, moderate and minimal periodontal lesions in young adults with rapidly progressive periodontitis. J Periodontal Res. 1995;30:66–72. doi: 10.1111/j.1600-0765.1995.tb01254.x. [DOI] [PubMed] [Google Scholar]
- 28.Moore WE, Holdeman LV, Smibert RM, Hash DE, Burmeister JA, Ranney RR. Bacteriology of severe periodontitis in young adult humans. Infect Immun. 1992;38:1137–1148. doi: 10.1128/iai.38.3.1137-1148.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dzink JL, Socransky SS, Haffajee AD. The predominant cultivable microbiota of active and inactive lesions of destructive periodontal diseases. J Clin Periodontol. 1988;15:316–323. doi: 10.1111/j.1600-051x.1988.tb01590.x. [DOI] [PubMed] [Google Scholar]
- 30.Cortelli JR, Cortelli SC, Jordan S, Haraszthy VI, Zambon JJ. Prevalence of periodontal pathogens in Brazilians with aggressive or chronic periodontitis. J Clin Periodontol. 2005;32:860–866. doi: 10.1111/j.1600-051X.2005.00777.x. [DOI] [PubMed] [Google Scholar]
- 31.Darby IB, Hodge PJ, Riggio MP, Kinane DF. Microbial comparison of smoker and non-smoker adult and early-onset periodontitis patients by polymerase chain reaction. J Clin Periodontol. 2000;27:417–424. doi: 10.1034/j.1600-051x.2000.027006417.x. [DOI] [PubMed] [Google Scholar]
- 32.Teles RP, Gursky LC, Faveri M, Rosa EA, Teles FR, Feres M, Socransky SS, Haffajee AD. Relationships between subgingival microbiota and GCF biomarkers in generalized aggressive periodontitis. J Clin Periodontol. 2010;37:313–323. doi: 10.1111/j.1600-051X.2010.01534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kolenbrander PE, Andersen RN, Moore LV. Coaggregation of Fusobacterium nucleatum, Selenomonas flueggei, Selenomonas infelix, Selenomonas noxia, and Selenomonas sputigena with strains from 11 genera of oral bacteria. Infect Immun. 1989;57:3194–3203. doi: 10.1128/iai.57.10.3194-3203.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Socransky SS, Tanner AC, Goodson JM, Haffajee AD, Walker CB, Ebersole JL, Sornberger GC. An approach to the definition of periodontal disease syndromes by cluster analysis. J Clin Periodontol. 1982;9:460–471. doi: 10.1111/j.1600-051x.1982.tb02107.x. [DOI] [PubMed] [Google Scholar]
- 35.Gmür R, Wyss C, Xue Y, Thurnheer T, Guggenheim B. Gingival crevice microbiota from Chinese patients with gingivitis or necrotizing ulcerative gingivitis. Eur J Oral Sci. 2004;112:33–41. doi: 10.1111/j.0909-8836.2004.00103.x. [DOI] [PubMed] [Google Scholar]
- 36.Haffajee AD, Socransky SS, Ebersole JL, Smith DJ. Clinical, microbiological and immunological features associated with the treatment of active periodontosis lesions. J Clin Periodontol. 1984;11:600–618. doi: 10.1111/j.1600-051x.1984.tb00913.x. [DOI] [PubMed] [Google Scholar]
- 37.Tanner A, Maiden MF, Macuch PJ, Murray LL, Kent RL., Jr Microbiota of health, gingivitis, and initial periodontitis. J Clin Periodontol. 1998;25:85–98. doi: 10.1111/j.1600-051x.1998.tb02414.x. [DOI] [PubMed] [Google Scholar]
- 38.Kumar PS, Griffen AL, Barton JA, Paster BJ, Moeschberger ML, Leys EJ. New bacterial species associated with chronic periodontitis. J Dent Res. 2003;82:338–344. doi: 10.1177/154405910308200503. [DOI] [PubMed] [Google Scholar]
- 39.Moore WE, Holdeman LV, Cato EP, Good IJ, Smith EP, Ranney RR, Palcanis KG. Variation in periodontal floras. Infect Immun. 1984;46:720–726. doi: 10.1128/iai.46.3.720-726.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dzink JL, Tanner AC, Haffajee AD, Socransky SS. Gram negative species associated with active destructive periodontal lesions. J Clin Periodontol. 1985;12:648–659. doi: 10.1111/j.1600-051x.1985.tb00936.x. [DOI] [PubMed] [Google Scholar]
- 41.Socransky SS. Microbiology of periodontal disease present status and future considerations. J Periodontol. 1977;48:497–504. doi: 10.1902/jop.1977.48.9.497. [DOI] [PubMed] [Google Scholar]
- 42.Kumada H, Watanabe K, Nakamu A, Haishima Y, Kondo S, Hisatsune K, Umemoto T. Chemical and biological properties of lipopolysaccharide from Selenomonas sputigena ATCC 33150. Oral Microbiol Immunol. 1997;12:162–167. doi: 10.1111/j.1399-302x.1997.tb00373.x. [DOI] [PubMed] [Google Scholar]
- 43.Li CL, Liang JP, Jiang YT. Association of uncultivated oral phylotypes AU126 and X112 with periodontitis. Oral Dis. 2006;12:371–374. doi: 10.1111/j.1601-0825.2005.01205.x. [DOI] [PubMed] [Google Scholar]
- 44.Preza D, Olsen I, Aas JA, Willumsen T, Grinde B, Paster BJ. Bacterial profiles of root caries in elderly patients. J Clin Microbiol. 2008;46:2015–2021. doi: 10.1128/JCM.02411-07. [DOI] [PMC free article] [PubMed] [Google Scholar]


