Abstract
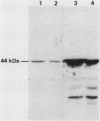
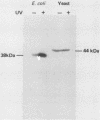
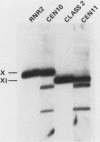
Ribonucleotide reductase catalyzes the first step in the pathway for the production of deoxyribonucleotides needed for DNA synthesis. The gene encoding the small subunit of ribonucleotide reductase was isolated from a Saccharomyces cerevisiae genomic DNA expression library in lambda gt11 by a fortuitous cross-reaction with anti-RecA antibodies. The cross-reaction was due to an identity between the last four amino acids of each protein. The gene has been named RNR2 and is centromere linked on chromosome X. The nucleotide sequence was determined, and the deduced amino acid sequence, 399 amino acids, shows extensive homology with other eucaryotic ribonucleotide reductases. Transplason mutagenesis was used to disrupt the RNR2 gene. A novel assay using colony color sectoring was developed to demonstrate visually that RNR2 is essential for mitotic viability. RNR2 encodes a 1.5-kilobase mRNA whose levels increase 18-fold after treatment with the DNA-damaging agent 4-nitroquinoline 1-oxide. CDC8 was also found to be inducible by DNA damage, but POL1 and URA3 were not inducible by 4-nitroquinoline 1-oxide. The expression of these genes defines a new mode of regulation for enzymes involved in DNA biosynthesis and sharpens our picture of the events leading to DNA repair in eucaryotic cells.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson S. Shotgun DNA sequencing using cloned DNase I-generated fragments. Nucleic Acids Res. 1981 Jul 10;9(13):3015–3027. doi: 10.1093/nar/9.13.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angulo J. F., Schwencke J., Moreau P. L., Moustacchi E., Devoret R. A yeast protein analogous to Escherichia coli RecA protein whose cellular level is enhanced after UV irradiation. Mol Gen Genet. 1985;201(1):20–24. doi: 10.1007/BF00397980. [DOI] [PubMed] [Google Scholar]
- Barker D. G., White J. H., Johnston L. H. The nucleotide sequence of the DNA ligase gene (CDC9) from Saccharomyces cerevisiae: a gene which is cell-cycle regulated and induced in response to DNA damage. Nucleic Acids Res. 1985 Dec 9;13(23):8323–8337. doi: 10.1093/nar/13.23.8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Botstein D., Falco S. C., Stewart S. E., Brennan M., Scherer S., Stinchcomb D. T., Struhl K., Davis R. W. Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene. 1979 Dec;8(1):17–24. doi: 10.1016/0378-1119(79)90004-0. [DOI] [PubMed] [Google Scholar]
- Burnette W. N. "Western blotting": electrophoretic transfer of proteins from sodium dodecyl sulfate--polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981 Apr;112(2):195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- Carle G. F., Olson M. V. An electrophoretic karyotype for yeast. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3756–3760. doi: 10.1073/pnas.82.11.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carle G. F., Olson M. V. Separation of chromosomal DNA molecules from yeast by orthogonal-field-alternation gel electrophoresis. Nucleic Acids Res. 1984 Jul 25;12(14):5647–5664. doi: 10.1093/nar/12.14.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dente L., Cesareni G., Cortese R. pEMBL: a new family of single stranded plasmids. Nucleic Acids Res. 1983 Mar 25;11(6):1645–1655. doi: 10.1093/nar/11.6.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström Y., Eriksson S., Jildevik I., Skog S., Thelander L., Tribukait B. Cell cycle-dependent expression of mammalian ribonucleotide reductase. Differential regulation of the two subunits. J Biol Chem. 1985 Aug 5;260(16):9114–9116. [PubMed] [Google Scholar]
- Eriksson S., Gräslund A., Skog S., Thelander L., Tribukait B. Cell cycle-dependent regulation of mammalian ribonucleotide reductase. The S phase-correlated increase in subunit M2 is regulated by de novo protein synthesis. J Biol Chem. 1984 Oct 10;259(19):11695–11700. [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Hieter P., Mann C., Snyder M., Davis R. W. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell. 1985 Feb;40(2):381–392. doi: 10.1016/0092-8674(85)90152-7. [DOI] [PubMed] [Google Scholar]
- Hieter P., Pridmore D., Hegemann J. H., Thomas M., Davis R. W., Philippsen P. Functional selection and analysis of yeast centromeric DNA. Cell. 1985 Oct;42(3):913–921. doi: 10.1016/0092-8674(85)90287-9. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L. M., Snyder M., Chang L. M., Davis R. W., Campbell J. L. Isolation of the gene encoding yeast DNA polymerase I. Cell. 1985 Nov;43(1):369–377. doi: 10.1016/0092-8674(85)90042-x. [DOI] [PubMed] [Google Scholar]
- Lammers M., Follmann H. Deoxyribonucleotide biosynthesis in yeast (Saccharomyces cerevisiae). A ribonucleotide reductase system of sufficient activity for DNA synthesis. Eur J Biochem. 1984 Apr 16;140(2):281–287. doi: 10.1111/j.1432-1033.1984.tb08099.x. [DOI] [PubMed] [Google Scholar]
- Larsson A., Sjöberg B. M. Identification of the stable free radical tyrosine residue in ribonucleotide reductase. EMBO J. 1986 Aug;5(8):2037–2040. doi: 10.1002/j.1460-2075.1986.tb04461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowdon M., Vitols E. Ribonucleotide reductase activity during the cell cycle of Saccharomyces cerevisiae. Arch Biochem Biophys. 1973 Sep;158(1):177–184. doi: 10.1016/0003-9861(73)90611-5. [DOI] [PubMed] [Google Scholar]
- Ludwig W., Follmann H. The specificity of ribonucleoside triphosphate reductase. Multiple induced activity changes and implications for deoxyribonucleotide formation. Eur J Biochem. 1978 Jan 16;82(2):393–403. doi: 10.1111/j.1432-1033.1978.tb12034.x. [DOI] [PubMed] [Google Scholar]
- McIntosh E. M., Haynes R. H. Sequence and expression of the dCMP deaminase gene (DCD1) of Saccharomyces cerevisiae. Mol Cell Biol. 1986 May;6(5):1711–1721. doi: 10.1128/mcb.6.5.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson T. A., Prakash L., Prakash S., Osley M. A., Reed S. I. Regulation of CDC9, the Saccharomyces cerevisiae gene that encodes DNA ligase. Mol Cell Biol. 1985 Jan;5(1):226–235. doi: 10.1128/mcb.5.1.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson G. W., Nicolet C. M., Kalainov D., Friedberg E. C. A yeast excision-repair gene is inducible by DNA damaging agents. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1842–1846. doi: 10.1073/pnas.83.6.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby S. W., Szostak J. W. Specific Saccharomyces cerevisiae genes are expressed in response to DNA-damaging agents. Mol Cell Biol. 1985 Jan;5(1):75–84. doi: 10.1128/mcb.5.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D. C., Cantor C. R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984 May;37(1):67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- Sjöberg B. M., Hahne S., Mathews C. Z., Mathews C. K., Rand K. N., Gait M. J. The bacteriophage T4 gene for the small subunit of ribonucleotide reductase contains an intron. EMBO J. 1986 Aug;5(8):2031–2036. doi: 10.1002/j.1460-2075.1986.tb04460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. E., Fisher P. A. Identification, developmental regulation, and response to heat shock of two antigenically related forms of a major nuclear envelope protein in Drosophila embryos: application of an improved method for affinity purification of antibodies using polypeptides immobilized on nitrocellulose blots. J Cell Biol. 1984 Jul;99(1 Pt 1):20–28. doi: 10.1083/jcb.99.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M., Elledge S., Davis R. W. Rapid mapping of antigenic coding regions and constructing insertion mutations in yeast genes by mini-Tn10 "transplason" mutagenesis. Proc Natl Acad Sci U S A. 1986 Feb;83(3):730–734. doi: 10.1073/pnas.83.3.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Standart N. M., Bray S. J., George E. L., Hunt T., Ruderman J. V. The small subunit of ribonucleotide reductase is encoded by one of the most abundant translationally regulated maternal RNAs in clam and sea urchin eggs. J Cell Biol. 1985 Jun;100(6):1968–1976. doi: 10.1083/jcb.100.6.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storms R. K., Ord R. W., Greenwood M. T., Mirdamadi B., Chu F. K., Belfort M. Cell cycle-dependent expression of thymidylate synthase in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Dec;4(12):2858–2864. doi: 10.1128/mcb.4.12.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelander L., Berg P. Isolation and characterization of expressible cDNA clones encoding the M1 and M2 subunits of mouse ribonucleotide reductase. Mol Cell Biol. 1986 Oct;6(10):3433–3442. doi: 10.1128/mcb.6.10.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelander L., Eriksson S., Akerman M. Ribonucleotide reductase from calf thymus. Separation of the enzyme into two nonidentical subunits, proteins M1 and M2. J Biol Chem. 1980 Aug 10;255(15):7426–7432. [PubMed] [Google Scholar]
- Thelander M., Gräslund A., Thelander L. Subunit M2 of mammalian ribonucleotide reductase. Characterization of a homogeneous protein isolated from M2-overproducing mouse cells. J Biol Chem. 1985 Mar 10;260(5):2737–2741. [PubMed] [Google Scholar]
- Wehland J., Schröder H. C., Weber K. Amino acid sequence requirements in the epitope recognized by the alpha-tubulin-specific rat monoclonal antibody YL 1/2. EMBO J. 1984 Jun;3(6):1295–1300. doi: 10.1002/j.1460-2075.1984.tb01965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J. H., Barker D. G., Nurse P., Johnston L. H. Periodic transcription as a means of regulating gene expression during the cell cycle: contrasting modes of expression of DNA ligase genes in budding and fission yeast. EMBO J. 1986 Jul;5(7):1705–1709. doi: 10.1002/j.1460-2075.1986.tb04414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis D. K., Uhlin B. E., Amini K. S., Clark A. J. Physical mapping of the srl recA region of Escherichia coli: analysis of Tn10 generated insertions and deletions. Mol Gen Genet. 1981;183(3):497–504. doi: 10.1007/BF00268771. [DOI] [PubMed] [Google Scholar]