Abstract
The mechanisms by which environmental influences lead to the development of complex neurode-generative diseases are largely unknown. It is known, however, that epigenetic mechanisms can mediate alterations in transcription due to environmental influences. In order to identify genes susceptible to regulation in the adult cortex by one type of epigenetic mechanism, histone, and protein acetylation, we treated mice with the histone deacetylase inhibitor Trichostatin A (TSA). After 1 week of treatment with TSA, RNA was extracted from the brain cortices of mice and gene expression differences were analyzed by microarray profiling. The altered genes were then compared with genes differentially expressed in microarray studies of disease by database and literature searches. Genes regulated by TSA were found to significantly overlap with differentially expressed genes in the Alzheimer’s disease (AD) brain. Several TSA-regulated genes involved in chromatin remodeling and epigenetic reprogramming including histone cluster 1, H4 h (Hist1H4 h), methionine adenosyltransferase II, alpha (Mat2a), and 5-methyltetrahydrofolate homocysteine reductase (Mtrr) overlapped with genes altered in early-stage AD in gray matter. We also show that the expression of hemoglobin, which has been shown to be altered in neurons in the AD brain, is regulated by TSA treatment. This analysis suggests involvement of epigenetic mechanisms in neurons in early stages of AD.
Keywords: Acetylation, Histone deacetylase inhibitors, Trichostatin A, Epigenetics, Alzheimer’s disease, Expression microarray
Introduction
Many neurodegenerative diseases including AD are complex diseases in that, except in rare familial forms of disease, the development of disease does not follow Mendelian rules of inheritance. Susceptibility is conferred by a combination of genetic and environmental influences, and twin concordance rates suggest a strong environmental component exists for the development of AD (Gatz et al. 2006; Brickell et al. 2007). While susceptibility alleles have been identified in familial and sporadic forms of AD (Goate et al. 1991; Corder et al. 1993; Selkoe 1996; Delacourte et al. 2002; Blacker et al. 2003), the environmental factors involved in disease development and progression have not been clearly defined. Differences in environmental exposure between individuals can lead to differences in gene expression through epigenetic mechanisms (Szyf et al. 2008). Epigenetic modifications allow for the modulation of gene expression without any change to the primary DNA sequence and provide the basis for differences in gene expression even in identical twins. These modifications can be mitotically and sometimes meiotically heritable and include chemical modifications to histones, methylation of the cytosine bases of DNA, and the synthesis of non-coding RNA (Mehler 2008).
During the development of the nervous system, epigenetic silencing orchestrated by the neuron restrictive silencing factor, alters chromatin structure, and shuts down transcription of non-neuronal genes in neurons (Ballas and Mandel 2005). The maintenance of these epigenetic modifications provides a basis for cell memory. Even in postmitotic cells, however, some genes continue to be regulated by modifications of chromatin structure due to environmental or physiological signals. Reports have shown that histone modifications are involved in the immune response and that inflammatory signals can modulate the repression of genes to allow for wound healing (Shaw and Martin 2009). Furthermore, epigenetic mechanisms can alter transcription in response to external environmental influences such as malnutrition and exposure to toxins as well as environmental stresses within a cell such as oxidative damage (van Vliet et al. 2007; Ryu et al. 2003; Cyr and Domann 2011). One type of epigenetic modification that can modulate transcription in response to an oxidative environment is histone acetylation (Shahbazian and Grunstein 2007). The balance between histone acetylation and deacetylation is maintained by two types of enzymes, histone acetyltransferases (HATs) and histone deacetylases (HDACs). Acetylation of lysines on the N-terminal tail of histones by HATs eliminates the positive charge on the lysine residue and therefore reduces the affinity of the histone for DNA (Gregory et al. 2001). As a result, the chromatin is in a more open state allowing transcriptional activators and coactivators access to promoter regions, thereby enhancing transcription. While histone hyperacetylation activates transcription in general, histone hypoacetylation tends to silence gene transcription. The balance of histone and protein acetylation can be tipped toward increased acetylation in an oxidative environment due to increased protein tyrosine nitration and other redox modifications to HDACs, which lead to decreased deacetylase activity (Ito et al. 2004; Doyle and Fitzpatrick 2010).
While evidence exists for an epigenetic basis in disease susceptibility and progression in complex neurodegenerative diseases such as AD (Chouliaras et al. 2010; Mastroeni et al. 2011), not much is known concerning the genes that are susceptible to regulation by epigenetic modifications including histone and protein acetylation in the adult CNS.Inthe present study, we identify genes and transcriptional pathways susceptible to regulation by an imbalance in acetylation in the cortex by treating mice with the histone deacetylase inhibitor TSA followed by expression microarray profiling.
Methods and materials
Treatment of mice with TSA and microarray profiling
Four-week-old, male C57Bl/6 mice were treated with either TSA (7.5 mg/kg/day) or vehicle (9:1 PBS to DMSO) for 1 week via daily intraperitoneal injections. After 1 week of treatment, mice were killed by cervical dislocation, and brain cortices were harvested and pooled into groups of three mice each and placed on ice. Total RNA was immediately extracted from harvested tissue using RNeasy Lipid Tissue Mini Kit (Qiagen, Maryland, USA) according to the manufacturer’s instructions and stored at −80 °C until used for hybridization to gene arrays. RNA quality was determined by formaldehyde agarose gel electrophoresis to visualize intact 28S and 18S RNA. Total RNA (100 mg) was converted to cDNA, labeled, and fragmented using an Affymetrix 3′ IVT Express Kit per manufacturer’s instructions (Affymetrix, Santa Clara, CA, USA). cDNA was fragmented and hybridized overnight at 42 °C to Affymetrix Mouse Genome 430A 2.0 arrays, which contains oligonucleotide probe sets representing 14,000 well-characterized genes. After hybridization, arrays were washed in a Genechip Fluidics Station and scanned on a GeneChip Scanner 3000 (both from Affymetrix) to generate cell intensity files (CEL files). After normalization and background correction by the Robust MultiChip Analysis (RMA) method with Affymetrix Expression Console software, statistical analysis of resulting CHP files was performed with Partek Genomics Suite 6.5 (Partek, St. Louis, MO, USA). A one-way ANOVA was performed, and genes were considered significantly changed if they had a fold change greater than ±1.2 and a p value less than 0.05.
In order to explore overlaps with disease states, human homologues of the changed genes were first determined using NetAffyx (www.affymetrix.com). Of the 544 probe sets differentially expressed in the mouse cortex due to TSA treatment, 369 human homologues were identified. Overlaps were computed for the human homologues with Gene Set Enrichment Analysis software (Subramanian et al. 2005) for gene sets in the C2 collection of the Molecular Signature Database (http://www.broadinstitute.org/gsea/MSigDB/index.jsp). This software determines significance of overlaps by a hypergeometric distribution test (p < 0.05 was considered significant), which corresponds to a one-tailed Fisher’s exact test. Literature searches of microarray results were also performed and significant genes regulated by TSA were compared for overlaps in previously published microarray studies analyzing postmortem AD brain tissue (Colangelo et al. 2002; Yao et al. 2003; Ricciarelli et al. 2004; Dunckley et al. 2006; Weeraratna et al. 2007; Liang et al. 2008; Williams et al. 2009; Qin et al. 2009; Tan et al. 2010; Bossers et al. 2010; Blalock et al. 2011; Ginsberg et al. 2012).
Protein extraction and western blotting for acetylated proteins
After brain cortices were extracted, the rest of the brain was then pooled and used to prepare nuclear protein fractions. The fractions were run on 4–12 % Bis–Tris polyacrylamide gels (Invitrogen, Carlsbad, CA), transferred to nitrocellulose, and were probed with either acetyllysine (1:1,000, Upstate Biotechnology, Lake Placid, NY, USA) or lamin B1 (1:200, Novus Biologicals, Littleton, CO, USA) primary antibodies. After primary antibody hybridization, blots were hybridized with either mouse or rabbit secondary antibodies (1:10,000 each, Santa Cruz Biotechnology, Santa Cruz, CA, USA) and developed using luminol reagent (Santa Cruz Biotechnology).
Quantitative RT-PCR to confirm differential gene expression
Quantitative RT-PCR (qRT-PCR) was performed in order to confirm changes in gene expression obtained by microarray profiling. qRT-PCR was performed on RNA isolated from the cortex of TSA-treated mice and control mice (vehicle-treated) from three groups (3 TSA treated and 3 control brains were pooled in each group). Statistical significance was determined by a Student’s t test from at least two separate experiments performed in triplicate. Primers were designed to target hemoglobin β chain (Hbb1 and Hbb2), S100a8, Mat2a, and Ung transcripts. To eliminate amplification from any contaminating genomic DNA, primers were designed to span an intron or RNA was treated with Turbo DNA-free (Invitrogen, Carlsbad, CA, USA). Beta-actin (Applied Biosystems, Carlsbad, CA, USA) was used as a control to normalize Ct values. qRT-PCR was performed per manufacturer’s specifications using Brilliant III Ultra-Fast SYBR Green qRT-PCR Master Mix and 2–10 nanograms of total RNA. Primers for Hbb1 were 5′ CCTGGGCA GGCTGCTGGTTG3′ and 5′ TGGCAAAGGTGCCCTTG AGGC 3′, for Hbb2, 5′ TGGGTAATCCCAAGGTGA AGGCCC 3′ and 5′ GCCCAGCACAATCACGATCGCA 3′, for S100a8, 5′ TCGAGGAGTTCCTTGCGATGGTG 3′ and 5′ GGACCCAGCCCTAGGCCA. GAA3′, for Mat2a, 5′ AACGGGCAGCTCAACGGCTTC 3′ and 5′ TGAAGGT GTGCATCA. AGGACAGCA 3′, and for Ung, 5′ CAGCAG TTTGGTGCCTGCTGC 3′ and 5′ AACCCAGGGAGAAC CCCGCC 3′.
Results
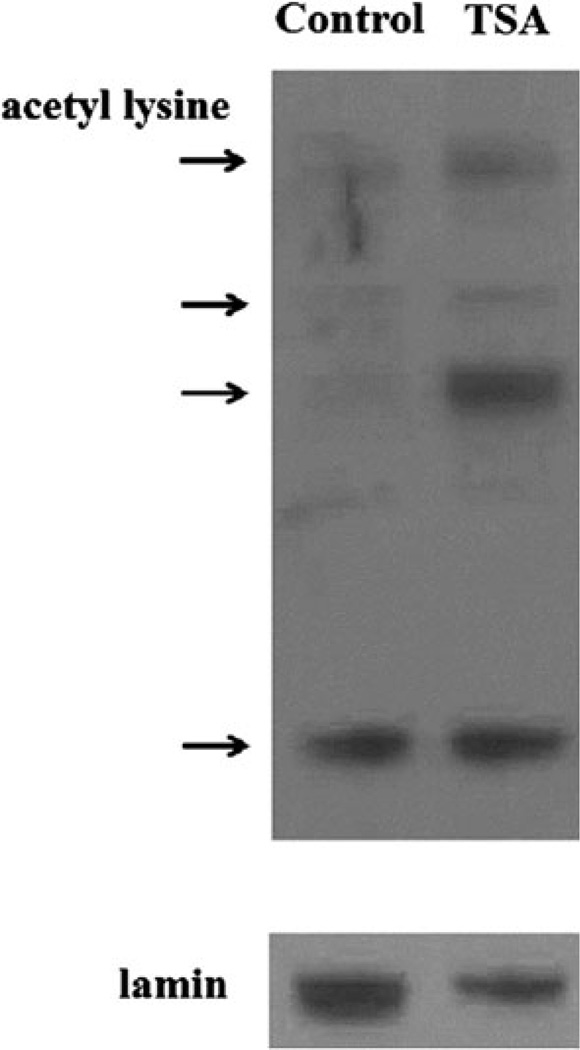
In order to evaluate the impact of TSA on acetylation patterns in the brain, we performed western blots with protein isolated from the brains of TSA- and vehicle-treated mice with an antibody to acetyl-lysine as shown in Fig. 1. We observed that mice treated with TSA for 1 week experienced increases in general protein acetylation in nuclear fractions in the brain. To more specifically identify targets of acetylation, we performed expression microarray profiling of total RNA isolated from the cortex of TSA- and vehicle-treated mice. After statistical analysis of the microarray data by a one-way ANOVA (Partek Genomics Suite Software 6.5), 369 genes with human homologues were found to be differentially expressed between TSA-and vehicle-treated mice with a fold change >±1.2 and p <0.05 (Supplementary Table 1). These genes were then analyzed with Gene Set Enrichment Analysis software to compute overlaps with previously published genes sets in the Molecular Signatures Database (http://www.broad.mit.edu/gsea/MSigDB/index.jsp).
Fig. 1.
Representative western blot showing increased acetyl-lysine immunoreactive proteins (denoted by arrows) in nuclear extracts isolated from the brains of TSA-treated compared to control (vehicle-treated) mice
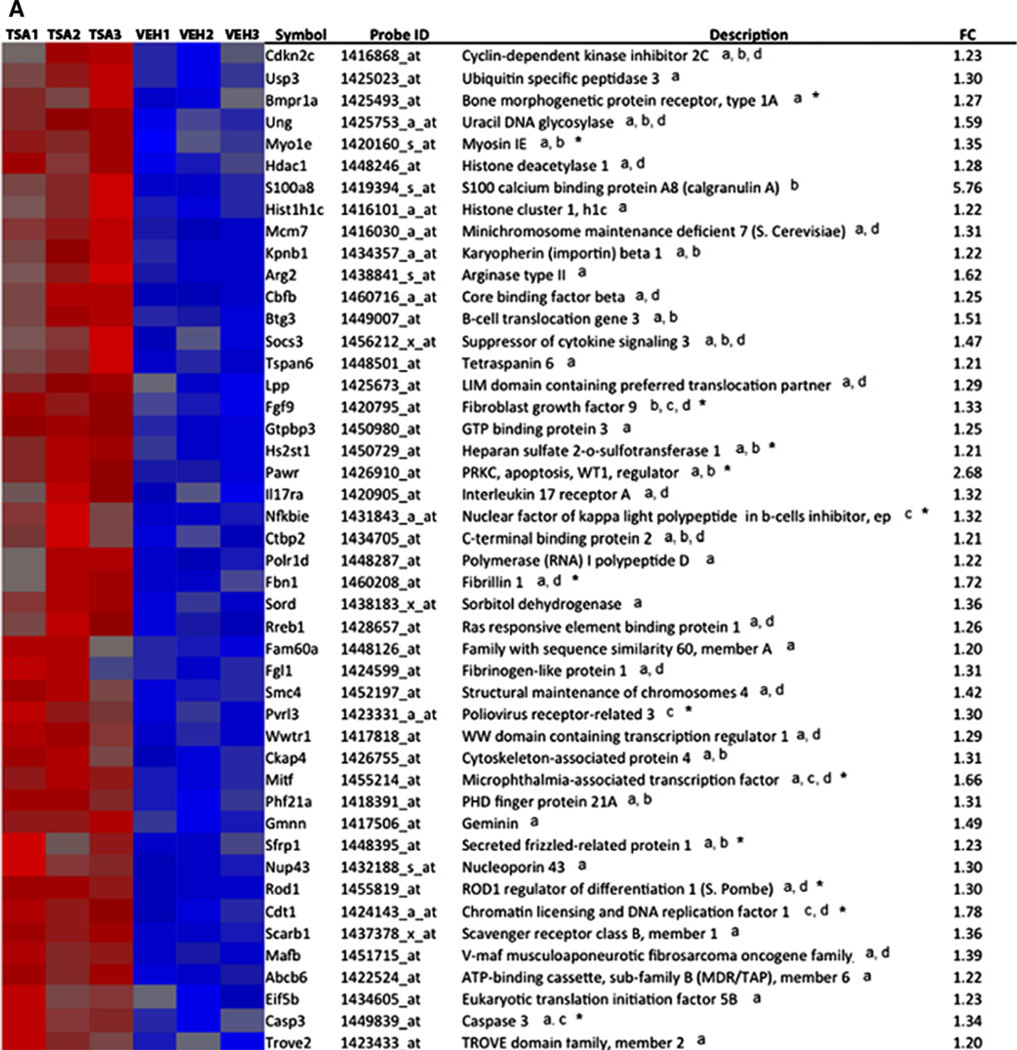
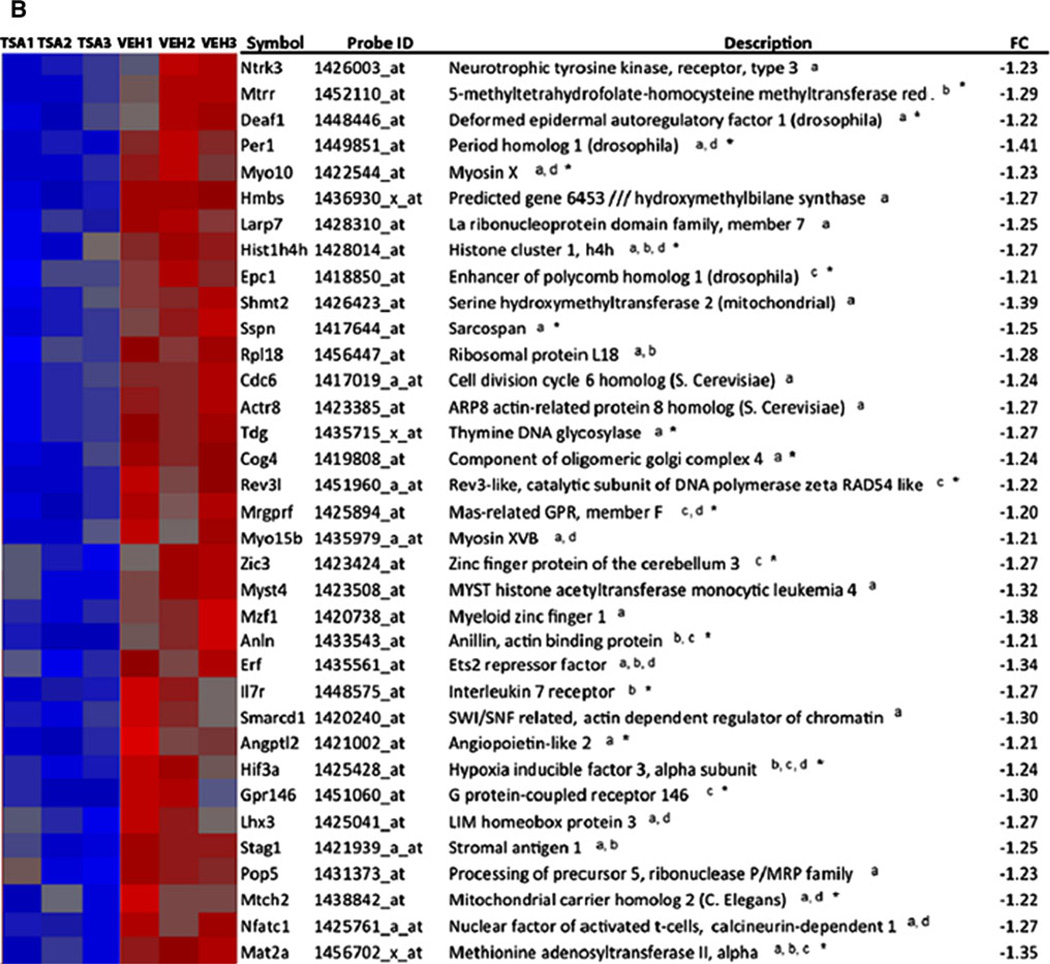
Interrogation of the Molecular Signatures Database with our TSA-regulated genes, we found significant overlap (p < 0.05) with studies which profiled gene expression in cancer (Lindgren et al. 2006), during the cell cycle (Ben-Porath et al. 2008), and in early-stage postmortem AD brain tissue from hippocampus (Blalock et al. 2004) shown in Table 1. Gene set enrichment analysis identified 65 genes of 369 TSA-regulated genes that overlapped with genes altered in AD. Overlap was also found in other microarray studies analyzing early-stage AD cortex and laser-captured hippocampal gray matter through literature searches (Bossers et al. 2010; Blalock et al., 2011). These studies analyzed early-stage disease defined by Braak stage (Braak and Braak 1991) or MiniMental Status Exam and neurofibrillary tangle density, respectively. Relative microarray expression levels, gene symbols, and gene descriptions for a total of 81 genes in our TSA microarrays found to overlap with genes altered in the AD brain in studies which analyzed early-stage disease are shown in Fig. 2. Thirty-two of the 81 genes were altered in early-stage disease denoted by an asterisk in Fig. 2, and 23 were altered in laser-dissected gray matter denoted by overlap in the Blalock et al. (2011) study. Another study examining gene expression in neocortex also showed overlap in 26 of the 81 genes shown in Fig. 2 (Tan et al. 2010), while studies that analyzed synaptoneurosomes and only later-stage disease did not show significant overlap with TSA-regulated genes (Colangelo et al. 2002; Yao et al. 2003; Ricciarelli et al. 2004; Dunckley et al. 2006; Weeraratna et al. 2007; Liang et al. 2008; Qin et al. 2009; Williams et al. 2009; Ginsberg et al. 2012).
Table 1.
Gene set enrichment analysis
| Gene set | # of genes in gene set | Description | # of genes in overlap | p value |
|---|---|---|---|---|
| BENPORATH_CYCLING_GENES | 648 | Genes showing cell cycle stage-specific expression |
39 | 5.71 E–12 |
| LINDGREN_BLADDER_CANCER_CLUSTER_3_UP | 325 | Genes whose expression profile is specific to cluster III of urothelial cell carcinoma (UCC) tumors |
25 | 2.96 E–10 |
| BLALOCK_ALZHEIMERS_DISEASE_UP | 1,720 | Genes upregulated in brain from patients with Alzheimer’s disease |
65 | 4.62 E–10 |
Gene set enrichment analysis was performed on previously published microarray studies in the Molecular Signatures Database (www.broadinstitute.org/gsea/MSigDB/index.jsp). The top three gene sets, ordered by significance of p values, overlapping with TSA-regulated genes include cell cycle genes (Ben-Porath et al. 2008), cancer genes (Lindgren et al. 2006), and AD genes (Blalock et al. 2004)
Fig. 2.
TSA-regulated genes overlap with genes differentially expressed in AD. Overlaps were identified by a combination of gene set enrichment analysis and literature searches. Heat maps show relative gene expression levels for TSA-regulated genes by expression microarray profiling. Red denotes increased expression and blue denotes decreased expression of genes in TSA-treated mice compared to controls (vehicle-treated, veh.). a Heat map of genes upregulated by TSA, which overlap with genes altered in AD. b Heat map of genes downregulated by TSA treatment, which overlap with genes altered in AD. aOverlap identified by gene set enrichment analysis (Blalock et al. 2004, which analyzed early and later stages of AD), boverlap in gray matter (Blalock et al. 2011, which analyzed early-stage disease in laser-captured hippocampal gray matter), coverlap with Bossers et al. (2010), which analyzed cortical gene expression in early-stage AD, doverlap with Tan et al. (2010), which analyzed neocortex. *TSA-regulated genes that overlap in early-stage AD. FC indicates fold change, TSA treated relative to control
Microarray expression levels, gene symbols, and descriptions for genes involved in epigenetic reprogramming which were differentially expressed (p < 0.05) by TSA treatment are shown in Supplemental Figure 1. This group includes 26 genes involved in chromatin remodeling, DNA repair, and methionine metabolism. Of particular interest are three genes, Mat2a, Mtrr, and Hist1H4h, which are regulated by TSA treatment, altered early in disease in gray matter in AD, and are involved in epigenetic reprogramming.
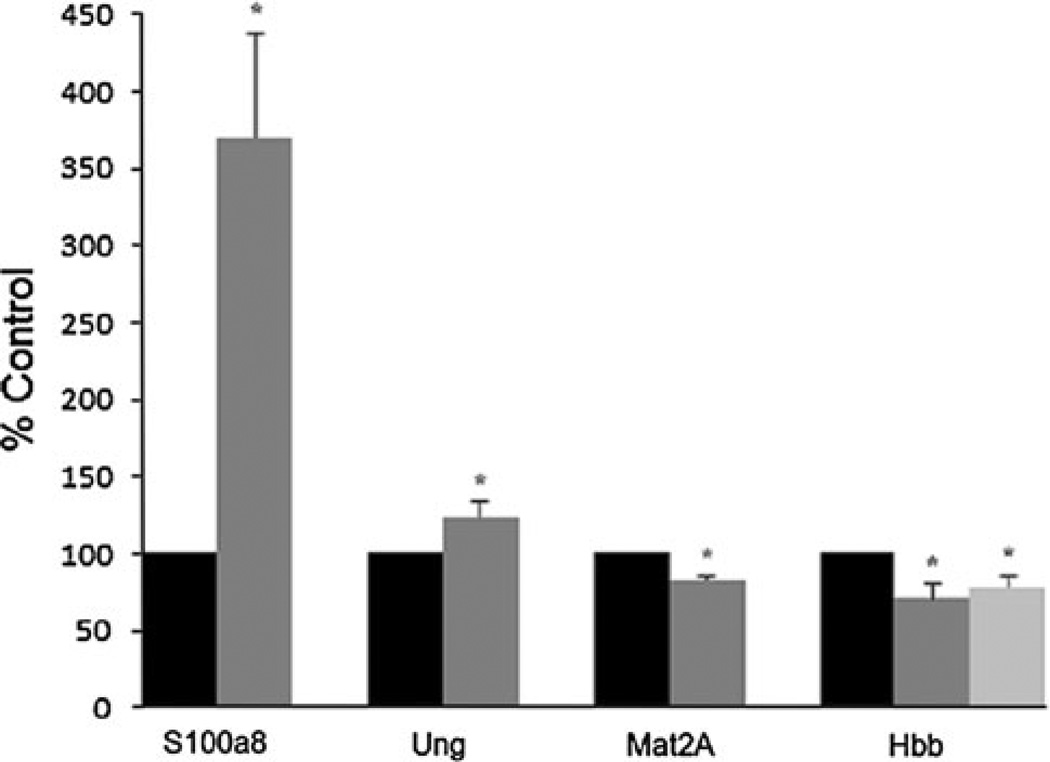
In order to confirm that the microarray expression level differences of down to 1.2-fold we detected were reliable, we confirmed differential gene expression in our TSA-treated mouse brains by qRT-PCR. We selected several genes for confirmation, which were altered at different levels by microarray from over fivefold down to 1.3-fold and overlapped in laser-captured gray matter in AD. These included S100a8 (upregulated 5.76-fold in TSA-treated mice by microarray), uracil DNA glycosylase (Ung) (upregulated 1.59-fold by microarray), and Mat2a (downregulated in TSA-treated mice 1.35-fold by microarray). By qRT-PCR, the expression of S100a8 was increased 3.70-fold in TSA-treated mice, Ung was increased by 1.29-fold, and Mat2a was decreased by 1.21-fold (p < 0.05 for all three genes) as shown in Fig. 3. Also of interest, we have observed that hemoglobin β is expressed in neurons (Broadwater et al. 2011) and its expression has been shown to be regulated by histone acetylation (Fathallah et al. 2008). While we did not detect differential expression of hemoglobin β in our microarray analysis (−1.3-fold, p value of 0.11), we did detect a significant decrease in the expression of hemoglobin β chain (Hbb1 and Hbb2) transcripts (−1.40-fold for Hbb1, −1.28-fold for Hbb2, p <0.05) in mRNA isolated from the cortex of TSA-treated mice compared to control mice by qRT-PCR as shown in Fig. 3.
Fig. 3.
Quantitative RT-PCR confirms altered expression of genes found to be differentially expressed due to TSA treatment by microarray profiling. By qRT-PCR of RNA isolated from the cortex of TSA-treated and control mice, the expression of S100a8 was increased 3.70-fold, Ung was increased 1.29-fold, Mat2a was decreased 1.21-fold, and the hemoglobin β1 and β2 transcripts (Hbb1 and Hbb2) were decreased by 1.40- and 1.28-fold, respectively. Black bars represent gene expression levels from control mice, and gray bars represent gene expression levels from TSA-treated mice. For Hbb, the dark gray bar represents Hbb1 expression and the lighter gray bar represents Hbb2 expression. Gene expression levels in TSA-treated mice are shown as percent of control from at least two experiments performed in triplicate. Error bars represent SEM, *p < 0.05
Discussion
We created an imbalance in histone acetylation in mice by treatment with the HDAC inhibitor TSA and then analyzed gene expression in the cortex by microarray profiling with the goal of identifying genes susceptible to epigenetic regulation in the adult brain. Our analysis identified 369 genes susceptible to regulation by acetylation, which could therefore potentially be modulated by environmental influences and contribute to the development of complex disease. Our data show both increases and decreases in transcription in response to HDAC inhibition in mouse cortex. This is consistent with the fact that increased acetylation in the brain can alter gene transcription by several mechanisms. HDAC inhibition can directly alter transcription through histone acetylation at specific sites leading to a more open chromatin conformation, which would lead to upregulation of gene expression. HDAC inhibition can also result in increased acetylation of transcriptional regulators, thereby altering their activity that has been demonstrated for transcription factors including Sp1 and HNF4a (Ryu et al. 2003; Yang et al. 2009). Indirect effects of HDAC inhibition can alter transcription through upregulation of transcription factors, coactivators, or repressors. It is most likely that a combination of these mechanisms is involved in the transcriptional response observed.
TSA is an HDAC inhibitor, which will lead to increased acetylation of histones and a more open chromatin structure so it is not surprising that we detected overlap between TSA-regulated genes and genes involved in chromatin remodeling including histone deacetylase 1 (HDAC1), SWI/SNF-related, actin-dependent regulator of chromatin(Smarcd1), structural maintenance of chromosomes 4 (Smc4), and MYST histone acetyltransferase 4 (Myst4) as shown in Supplementary Figure 1. We also found overlap with genes involved in DNA repair such as Ung and thymine DNA glycosylase (Tdg) and genes involved in methionine or one carbon metabolism including Mat2A and Mtrr. Both Mat2A and Mtrr are involved in reactions that generate S-adenosyl methionine (SAM), which is the methyl donor for most methylation reactions in cells including cytosine-guanine dinucleotide (CpG) methylation. The fact that we see overlap between DNA repair and methionine metabolism genes and genes regulated by TSA treatment suggests that these genes together with genes involved in chromatin structure are a part of an overall program involved in epigenetic reprogramming.
The top two gene groups from the Molecular Signatures Database that overlapped with genes regulated by TSA in our gene set enrichment analysis were cancer and cell cycle genes (Ben-Porath et al. 2008; Lindgren et al. 2006) (Table 1). The third gene set that overlapped with our TSA microarray study was a microarray analysis of postmortem hippocampus in early-stage and late-stage AD brain (Blalock et al. 2004). We expected to see overlap with cancer and cell cycle gene sets due to previous studies, which have shown that treatment with HDAC inhibitors is beneficial in cancer, inhibiting proliferation, preventing angiogenesis, and inducing cell death pathways (Lane and Chabner 2009). Previous studies have implicated acetylation-mediated mechanisms in neurodegenerative disease as well and support our data showing overlap in AD. These include studies showing that HDAC inhibition in mouse models of neurodegenerative disease, including Huntington’s disease mice and the experimental autoimmune encephalomyelitis mouse model of multiple sclerosis, can provide neuroprotection (Ferrante et al. 2003; Gardian et al. 2005; Camelo et al. 2005). Studies have also reported that HDAC inhibition in AD mouse models reverses memory impairment (Green et al. 2008; Ricobaraza et al. 2009). An imbalance in histone acetylation in AD would be expected to lead to alterations in transcription and in fact, microarray profiling studies that have analyzed postmortem AD brain tissue have shown that transcription is altered in AD and that cortical areas are particularly vulnerable (Wang et al. 2010). Our study has identified a subset of genes altered in AD, shown in Fig. 2, which are susceptible to regulation by an imbalance in acetylation.
Interestingly, another study has shown that resveratrol, which activates the sirtuin class III HDACs, has been shown to slow the development of AD pathology (Anekonda and Reddy 2006). This study also suggests an imbalance in acetylation in AD. The balance between histone and protein acetylation and deacetylation may be altered in neurodegenerative diseases such as AD as a result of mitochondrial dysfunction and increased reactive oxygen species (ROS), which has been reported in the AD brain (Smith et al. 1997; Hirai et al. 2001; Moreira et al. 2008). AD is an age-related form of dementia marked by decreased neuronal and synaptic activity and cognitive impairment (Selkoe 2004). The central AD pathologies include extracellular plaques resulting from aberrant processing of amyloid precursor protein (APP) into beta-amyloid and intracellular neurofibrillary tangles containing aggregated tau protein. In the AD brain, microglia are activated by contact with fibrillar beta-amyloid in extracellular plaques. This contact activates microglial NADPH oxidase, which generates damaging ROS (Qin et al. 2006; Wilkinson and Landreth 2006). In addition, activated microglia release nitric oxide, which can react with NADPH-generated ROS to create highly reactive peroxynitrite, which damages cells by oxidizing proteins and lipids and can lead to neuronal apoptosis (Combs et al. 2001). This oxidative environment can influence both acetylation and methylation reactions in cells (Cyr and Domann 2011), leading to epigenetic reprogramming and altered transcription. In support of this notion, Drake et al. (2004) demonstrated a link between mitochondrial dysfunction, increased ROS, and altered histone modifications. In this study, 4-hydroxynonenol, which is an end product of lipid peroxidation, has been shown to be increased in the AD brain and to bind to histones. This interaction has been suggested to alter transcription by interfering with the ability of histones to bind to DNA in the AD brain. Other studies have shown a direct involvement of proteins involved in AD pathology including tau and beta-amyloid in mediating altered acetylation of histones H3 and H4, and β-tubulin (Perez et al. 2009; Lithner et al. 2009). Studies have now also directly linked alterations in acetylation of histones to gene expression involved in learning and memory and cognitive function (Stilling and Fischer 2011). Peleg et al. (2010) have reported that acetylation of histone H4 is altered in aged mice and leads to deficits in memory consolidation. Another study has provided evidence of a link between histone H4 acetylation and cognitive dysfunction more specifically related to AD (Francis et al. 2009). This study showed that levels of histone H4 acetylation were decreased in the APP/PS1 mouse model of AD compared to wild-type littermates after fear conditioning training, which is a test of associative memory. TSA treatment of the APP/PS1 mice prior to fear conditioning prevented histone H4 deacetylation and restored contextual fear behavior and hippocampal long-term potentiation.
In the present study, HDAC inhibition in mice with TSA resulted in altered expression of 81 genes, which overlapped with differentially expressed genes in the AD brain. It is important to note that in AD, the direction of the alteration of the 81 overlapping genes is not always the same as in the TSA-treated mice. For example, of the 65 genes that overlapped in the Blalock et al. (2004) study identified by gene set enrichment analysis, all 65 genes were upregulated in AD, while in TSA-treated mice, the expression of 41 of these genes was increased and the expression of 24 was decreased. These results may explain contradictory reports with regard to HDAC activity and HDAC inhibition in the literature. In some studies, increased histone and protein acetylation or inhibition of HDACs through oxidative modifications has been implicated in inflammatory processes and in AD pathology, suggesting that treatment with HDAC inhibitors would mimic the disease and be deleterious (Ito et al. 2004; Lithner et al. 2009; Doyle and Fitzpatrick 2010). However, evidence from studies in mouse models of AD and neurodegenerative disease shows that HDAC inhibitors reverse memory impairment and provide neuroprotection (Ferrante et al. 2003; Gardian et al. 2005; Camelo et al. 2005; Green et al. 2008; Ricobaraza et al. 2009). The finding that HDAC inhibition seems to be involved in disease pathology, on one hand, but that HDAC inhibitors can be therapeutic in neurodegenerative disease mouse models can be explained by our results, which show that for a third of the overlapping genes identified in our study, the direction of transcriptional regulation by TSA was reversed compared to AD. This suggests that in AD, there exists an imbalance in acetylation due to a combination of decreased HDAC activity (or increased HAT activity) for some genes and increased HDAC activity (or decreased HAT activity) for others. These results indicate the necessity for drugs targeted to specific HDACs and HATs in the treatment for complex neurodegenerative diseases such as AD.
A potential candidate HDAC that may provide a more specific therapeutic target in AD is HDAC2. While studies have linked altered acetylation of specific histones, histone H4 in particular, to AD pathology (Francis et al. 2009), other studies point to involvement of specific HDACs including HDAC2 in cognitive impairment and in AD. HDAC2 has been shown to be involved in learning and memory by regulating the expression of genes involved in synaptic plasticity in a study by Guan et al. (2009). In this study, HDAC2 knockout mice display increased acetylation of histone H4 on lysine 12 (H4K12) in the hippocampus and enhanced learning and memory. Another study has reported that knocking down HDAC2 in the hippocampus by short hairpin RNA repairs cognitive function in an AD mouse model (Gräff et al. 2012). Another specific HDAC implicated in AD pathology is HDAC6. A study by Ding et al. (2008) reported increased HDAC6 in AD postmortem brain samples. Rather than altering transcription in the nucleus by deacetylating histones, HDAC6 acts primarily in the cytoplasm where it deacetylates β-tubulin. Aberrant HDAC6 activity has been suggested to contribute to neurodegeneration by decreasing acetylation of β-tubulin resulting in disruption of intracellular transport (Li et al. 2011). While TSA inhibits both HDAC2 and HDAC6, our study was designed to identify alterations in transcription through inhibition of HDACs that act in the nucleus, such as HDAC2, and would not have detected the effects of HDAC6 inhibition.
The fact that 81 genes regulated by TSA overlap in the Blalock et al. (2004, 2011) and Bossers et al. (2010) studies, which analyzed early-stage AD brains, suggests that epigenetic mechanisms may play an early role in the development of AD. Interestingly, these results also suggest that an imbalance in acetylation can lead to alterations in one carbon metabolism and potentially methylation reactions. This is shown by the downregulation of genes involved in methionine or one carbon metabolism, Mat2a and Mtrr, by TSA in our study. Mtrr reduces methionine synthase, which then catalyzes the reaction converting homocysteine to methionine. Mat2a catalyzes the next step in the one carbon cycle and converts methionine to SAM, the methyl donor for most methylation reactions in cells. The fact that these genes overlap with genes altered in early-stage disease in laser-captured gray matter suggests a role for altered DNA or histone methylation in neuronal pathology in early stages of AD. These results also reflect the complexity and crosstalk between acetylation and methylation in chromatin remodeling (Berger 2007). Our data are consistent with other studies, which have shown that alterations in transcription in AD may be mediated by epigenetic modifications involving methylation. It has been reported that the methionine metabolism intermediate S-adenosyl homocysteine (SAH) is increased in postmortem AD brain and inhibits methyltransferases (Kennedy et al. 2004). Other studies have shown that intermediates of methionine metabolism, both SAM and SAH, can modulate the transcription of genes involved in AD pathology. In AD transgenic mice, the level of these intermediates has been manipulated by folate and vitamin B deficiency. These deficiencies were shown to lead to an accumulation of SAH, inhibition of methyltransferases, and upregulation of the expression of presenilin (PS1) and betaamyloid cleaving enzyme (Fuso et al. 2008). In a subsequent study, the expression of PS1 was found to be regulated by increased SAH due to altered levels of methylated DNA in gene regulatory regions (Fuso et al. 2011).
Our data suggest that hemoglobin may be an additional target of epigenetic reprogramming in neurons in AD. Expression of the hemoglobin β and β chains has been reported in neurons in both human and mouse brains (Biagioli et al. 2009; Richter et al., 2009; Schelshorn et al., 2009; Broadwater et al. 2011) where it has been proposed to play a role in neuronal respiration. Hemoglobin expression has been shown to be decreased in the AD brain but the significance of this observation to AD pathology is still not clear (Ferrer et al. 2011). Studies have shown that hemoglobin expression can be regulated by both histone acetylation and CpG methylation of regulatory regions in erythroid cells (Fathallah et al. 2008; Lathrop et al. 2009), but not much is known concerning its regulation in neurons. We have shown that hemoglobin β transcription is susceptible to regulation by TSA treatment in the brain providing evidence for epigenetic mechanisms in the regulation of hemoglobin expression in neurons as well.
This study has identified genes susceptible to regulation by an imbalance in acetylation in the adult CNS. It has provided evidence for epigenetic reprogramming in mediating transcriptional alterations in early stages of AD in gray matter. Future studies will be required to understand the types of environmental influences and mechanisms that initiate these processes in AD.
Supplementary Material
Acknowledgments
This study was funded in part by NIH Grant NS058921 (JM) and by a Grant from the Ohio Board of Regents.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00221-012-3172-y) contains supplementary material, which is available to authorized users.
References
- Anekonda TS, Reddy PH. Neuronal protection by sirtuins in Alzheimer’s disease. J Neurochem. 2006;96:305–313. doi: 10.1111/j.1471-4159.2005.03492.x. [DOI] [PubMed] [Google Scholar]
- Ballas N, Mandel G. The many faces of REST oversee epigenetic programming of neuronal genes. Curr Opin Neurobiol. 2005;15:500–506. doi: 10.1016/j.conb.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Biagioli M, Pinto M, Cesselli D, et al. Unexpected expression of α- and β- globin in mesencephalic dopaminergic neurons and glial cells. Proc Natl Acad Sci USA. 2009;106:15454–15459. doi: 10.1073/pnas.0813216106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacker D, Bertram L, Saunders AJ, et al. Results of a high-resolution genome screen of 437 Alzheimer’s disease families. Hum Mol Genet. 2003;12:23–32. doi: 10.1093/hmg/ddg007. [DOI] [PubMed] [Google Scholar]
- Blalock EM, Geddes JW, Chen KC, et al. Incipient Alzheimer’s disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc Natl Acad Sci USA. 2004;101:2173–2178. doi: 10.1073/pnas.0308512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blalock EM, Buechel HM, Popovic J, Geddes JW, Landfield PW. Microarray analyses of laser-captured hippocampus reveal distinct gray and white matter signatures associated with incipient Alzheimer’s disease. J Chem Neuroanat. 2011;42:118–126. doi: 10.1016/j.jchemneu.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossers K, Wirz KTS, Meerhoff GF, et al. Concerted changes in transcripts in the prefrontal cortex precede neuropathology in Alzheimer’s disease. Brain. 2010;133:3699–3723. doi: 10.1093/brain/awq258. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. (review) [DOI] [PubMed] [Google Scholar]
- Brickell KL, Leverenz JB, Steinbart EJ, et al. Clinicopathological concordance and discordance in three monozygotic twin pairs with familial Alzheimer’s disease. J Neurol Neurosurg Psychiatr. 2007;78:1050–1055. doi: 10.1136/jnnp.2006.113803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadwater L, Pandit A, Azzam S, Clements R, Vadnal J, Yong VW, Freeman EJ, Gregory RB, McDonough J. Analysis of the mitochondrial proteome in multiple sclerosis cortex. BBA Mol Bas Dis. 2011;1812:630–641. doi: 10.1016/j.bbadis.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camelo S, Iglesias AH, Hwang D, et al. Transcriptional therapy with the histone deacetylase inhibitor trichostatin A ameliorates experimental autoimmune encephalomyelitis. J Neuroimmunol. 2005;164:10. doi: 10.1016/j.jneuroim.2005.02.022. [DOI] [PubMed] [Google Scholar]
- Chouliaras L, Rutten BPF, Kenis G, et al. Epigenetic regulation in the pathophysiology of Alzheimer’s disease. Prog Neurobiol. 2010;90:498–510. doi: 10.1016/j.pneurobio.2010.01.002. [DOI] [PubMed] [Google Scholar]
- Colangelo V, Schurr J, Ball MJ, Pelaez RP, Bazan NG, Lukiw WJ. Gene expression profiling of 12633 genes in Alzheimer hippocampal CA1: transcription and neurotrophic factor downregulation and up-regulation of apoptotic and pro-inflammatory signaling. J Neurosci Res. 2002;70:462–473. doi: 10.1002/jnr.10351. [DOI] [PubMed] [Google Scholar]
- Combs CK, Karlo JC, Kao SC, Landreth GE. beta-Amyloid stimulation of microglia and monocytes results in TNFalpha-dependent expression of inducible nitric oxide synthase and neuronal apoptosis. J Neurosci. 2001;21:1179–1188. doi: 10.1523/JNEUROSCI.21-04-01179.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- Cyr AR, Domann FE. The redox basis of epigenetic modifications: from mechanisms to functional consequences. Antioxid Redox Signal. 2011;15:551–589. doi: 10.1089/ars.2010.3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacourte A, Sergeant N, Champain D, et al. Nonoverlapping but synergetic tau and APP pathologies in sporadic Alzheimer’s disease. Neurology. 2002;59:398–407. doi: 10.1212/wnl.59.3.398. [DOI] [PubMed] [Google Scholar]
- Ding H, Dolan PJ, Johnson GV. Histone deacetylase 6 interacts with the microtubule-associated protein tau. J Neurochem. 2008;106:2119–2130. doi: 10.1111/j.1471-4159.2008.05564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle K, Fitzpatrick FA. Redox signaling, alkylation (carbonylation) of conserved cysteines inactivates class I histone deacetylases 1, 2, and 3 and antagonizes their transcriptional repressor function. J Biol Chem. 2010;285:17417–17424. doi: 10.1074/jbc.M109.089250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake J, Petroze R, Castegna A, et al. 4-Hydroxynonenal oxidatively modifies histones: implications for Alzheimer’s disease. Neurosci Lett. 2004;356:155–158. doi: 10.1016/j.neulet.2003.11.047. [DOI] [PubMed] [Google Scholar]
- Dunckley T, Beach TG, Ramsey KE, et al. Gene expression correlates of neurofibrillary tangles in Alzheimer’s disease. Neurobiol Aging. 2006;27:1359–1371. doi: 10.1016/j.neurobiolaging.2005.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fathallah H, Portnoy G, Atweh GF. Epigenetic analysis of the human alpha- and beta-globin gene clusters. Blood Cells Mol Dis. 2008;40:166–173. doi: 10.1016/j.bcmd.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante RJ, Kubilus JK, Lee J, et al. Histone deacetylase inhibition by sodium butyrate chemotherapy ameliorates the neurodegenerative phenotype in Huntington’s Disease mice. J Neurosci. 2003;23:9418–9427. doi: 10.1523/JNEUROSCI.23-28-09418.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer I, Go´mez A, Carmona M, Huesa G, Porta S, Riera-Codina M, Biagioli M, Gustincich S, Aso E. Neuronal hemoglobin is reduced in Alzheimer’s disease, argyrophilic grain disease, Parkinson’s disease, and dementia with Lewy bodies. J Alzheimers Dis. 2011;23:537–550. doi: 10.3233/JAD-2010-101485. [DOI] [PubMed] [Google Scholar]
- Francis YI, Fà M, Ashraf H, Zhang H, Staniszewski A, Latchman DS, Arancio O. Dysregulation of histone acetylation in the APP/PS1 mouse model of Alzheimer’s disease. J Alzheimers Dis. 2009;18:131–139. doi: 10.3233/JAD-2009-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuso A, Nicolia V, Cavallaro RA, Ricceri L, D’Anselmi F, Coluccia P, Calamandrei G, Scarpa S. B-vitamin deprivation induces hyperhomocysteinemia and brain S-adenosylhomocysteine, depletes brain S-adenosylmethionine, and enhances PS1 and BACE expression and amyloid-beta deposition in mice. Mol Cell Neurosci. 2008;37:731–746. doi: 10.1016/j.mcn.2007.12.018. [DOI] [PubMed] [Google Scholar]
- Fuso A, Nicolia V, Pasqualato A, Fiorenza MT, Cavallaro RA, Scarpa S. Changes in Presenilin 1 gene methylation pattern in diet-induced B vitamin deficiency. Neurobiol Aging. 2011;32:187–199. doi: 10.1016/j.neurobiolaging.2009.02.013. [DOI] [PubMed] [Google Scholar]
- Gardian G, Browne SE, Choi DK, et al. Neuroprotective effects of phenylbutyrate in the N171-82Q transgenic mouse model of Huntington’s disease. J Biol Chem. 2005;280:556–563. doi: 10.1074/jbc.M410210200. [DOI] [PubMed] [Google Scholar]
- Gatz M, Reynolds CA, Fratiglioni L, et al. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatr. 2006;63:168–174. doi: 10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- Ginsberg SD, Alldred MJ, Che S. Gene expression levels assessed by CA1 pyramidal neuron and regional hippocampal dissections in Alzheimer’s disease. Neurobiol Dis. 2012;45:99–107. doi: 10.1016/j.nbd.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goate A, Chartier-Harlin MC, Mullan M, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature. 1991;349:704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- Gräff J, Rei D, Guan JS, et al. An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature. 2012;483:222–226. doi: 10.1038/nature10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green KN, Steffan JS, Martinez-Coria H, et al. Nicotinamide restores cognition in Alzheimer’s disease transgenic mice via a mechanism involving sirtuin inhibition and selective reduction of Thr231-phosphotau. J Neurosci. 2008;28:11500–11510. doi: 10.1523/JNEUROSCI.3203-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory PD, Wagner K, Hörz W. Histone acetylation and chromatin remodeling. Exp Cell Res. 2001;265:195–202. doi: 10.1006/excr.2001.5187. [DOI] [PubMed] [Google Scholar]
- Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, Nieland TJ, Zhou Y, Wang X, Mazitschek R, Bradner JE, DePinho RA, Jaenisch R, Tsai LH. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459(7243):55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai K, Aliev G, Nunomura A, et al. Mitochondrial abnormalities in Alzheimer’s disease. J Neurosci. 2001;21:3017–3023. doi: 10.1523/JNEUROSCI.21-09-03017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Hanazawa T, Tomita K, et al. Oxidative stress reduces histone deacetylase 2 activity and enhances IL-8 gene expression: role of tyrosine nitration. Biochem Biophys Res Commun. 2004;315:240–245. doi: 10.1016/j.bbrc.2004.01.046. [DOI] [PubMed] [Google Scholar]
- Kennedy BP, Bottiglieri T, Arning E, Ziegler MG, Hansen LA, Masliah E. Elevated S-adenosylhomocysteine in Alzheimer brain: influence on methyltransferases and cognitive function. J Neural Transm. 2004;111:547–567. doi: 10.1007/s00702-003-0096-5. [DOI] [PubMed] [Google Scholar]
- Lane AA, Chabner BA. Histone deacetylase inhibitors in cancer therapy. J Clin Oncol. 2009;10(27):5459–5468. doi: 10.1200/JCO.2009.22.1291. [DOI] [PubMed] [Google Scholar]
- Lathrop MJ, Hsu M, Richardson CA, Olivier EN, Qiu C, Bouhassira EE, Fiering S, Lowrey CH. Developmentally regulated extended domains of DNA hypomethylation encompass highly transcribed genes of the human beta-globin locus. Exp Hematol. 2009;37:807–813. doi: 10.1016/j.exphem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Jiang H, Chang M, Xie H, Hu L. HDAC6 α-tubulin deacetylase: a potential therapeutic target in neurodegenerative diseases. J Neurol Sci. 2011;304(1–2):1–8. doi: 10.1016/j.jns.2011.02.017. [DOI] [PubMed] [Google Scholar]
- Liang WS, Dunckley T, Beach TG, et al. Altered neuronal gene expression in brain regions differentially affected by Alzheimer’s disease: a reference data set. Physiol Genomics. 2008;33:240–256. doi: 10.1152/physiolgenomics.00242.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren D, Liedberg F, Andersson A, et al. Molecular characterization of early-stage bladder carcinomas by expression profiles, FGFR3 mutation status, and loss of 9q. Oncogene. 2006;25:2685–2696. doi: 10.1038/sj.onc.1209249. [DOI] [PubMed] [Google Scholar]
- Lithner CU, Hernandez CM, Nordberg A, Sweatt JD. Epigenetic changes related to beta-amyloid-implications for Alzheimer’s disease. Alzheimer’s Dementia. 2009;5:304. [Google Scholar]
- Mastroeni D, Grover A, Delvaux E, Whiteside C, Coleman PD, Rogers J. Epigenetic mechanisms in Alzheimer’s disease. Neurobiol Aging. 2011;32:1161–1180. doi: 10.1016/j.neurobiolaging.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehler MF. Epigenetic principles and mechanisms underlying nervous system functions in health and disease. Prog Neurobiol. 2008;86:305. doi: 10.1016/j.pneurobio.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira PI, Santos MS, Oliveira CR, et al. Alzheimer disease and the role of free radicals in the pathogenesis of the disease. CNS Neurol Disord: Drug Targets. 2008;7:3–10. doi: 10.2174/187152708783885156. [DOI] [PubMed] [Google Scholar]
- Peleg S, Sananbenesi F, Zovoilis A, Burkhardt S, Bahari-Javan S, Agis-Balboa RC, Cota P, Wittnam JL, Gogol-Doering A, Opitz L, Salinas-Riester G, Dettenhofer M, Kang H, Farinelli L, Chen W, Fischer A. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328:753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- Perez M, Santa-Maria I, Gomez de Barreda E, et al. Tau-an inhibitor of deacetylase HDAC6 function. J Neurochem. 2009;109:1756–1766. doi: 10.1111/j.1471-4159.2009.06102.x. [DOI] [PubMed] [Google Scholar]
- Qin B, Cartier L, Dubois-Dauphin M, et al. A key role for the microglial NADPH oxidase in APP-dependent killing of neurons. Neurobiol Aging. 2006;27:1577–1587. doi: 10.1016/j.neurobiolaging.2005.09.036. [DOI] [PubMed] [Google Scholar]
- Qin W, Haroutunian V, Katsel P, Cardozo CP, Ho L, Buxbaum JD, Pasinetti GM. PGC-1alpha expression decreases in the Alzheimer disease brain as a function of dementia. Arch Neurol. 2009;66:352–361. doi: 10.1001/archneurol.2008.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricciarelli R, d’Abramo C, Massone S, Marinari U, Pronzato M, Tabaton M. Microarray analysis in Alzheimer’s disease and normal aging. IUBMB Life. 2004;56:349–354. doi: 10.1080/15216540412331286002. [DOI] [PubMed] [Google Scholar]
- Richter F, Meurers BH, Zhu C, Medvedeva VP, Chesselet M-F. Neurons express hemoglobin α- and β-chains in rat and human brains. J Comp Neurol. 2009;515:538–547. doi: 10.1002/cne.22062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricobaraza A, Cuadrado-Tejedor M, Pérez-Mediavilla A, et al. Phenylbutyrate ameliorates cognitive deficit and reduces tau pathology in an Alzheimer’s disease mouse model. Neuropsychopharmacology. 2009;34:1721–1732. doi: 10.1038/npp.2008.229. [DOI] [PubMed] [Google Scholar]
- Ryu H, Lee J, Olofsson BA, et al. Histone deacetylase inhibitors prevent oxidative neuronal death independent of expanded polyglutamine repeats via an Sp1-dependent pathway. Proc Natl Acad Sci. 2003;100:4281–4286. doi: 10.1073/pnas.0737363100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelshorn DW, Schneider A, Kuschinsky AW, Weber D, Kruger C, Dittgen T, Burgers HF, Sabouri F, Gassler N, Bach A, Martin H, Maurer MH. Expression of hemoglobin in rodent neurons. J Cereb Blood Flow Metabol. 2009;29:585–595. doi: 10.1038/jcbfm.2008.152. [DOI] [PubMed] [Google Scholar]
- Selkoe DJ. Amyloid beta-protein and the genetics of Alzheimer’s disease. J Biol Chem. 1996;271:18295–18298. doi: 10.1074/jbc.271.31.18295. [DOI] [PubMed] [Google Scholar]
- Selkoe DJ. Cell biology of protein misfolding: the examples of Alzheimer’s and Parkinson’s diseases. Nat Cell Biol. 2004;6:1054–1061. doi: 10.1038/ncb1104-1054. [DOI] [PubMed] [Google Scholar]
- Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. (review) [DOI] [PubMed] [Google Scholar]
- Shaw T, Martin P. Epigenetic reprogramming during wound healing: loss of polycomb-mediated silencing may enable upregulation of repair genes. EMBO Rep. 2009;10:881–886. doi: 10.1038/embor.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MA, Richey Harris PL, Sayre LM, et al. Widespread peroxynitrite-mediated damage in Alzheimer’s disease. J Neurosci. 1997;17:2653–2657. doi: 10.1523/JNEUROSCI.17-08-02653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stilling RM, Fischer A. The role of histone acetylation in age-associated memory impairment and Alzheimer’s disease. Neurobiol Learn Mem. 2011;96:19–26. doi: 10.1016/j.nlm.2011.04.002. (review) [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyf M, McGowan P, Meaney MJ. The social environment and the epigenome. Environ Mol Mut. 2008;49:46–60. doi: 10.1002/em.20357. [DOI] [PubMed] [Google Scholar]
- Tan MG, Chua W-T, Esiri MM, et al. Genome wide profiling of altered gene expression in the neocortex of Alzheimer’s disease. J Neurosci Res. 2010;88:1157–1169. doi: 10.1002/jnr.22290. [DOI] [PubMed] [Google Scholar]
- van Vliet J, Oates N, Whitelaw E. Epigenetic mechanisms in the context of complex diseases. Cell Mol Life Sci. 2007;64:1531–1538. doi: 10.1007/s00018-007-6526-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Michaelis ML, Michaelis EK. Functional genomics of brain aging and Alzheimer’s disease: focus on selective neuronal vulnerability. Curr Genomics. 2010;11:618–633. doi: 10.2174/138920210793360943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeraratna AT, Kalehua A, Deleon I, et al. Alterations in immunological and neurological gene expression patterns in Alzheimer’s disease tissues. Exp Cell Res. 2007;313:450–461. doi: 10.1016/j.yexcr.2006.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson BL, Landreth GE. The microglial NADPH oxidase complex as a source of oxidative stress in Alzheimer’s disease. J Neuroinflamm. 2006;3:30. doi: 10.1186/1742-2094-3-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C, Mehrian Shai R, Wu Y, et al. Transcriptome analysis of synaptoneurosomes identifies neuroplasticity genes overexpressed in incipient Alzheimer’s disease. PLoS One. 2009;4:e4936. doi: 10.1371/journal.pone.0004936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Kong X, Martins-Santos ME, Aleman G, Chaco E, Liu GE, Wu SY, Samols D, Hakimi P, Chiang CM, Hanson RW. Activation of SIRT1 by resveratrol represses transcription of the gene for the cytosolic form of phosphoenolpyruvate carboxykinase (GTP) by deacetylating hepatic nuclear factor 4alpha. J Biol Chem. 2009;284(40):27042–27053. doi: 10.1074/jbc.M109.047340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao PJ, Zhu M, Pyun EI, Brooks AI, Therianos S, Meyers VE, Coleman PD. Defects in expression of genes related to synaptic vesicle trafficking in frontal cortex of Alzheimer’s disease. Neurobiol Dis. 2003;12:97–109. doi: 10.1016/s0969-9961(02)00009-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.