Abstract
The fruit fly, Drosophila melanogaster, has been used to study genetics, development, and signaling for nearly a century but only over the past few decades has this tremendous resource been the focus of cardiovascular research. Fly genetics offers sophisticated transgenic systems, molecularly-defined genomic deficiencies, genome-wide transgenic RNAi lines, and numerous curated mutants to perform genetic screens. As a genetically-tractable model, the fly facilitates gene discovery and can complement mammalian models of disease. The circulatory system in the fly is comprised of well-defined sets of cardiomyocytes and methodological advances have permitted accurate characterization of cardiac morphology and function. Thus, fly genetics and genomics offers new approaches for gene discovery of adult cardiac phenotypes to identify evolutionarily conserved molecular signals that drive cardiovascular disease.
Keywords: Drosophila, Cardiomyopathy, Genomics
Introduction
“…the small and totally harmless fruit fly, Drosophila. This animal has been extremely cooperative in our hands - and has revealed to us some of its innermost secrets and tricks for developing from a single celled egg to a complex living being of great beauty and harmony.”
Christiane Nüsslein-Volhard, Nobel Banquet Speech, 10 Dec 1995
Advantages of Drosophila as a Model System
Over the past century, the study of Drosophila melanogaster has yielded insight into fundamental concepts that underlie basic biology. The initial work by Morgan, Sturtevant, Bridges, and Muller formed that basis for modern genetics and built a framework that facilitated investigations into the concepts of the gene, mutagenesis, chromosomal structure, the inheritance of complex traits, evolutionary development of organisms, and population genetics.1
Fruit fly genetics offers a set of resources currently unavailable in other models systems that are uniquely advantageous for gene discovery. First, the fly has a short life cycle of ~ 10 days from mating of adult flies to deposition of fertilized eggs, embryo development to first instar larvae that proceed to second and third instar larvae, pupal formation and subsequent eclosion to produce the next generation of adults.2, 3 Adults achieve reproductive maturity within a few hours of emerging from the pupal case and have a lifespan of 60 to 100 days under standard laboratory conditions. Therefore, genetic crosses and the establishment of inbred lines requires considerably less time compared to mammalian models. Second, fly genetics has the unique advantage of balancers chromosomes that contain multiple inversions and suppress meiotic recombination with a corresponding non-rearranged chromosome.4 Recessive, often times lethal, mutations can be maintained in the presence of balancer chromosomes as stable stocks and followed in subsequent genetic crosses. Third, the presence of specific mutations, transgenes, and balancers can be followed by easily observed physical traits. For example, the presence of a transgene is usually accompanied by a mini-white gene cassette that results in an eye color change from white to red.5 Lastly, the Drosophila melanogaster genome is significantly smaller that mammalian genomes thereby decreasing the time and increasing the efficiency of screening. The genome is ~5% the size of the human genome, comprised of five chromosomes (X, Y, 2L/R, 3L/R, and 4) that encode ~125 million base pairs of DNA comprised of ~13,000 predicted gene products (Figure 1)6–8. Although smaller than that of the mouse and human, the fly genome efficiently encodes genes that have multiple spliced isoforms, use different promoter start sites, and genes are sometimes contained within the intronic sequences of other genes. Thus, the compact fly genome encodes similar gene products that are present in higher vertebrates. In fact, analyses of Drosophila and human genomes have shown ~80% of human diseases in which the disease-related gene has been identified have an orthologue in Drosophila.9–11
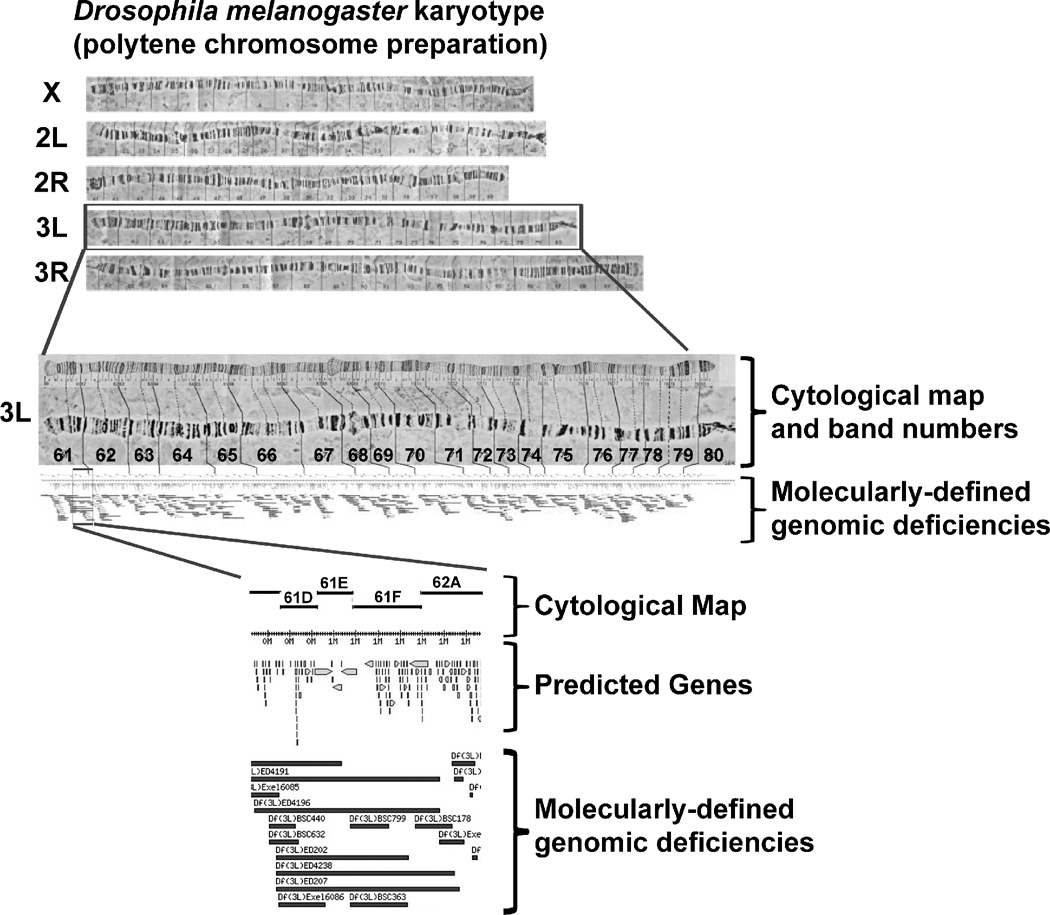
Figure 1. The Drosophila melanogaster genome.
(A) The karyotyping of D. melanogaster prepared from the salivary gland showing chromosomes X, 2L/R, and 3L/R. The Y and fourth chromosomes are not depicted. (B) An enlarged view of the 3L chromosome with cytological map showing bands 61 through 80 (above) and molecularly-defined deficiencies (below). (C) An enlarged view of the 61D through 62A cytological map showing predicted genes encoded within the region and corresponding molecularly-defined genomic deficiencies from the Exelixis and DrosDel collections.60–63 Figures adapted from http://flystocks.bio.indiana.edu.
Genetic Engineering in Drosophila
Several accomplishments in fly transgenics over the past several years have further advanced the use of the Drosophila as a model system. Ectopic transgene expression in the fly is usually achieved using the bipartite Gal4-upstream activating sequence (UAS) system derived from gene expression for galactose metabolism in yeast (Figure 2A).5, 12 Transgenic flies that are designated GAL4 expression flies harbor promoters of interest and control the tissue-specific expression of the yeast Gal4 transcription factor or designated UAS-target gene lines harbor transgenes of interest downstream of a specific UAS sequence. Typically, different Gal4-driver lines are crossed with UAS-target gene lines and the effects of tissue-specific gene expression are examined in the progeny. The Gal4-UAS system provides a number of distinct advantages including: promoter expression patterns when using UAS-beta-galactosidase (UAS-lacZ) or UAS-Green Fluorescent Protein (GFP) as a tissue marker; ectopic expression in a variety of tissues; and the tissue-specific effects of specific gene knockdown using UAS-RNAi lines (http://www.flyrnai.org/DRSC-OVR.html). 13, 14 For example, tinC-GAL4 can be used to drive cardiac-specific transgenes to express recombinant cDNA to examine ectopic protein expression or RNAi to examine specific gene knockdown in the heart.15 Compared to strategies to generate cardiac specific transgene expression in the mouse, transgenic flies can be engineered in a timeframe of 6–8 weeks. Further refinements in transgenic expression in the fly include PhiC31 site-specific integrase that uses attB and attP sites to achieve targeted integration of transgenes into the fly genome.16, 17 This approach controls for positional effects due to transgene location.
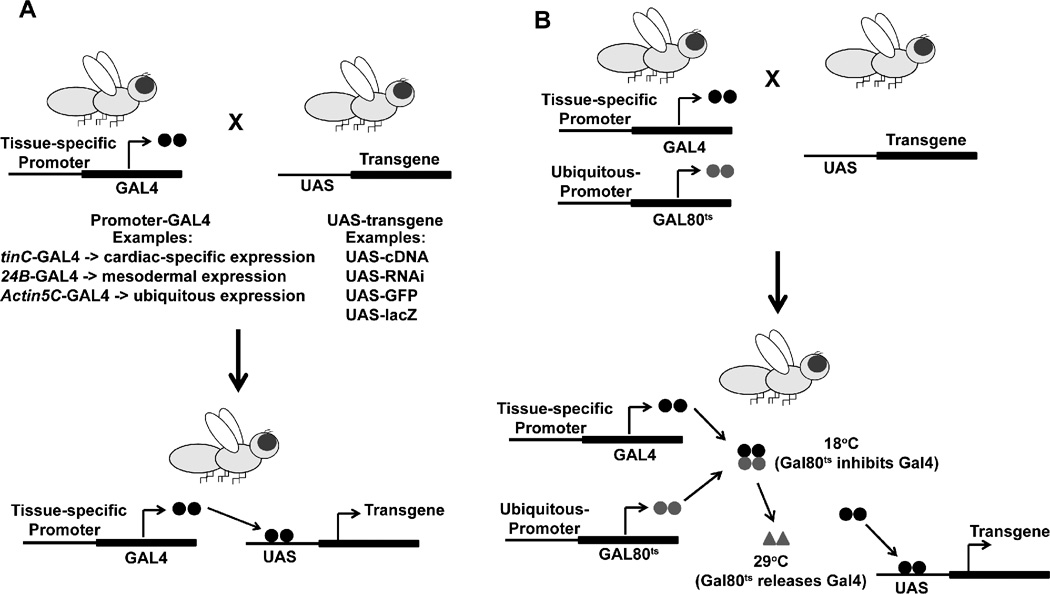
Figure 2. Transgenic-Expression Systems in Drosophila.
(A) The Gal4-UAS bipartite transgene expression approach relies on breeding transgenic flies that harbor either a tissue-specific promoter that drives Gal4 production or UAS-transgene constructs. The progeny possess one copy of the Gal4 and UAS constructs and express the transgene of interest under the control of the promoter of interest. This system allows versatility by using different promoters to control transgene expression. (B) The ubiquitous expression of a temperature-sensitive Gal80 (Gal80ts) is incorporated into the Gal4-UAS system to add temporal control of transgene expression using a shift from a restrictive to permissive temperature. Gal80ts reversibly suppresses Gal4 activity at 18°C and permits Gal4 binding to UAS at 29°C.
Gal4-driver lines also can be engineered to include the ubiquitous expression of a temperature-sensitive Gal80 (Gal80ts) that reversibly suppresses Gal4 activity at 18°C and permits Gal4 binding to UAS at 29°C thereby permitting temporal control in the context of tissue-specific transgenes expression (Figure 2B).18, 19 Other systems based on hormone or tetracycline-sensitive Gal4 based systems have been described that allow for inducible transgene expression.12, 20
Strategies to achieve homologous recombination for gene knockout (“ends-out” targeting) or replacement (“ends-in” targeting) in the fly based on FLP site-specific recombinase, and its target site, FRT, have been developed by Golic and colleagues (Figure 3A and B).21–28 The “ends-out” approach is based on using a transgenic fly harboring a mini-white cassette that is flanked by genomic targeting sequence along with unique I-SceI recognition sites within two FRT sites (Figure 3A). The genomic targeting construct undergoes mobilization and linearization for in vivo homologous recombination in progeny that are bred in to a fly line that harbors hsp70-FLP and hsp70-SceI. Progeny are then screened for potential homologous recombination and target gene disruption by insertion of the mini-white cassette into the endogenous gene locus. The ‘ends-in” approach is similar but results in a tandem gene duplication with the targeted construct near the endogenous gene locus and subsequent reduction to produce the targeted replacement of the gene of interest (Figure 3B). One of the main differences between gene targeting strategies in the mouse and fly is that homologous recombination using the mouse is monitored in embryonic stem cells prior to injecting mouse blastocysts while homologous recombination using the fly occurs in vivo and the progeny produced from fly crosses are screened.21 The timeframe for generating transgenic flies using the “ends-in” or “ends-out” strategies is on the order of a few months depending on the culture conditions. Additional strategies based on recombination-mediated genetic engineering, referred to as recombineering, have been developed to create large genomic DNA insertions based on recombination with bacterial artificial chromosomes.17, 29 These strategies can be combined with site-directed mutagenesis to perform gene product structure-function studies.
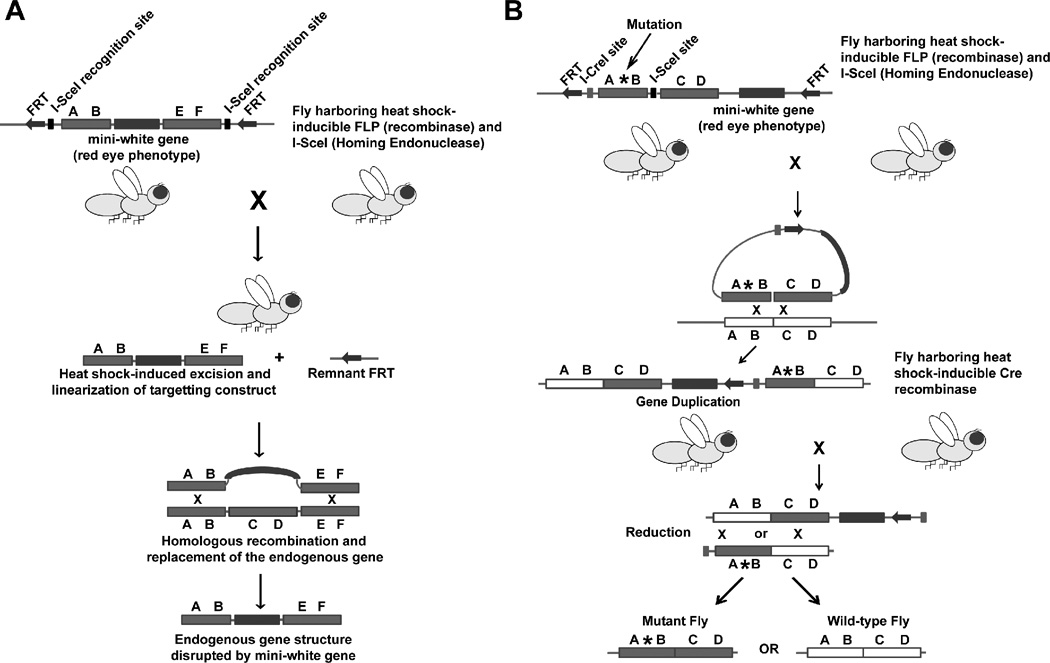
Figure 3. Targeted gene knockout and gene replacement in Drosophila.
(A) The “ends-out” approach for gene knockout. A fly harboring a transgenic construct containing the mini-white gene within the genomic sequence of a gene targeted for replacement is bred with a fly that harbors an inducible FLP recombinase and I-SceI homing enzyme. In a subset of progeny, the targeting construct is excised by the FLP recombinase, linearized by I-SceI, and undergoes homologous recombination with the endogenous gene. The endogenous gene is disrupted by replaced with the mini-white gene. (B) The “ends-out” approach for gene replacement. A fly harboring a transgenic construct that contains an engineered mutation (asterisk) and the mini-white gene within the genomic sequence of a gene targeted for replacement is bred with a fly that harbors an inducible FLP recombinase and I-SceI homing enzyme. In a subset of the first generation, the targeting construct is excised by FLP recombinase, linearized by I-SceI, and undergoes homologous recombination with the endogenous gene locus to produce tandem gene duplication. Then, the fly containing the gene duplication is bred with a fly that harbors an inducible Cre recombinase that recognizes the I-CreI site that is engineered in the targeting construct. In a subset of the progeny, the tandem genes undergo homologous recombination and reduction of gene copy number thereby producing flies that harbor either a wild-type gene or the mutant gene. The figure is based on Maggert, K.A. et al122
Drosophila Genetic Resources
To promote advancement of discovery, the community of Drosophila researchers has complied and shared mutants and reagents leading to the creation and maintenance of stock collections including Bloomington, Vienna, and Kyoto (http://flystocks.bio.indiana.edu/, http://stockcenter.vdrc.at/control/main, and http://www.dgrc.kit.ac.jp/en/index.html). Information pertaining to the Bloomington Stock collection was initially curated and described in the “Red Book” (“Genetic Variations of Drosophila melanogaster” (1967) and later “The Genome of Drosophila melanogaster” (1992)) that contributed significantly to the Flybase consortium (http://flybase.org).30, 31 As of 2010, the Bloomington Stock center maintained 30,810 stocks and distributed 196,930 subcultures to the scientific community (http://flystocks.bio.indiana.edu). Additional resources include a searchable database of high-throughput in situ hybridization studies that contains >100,000 images of expression from >4000 genes (FlyExpress.net). This platform provides a resource to examine the spatiotemporal expression patterns of genes expressed during Drosophila embryogenesis.
Recently, the Drosophila model organism ENCyclopedia Of DNA Elements (modENCODE) project has produced large data sets of transcript profiles, histone modifications, transcription factors, and replication programs in cell lines, isolated tissues, and whole organisms across several developmental stages.32 This powerful resource is designed to provide insight into potential new functions for genes, a better understanding of developmental- and tissue-specific gene regulation, and integration of functional changes in the transcriptome with the genome.
Efforts to systematically examine the effects of gene knockdown have led to the creation of large collections of transgenic flies that harbor specific RNAi under the control of UAS have been generated by Vienna Drosophila RNAi Center (http://stockcenter.vdrc.at/control/main) and the Transgenic RNAi Project (TRiP) at Harvard Medical School http://www.flyrnai.org/TRiP-HOME.html).13, 14 In combination with tissue specific promoters that drive Gal4 expression, these UAS-RNAi lines have been used to conduct large scale genome-wide screens of gene function in a variety of contexts. For example, RNAi screens have been performed to examine muscle development, cardiac function, and extracellular signal-regulated kinase (ERK) signaling (see below).15, 33, 34
Collectively, the resources available in the field of Drosophila genetics and genomics have advanced the identification of genes involved in a variety of basic biological processes including signal transduction, cell differentiation, and organ development. While other models, including mouse and large animals, are well suited to investigate the pathophysiology underlying human disease, Drosophila research provides a genetically-tractable model system to identify new genes and signaling pathways. Therefore, applying the unique set of resources available in the fields of fly genetics and genomics has the potential to further the understanding of human cardiovascular disease.
General Screening Strategies in Drosophila
The major strength of the fruit fly as a model of cardiovascular disease relies on the remarkable, diverse resources that facilitate large-scale genetic screens, otherwise currently unattainable in mouse models. The strategies to identify genes that impact fly cardiac function can be separated into two general concepts: (1) genes that directly cause cardiac abnormalities or (2) genes that suppress or enhance abnormal cardiac phenotypes, so called suppressor or enhancer screens. In the first case, wild-type fly lines are mutagenized and then screened for the occurrence of new measurable phenotypes. This powerful approach has been the cornerstone of genetic screens in the fly and has led to new insights into signaling pathways that are conserved among multiple species, including mammals. Mutagenesis screens based on development of the fly eye or wing vein morphology have identified key components of receptor tyrosine kinase signaling pathways including epidermal growth factor receptor (EGFR), Ras, Raf, Sos, Grb2, and PTP11N.35–45 Additionally, fly screens have identified components in pathways involving Notch, Wnt, bone morphogenetic protein (BMP), Smad, Salvador/Warts/Hippo and G protein-coupled receptor (GPCR) signaling in a variety of contexts.46–55
In a suppressor or enhancer screen, sensitized lines that express particular phenotypes are mutagenized or bred into another mutagenized strain to identify suppressors and enhancers of a particular phenotype or phenotypic trait. The literature in the fly genetics field provides a wealth of information pertaining to mutagenesis screens and describes the identification of key components in many signaling pathways. Mutagenesis approaches include: chemical, irradiation, P-element, and genomic deficiencies. For example, genetic screens performed in the context of Ras or Raf mutations have identified dominant suppressors and enhancers of the Ras-Raf-ERK pathway.38
Chemical/Irradiation Mutagenesis
The wealth of resources that are available in the fly genetics and genomics community provides multiple approaches to examine gene function in relationship to a particular measurable trait. Traditionally, gene function has been elucidated by examining nulls (complete loss of function allele), hypomorphs (partial loss of function allele), hypermorphs (gain of function allele), or antimorphs (antagonist of wild-type allele) that are generated by chemical or radiation-induced mutagenesis.2, 3 When combined with a high-throughput phenotyping strategy, chemical mutagenesis can generate a number of interesting mutants that can be mapped using several strategies to identify candidate genes.56
P-element Mutagenesis
Mutagenesis strategies based on P-element inserts are unique to fly genetics. P-elements are foreign pieces of engineered DNA that insert throughout the fly genome and potentially disrupt gene function by physically altering gene structure. Large collections of mutants have been generated in which the exact positions of the P-element insertions in the genome have been mapped and sequenced to studying gene function.57–59 In some cases, the P-elements have been engineered to encode GFP (i.e., P(GawP)) or beta-galactosidase (i.e., P(lacW or P(PZ)) to examine the expression patterns of genes. P(EP) and P(XP) P-elements harbor UAS sequences designed to potentially amplify gene transcription near the P-element insertion.
Molecularly-defined Genomic Deficiencies
More recently, large collections of molecularly-defined genomic deficiencies have been engineered by two groups, Exelixis and DrosDel.60–63 The deficiencies were created using FLP recombinase strategies. Collections of flies were generated that harbored P-elements containing FRT sites that are recognized by FLP recombinase. Deficiencies were created using FLP recombinase mediated excision of genomic DNA flanked by two neighboring in trans P-elements (i.e., Piggybac and P(RS3)/P(XP) P-elements) (Figure 1).60, 62, 63 These deficiency mutants, maintained in a haploinsufficient state in the context of balancer chromosomes and in isogenic backgrounds, are powerful resources to examine the phenotypic effects of gene deletion. These large collections also provide a unique approach to identify and map candidate genes that affects particular traits. Using these methods, the combination of the Exelixis and DrosDel P-element collections can be used to construct ~500,000 theoretical molecularly-defined genomic deficiencies (http://www.drosdel.org.uk/fdd/fdd_info.php).60–63
The Drosophila Circulatory System
The Embryonic Heart (Dorsal Vessel)
While the fly circulatory system is simple compared to higher vertebrates, investigations of the Drosophila heart have yielded new insights into mammalian cardiac development and function. The fly circulatory system develops from an orchestrated set of fundamental signals that occur in the mesoderm.64–67 Secreted ligands including decapentaplegia (dpp) of the bone morphogenetic protein family regulate embryonic heart development and trigger cell fate specification in the developing mesoderm. A set of cardiac precursor cells that expresses specific transcription factors including tin, the fly othologue of Nkx2.5 and GATA family members arise in the embryo, migrate along the mesoderm, and form the recognizable single layered dorsal vessel at stage 16 of embryonic development.68 The dorsal vessel is divided into the heart proper and the anterior and posterior aorta (Figure 4A). Many of the temporal and tissue specific signals required for dorsal vessel formations are evolutionarily conserved among mammals.69 For example, genetic studies have identified tin as a critical component in heart development and mutations that remove tin cause a complete absence of heart cells.68 Mutations in human Nkx2.5 are associated with congenital heart disease including atrial septal defects and arrhythmias.70
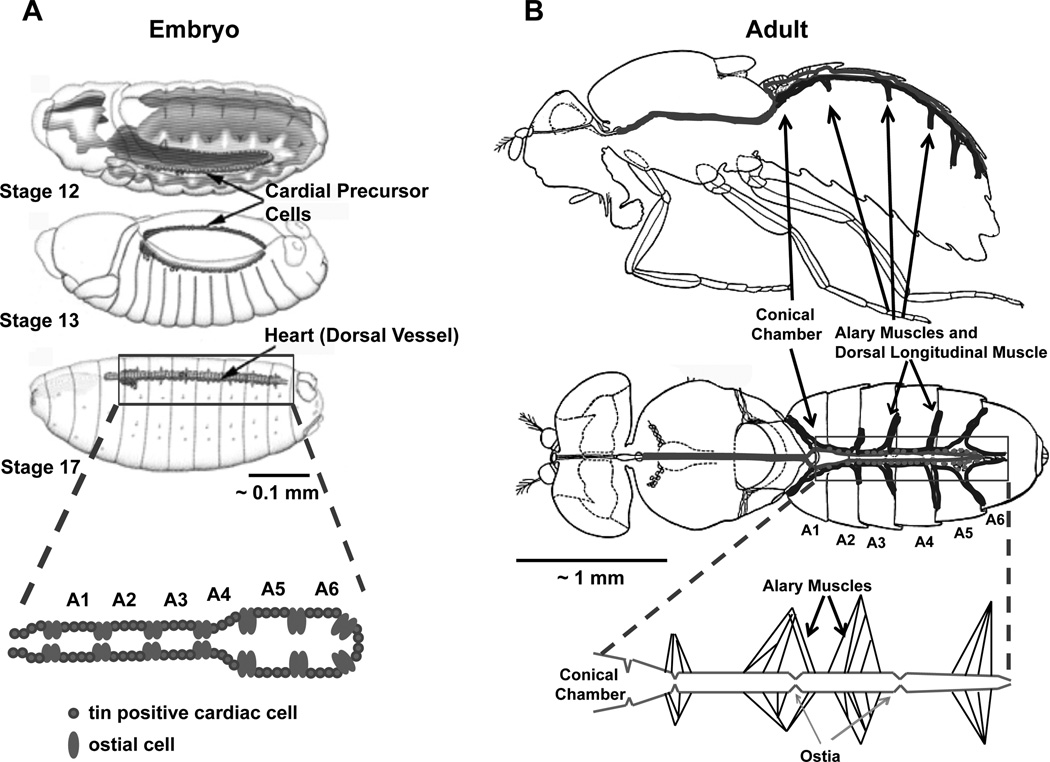
Figure 4. The embryonic and adult Drosophila circulatory system.
(A) The developing embryonic circulatory system arises from cardial precursor cells that migrate to form the dorsal vessel at Stage 16. Stages 12, 13, and 17 are shown. The figure is adapted from Fly Embryo RNAi Project (http://flyembryo.nhlbi.nih.gov). (B) The adult fly circulatory system consists of an open system with the main conical chamber, heart, located along the dorsal aspect of the A1 abdominal segment. Suspensory muscles including the Alary and Ventral Longitudinal Muscle also referred to as the Dorsal Diaphragm. Pericardial cells are closely juxtaposed along the length of the abdominal portion of the circulatory system. The figure is adapted from Miller.72 A 1 mm scale bars are shown for comparison.
Each side of the embryonic dorsal vessel is arranged in sets of four tin positive cardioblasts that form the contractile cells of the heart and are separated by pairs of Svp-expressing ostial cells (Figure 4). Pericardial cells are non-contractile cells that surround the heart and have been implicated in detoxification of the hemolymph.71, 72 Additionally, the dorsal vessel is surrounded by an extracellular matrix composed of pericardin, a type IV collagen-like protein.71
The Larval and Adult Heart
As the larva develops from the embryo, the dorsal vessel maintains its general morphology. Then, during the formation of the pupal stage, when the larva transitions into the adult, the fly circulatory system undergoes tremendous morphological change.73, 74 During morphogenesis, the heart proper of the dorsal vessel involutes and the posterior aorta in the first abdominal segment becomes the main pumping chamber of the adult heart.73–75 The adult heart consists of a single layer of cardiomyocytes that have circumferentially oriented myofibers and is closely juxtaposed to the ventral longitudinal muscle (also referred to as the “dorsal diaphragm”) and sets of suspensory muscles arising from the dorsal cuticle (called alary muscles) (Figure 4B). The ventral longitudinal muscle belongs to a distinct non-cardiac muscle type that does not express cardiomyocyte-specific tin and arises from lymph cells via trans-differentiation during the pupal stage of development.76 The larval and adult hearts continue to possess ostial cells derived from the Svp-expressing cells of the embryonic dorsal vessel.77 The ostial cells function as valves that facilitate the entry of hemolymph into the circulatory system. Additionally, pericardial cells are also maintained in close proximity to the larval and adult hearts.72
The embryonic and larval dorsal vessel lack innervation and generate a completely myogenic cardiac impulse. However, pairs of transverse glutamergic nerves innervate the lateral edges of the cardiac chamber and abdominal segments of circulatory system.78 The adult fly heart has anterior (rostral) and posterior (caudal) pacemakers that generate measurable retrograde and anterograde propagating calcium transients.79 Consistent with these observations, earlier work by Wasserthal using a linear optosensor chip and an IR-light beam demonstrated anterograde and retrograde pulses of hemolymph.80 During anterograde beating, the hemolymph enters the heart through all inflow ostia and is propelled through the narrow aorta located in the thorax. During retrograde beating, the hemolymph enters the heart through the first ostia and moves toward the rostral end. Thus, hemolymph circulates through sets of valves along the caudal circulatory system into the lumen of the tube and is propelled in a rostral or caudal direction.
Heart rate in the fly heart responds to agonist stimulation suggesting a level of neurohormonal control.81–83 For example, the fly heart responds to several bioactive amines including octopamine, serotonin, norepinephrine, and dopamine as well as peptide hormones, including crustacean cardioaccelerator peptide (CCP).81
The dissected fly heart also facilitates the examination of myocardial calcium handling using cardiac-specific genetically encoded calcium-dependent fluorescent proteins (Figure 5A).79 Recently, flies harboring GCaMP2 under the control of the cardiac-specific tinC driver have been used to characterize myocardial calcium transients in adult flies.79 The calcium transients in adult flies share several properties observed in with mammalian hearts. The propagating calcium transients in the fly heart are dependent on extracellular sodium, inhibited by chelation of extracellular calcium or pharmacological inhibition of L-type calcium channels, and are sensitive to thapsigargin-mediated depletion of intracellular calcium stores. Moreover, a troponin-I mutant that has impaired cardiac function and an enlarged heart chamber demonstrated abnormalities in myocardial calcium handling including a decreased response to caffeine-augmented calcium release, consistent with some observations in human cardiomyopathies.79
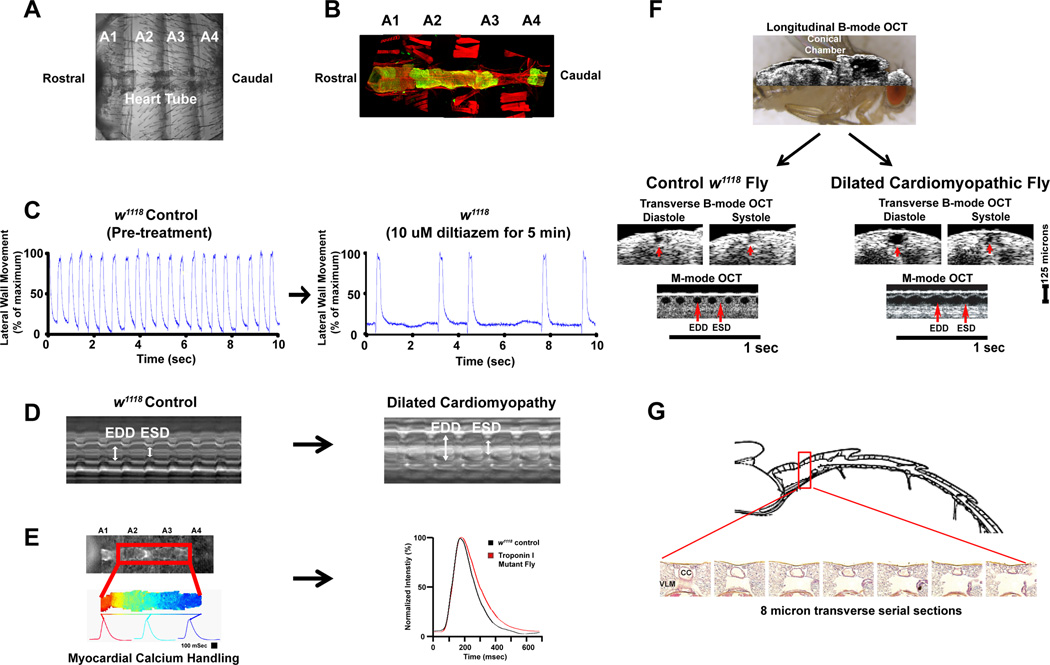
Figure 5. Strategies to assess adult Drosophila cardiac morphology and function.
(A) A dissected adult fly heart perfused in artificial hemolymph is shown. Abdominal segments are denoted as A1, A2, A3, and A4. This specimen preparation can be used to assess heart rate and rhythm, myocardial calcium handling, cardiac morphology, and cardiac function. (B) An example of cardiac morphology in a tinC-GFP transgenic fly depicting the cardiac tube (green) and actin stained with phalloidin (red) using confocal microscopy with Z-stack reconstruction as previously described.119(C) Examples of heart rate and rhythm obtained using a Leica 165FC steromicroscope equipped with an Andor iXon high-speed camera. Heart rate was measured by examining movement of the lateral wall as described by Wessells and Bodmer.105, 109 An example of the effects of Diltiazem, a calcium-channel blocker, is shown. (D) Representative m-mode of cardiac function was obtained using high-speed brightfield imaging similar to methods as described.123 M-modes showing w1118 (control) and a dilated cardiomyopathic mutant are shown for comparison. (E) An example of myocardial calcium transients measured in adult transgenic flies that had cardiac-specific expression of GCaMP2 in a w1118 background (control) or in the presence of hdp2, a mutation in Troponin-I, that has a dilated cardiomyopathy phenotype are shown.79(F) Longitudinal B-mode OCT image during diastole superimposed on a live adult fly (left) and transverse B-mode OCT during diastole and systole with representative M-mode OCT showing cardiac chamber size and function in w1118 (control) and a mutant that has dilated cardiomyopathy as described by Wolf et. al.106(G) Example of transverse sectioning and hemotylin/eosin staining of a fixed adult fly. The cardiac chamber (CC) and surrounding tissues including the ventral longitudinal muscle (VML) are shown. The image is adapted from Yu et. al.119
It should be noted that unlike mammals, oxygen transport occurs through diffusion from spiracles that invaginate from the cuticle into the interior of the adult fly.72 Thus, the flow of hemolymph serves to transport immune cells, nutrients, and molecules that are necessary to maintain homeostasis. The fly is emerging as a model to examine metabolomics, lipidomics, and mitochondrial function.84–87 The major fuel sources in Drosophila flight muscle are glycogen, free glucose, and trehalose, a disaccharide composed of two glucose molecules. A model of tissue hypoxia tolerance has been used to examine changes in metabolic profiles using NMR-based measurements and principle component analyses.88, 89 An age-dependent reduction in the hypoxia tolerance was attributed to a decrease in ATP production during reoxygenation due to reduced recovery of mitochondrial respiratory pathways.88
Similar to protein and carbohydrate metabolism, flies and mammals demonstrate a conservation among genes that encodes proteins that are involved in lipid metabolism. Mass-spectrometry based analyses of ~400 different lipids from Drosophila showed similarities between fly and mammalian lipidomes.90 Additionally, fly mutants have been isolated that encode genes involved in the metabolism of ceramides, sphingolipids, cholesterol esters, fatty acids, glycerophospholipids, triacylglycerides, and lipid droplets.85, 90–99 The unique abilities to combine fly genetics and metabolic profiling in different tissues suggests a unique approach to examine myocardial metabolomics in a genetically-tractable model system.
Evaluating Cardiac Structure and Function in Drosophila
Genetic Screens Using the Embryonic and Larval Heart
A number of approaches have been developed to characterize the fly circulatory system. In general, these approaches are based on an examination of cardiac morphology or function. Initial screens of fly mesodermal and heart development, conducted by Bodmer and Frasch, were based on the morphologic examination of the embryonic dorsal vessel.64, 66–68 These seminal studies identified a number of transcription factors that were critical for cardiac development and are highly conserved throughout mammals, including humans. Skeath and coworkers have examined the cell lineage of all heart cells in the fly embryo and identified the ETS-transcription factor, pointed, and the GATA transcription factor, pannier, as important components from a screen of ~2000 3rd chromosome P-element lines.100 The identification of tin and subsequent understanding of the regulatory networks including dMef, Hand, Svp, ladybird, eve, pannier, and pointed have contributed significantly to the understanding of cardiac development in humans.66–69
An interesting screen was conducted to examine modifiers of heart cell fate using a P-element mediated gain-of-function strategy by Bidet and coworkers.101 Fly lines that harbor single P-element insertions encoding a UAS promoter were bred with the pan-mesodermal driver 24B–Gal4 fly line and dorsal vessel cells were examined for abnormalities in heart morphology. Using this strategy, the effects of over-expression of genes near each P-element insertion was correlated with alterations in dorsal vessel cell number and suggested that alteration in the levels of rhomboid, the transcription co-factor yan, and the Rho-GTPase Rac2 influenced cardiac development.101
Additional genetic screens based on the examination of the embryonic heart have led to new insights into signaling pathways that control cardiac development. For example, a P-element screen identified mutants in which the pericardial cells dissociated from the cardioblasts during dorsal vessel development.102, 103 The mutations were subsequently mapped to 3-hydroxy-3-methylglutaryl-coenzyme A reductase, the mevalonate pathway, and the G-protein gamma subunit 1 that requires geranylgeranylation for proper function. These findings, in conjunction with an examination of septate junction proteins, suggest that G-protein function is necessary to maintain cardiac integrity during fly heart development.
Genetic Screens Based on Heart Rate and Rhythm in Pupae and Intact Adults
During the past few years, attention has been directed towards the adult fly heart as a model of human cardiac disease.104–107 Several approaches have been developed to characterize the pupal and adult cardiac function and structure (Figure 5B and C). Pasternostro and co-workers initially described an age-associated decline in cardiac function in adult Drosophila.108 Wessells and Bodmer developed methods to measure heart rate frequency in early pupae that retain the translucent properties of larvae or in adults using high-speed image capture of cardiac chamber movement under bright field microscopy.109 These strategies have also been used to monitor adult heart rate responses to externally applied electrical pacing to characterize the effects of aging on cardiac parameters in adult flies. For example, an age-related decline in cardiac function, manifest as changes in basal heart rate and altered heart rate response to external pacing, were minimized in flies that had alterations in Insulin-like growth factor (IGF) receptor signaling. Furthermore, cardiac-specific expression of dPTEN, a phosphatase that inhibits insulin-receptor dependent PI3K signaling, rescued the age-dependent changes in cardiac function attributed to insulin-IGF receptor signaling.109 The cardiac-specific expression of the forkhead transcription factor dFOXO or reductions in dTOR rescued or attenuated age-dependent changes in cardiac function attributed to insulin-IGF receptor signaling.110
Furthermore, eukaryotic translation initiation factor 4E (Eif4e) binding protein (Eif4e–BP) acts downstream on dTOR and dFOXO and can modulate the age-associated changes in cardiac function attributed to IGF signaling abnormalities.111 In flies, the FOXO/Eif4e–BP pathway has been shown to control protein homeostatsis (called “proteostasis”) by regulating autophagy.112 In transgenic mice, the cardiac-specific ablation of TOR resulted in a dilated cardiomyopathy that was characterized by apoptosis, autophagy, altered mitochondrial structure, and accumulation of Eif4e–BP.113
Genetic Screens Based on Heart Rate, Rhythm, and Function in Dissected Heart Preparations
Adult fly cardiac function can be studied using methods based on the dissection of the surrounding tissues from the fly heart thus providing a specimen that is attached to the dorsal cuticle and can be maintained by perfusion in artificial hemolymph.114–116 This approach provides a platform for multiple phenotyping strategies. Bright field or fluorescence microscopy with high-speed image capture is used to evaluate heart rate, rhythm, and cardiac function by measuring the movement of the lateral walls of the in the second and third abdominal segments. Using these methods, potential mechanisms that contribute to age-related changes in cardiac function have been examined. Examples include: mutations in KCNQ potassium channels mutations have been associated with cardiac arrhythmias mimic the effects of aging in the fly and investigations of sestrin as a feedback inhibitor of TOR that prevents age-related pathologies.115, 117 Recently, Bodmer and Penninger performed a global in vivo RNAi screen using the Vienna Drosophila RNAi collection to identify genes related to cardiac dysfunction.15 Transgenic flies harboring UAS-RNAi were crossed to the cardiac-specific tinCdelta-4-Gal4 driver lines and the progeny were examined. Developmental and adult lethality at 25°C and temperature-induced stress at 29°C in adults were examined as the primary phenotypic screen. After multiple rounds of screening, 7116 (89.2%) viable adult transformants, 365 (4.6%) lethal transformants, and 490 (6.2%) transformants that were considered to have an adult heart defect were identified. From these studies, a global network of heart function was generated and the silencing of the CCR-NOT pathway was found to cause a dilated cardiomyopathy.
Genetic Screens Based on Cardiac Function in Awake, Adult Flies
To examine cardiac chamber size and function in awake, adult Drosophila an approach has been developed based optical coherent tomography (OCT).18, 106, 118, 119 OCT uses 1310 nm near-infrared light to provide non-invasive, non-destructive images that are analogous to transthoracic echocardiography in humans and mice (Figure 5B). Therefore, OCT imaging facilitates relatively fast analyses of cardiac function in adult flies without the cardiovascular effects associated with prolonged exposure to anesthesia or damage to surrounding tissues during dissection. Moreover, due to the non-invasive properties of OCT, the effects of temporal, cardiac-specific transgene expression can be examined serially in individual flies.18 Genetic screens based on OCT imaging of heart chamber size and function from stocks of molecularly-defined genomic deficiencies from the Exelixis and DrosDel collections have led to the identification of genes that cause cardiomyopathies in the fly model. An examination of deficiencies of the 3rd chromosome identified mutants in rhomboid-3 that had enlarged cardiac chambers.119 Rhomboid-3 is a member of an evolutionarily conserved family of seven-transmembrane serine proteases that hydrolyze membrane bound Spitz, the fly orthologue of epidermal growth factor (EGF).120, 121 In fact, the post-developmental inhibition of EGFR by means of cardiac-specific expression of dominant-negative EGFR results in progressive deterioration of cardiac function suggesting that proper EGFR signaling is required to maintain adult cardiac function.119 Importantly, these findings suggest that the fly may model cardiovascular-specific aspects of EGFR signaling and provide insights into the mechanisms that contribute to chemotherapy-induced cardiomyopathies.
The screen of genomic deficiencies has also identified a new gene, weary (wry), that is important for the maintenance of normal heart function in the adult fly.118 Wry encodes a protein that has a similar structure to members of the Notch family but lacks a DSL (Delta-Serrate-Lag) domain commonly found in other Drosophila Notch ligands.118 Cell aggregation assays and gamma-secretase inhibitors demonstrate that Wry can mediate cellular adhesion with Notch expressing cells and transactivate Notch to promote signaling and nuclear transcription. Interestingly, deficiencies in wry produced by genomic deficiencies, P-element insertions, or cardiac-specific RNAi knockdown results in a dilated cardiomyopathic phenotype. Mutations in human Notch signaling and mutant mouse models of Notch signaling have been implicated in congenital heart disease, however the involvement of Notch signaling in adult mammalian cardiac disease remains unclear. Notch ligands appear important to maintain cardiac function in adult flies and gene orthologs involved in Notch signaling may be important in pathogenesis of mammalian dilated cardiomyopathy. Moreover, Notch signaling components may represent therapeutic targets for the treatment of dilated cardiomyopathies.
Comparison of Drosophila to other model systems and limitations of the fly model of cardiovascular disease
It is clear that Drosophila is an excellent discovery engine that continues to provide a tremendous resource to identify new genes in cardiovascular biology. Despite the advantages of the fly as a genetically-tractable system, there are obvious limitations compared to models that use Zebrafish, mice, or large animals (Table I). For example, the fly lacks genetic redundancy of more complex organisms, thereby providing the possibility of more efficient evaluation of candidate genes since fewer genes need to be targeted and less time may be required for analyses. However, the lack of genetic redundancy may represent a limitation since specific gene regulatory mechanisms that are present in mammalian systems may not be present in the fly.
Table I. Comparison of Model Systems of Cardiovascular Diseases.
The strengths and limitations of fly, zebra fish, mouse, and large animal models of cardiovascular diseases are shown.
| Model | Advantages | Disadvantages |
|---|---|---|
| Fly |
|
|
| Zebrafish |
|
|
| Mouse |
|
|
| Large Animal (pig/rabbit/sheep/dog) |
|
|
The translation of fly cardiac physiology to mammals is difficult due to a single chamber open circulatory system compared to Zebrafish and mammals. Ischemia-reperfusion studies cannot be conducted in the fly because the fly does not have a coronary circulation and relies on oxygen transport by diffusion. The cardiac conduction system of the fly is distinctly different from the two chambered heart of the Zebrafish or the four chambered heart of mammals. The presence of two pacemakers, anterograde and retrograde pulses, and irregularities in adult heart rate can make assessment of arrhythmia difficult in intact flies. Importantly, the wealth of resources available to Drosophila research and the evolutionary conservation of signaling mechanisms make the fly a valuable model to identify genes that cause or contribute to human cardiovascular diseases
Conclusions
The rich history and tremendous resources available in fly genetics continues to offer new avenues to identify signaling pathways that are important in many biological processes. In conjunction with recent advances in cardiac phenotyping, the fly provides a genetically-tractable approach to understand the complex signals that lead to cardiac dysfunction. Thus, continued studies based on fly genetic and genomics has the potential to advance our understanding of human cardiovascular diseases.
Acknowledgments
Sources of Funding:
This work was supported by the National Institutes of Health through a grant (K08HL085072) to M.J.W. and HL-083065 to H.A.R.
Non-standard Abbreviations and Acronyms
- BMP
Bone morphogenetic protein
- dFOXO
Drosophila O subclass of the forkhead family of transcription factors
- dTOR
Drosophila target of rapamycin
- EGFR
Epidermal Growth Factor Receptor
- ERK
Extracellular signal-regulated kinases
- FLP
recombinase that recognizes FRT site
- FLP
recombinase that recognizes FRT site
- GAL4
transcriptional activator from yeast
- Gal80ts
temperature-sensitive protein that interact with GAL4
- GCaMP2
circularly permutated enhanced green fluorescent protein fused with M13 helix of myosin light chain kinase at the N terminus and calmodulin at the C terminus
- GFP
green fluorescent protein
- GPCR
G-protein coupled receptor
- KCNQ
potassium channel gene
- modENCODE
model organism ENCyclopedia Of DNA Elements
- OCT
optical coherence tomography
- RNAi
small interference RNA
- tinC
DNA sequence within the tinman gene used to drive cardiac-specific transgene expression
- TRiP
Transgenic RNAi Project at Harvard Medical School
- UAS
Upstream Activating Sequence from yeast
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures None
References
- 1.Kohler RE. Lords of the fly : Drosophila genetics and the experimental life. Chicago: University of Chicago Press; 1994. [Google Scholar]
- 2.Ashburner M, Golic KG, Hawley RS. Drosophila : A laboratory handbook. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 2005. [Google Scholar]
- 3.Greenspan RJ. Fly pushing : The theory and practice of drosophila genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2004. [Google Scholar]
- 4.Dahmann C. Drosophila : Methods and protocols. Totowa, N.J.: Humana Press; 2008. [Google Scholar]
- 5.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 6.Celniker SE, Dillon LA, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, Micklem G, Piano F, Snyder M, Stein L, White KP, Waterston RH. Unlocking the secrets of the genome. Nature. 2009;459:927–930. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Celniker SE, Rubin GM. The drosophila melanogaster genome. Annu Rev Genomics Hum Genet. 2003;4:89–117. doi: 10.1146/annurev.genom.4.070802.110323. [DOI] [PubMed] [Google Scholar]
- 8.Rubin GM, Yandell MD, Wortman JR, Gabor Miklos GL, Nelson CR, Hariharan IK, Fortini ME, Li PW, Apweiler R, Fleischmann W, Cherry JM, Henikoff S, Skupski MP, Misra S, Ashburner M, Birney E, Boguski MS, Brody T, Brokstein P, Celniker SE, Chervitz SA, Coates D, Cravchik A, Gabrielian A, Galle RF, Gelbart WM, George RA, Goldstein LS, Gong F, Guan P, Harris NL, Hay BA, Hoskins RA, Li J, Li Z, Hynes RO, Jones SJ, Kuehl PM, Lemaitre B, Littleton JT, Morrison DK, Mungall C, O'Farrell PH, Pickeral OK, Shue C, Vosshall LB, Zhang J, Zhao Q, Zheng XH, Lewis S. Comparative genomics of the eukaryotes. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chien S, Reiter LT, Bier E, Gribskov M. Homophila: Human disease gene cognates in drosophila. Nucleic Acids Res. 2002;30:149–151. doi: 10.1093/nar/30.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fortini ME, Skupski MP, Boguski MS, Hariharan IK. A survey of human disease gene counterparts in the drosophila genome. J Cell Biol. 2000;150:F23–30. doi: 10.1083/jcb.150.2.f23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reiter LT, Potocki L, Chien S, Gribskov M, Bier E. A systematic analysis of human disease-associated gene sequences in drosophila melanogaster. Genome Res. 2001;11:1114–1125. doi: 10.1101/gr.169101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elliott DA, Brand AH. The gal4 system : A versatile system for the expression of genes. Methods Mol Biol. 2008;420:79–95. doi: 10.1007/978-1-59745-583-1_5. [DOI] [PubMed] [Google Scholar]
- 13.Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ. A genome-wide transgenic rnai library for conditional gene inactivation in drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 14.Haley B, Hendrix D, Trang V, Levine M. A simplified mirna-based gene silencing method for drosophila melanogaster. Dev Biol. 2008;321:482–490. doi: 10.1016/j.ydbio.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neely GG, Kuba K, Cammarato A, Isobe K, Amann S, Zhang L, Murata M, Elmen L, Gupta V, Arora S, Sarangi R, Dan D, Fujisawa S, Usami T, Xia CP, Keene AC, Alayari NN, Yamakawa H, Elling U, Berger C, Novatchkova M, Koglgruber R, Fukuda K, Nishina H, Isobe M, Pospisilik JA, Imai Y, Pfeufer A, Hicks AA, Pramstaller PP, Subramaniam S, Kimura A, Ocorr K, Bodmer R, Penninger JM. A global in vivo drosophila rnai screen identifies not3 as a conserved regulator of heart function. Cell. 2010;141:142–153. doi: 10.1016/j.cell.2010.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic drosophila by using the site-specific integrase from phage phic31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Venken KJ, Bellen HJ. Emerging technologies for gene manipulation in drosophila melanogaster. Nat Rev Genet. 2005;6:167–178. doi: 10.1038/nrg1553. [DOI] [PubMed] [Google Scholar]
- 18.Kim IM, Wolf MJ. Serial examination of an inducible and reversible dilated cardiomyopathy in individual adult drosophila. PLoS One. 2009;4:e7132. doi: 10.1371/journal.pone.0007132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roman G, Endo K, Zong L, Davis RL. P[switch], a system for spatial and temporal control of gene expression in drosophila melanogaster. Proc Natl Acad Sci U S A. 2001;98:12602–12607. doi: 10.1073/pnas.221303998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bello B, Resendez-Perez D, Gehring WJ. Spatial and temporal targeting of gene expression in drosophila by means of a tetracycline-dependent transactivator system. Development. 1998;125:2193–2202. doi: 10.1242/dev.125.12.2193. [DOI] [PubMed] [Google Scholar]
- 21.Golic KG. Site-specific recombination between homologous chromosomes in drosophila. Science. 1991;252:958–961. doi: 10.1126/science.2035025. [DOI] [PubMed] [Google Scholar]
- 22.Golic KG. Local transposition of p elements in drosophila melanogaster and recombination between duplicated elements using a site-specific recombinase. Genetics. 1994;137:551–563. doi: 10.1093/genetics/137.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Golic KG, Lindquist S. The flp recombinase of yeast catalyzes site-specific recombination in the drosophila genome. Cell. 1989;59:499–509. doi: 10.1016/0092-8674(89)90033-0. [DOI] [PubMed] [Google Scholar]
- 24.Golic MM, Rong YS, Petersen RB, Lindquist SL, Golic KG. Flp-mediated DNA mobilization to specific target sites in drosophila chromosomes. Nucleic Acids Res. 1997;25:3665–3671. doi: 10.1093/nar/25.18.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gong WJ, Golic KG. Ends-out, or replacement, gene targeting in drosophila. Proc Natl Acad Sci U S A. 2003;100:2556–2561. doi: 10.1073/pnas.0535280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rong YS, Golic KG. Gene targeting by homologous recombination in drosophila. Science. 2000;288:2013–2018. doi: 10.1126/science.288.5473.2013. [DOI] [PubMed] [Google Scholar]
- 27.Rong YS, Golic KG. A targeted gene knockout in drosophila. Genetics. 2001;157:1307–1312. doi: 10.1093/genetics/157.3.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rong YS, Titen SW, Xie HB, Golic MM, Bastiani M, Bandyopadhyay P, Olivera BM, Brodsky M, Rubin GM, Golic KG. Targeted mutagenesis by homologous recombination in d Melanogaster. Genes Dev. 2002;16:1568–1581. doi: 10.1101/gad.986602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Venken KJ, Kasprowicz J, Kuenen S, Yan J, Hassan BA, Verstreken P. Recombineering-mediated tagging of drosophila genomic constructs for in vivo localization and acute protein inactivation. Nucleic Acids Res. 2008;36:e114. doi: 10.1093/nar/gkn486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindsley DL, Grell EH, Bridges CB. Genetic variations of drosophila melanogaster. Washington?: 1967. [Google Scholar]
- 31.Lindsley DL, Zimm GG. The genome of drosophila melanogaster. San Diego: Academic Press; 1992. [Google Scholar]
- 32.Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L, Lin MF, Washietl S, Arshinoff BI, Ay F, Meyer PE, Robine N, Washington NL, Di Stefano L, Berezikov E, Brown CD, Candeias R, Carlson JW, Carr A, Jungreis I, Marbach D, Sealfon R, Tolstorukov MY, Will S, Alekseyenko AA, Artieri C, Booth BW, Brooks AN, Dai Q, Davis CA, Duff MO, Feng X, Gorchakov AA, Gu T, Henikoff JG, Kapranov P, Li R, Macalpine HK, Malone J, Minoda A, Nordman J, Okamura K, Perry M, Powell SK, Riddle NC, Sakai A, Samsonova A, Sandler JE, Schwartz YB, Sher N, Spokony R, Sturgill D, van Baren M, Wan KH, Yang L, Yu C, Feingold E, Good P, Guyer M, Lowdon R, Ahmad K, Andrews J, Berger B, Brenner SE, Brent MR, Cherbas L, Elgin SC, Gingeras TR, Grossman R, Hoskins RA, Kaufman TC, Kent W, Kuroda MI, Orr-Weaver T, Perrimon N, Pirrotta V, Posakony JW, Ren B, Russell S, Cherbas P, Graveley BR, Lewis S, Micklem G, Oliver B, Park PJ, Celniker SE, Henikoff S, Karpen GH, Lai EC, Macalpine DM, Stein LD, White KP, Kellis M. Identification of functional elements and regulatory circuits by drosophila modencode. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friedman A, Perrimon N. A functional rnai screen for regulators of receptor tyrosine kinase and erk signalling. Nature. 2006;444:230–234. doi: 10.1038/nature05280. [DOI] [PubMed] [Google Scholar]
- 34.Schnorrer F, Schonbauer C, Langer CC, Dietzl G, Novatchkova M, Schernhuber K, Fellner M, Azaryan A, Radolf M, Stark A, Keleman K, Dickson BJ. Systematic genetic analysis of muscle morphogenesis and function in drosophila. Nature. 2010;464:287–291. doi: 10.1038/nature08799. [DOI] [PubMed] [Google Scholar]
- 35.Engstrom L, Noll E, Perrimon N. Paradigms to study signal transduction pathways in drosophila. Curr Top Dev Biol. 1997;35:229–261. doi: 10.1016/s0070-2153(08)60261-6. [DOI] [PubMed] [Google Scholar]
- 36.Halfon MS, Carmena A, Gisselbrecht S, Sackerson CM, Jimenez F, Baylies MK, Michelson AM. Ras pathway specificity is determined by the integration of multiple signal-activated and tissue-restricted transcription factors. Cell. 2000;103:63–74. doi: 10.1016/s0092-8674(00)00105-7. [DOI] [PubMed] [Google Scholar]
- 37.Johnson Hamlet MR, Perkins LA. Analysis of corkscrew signaling in the drosophila epidermal growth factor receptor pathway during myogenesis. Genetics. 2001;159:1073–1087. doi: 10.1093/genetics/159.3.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karim FD, Chang HC, Therrien M, Wassarman DA, Laverty T, Rubin GM. A screen for genes that function downstream of ras1 during drosophila eye development. Genetics. 1996;143:315–329. doi: 10.1093/genetics/143.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu X, Melnick MB, Hsu JC, Perrimon N. Genetic and molecular analyses of mutations involved in drosophila raf signal transduction. EMBO J. 1994;13:2592–2599. doi: 10.1002/j.1460-2075.1994.tb06549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perrimon N, Lu X, Hou XS, Hsu JC, Melnick MB, Chou TB, Perkins LA. Dissection of the torso signal transduction pathway in drosophila. Mol Reprod Dev. 1995;42:515–522. doi: 10.1002/mrd.1080420421. [DOI] [PubMed] [Google Scholar]
- 42.Simon MA. Receptor tyrosine kinases: Specific outcomes from general signals. Cell. 2000;103:13–15. doi: 10.1016/s0092-8674(00)00100-8. [DOI] [PubMed] [Google Scholar]
- 43.Simon MA, Bowtell DD, Dodson GS, Laverty TR, Rubin GM. Ras1 and a putative guanine nucleotide exchange factor perform crucial steps in signaling by the sevenless protein tyrosine kinase. Cell. 1991;67:701–716. doi: 10.1016/0092-8674(91)90065-7. [DOI] [PubMed] [Google Scholar]
- 44.Therrien M, Chang HC, Solomon NM, Karim FD, Wassarman DA, Rubin GM. Ksr, a novel protein kinase required for ras signal transduction. Cell. 1995;83:879–888. doi: 10.1016/0092-8674(95)90204-x. [DOI] [PubMed] [Google Scholar]
- 45.Wassarman DA, Therrien M, Rubin GM. The ras signaling pathway in drosophila. Curr Opin Genet Dev. 1995;5:44–50. doi: 10.1016/s0959-437x(95)90052-7. [DOI] [PubMed] [Google Scholar]
- 46.Artavanis-Tsakonas S, Muskavitch MA. Notch: The past, the present, and the future. Curr Top Dev Biol. 2010;92:1–29. doi: 10.1016/S0070-2153(10)92001-2. [DOI] [PubMed] [Google Scholar]
- 47.Baker NE. Molecular cloning of sequences from wingless, a segment polarity gene in drosophila: The spatial distribution of a transcript in embryos. EMBO J. 1987;6:1765–1773. doi: 10.1002/j.1460-2075.1987.tb02429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grimwade BG, Muskavitch MA, Welshons WJ, Yedvobnick B, Artavanis-Tsakonas S. The molecular genetics of the notch locus in drosophila melanogaster. Dev Biol. 1985;107:503–519. doi: 10.1016/0012-1606(85)90331-8. [DOI] [PubMed] [Google Scholar]
- 49.Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 50.Raftery LA, Twombly V, Wharton K, Gelbart WM. Genetic screens to identify elements of the decapentaplegic signaling pathway in drosophila. Genetics. 1995;139:241–254. doi: 10.1093/genetics/139.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sekelsky JJ, Newfeld SJ, Raftery LA, Chartoff EH, Gelbart WM. Genetic characterization and cloning of mothers against dpp, a gene required for decapentaplegic function in drosophila melanogaster. Genetics. 1995;139:1347–1358. doi: 10.1093/genetics/139.3.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siegfried E, Wilder EL, Perrimon N. Components of wingless signalling in drosophila. Nature. 1994;367:76–80. doi: 10.1038/367076a0. [DOI] [PubMed] [Google Scholar]
- 53.Spencer FA, Hoffmann FM, Gelbart WM. Decapentaplegic: A gene complex affecting morphogenesis in drosophila melanogaster. Cell. 1982;28:451–461. doi: 10.1016/0092-8674(82)90199-4. [DOI] [PubMed] [Google Scholar]
- 54.Vinson CR, Conover S, Adler PN. A drosophila tissue polarity locus encodes a protein containing seven potential transmembrane domains. Nature. 1989;338:263–264. doi: 10.1038/338263a0. [DOI] [PubMed] [Google Scholar]
- 55.Wu S, Huang J, Dong J, Pan D. Hippo encodes a ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- 56.St Johnston D. The art and design of genetic screens: Drosophila melanogaster. Nat Rev Genet. 2002;3:176–188. doi: 10.1038/nrg751. [DOI] [PubMed] [Google Scholar]
- 57.Bellen HJ, Levis RW, Liao G, He Y, Carlson JW, Tsang G, Evans-Holm M, Hiesinger PR, Schulze KL, Rubin GM, Hoskins RA, Spradling AC. The bdgp gene disruption project: Single transposon insertions associated with 40% of drosophila genes. Genetics. 2004;167:761–781. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bellen HJ, O'Kane CJ, Wilson C, Grossniklaus U, Pearson RK, Gehring WJ. P-element-mediated enhancer detection: A versatile method to study development in drosophila. Genes Dev. 1989;3:1288–1300. doi: 10.1101/gad.3.9.1288. [DOI] [PubMed] [Google Scholar]
- 59.Hummel T, Klambt C. P-element mutagenesis. Methods Mol Biol. 2008;420:97–117. doi: 10.1007/978-1-59745-583-1_6. [DOI] [PubMed] [Google Scholar]
- 60.Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, Deal-Herr ME, Grant D, Marcinko M, Miyazaki WY, Robertson S, Shaw KJ, Tabios M, Vysotskaia V, Zhao L, Andrade RS, Edgar KA, Howie E, Killpack K, Milash B, Norton A, Thao D, Whittaker K, Winner MA, Friedman L, Margolis J, Singer MA, Kopczynski C, Curtis D, Kaufman TC, Plowman GD, Duyk G, Francis-Lang HL. Systematic generation of high-resolution deletion coverage of the drosophila melanogaster genome. Nat Genet. 2004;36:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- 61.Ryder E, Ashburner M, Bautista-Llacer R, Drummond J, Webster J, Johnson G, Morley T, Chan YS, Blows F, Coulson D, Reuter G, Baisch H, Apelt C, Kauk A, Rudolph T, Kube M, Klimm M, Nickel C, Szidonya J, Maroy P, Pal M, Rasmuson-Lestander A, Ekstrom K, Stocker H, Hugentobler C, Hafen E, Gubb D, Pflugfelder G, Dorner C, Mechler B, Schenkel H, Marhold J, Serras F, Corominas M, Punset A, Roote J, Russell S. The drosdel deletion collection: A drosophila genomewide chromosomal deficiency resource. Genetics. 2007;177:615–629. doi: 10.1534/genetics.107.076216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ryder E, Blows F, Ashburner M, Bautista-Llacer R, Coulson D, Drummond J, Webster J, Gubb D, Gunton N, Johnson G, O'Kane CJ, Huen D, Sharma P, Asztalos Z, Baisch H, Schulze J, Kube M, Kittlaus K, Reuter G, Maroy P, Szidonya J, Rasmuson-Lestander A, Ekstrom K, Dickson B, Hugentobler C, Stocker H, Hafen E, Lepesant JA, Pflugfelder G, Heisenberg M, Mechler B, Serras F, Corominas M, Schneuwly S, Preat T, Roote J, Russell S. The drosdel collection: A set of p-element insertions for generating custom chromosomal aberrations in drosophila melanogaster. Genetics. 2004;167:797–813. doi: 10.1534/genetics.104.026658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, Buchholz R, Demsky M, Fawcett R, Francis-Lang HL, Ryner L, Cheung LM, Chong A, Erickson C, Fisher WW, Greer K, Hartouni SR, Howie E, Jakkula L, Joo D, Killpack K, Laufer A, Mazzotta J, Smith RD, Stevens LM, Stuber C, Tan LR, Ventura R, Woo A, Zakrajsek I, Zhao L, Chen F, Swimmer C, Kopczynski C, Duyk G, Winberg ML, Margolis J. A complementary transposon tool kit for drosophila melanogaster using p and piggybac. Nat Genet. 2004;36:283–287. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- 64.Bodmer R, Venkatesh TV. Heart development in drosophila and vertebrates: Conservation of molecular mechanisms. Dev Genet. 1998;22:181–186. doi: 10.1002/(SICI)1520-6408(1998)22:3<181::AID-DVG1>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 65.Cripps RM, Olson EN. Control of cardiac development by an evolutionarily conserved transcriptional network. Dev Biol. 2002;246:14–28. doi: 10.1006/dbio.2002.0666. [DOI] [PubMed] [Google Scholar]
- 66.Zaffran S, Frasch M. Early signals in cardiac development. Circ Res. 2002;91:457–469. doi: 10.1161/01.res.0000034152.74523.a8. [DOI] [PubMed] [Google Scholar]
- 67.Zaffran S, Xu X, Lo PC, Lee HH, Frasch M. Cardiogenesis in the drosophila model: Control mechanisms during early induction and diversification of cardiac progenitors. Cold Spring Harb Symp Quant Biol. 2002;67:1–12. doi: 10.1101/sqb.2002.67.1. [DOI] [PubMed] [Google Scholar]
- 68.Bodmer R. The gene tinman is required for specification of the heart and visceral muscles in drosophila. Development. 1993;118:719–729. doi: 10.1242/dev.118.3.719. [DOI] [PubMed] [Google Scholar]
- 69.Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schott JJ, Benson DW, Basson CT, Pease W, Silberbach GM, Moak JP, Maron BJ, Seidman CE, Seidman JG. Congenital heart disease caused by mutations in the transcription factor nkx2–5. Science. 1998;281:108–111. doi: 10.1126/science.281.5373.108. [DOI] [PubMed] [Google Scholar]
- 71.Chartier A, Zaffran S, Astier M, Semeriva M, Gratecos D. Pericardin, a drosophila type iv collagen-like protein is involved in the morphogenesis and maintenance of the heart epithelium during dorsal ectoderm closure. Development. 2002;129:3241–3253. doi: 10.1242/dev.129.13.3241. [DOI] [PubMed] [Google Scholar]
- 72.Demerec M. The biology of drosophila. Plainview, N.Y.: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 73.Curtis NJ, Ringo JM, Dowse HB. Morphology of the pupal heart, adult heart, and associated tissues in the fruit fly, drosophila melanogaster. J Morphol. 1999;240:225–235. doi: 10.1002/(SICI)1097-4687(199906)240:3<225::AID-JMOR2>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 74.Monier B, Astier M, Semeriva M, Perrin L. Steroid-dependent modification of hox function drives myocyte reprogramming in the drosophila heart. Development. 2005;132:5283–5293. doi: 10.1242/dev.02091. [DOI] [PubMed] [Google Scholar]
- 75.Zeitouni B, Senatore S, Severac D, Aknin C, Semeriva M, Perrin L. Signalling pathways involved in adult heart formation revealed by gene expression profiling in drosophila. PLoS Genet. 2007;3:1907–1921. doi: 10.1371/journal.pgen.0030174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shah AP, Nongthomba U, Tanaka KK, Denton ML, Meadows SM, Bancroft N, Molina MR, Cripps RM. Cardiac remodeling in drosophila arises from changes in actin gene expression and from a contribution of lymph gland-like cells to the heart musculature. Mech Dev. 2011 doi: 10.1016/j.mod.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Molina MR, Cripps RM. Ostia, the inflow tracts of the drosophila heart, develop from a genetically distinct subset of cardial cells. Mech Dev. 2001;109:51–59. doi: 10.1016/s0925-4773(01)00509-3. [DOI] [PubMed] [Google Scholar]
- 78.Dulcis D, Levine RB. Innervation of the heart of the adult fruit fly, drosophila melanogaster. J Comp Neurol. 2003;465:560–578. doi: 10.1002/cne.10869. [DOI] [PubMed] [Google Scholar]
- 79.Lin N, Badie N, Yu L, Abraham D, Cheng H, Bursac N, Rockman HA, Wolf MJ. A method to measure myocardial calcium handling in adult drosophila. Circ Res. 2011;108:1306–1315. doi: 10.1161/CIRCRESAHA.110.238105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wasserthal LT. Drosophila flies combine periodic heartbeat reversal with a circulation in the anterior body mediated by a newly discovered anterior pair of ostial valves and 'venous' channels. J Exp Biol. 2007;210:3707–3719. doi: 10.1242/jeb.007864. [DOI] [PubMed] [Google Scholar]
- 81.Johnson E, Ringo J, Dowse H. Modulation of drosophila heartbeat by neurotransmitters. J Comp Physiol B. 1997;167:89–97. doi: 10.1007/s003600050051. [DOI] [PubMed] [Google Scholar]
- 82.Johnson E, Ringo J, Dowse H. Native and heterologous neuropeptides are cardioactive in drosophila melanogaster. J Insect Physiol. 2000;46:1229–1236. doi: 10.1016/s0022-1910(00)00043-3. [DOI] [PubMed] [Google Scholar]
- 83.Johnson E, Sherry T, Ringo J, Dowse H. Modulation of the cardiac pacemaker of drosophila: Cellular mechanisms. J Comp Physiol B. 2002;172:227–236. doi: 10.1007/s00360-001-0246-8. [DOI] [PubMed] [Google Scholar]
- 84.Acehan D, Khuchua Z, Houtkooper RH, Malhotra A, Kaufman J, Vaz FM, Ren M, Rockman HA, Stokes DL, Schlame M. Distinct effects of tafazzin deletion in differentiated and undifferentiated mitochondria. Mitochondrion. 2009;9:86–95. doi: 10.1016/j.mito.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dobrosotskaya IY, Seegmiller AC, Brown MS, Goldstein JL, Rawson RB. Regulation of srebp processing and membrane lipid production by phospholipids in drosophila. Science. 2002;296:879–883. doi: 10.1126/science.1071124. [DOI] [PubMed] [Google Scholar]
- 86.Dorn GW, 2nd, Clark CF, Eschenbacher WH, Kang MY, Engelhard JT, Warner SJ, Matkovich SJ, Jowdy CC. Marf and opa1 control mitochondrial and cardiac function in drosophila. Circ Res. 2011;108:12–17. doi: 10.1161/CIRCRESAHA.110.236745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lim HY, Wang W, Wessells RJ, Ocorr K, Bodmer R. Phospholipid homeostasis regulates lipid metabolism and cardiac function through srebp signaling in drosophila. Genes Dev. 2011;25:189–200. doi: 10.1101/gad.1992411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Coquin L, Feala JD, McCulloch AD, Paternostro G. Metabolomic and flux-balance analysis of age-related decline of hypoxia tolerance in drosophila muscle tissue. Mol Syst Biol. 2008;4:233. doi: 10.1038/msb.2008.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Feala JD, Coquin L, McCulloch AD, Paternostro G. Flexibility in energy metabolism supports hypoxia tolerance in drosophila flight muscle: Metabolomic and computational systems analysis. Mol Syst Biol. 2007;3:99. doi: 10.1038/msb4100139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Parisi M, Li R, Oliver B. Lipid profiles of female and male drosophila. BMC Res Notes. 2011;4:198. doi: 10.1186/1756-0500-4-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Arrese EL, Patel RT, Soulages JL. The main triglyceride-lipase from the insect fat body is an active phospholipase a(1): Identification and characterization. J Lipid Res. 2006;47:2656–2667. doi: 10.1194/jlr.M600161-JLR200. [DOI] [PubMed] [Google Scholar]
- 92.Bauer R, Voelzmann A, Breiden B, Schepers U, Farwanah H, Hahn I, Eckardt F, Sandhoff K, Hoch M. Schlank, a member of the ceramide synthase family controls growth and body fat in drosophila. EMBO J. 2009;28:3706–3716. doi: 10.1038/emboj.2009.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Beller M, Riedel D, Jansch L, Dieterich G, Wehland J, Jackle H, Kuhnlein RP. Characterization of the drosophila lipid droplet subproteome. Mol Cell Proteomics. 2006;5:1082–1094. doi: 10.1074/mcp.M600011-MCP200. [DOI] [PubMed] [Google Scholar]
- 94.DiAngelo JR, Birnbaum MJ. Regulation of fat cell mass by insulin in drosophila melanogaster. Mol Cell Biol. 2009;29:6341–6352. doi: 10.1128/MCB.00675-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gronke S, Mildner A, Fellert S, Tennagels N, Petry S, Muller G, Jackle H, Kuhnlein RP. Brummer lipase is an evolutionary conserved fat storage regulator in drosophila. Cell Metab. 2005;1:323–330. doi: 10.1016/j.cmet.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 96.Gronke S, Muller G, Hirsch J, Fellert S, Andreou A, Haase T, Jackle H, Kuhnlein RP. Dual lipolytic control of body fat storage and mobilization in drosophila. PLoS Biol. 2007;5:e137. doi: 10.1371/journal.pbio.0050137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Horne I, Haritos VS, Oakeshott JG. Comparative and functional genomics of lipases in holometabolous insects. Insect Biochem Mol Biol. 2009;39:547–567. doi: 10.1016/j.ibmb.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 98.Nohturfft A, Losick RCell biology. Fats, flies, and palmitate. Science. 2002;296:857–858. doi: 10.1126/science.1072154. [DOI] [PubMed] [Google Scholar]
- 99.Ugrankar R, Liu Y, Provaznik J, Schmitt S, Lehmann M. Lipin is a central regulator of adipose tissue development and function in drosophila melanogaster. Mol Cell Biol. 2011;31:1646–1656. doi: 10.1128/MCB.01335-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Alvarez AD, Shi W, Wilson BA, Skeath JB. Pannier and pointedp2 act sequentially to regulate drosophila heart development. Development. 2003;130:3015–3026. doi: 10.1242/dev.00488. [DOI] [PubMed] [Google Scholar]
- 101.Bidet Y, Jagla T, Da Ponte JP, Dastugue B, Jagla K. Modifiers of muscle and heart cell fate specification identified by gain-of-function screen in drosophila. Mech Dev. 2003;120:991–1007. doi: 10.1016/s0925-4773(03)00182-5. [DOI] [PubMed] [Google Scholar]
- 102.Yi P, Han Z, Li X, Olson EN. The mevalonate pathway controls heart formation in drosophila by isoprenylation of ggamma1. Science. 2006;313:1301–1303. doi: 10.1126/science.1127704. [DOI] [PubMed] [Google Scholar]
- 103.Yi P, Johnson AN, Han Z, Wu J, Olson EN. Heterotrimeric g proteins regulate a noncanonical function of septate junction proteins to maintain cardiac integrity in drosophila. Dev Cell. 2008;15:704–713. doi: 10.1016/j.devcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bier E, Bodmer R. Drosophila, an emerging model for cardiac disease. Gene. 2004;342:1–11. doi: 10.1016/j.gene.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 105.Wessells RJ, Bodmer R. Screening assays for heart function mutants in drosophila. Biotechniques. 2004;37:58–60. doi: 10.2144/04371ST01. 62, 64 passim. [DOI] [PubMed] [Google Scholar]
- 106.Wolf MJ, Amrein H, Izatt JA, Choma MA, Reedy MC, Rockman HA. Drosophila as a model for the identification of genes causing adult human heart disease. Proc Natl Acad Sci U S A. 2006;103:1394–1399. doi: 10.1073/pnas.0507359103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wolf MJ, Rockman HA. Drosophila melanogaster as a model system for genetics of postnatal cardiac function. Drug Discov Today Dis Models. 2008;5:117–123. doi: 10.1016/j.ddmod.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Paternostro G, Vignola C, Bartsch DU, Omens JH, McCulloch AD, Reed JC. Age-associated cardiac dysfunction in drosophila melanogaster. Circ Res. 2001;88:1053–1058. doi: 10.1161/hh1001.090857. [DOI] [PubMed] [Google Scholar]
- 109.Wessells RJ, Fitzgerald E, Cypser JR, Tatar M, Bodmer R. Insulin regulation of heart function in aging fruit flies. Nat Genet. 2004;36:1275–1281. doi: 10.1038/ng1476. [DOI] [PubMed] [Google Scholar]
- 110.Luong N, Davies CR, Wessells RJ, Graham SM, King MT, Veech R, Bodmer R, Oldham SM. Activated foxo-mediated insulin resistance is blocked by reduction of tor activity. Cell Metab. 2006;4:133–142. doi: 10.1016/j.cmet.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 111.Wessells R, Fitzgerald E, Piazza N, Ocorr K, Morley S, Davies C, Lim HY, Elmen L, Hayes M, Oldham S, Bodmer R. D4ebp acts downstream of both dtor and dfoxo to modulate cardiac functional aging in drosophila. Aging Cell. 2009;8:542–552. doi: 10.1111/j.1474-9726.2009.00504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Demontis F, Perrimon N. Foxo/4e–bp signaling in drosophila muscles regulates organism-wide proteostasis during aging. Cell. 2010;143:813–825. doi: 10.1016/j.cell.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang D, Contu R, Latronico MV, Zhang J, Rizzi R, Catalucci D, Miyamoto S, Huang K, Ceci M, Gu Y, Dalton ND, Peterson KL, Guan KL, Brown JH, Chen J, Sonenberg N, Condorelli G. Mtorc1 regulates cardiac function and myocyte survival through 4e–bp1 inhibition in mice. J Clin Invest. 2010;120:2805–2816. doi: 10.1172/JCI43008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ocorr K, Fink M, Cammarato A, Bernstein S, Bodmer R. Semi-automated optical heartbeat analysis of small hearts. J Vis Exp. 2009 doi: 10.3791/1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ocorr K, Reeves NL, Wessells RJ, Fink M, Chen HS, Akasaka T, Yasuda S, Metzger JM, Giles W, Posakony JW, Bodmer R. Kcnq potassium channel mutations cause cardiac arrhythmias in drosophila that mimic the effects of aging. Proc Natl Acad Sci U S A. 2007;104:3943–3948. doi: 10.1073/pnas.0609278104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vogler G, Ocorr K. Visualizing the beating heart in drosophila. J Vis Exp. 2009 doi: 10.3791/1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lee JH, Budanov AV, Park EJ, Birse R, Kim TE, Perkins GA, Ocorr K, Ellisman MH, Bodmer R, Bier E, Karin M. Sestrin as a feedback inhibitor of tor that prevents age-related pathologies. Science. 2010;327:1223–1228. doi: 10.1126/science.1182228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kim IM, Wolf MJ, Rockman HA. Gene deletion screen for cardiomyopathy in adult drosophila identifies a new notch ligand. Circ Res. 2010;106:1233–1243. doi: 10.1161/CIRCRESAHA.109.213785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yu L, Lee T, Lin N, Wolf MJ. Affecting rhomboid-3 function causes a dilated heart in adult drosophila. PLoS Genet. 2010;6:e1000969. doi: 10.1371/journal.pgen.1000969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lemberg MK, Freeman M. Functional and evolutionary implications of enhanced genomic analysis of rhomboid intramembrane proteases. Genome Res. 2007;17:1634–1646. doi: 10.1101/gr.6425307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Schweitzer R, Shaharabany M, Seger R, Shilo BZ. Secreted spitz triggers the der signaling pathway and is a limiting component in embryonic ventral ectoderm determination. Genes Dev. 1995;9:1518–1529. doi: 10.1101/gad.9.12.1518. [DOI] [PubMed] [Google Scholar]
- 122.Maggert KA, Gong WJ, Golic KG. Methods for homologous recombination in drosophila. Methods Mol Biol. 2008;420:155–174. doi: 10.1007/978-1-59745-583-1_9. [DOI] [PubMed] [Google Scholar]
- 123.Fink M, Callol-Massot C, Chu A, Ruiz-Lozano P, Izpisua Belmonte JC, Giles W, Bodmer R, Ocorr K. A new method for detection and quantification of heartbeat parameters in drosophila, zebrafish, and embryonic mouse hearts. Biotechniques. 2009;46:101–113. doi: 10.2144/000113078. [DOI] [PMC free article] [PubMed] [Google Scholar]