Abstract
The measurement of nitric oxide in lipopolysaccharide (LPS)-stimulated RAW 264.7 cells is used as a model for evaluating the anti-inflammatory or chemopreventive potential of substances. Thienodolin, isolated from a Streptomyces sp. derived from Chilean marine sediment, inhibited nitric oxide production in LPS-stimulated RAW 264.7 cells (IC50 = 17.2 ± 1.2 μM). At both the mRNA and protein levels, inducible nitric oxide synthase (iNOS) was suppressed in a dose-dependent manner. Mitogen-activated protein kinases (MAPKs), one major upstream signaling pathway involved in the transcription of iNOS, were not affected by treatment of thienodolin. However, the compound blocked the degradation of IκBα resulting in inhibition of NF-κB p65 nuclear translocation, and inhibited the phosphorylation of signal transducers and activators of transcription 1 (STAT1) at Tyr701. This study supports further exploration of thienodolin as a potential therapeutic agent with a unique mechanistic activity.
Keywords: Nitric oxide, Inducible nitric oxide synthase, Macrophage, Thienodolin, Inflammation, Cancer prevention
Nitric oxide synthases (NOS) catalyze the bioconversion of l-arginine to l-citrulline, and one a reactive nitrogen species, nitric oxide (NO). Three major isoforms of NOS have been identified, including endothelial NOS (eNOS or NOS I), inducible NOS (iNOS or NOS II), and neuronal NOS (nNOS or NOS III) [1]. In addition, other variants have been identified, including mitochondrial NOS, which participates in mitochondrial respiration [2]. eNOS and nNOS help to maintain homeostasis in the cardiovascular and nervous systems, respectively, whereas iNOS is involved in a host of defense mechanisms with high expression levels induced by various exogenous or endogenous stimuli, mainly in macrophages and in smooth muscle and liver [3,4]. Consequently, relative to other isoforms [5,6], over a thousand-fold more NO is generated by iNOS under inflammatory conditions.
In spite of the physiological roles of NO, the aberrant existence of NO primarily due to the overexpression of iNOS has been reported to play a pathophysiological role in chronic diseases and septic (endotoxin) shock. In fact, excessive NO production can cause post-ischemic stroke damage, septic shock, seizures, schizophrenia, migraine headaches, Alzheimer's disease, tolerance to and dependence on morphine, development of colitis, tissue damage and inflammation, overproduction of osteoclasts leading to osteoporosis, Paget's disease and rheumatoid arthritis, destruction of photoreceptors in the retina, and long-term depression [7].
Once iNOS is induced, it can sustainably produce excessive amounts of NO for a prolonged period of time [8]. As a result, several chronic inflammatory diseases have been associated with continuous iNOS expression. As examples, iNOS expression was observed in rheumatoid arthritis, multiple sclerosis, Sjögren's syndrome, asthma, bronchioctasis, idiopathic pulmonary fibrosis, atherosclerotic plaques, inflammatory bowel diseases (ulcerative colitis, Crohn's disease, necrotizing enterocolitis, and coeliac disease), glomerulonephritis, dilated cardiomyopathy, psoriasis, cutaneous lupus, systemic lupus, systemic sclerosis, dermatitis, and periapical periodonitis [9]. In addition, some chronic inflammatory conditions are positively correlated with neoplastic transformation [10]. It was observed that injection of Escherichia coli into the bladders of rats resulted in inflammation, papillary hyperplasia, and eventually squamous metaplasia [11]. In accord with these observations, iNOS, which is frequently expressed in chronic inflammatory lesions, has been detected in malignant tumors of breast, brain, lung, prostate, colon, pancreas, and skin. Furthermore, it was found that patients with iNOS-expressing melanomas show significantly shorter survival rates than iNOS-negative counterparts [12]. In this respect, the discovery of iNOS inhibitors is important for the treatment of inflammatory diseases, as well as the prevention of cancers.
During the course of our search for bioactive natural products from marine-derived actinomycete strains, the crude extract of our strain, CNY-325, exhibited significant activity in screens associated with tumor induction. This strain, isolated from a Chilean marine sediment, was identified as a Streptomyces sp. based on 16S rDNA gene sequence analysis. Bioassay-guided separation of the crude extract using diverse chromatographic methods yielded dechlorothienodolin (1) and thienodolin (2) (Figure 1).
Figure 1.
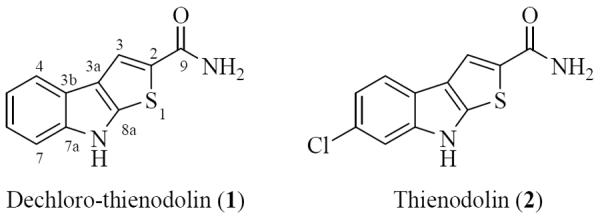
Chemical structures of dechloro-thienodolin (1) and thienodolin (2).
The molecular formula of dechloro-thienodolin (1) was assigned as C11H8N2OS by interpretation of combined HRESIMS and 13C NMR spectral data. The IR spectrum of 1 showed an absorption band at 1650 cm−1, which suggested the presence of an amide group. The distinct chemical shifts and coupling constants of four aromatic proton signals (H-4~H-7; δ 7.74, dd, J = 8.2, 1.3 Hz, 7.14, ddd, J = 8.2, 8.2, 1.3 Hz, 7.23, ddd, J = 8.2, 8.2, 1.3 Hz, 7.48, dd, J = 8.2, 1.3 Hz, respectively) in the 1H NMR spectrum illustrated the presence of a 1,2 disubstituted benzene ring. The 1H NMR spectrum of 1 displayed an olefinic proton H-4, which showed an HMBC correlation to a quaternary olefinic carbon (C-3a, δ 123.7). A long range HMBC correlation from H-3 to three quaternary olefinic carbons (C-3a, δ 123.7; C-8a, δ 144.3; C-2, δ 131.5), and to a primary amide carbonyl carbon (C-9, δ 164.3), were also observed. These data, in conjunction with the molecular formula, revealed the structure of 1 as dechloro-thienodolin. This assignment was confirmed by comparison of previously reported spectroscopic data. Thienodolin (2) was reported as a plant growth-regulating substance from Streptomyces albogriseolus [13]. In 2004, Engqvist et al. reported a total synthesis of thienodolin (2), as well as its unsubstituted analogue dechloro-thienodolin (1). Dechloro-thienodolin (1) was isolated in this study for the first time as a natural product [14]. In the present study, we investigated the inhibitory effects of dechloro-thienodolin and thienodolin on NO production and iNOS expression, as well as underlying mechanisms in lipopolysaccharide (LPS)-stimulated RAW 264.7 cells.
First, we examined the influence of dechloro-thienodolin and thienodolin on NO production in LPS-stimulated RAW 264.7 cells. We measured nitrite as the stable end-production of NO. It was observed that the expression levels of eNOS, iNOS and nNOS in resting RAW 264.7 cells are barely detectable [15]. In accordance with this, the basal level of nitrite in media of cultured RAW 264.7 cells was 0.4 ± 0.2 μM, whereas incubation with LPS (1 μg/mL) for 20 h increased the nitrite production to 23.5 ± 0.8 μM. However, the pretreatment with compounds for 15 min prior to LPS exposure inhibited the nitrite production in comparison with LPS-treated control cells in a dose-dependent manner with a significant difference at 25 μM and 50 μM. Notably, thienodolin inhibited the nitrite production more than dechloro-thienodolin at each concentration with greater potency, yielding an IC50 value of 17.2 μM. As determined by the MTT assay, dechloro-thienodolin and thienodolin did not affect cell survival under any of these experimental conditions (Figure 2).
Figure 2.
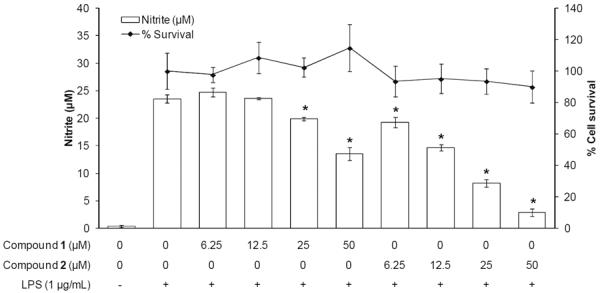
Effect of dechloro-thienodolin (1) and thienodolin (2) on nitrite production by LPS-stimulated RAW 264.7 cells for 20 h. RAW 264.7 cells were pretreated with or without compounds for 15 min prior to LPS treatment for 20 h. The levels of nitrite in cultured media were measured by Griess reagent. Data are expressed as mean±SEM. *p<0.05.
Several parameters are known to affect NO synthesis including the amount or availability of l-arginine as a substrate and iNOS proteins as an enzyme. More specifically, the cellular level of l-arginine is dependent on the expression level of cationic amino acid transporters and arginase, and the enzyme activity of iNOS is dependent on iNOS mRNA and protein expression levels, iNOS-specific cofactors, post-translational modification, and existence of endogenous antagonists [9]. The burst of NO upon LPS-stimulation in RAW 264.7 macrophages primarily results from the drastic increase in iNOS expression. Therefore, we examined the effect of thienodolin on the expression levels of iNOS. For this purpose, RAW 264.7 cells were pretreated with various concentrations of thienodolin for 15 min and further incubated with LPS (1 μg/mL) for either 18 h or 5 h, in order to evaluate protein expression and mRNA expression, respectively. As shown in Figure 3, thienodolin suppressed the expression of iNOS dose-dependently at both the protein and mRNA levels. The expression of one of the housekeeping genes, β-actin, was examined as a loading control, but no apparent changes were observed.
Figure 3.
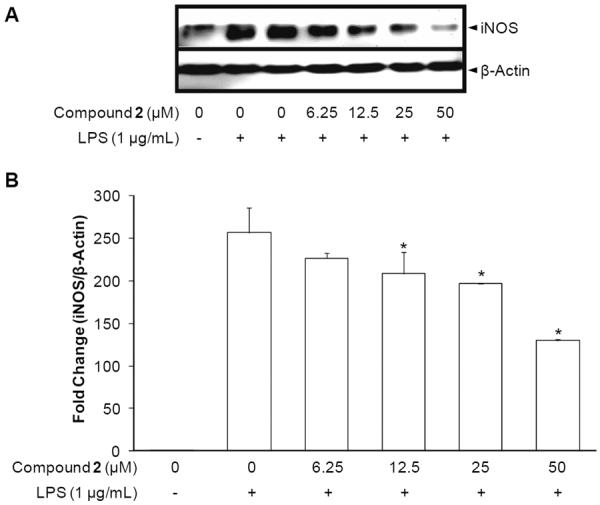
Effect of thieonodolin on iNOS protein expression and mRNA levels. A, Inhibitory effect of thienodolin (2) on iNOS protein expression. RAW 264.7 cells were pretreated with the indicated concentrations of thienodolin (0–50 μM) for 15 min, and then stimulated with LPS (1 μg/mL). After an 18 h incubation period, cells were lysed, and 15 μg protein was applied on a 9% SDS-polyacrylamide gel. The levels of iNOS protein expression were examined by Western blot analyses using antibodies against iNOS. β-Actin was used as an internal standard. B, Inhibitory effect of thienodolin (2) on iNOS mRNA levels. RAW 264.7 cells were pretreated with the indicated concentrations of thienodolin (0–50 μM) for 15 min, and then stimulated with 1 μg/mL LPS. After a 5 h incubation period, RNA was extracted and quantified. Total RNA (1 μg) was used in the RT-PCR reaction. After normalization to the internal standard, β-actin, fold-changes relative to vehicle-treated control are presented (n = 3). An asterisk (*) indicates a significant difference with P values less than 0.05.
Given this result, we investigated essential molecules in upstream signaling pathways, which mediate iNOS expression. In this cell-line based system, LPS, one of endotoxins located in the outer membrane of Gram-negative bacteria, which can lead to endotoxin shock, was used to activate the signaling pathways. Upon LPS exposure, plasma membrane-bound Toll-like receptor 4 (TLR4) recognizes it and propagates activation signals to two major intracellular pathways including the myeloid differentiation factor 88 (MyD88)-dependent and Toll/IL-1 receptor domain-containing adapter inducing interferon-β (TRIF)-dependent pathways. The activation of mitogen-activated protein kinases (MAPKs) and nuclear factor κB (NF-κB) take place as downstream signaling events, while the activation of signal transducer and activator of transcription 1 (STAT1) occurs in the TRIF-dependent pathway [16]. Eventually, those signaling molecules mentioned above either activate transcriptional factors or act as transcriptional factors. It has been reported that NF-κB, interferon regulatory factor-1 (IRF-1), signal transducer and activator of transcription-1α (STAT-1α), cAMP-induced transcription factors; cAMP-responsive element binding protein (CREB), CCAAT-enhancer box binding protein (C/EBP), and activating protein-1 (AP-1) promote the expression of iNOS [17].
Therefore, to further examine the molecular mechanism underlying thienodolin-mediated inhibition of iNOS expression, cellular levels of upstream signaling molecules, mitogen-activated protein kinases (MAPKs) were determined by Western blot analysis. RAW 264.7 cells were pretreated with thienodolin for 15 min, and exposed to LPS (1 μg/mL) for 30 min. As shown in Figure 4, LPS treatment resulted in the induced phosphorylation of MAPKs, including p-p38 MAPK, p-ERK1/2, and p-SAPK/JNK. However, thienodolin did not affect either the total or phosphorylated forms of MAPKs.
Figure 4.
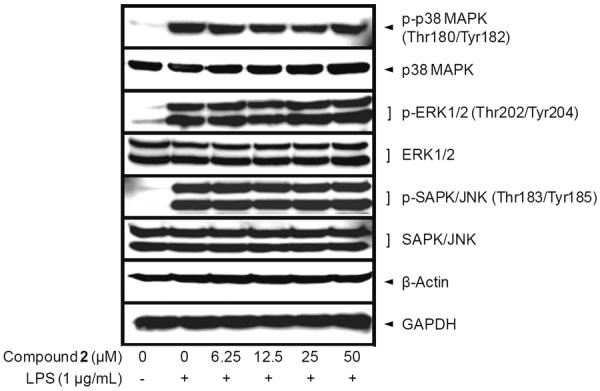
Effect of thienodolin on LPS-induced MAPKs activation in cultured RAW 264.7 cells. RAW 264.7 cells were pretreated with various concentrations up to 50 μM of thienodolin (2) for 15 min, and then incubated with LPS (1 μg/mL) for 15 min. Total cell lysate was prepared and the levels of p-p38 MAPK, total p38 MAPK, p-ERK1/2, total ERK1/2, p-SAPK/JNK, and SAPK/JNK were analyzed by Western blotting.
NF-κB is another key regulator of iNOS expression in inflammation [18]. Therefore, we examined the effect of thienodolin on the NF-κB pathway. In resting macrophages, NF-κB subunits are sequestered in the cytoplasm by interacting with inhibitor of κB (IκB) proteins. However, in LPS-driven activation, IκB is phosphorylated by IκB kinases (IKKs), and degraded in an ubiquitin-dependent manner, leading to the nuclear translocation of NF-κB. So far, several mammalian IκB family members have been identified, including IκBγ1, IκBγ2, IκBδ, IκBε, IκBR, IκBL, p100, p105, Bcl-3, and IκBζ [19]. Since it has been reported that the degradation of IκBα is crucial for NF-κB activation, and IκBα is the most well-characterized member [20], and introduction of a vector encoding mutant IκBα which is tolerant to the phosphorylation and degradation results in significant inhibition of nitrite production upon treatment with stimuli [21], the effect of thienodolin on protein degradation of IκBα was examined.
From our previous report [22], IκBα degradation reaches maximal levels from 10 to 30 min following LPS treatment. Based on this, RAW 264.7 cells were incubated with thienodolin for 15 min prior to LPS (1 μg/mL), and then for an additional 15 min. As shown in Figure 5A, treatment with LPS led to near complete IκBα degradation, however, it was blocked by thienodolin treatment in a concentration-dependent manner. Next, it was determined whether thienodolin inhibited the translocation of NF-κB to the nucleus. NF-κB exists as hetero- or homodimers of subunits including RelA (p65), RelB, cRel, p50 and p52 [18]. NF-κB p65 is known as a critical transactivation subunit for NF-κB [23]. Along with this, the effect of thienodolin on the nuclear localization of NF-κB p65 in RAW 264.7 cells was examined. As shown in Figure 5B, the NF-κB p65 subunit in the nuclear compartment was increased after the incubation with LPS (1 μg/mL) for 1 h in RAW 264.7 cells. However, thienodolin treatment prior to LPS exposure attenuated NF-κB p65 accumulation in the nucleus. Lamin A/C was used as a nuclear loading control. Protein expression of NF-κB p65 in the total cell lysate was not changed by the treatment of LPS or thienodolin (Figure 5C).
Figure 5.
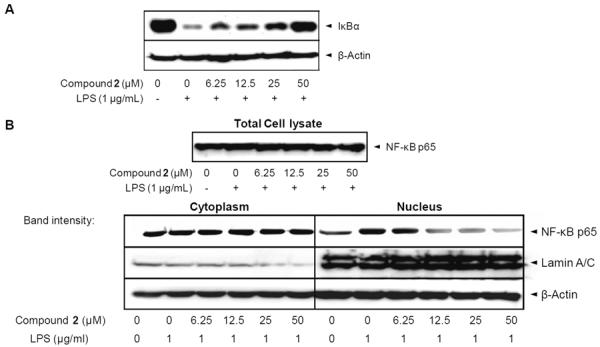
Effect of thienodolin on IκBα degradation, and NF-κB nuclear translocation, in cultured RAW 264.7 cells. A, Cells were pretreated with the indicated concentrations of thienodolin (2) for 15 min, followed by incubation with LPS (1 μg/mL) for 15 min. Cells were then lysed and IκBα protein levels were analyzed by Western blotting B, Cells were pretreated with the indicated concentrations of thienodolin (2) for 15 min, incubated with LPS for 1 h, and nuclear fractions were extracted and analyzed by Western blotting.
In the TRIF-dependent pathway of LPS signaling, interferon regulatory factor 3 (IRF3) is activated by phosphorylation, followed by induction of interferon-β (IFN-β), which eventually leads to the phosphorylation of STAT1 at Tyr701 [24,25]. In fact, it was reported that the treatment of IFN-β without LPS resulted in the phosphorylation of STAT1 at Tyr701 in RAW 264.7 cells [26]. STAT1 is one of the crucial transcriptional factors for iNOS expression as described above. Notably, the knock-out model of STAT1 has been reported for the down-regulated expression of iNOS in response to LPS in RAW 264.7 cells [27]. Focusing on the effect of STAT1 on iNOS expression, we examined the phosphorylation level of STAT1, which represents the activation of STAT1. As shown in Figure 6, the phosphorylation level of STAT1 at Tyr701 reached a maximum after 4–6 h of LPS exposure. However, pretreatment with thienodolin (50 μM) prior to LPS exposure for 4 h strongly reduced the phosphorylation level of STAT1 at Tyr701.
Figure 6.
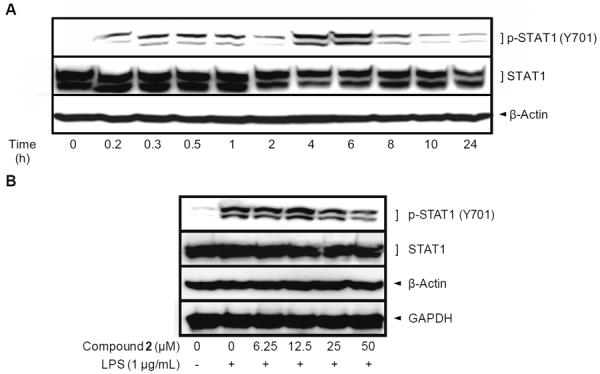
Effect of thienodolin (2) on phosphorylation of STAT1 at Tyr701 in cultured RAW 264.7 cells. A, Time course study of STAT1 phosphorylation at Tyr701 by LPS in RAW 264.7 cells. RAW 264.7 cells were treated with 1 μg/mL LPS for the indicated time periods and lysed to determine the protein expression levels of p-STAT1 (Tyr701) using Western blot analysis. B, Inhibitory effect of thienodolin (2) on the level of p-STAT1 (Tyr701). RAW 264.7 cells were pretreated with the indicated concentrations of thienodolin for 15 min followed by incubation with LPS (1 μg/mL) for 4 h. Cells were then lysed and p-STAT1 (Tyr701) levels were determined by Western blotting.
NO and its stable end products have been suggested to be involved in carcinogenesis as reactive nitrogen species by inducing DNA damage and mutation. LPS, as an endotoxin, can cause septic shock by generating high levels of NO production and iNOS induction [28]. Also, LPS might induce carcinogenic transformation in prostate epithelial cells [29]. Herein, it was observed that dechlorothienodolin and thienodolin inhibited NO production in LPS-stimulated RAW 264.7 cells, and therefore may represent promising candidates for anti-inflammation and cancer chemoprevention.
Since thienodolin showed more potent activity than dechlorothienodolin, further mechanistic studies were performed with this compound. Pretreatment with thienodolin prior to LPS exposure attenuated the induction of iNOS at the protein and mRNA levels. Since gene expression of iNOS is controlled by upstream signaling molecules, we examined the effect of thienodolin on MAPKs, NF-κB and STAT1. Collectively, thienodolin inhibited the degradation of IκBα, subsequent nuclear translocation of NF-κB p65, and phosphorylation levels of STAT1 at Tyr701.
There are various compounds capable of exhibiting similar effects. For example, some flavonoids, including genistein, kaempferol, quercetin, and daidzein, inhibit NO production and iNOS expression via down-regulating STAT-1 and NF-κB in LPS-stimulated cells [30]. Also, selenium, and stilbenoids including resveratrol and pinosylvin, have been reported to inhibit not only the NF-κB pathway but also the TRIF-dependent pathway [31–33].
Similar to other dietary phytochemicals, thienodolin showed multiple inhibitory mechanisms in LPS-induced iNOS expression.
In general, halogenation by chemical reactions requires harsh conditions and generates unnecessary byproducts, while biohalogenation does not. Therefore, naturally occurring halogenated compounds can be attractive as specifically attainable drug candidates [34]. Halogenated secondary metabolites are generated from tryptophan, pyrrole derivatives, phenol derivatives, and activated aliphatic compounds by flavin-dependent halogenases in microorganisms. Thienodolin, an indole ring chlorinated in the 6-position, is one of the halogenated secondary metabolites, and believed to be biosynthesized from tryptophan. Although the entire process has yet to be elucidated, it was found that tryptophan 6-halogenase is involved in catalyzing the initial steps of thienodolin biosynthesis in Streptomyces albogriseolus [13,34,35].
Tryptophan is an essential amino acid. It was reported that 3-hydroxyanthranilic acid, one of the tryptophan metabolites in human beings, significantly enhanced both indoleamine 2,3-dioxygenase (IDO) expression and IDO activity, and inhibited NO production and iNOS expression via its enhancement of heme oxygenase-1 expression. [36]. In this study, we described for the first time, the inhibitory effects of plausible halogenated tryptophan analogues on iNOS expression and NO production via down-regulation of upstream signaling molecules including NF-κB and STAT1. In summary, thienodolin can be considered as a potential drug candidate for anti-inflammation and cancer chemoprevention with a unique mechanism of action.
Experimental
General experimental procedures
IR, Perkin-Elmer 1600 FT-IR spectrometer; UV, Varian Cary UV-visible spectrophotometer; NMR, Varian Inova spectrometers; Low-resolution LC-MS, (Hewlett-Packard series 1100 LC/MS system), and high resolution mass TOF spectral data were acquired at the Scripps Research Institute, La Jolla, CA.
Collection and phylogenetic analysis of strain CNY-325
The marine-derived Streptomyces strain CNY-325 was isolated from a Chilean marine sediment (Valparaiso, V region, Chile). NCBI blast analysis of the partial (1480 base pairs) 16S rDNA gene sequence of CNY-325 indicated that this strain shared only 93% sequence identity to Streptomyces sp. GZ33, and thus is likely to be a new species.
Cultivation and extraction
Streptomyces sp. (strain CNY325) was cultured in 2.8 L Fernbach flasks each containing 1 L of a seawater-based medium (10 g starch, 4 g yeast extract, 2 g peptone, 1 g CaCO3, 40 mg Fe2(SO4)3-4H2O, 100 mg KBr) and shaken at 230 rpm at 27°C. After 7 days of cultivation, sterilized XAD-16 resin (20 g/L) was added to absorb the organic products, and the culture and resin were shaken at 215 rpm for 2 h. The resin was filtered through cheesecloth and washed with deionized water, and eluted with acetone. The acetone was removed under reduced pressure, and the resulting aqueous crude material was extracted with ethyl acetate (2 × 300 mL). The ethyl acetate-soluble fraction was dried in vacuo to yield 1.3 g of crude material from a 10 L culture.
Isolation of dechloro-thienodolin (1)
The crude extract (1.3 g) from strain CNH-365 was subjected to silica normal-phase vacuum flash chromatography using sequential mixtures of CH2Cl2 and methanol as eluents (elution order: 1% methanol, 2%, 5%, 10%, 50% methanol in CH2Cl2, 100% methanol). Based on LC-MS analysis, an aliquot (dry wt, 130 mg) of the fraction eluted with 10% methanol in CH2Cl2 from flash chromatography was fractionated by semi-preparative HPLC (Phenomenex C18, 250 × 100 mm, 5 μm, 70% aq. CH3CN, 3.0 mL/min flow rate, UV 210 nm) to yield, in order of elution, dechloro-thienodolin and 2 as dark-yellow solids. Final purification of the individual compounds was then accomplished by HPLC (Phenomenex C18, 250 × 100 mm, 5 μm, 80% aq. CH3CN, 2.5 mL/min flow rate, UV 210 nm) to afford 15.8 and 3.2 mg of dechloro-thiendolin (1) and thienodolin (2), respectively.
Dechloro-thienodolin (1)
Dark-yellow solid.
IR (KBr): νmax 3100, 1685, 1650 cm−1.
UV (MeOH): λmax (log ε) 232 (3.32), 260 (2.85), 285 (2.88), 340 (2.98) nm.
1H NMR (500 MHz, DMSO-d6): 11.87 (s, 2H, NH-8), 8.13 (s, 1H, H-3), 7.93 (br s, 1H, NH-9), 7.74 (dd, J = 8.2, 1.3, 1H, H-4), 7.48 (dd, J = 8.2, 1.3, 1H, H-7) 7.27 (br s, 1H, NH-9), 7.23 (ddd, J = 8.2, 8.2, 1.3, 1H, H-6), 7.14 (ddd, J = 8.2, 8.2, 1.3, 1H, H-5).
13C NMR (75 MHz, DMSO-d6): δ 164.3 (C, C-9), 144.3 (C, C-8a), 141.9 (C, C-7a), 131.5 (C, C-2), 123.7 (C, C-3a), 122.6 (CH, C-6), 121.7 (C, C-3b), 120.1 (CH, C-3), 119.6 (CH, C-5), 118.8 (CH, C-4), 111.9 (CH, C-7).
HR ESI-TOF m/z 215.0285 [M-H]+ (calcd for C11H7N2OS, 215.0285).
Thienodolin (2)
1H NMR (500 MHz, DMSO-d6): 8.11 (s, 1H, H-3), 8.00 (br s, 1H, NH-9), 7.75 (d, J = 8.5, 1H, H-4), 7.58 (s, 1H, H-7), 7.30 (br s, 1H, NH-9), 7.15 (d, J = 8.5, 1H, H-5).
13C NMR (75 MHz, DMSO-d6): δ 164.0 (C, C-9), 145.0 (C, C-8a), 142.2 (C, C-7a), 132.4 (C, C-2), 127.0 (C, C-6), 123.3 (C, C-3a), 120.5 (C, C-3b), 120.0 (CH, C-4), 119.9 (CH, C-3), 119.7 (CH, C-5), 111.6 (CH, C-7).
HR ESI-TOF m/z 272.9858 [M+Na]+ (calcd for C11H7ClN2OSNa, 272.9860).
Reagents
Lipopolysaccharide (LPS), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT), and bromochloro-propane (BCP) were purchased from Sigma-Aldrich Co. (St. Louis, MO, USA). Cell lysis buffer (10×), rabbit monoclonal antibodies against GAPDH, phospho (p)-p44/42 MAPK (ERK1/2) (Thr202/Tyr204), ERK1/2, p-p38 MAPK (Thr180/Tyr182), p-SAPK/JNK (Thr183/Tyr185), SAPK/JNK, rabbit polyclonal antibodies against β-actin, COX-2, IκB-κ, p38 MAPK, p-Stat1 (Tyr701), STAT1, lamin A/C, NF-κB p65, anti-rabbit IgG, and HRP-linked antibody were purchased from Cell Signaling Technology (Beverly, MA, USA). Rabbit polyclonal antibody against lamin A/C was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit polyclonal anti-iNOS antibody was purchased from Abcam Inc. (Cambridge, MA, USA). An enhanced chemiluminescence (ECL) detection kit was purchased from Amersham Biosciences (Piscataway, NJ, USA). Dulbecco's Modified Eagle's Medium (DMEM), fetal bovine serum (FBS), antibiotics-antimycotics (100 U/mL penicillin G sodium, 100 μg/mL streptomycin sulfate and 0.25 μg/mL amphotericin B), oligonucleotide PCR primers (Table 1), and Tri reagent® solution were purchased from Invitrogen™ (Carlsbad, CA, USA). RT2 First Strand Kit (Reagents for Twelve (12) First Strand cDNA Synthesis Reactions) was purchased from SABiosciences™ (Frederick, MD, USA). PerfeCTa™ SyBR® Green FastMix™, and ROX were purchased from Quanta biosciences™ (Gaithersburg, MD, USA). NE-PER® nuclear and cytoplasmic extraction reagents, and Lane Marker Reducing Sample Buffer (5×) were purchased from Thermo Fisher Scientific (Rockford, IL, USA).
Cell culture
RAW 264.7 murine macrophage cells were maintained in DMEM supplemented with 10% heat-inactivated FBS, 100 units/mL penicillin, and 100 μg/mL streptomycin at 37°C in a 5% CO2 humidified incubator.
Measurement of nitrite production
The level of nitrite, the stable end product of NO, was measured as described previously [37]. In brief, RAW 264.7 cells (1×104 cells per well) were seeded and incubated in 96-well culture plates at 37°C, and 5% CO2 in humidified air for 24 h. Then, complete medium was replaced with phenol red-free medium containing various concentrations of thienodolin, followed by treatment with 1 μg/mL of LPS for 20 h. The nitrite released in the culture media (100 μL) was reacted with Griess reagent and the absorbance was measured at 540 nm. The amount of nitrite was calculated using a standard curve of the known nitrite concentration versus absorbance at 540 nm. Under the same experimental conditions, cell viability was evaluated by conducting MTT assay. MTT assays were performed to evaluate the cytotoxic effect of tested compounds toward RAW 264.7 cells [38]. Briefly, after transferring 100 uL of media, MTT solution dissolved in PBS was added to each well at a final concentration of 500 μg/mL, and cells were further incubated at 37°C for 4 h. The supernatant was decanted from cells, and formazan crystals on the bottom of the wells were solubilized in 100% DMSO. The OD of each well was measured at 540 nm using a microplate reader. Absorbance (MTT values) of treatment groups was normalized to the LPS-treated control, and expressed as a percentage.
Western blot analyses
The effect of thienodolin on protein expression and subcellular localization in RAW 264.7 cells was evaluated by Western blot analysis. After incubation for the indicated time, cells were washed 3 times with PBS, harvested by using a cell scraper, and collected by centrifugation (×500 g, 3 min, 4°C). The cell pellets were resuspended with either 1× cell lysis buffer for the whole cell lysate, or with NE-PER® nuclear and cytoplasmic extraction reagents for fractionation of cytoplasmic and nuclear proteins, according to the manufacturer's instructions. The cell lysate was stored at −80°C until use. After the quantification of protein using Bradford reagent [39], equal amounts of cellular protein in each sample (10–30 μg) were resolved using 8–11% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and electrically transferred onto FluoroTrans® PVDF transfer membranes (Pall Life Sciences, Ann Arbor, MI, USA). The membranes were incubated with 5% skimmed milk in Tris-buffered saline containing 0.1% Tween 20 (TBST) for 1 h at room temperature to block non-specific protein binding. Then, membranes were incubated with corresponding primary antibodies (1:1000–1:2000) in 3% skimmed milk in TBS overnight at 4°C. After washing 3 times with TBST, membranes were further incubated with horseradish peroxidase-conjugated secondary antibodies for 1–3 h at room temperature. Membranes were washed 3 times with TBST, and the immune complexes were visualized using an ECL detection kit. Images were captured using a Geliance 1000 imager (Perkin Elmer, Inc., Waltham, MA, USA).
Quantitative real time-polymerase chain reaction (RT-PCR) analysis
Total RNA was isolated from RAW 264.7 cells using Trizol® reagent solution according to the manufacturer's instructions. RNA was dissolved in RNase-free water and the concentration and purity of RNA was determined by measuring the absorbances at 230, 260, and 280 nm using a BioSpec-nano spectrophotometer (Shimadzu Scientific Instruments, Inc, Columbia, MD, USA). An equal amount of RNA (1 μg) was reverse transcribed to cDNA using a RT2 First Strand Kit (C-03) at 42°C for 15 min and 95°C for 5 min in an ABI 7300 thermocycler (Applied Biosystems Inc., Foster City, CA, USA). Amplification of specific target cDNA was achieved by using 2× PerfeCTa™ SyBR® Green FastMix™, ROX reagent, with corresponding primer pairs as follows: iNOS: 5'-GGA GCG AGT TGT GGA TTG TC- 3' (sense) and 5'-GTG AGG GCT TGG CTG AGT GAG-3' (antisense); COX-2: 5'-GAA GTC TTT GGT CTG GTG CCT G-3' (sense) and 5'-GTC TGC TGG TTT GGA ATA GTT GC-3' (antisense); β-Actin: 5'-GCT ACA GCT TCA CCA CCA CAG-3' and 5'-GGT CTT TAC GGA TGT CAA CGT C-3' (antisense). PCR was conducted under the following 50-time thermal cycling conditions after the initial denaturation at 94°C for 3 min: Denaturation at 94°C for 30 s, annealing at 57°C for 30 s, and elongation at 72°C for 30 s [40]. The signal was collected at the end of each cycle and fold changes were calculated based on the 2−ΔΔCT method.
Statistical analysis
Data were expressed as means ± standard deviation (SD) for the indicated number of independently performed experiments. Statistical significance between treated and control groups was determined by one-way analysis of variance (ANOVA) using Microsoft Excel 2007 software (Microsoft Corporation, Redmond, Washington). P values less than 0.05 (p < 0.05) were considered statistically significant. The IC50 values were determined using TableCurve software 2D (version 4) curve-fitting software (Jandel Scientific, Corte Madera, CA).
Acknowledgments
This work is the result of financial support from the NIH under grants R37 CA044848 and P01 CA48112.
References
- [1].Mayer B, Hemmens B. Biosynthesis and action of nitric oxide in mammalian cells. Trends in Biochemical Sciences. 1997;22:477–481. doi: 10.1016/s0968-0004(97)01147-x. [DOI] [PubMed] [Google Scholar]
- [2].Bates TE, Loesch A, Burnstock G, Clark JB. Immunocytochemical evidence for a mitochondrially located nitric oxide synthase in brain and liver. Biochemical and Biophysical Research Communications. 1995;213:896–900. doi: 10.1006/bbrc.1995.2213. [DOI] [PubMed] [Google Scholar]
- [3].Liu VW, Huang PL. Cardiovascular roles of nitric oxide: a review of insights from nitric oxide synthase gene disrupted mice. Cardiovascular Research. 2008;77:19–29. doi: 10.1016/j.cardiores.2007.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Aktan F. iNOS-mediated nitric oxide production and its regulation. Life Sciences. 2004;75:639–653. doi: 10.1016/j.lfs.2003.10.042. [DOI] [PubMed] [Google Scholar]
- [5].Bricker S. The Anaesthesia Science Viva Book. Cambridge University Press; Cambridge: 2004. p. 135. [Google Scholar]
- [6].Hiraku Y, Kawanishi S, Ichinose T, Murata M. The role of iNOS-mediated DNA damage in infection- and asbestos-induced carcinogenesis. Annals of the New York Academy of Sciences. 2010;1203:15–22. doi: 10.1111/j.1749-6632.2010.05602.x. [DOI] [PubMed] [Google Scholar]
- [7].Salerno L, Sorrenti V, Di Giacomo C, Romeo G, Siracusa MA. Progress in the development of selective nitric oxide synthase (NOS) inhibitors. Current Pharmaceutical Design. 2002;3:177–200. doi: 10.2174/1381612023396375. [DOI] [PubMed] [Google Scholar]
- [8].Pilon G, Charbonneau A, White PJ, Dallaire P, Perreault M, Kapur S, Marette A. Endotoxin mediated-iNOS induction causes insulin resistance via ONOO−induced tyrosine nitration of IRS-1 in skeletal muscle. PLoS One. 2010;5:e15912. doi: 10.1371/journal.pone.0015912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Suschek CV, Schnorr O, Kolb-Bachofen V. The role of iNOS in chronic inflammatory processes in vivo: is it damage-promoting, protective, or active at all? Current Molecular Medicine. 2004;4:763–775. doi: 10.2174/1566524043359908. [DOI] [PubMed] [Google Scholar]
- [10].Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chang AH, Parsonnet J. Role of bacteria in oncogenesis. Clinical Microbiology Reviews. 2010;23:837–857. doi: 10.1128/CMR.00012-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Singh S, Gupta AK. Nitric oxide: role in tumour biology and iNOS/NO-based anticancer therapies. Cancer Chemotherapy and Pharmacology. 2011;67:1211–1224. doi: 10.1007/s00280-011-1654-4. [DOI] [PubMed] [Google Scholar]
- [13].Kanbe K, Naganawa H, Nakamura KT, Okami Y, Takeuchi T. Thienodolin, a new plant growth-regulating substance produced by a Streptomycete strain: II. Structure of thienodolin. Bioscience Biotechnology and Biochemistry. 1993;57:636–637. [Google Scholar]
- [14].Engqvist R, Javaid A, Bergman J. Synthesis of thienodolin. European Journal of Organic Chemistry. 2004:2589–2592. [Google Scholar]
- [15].Nicolin V, Ponti C, Narducci P, Grill V, Bortul R, Zweyer M, Vaccarezza M, Zauli G. Different levels of the neuronal nitric oxide synthase isoform modulate the rate of osteoclastic differentiation of TIB-71 and CRL-2278 RAW 264.7 murine cell clones. The Anatomical Record. Part A, Discoveries in Molecular, Cellular, and Evolutionary Biology. 2005;286:945–954. doi: 10.1002/ar.a.20239. [DOI] [PubMed] [Google Scholar]
- [16].Lu YC, Yeh WC, Ohashi PS. LPS/TLR4 signal transduction pathway. Cytokine. 2008;42:145–151. doi: 10.1016/j.cyto.2008.01.006. [DOI] [PubMed] [Google Scholar]
- [17].Kleinert H, Pautz A, Linker K, Schwarz PM. Regulation of the expression of inducible nitric oxide synthase. European Journal of Pharmacology. 2004;500:255–266. doi: 10.1016/j.ejphar.2004.07.030. [DOI] [PubMed] [Google Scholar]
- [18].Luqman S, Pezzuto JM. NFkappaB:a promising target for natural products in cancer chemoprevention. Phytotherapy Research. 2010;24:949–963. doi: 10.1002/ptr.3171. [DOI] [PubMed] [Google Scholar]
- [19].Majdalawieh A, Ro HS. Regulation of IkappaBalpha function and NF-kappaB signaling: AEBP1 is a novel proinflammatory mediator in macrophages. Mediators of Inflammation. 2010;2010:1–27. doi: 10.1155/2010/823821. article ID: 823821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Henkel T, Machleidt T, Alkalay I, Krönke M, Ben-Neriah Y, Baeuerle PA. Rapid proteolysis of I kappa B-alpha is necessary for activation of transcription factor NF-kappa B. Nature. 1993;365:182–185. doi: 10.1038/365182a0. [DOI] [PubMed] [Google Scholar]
- [21].Morris KR, Lutz RD, Choi HS, Kamitani T, Chmura K, Chan ED. Role of the NF-kappaB signaling pathway and kappaB cis-regulatory elements on the IRF-1 and iNOS promoter regions in mycobacterial lipoarabinomannan induction of nitric oxide. Infection and Immunity. 2003;71:1442–1452. doi: 10.1128/IAI.71.3.1442-1452.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Park EJ, Cheenpracha S, Chang LC, Kondratyuk TP, Pezzuto JM. Inhibition of lipopolysaccharide-induced cyclooxygenase-2 and inducible nitric oxide synthase expression by 4-[(2'-O-acetyl-α-L-rhamnosyloxy)benzyl]isothiocyanate from Moringa oleifera. Nutrition and Cancer. 2011;63:971–982. doi: 10.1080/01635581.2011.589960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Schmitz ML, Baeuerle PA. The p65 subunit is responsible for the strong transcription activating potential of NF-kappa B. EMBO Journal. 1991;10:3805–3817. doi: 10.1002/j.1460-2075.1991.tb04950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Fujihara M, Muroi M, Tanamoto K, Suzuki T, Azuma H, Ikeda H. Molecular mechanisms of macrophage activation and deactivation by lipopolysaccharide: roles of the receptor complex. Pharmacology & Therapeutics. 2003;100:171–194. doi: 10.1016/j.pharmthera.2003.08.003. [DOI] [PubMed] [Google Scholar]
- [25].Thomas KE, Galligan CL, Newman RD, Fish EN, Vogel SN. Contribution of interferon-beta to the murine macrophage response to the toll-like receptor 4 agonist, lipopolysaccharide. Journal of Biological Chemistry. 2006;281:31119–31130. doi: 10.1074/jbc.M604958200. [DOI] [PubMed] [Google Scholar]
- [26].Jacobs AT, Ignarro LJ. Lipopolysaccharide-induced expression of interferon-beta mediates the timing of inducible nitric-oxide synthase induction in RAW 264.7 macrophages. Journal of Biological Chemistry. 2001;276:47950–47957. doi: 10.1074/jbc.M106639200. [DOI] [PubMed] [Google Scholar]
- [27].Tsoyi K, Nizamutdinova IT, Jang HJ, Mun L, Kim HJ, Seo HG, Lee JH, Chang KC. Carbon monoxide from CORM-2 reduces HMGB1 release through regulation of IFN-β/JAK2/STAT-1/INOS/NO signaling but not COX-2 in TLR-activated macrophages. Shock. 2010;34:608–614. doi: 10.1097/SHK.0b013e3181e46f15. [DOI] [PubMed] [Google Scholar]
- [28].Yamashita T, Kawashima S, Ohashi Y, Ozaki M, Ueyama T, Ishida T, Inoue N, Hirata K, Akita H, Yokoyama M. Resistance to endotoxin shock in transgenic mice overexpressing endothelial nitric oxide synthase. Circulation. 2000;101:931–937. doi: 10.1161/01.cir.101.8.931. [DOI] [PubMed] [Google Scholar]
- [29].Kundu SD, Lee C, Billips BK, Habermacher GM, Zhang Q, Liu V, Wong LY, Klumpp DJ, Thumbikat P. The toll-like receptor pathway: a novel mechanism of infection-induced carcinogenesis of prostate epithelial cells. Prostate. 2008;68:223–229. doi: 10.1002/pros.20710. [DOI] [PubMed] [Google Scholar]
- [30].Hämäläinen M, Nieminen R, Vuorela P, Heinonen M, Moilanen E. Anti-inflammatory effects of flavonoids: genistein, kaempferol, quercetin, and daidzein inhibit STAT-1 and NF-kappaB activations, whereas flavone, isorhamnetin, naringenin, and pelargonidin inhibit only NF-kappaB activation along with their inhibitory effect on iNOS expression and NO production in activated macrophages. Mediators of Inflammation. 2007;2007:1–10. doi: 10.1155/2007/45673. article ID: 45673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Youn HS, Lee JY, Fitzgerald KA, Young HA, Akira S, Hwang DH. Specific inhibition of MyD88-independent signaling pathways of TLR3 and TLR4 by resveratrol: molecular targets are TBK1 and RIP1 in TRIF complex. Journal of Immunology. 2005;175:3339–3346. doi: 10.4049/jimmunol.175.5.3339. [DOI] [PubMed] [Google Scholar]
- [32].Youn HS, Lim HJ, Choi YJ, Lee JY, Lee MY, Ryu JH. Selenium suppresses the activation of transcription factor NF-kappa B and IRF3 induced by TLR3 or TLR4 agonists. International Journal of Immunopharmacology. 2008;8:495–501. doi: 10.1016/j.intimp.2007.12.008. [DOI] [PubMed] [Google Scholar]
- [33].Park EJ, Min HY, Chung HJ, Ahn YH, Pyee JH, Lee SK. Pinosylvin suppresses LPS-stimulated inducible nitric oxide synthase expression via the MyD88-independent, but TRIF-dependent downregulation of IRF-3 signaling pathway in mouse macrophage cells. Cellular Physiology and Biochemistry. 2011;27:353–362. doi: 10.1159/000327961. [DOI] [PubMed] [Google Scholar]
- [34].Wagner C, El Omari M, König GM. Biohalogenation: nature's way to synthesize halogenated metabolites. Journal of Natural Products. 2009;72:540–553. doi: 10.1021/np800651m. [DOI] [PubMed] [Google Scholar]
- [35].Seibold C, Schnerr H, Rumpf J, Kunzendorf A, Hatscher C, Wage T, Ernyei AJ, Dong C, Naishmith JH, Van Pe'e KH. A flavin-dependent tryptophan 6-halogenase and its use in modification of pyrrolnitrin biosynthesis. Biocatalysis and Biotransformation. 2006;24:401–408. [Google Scholar]
- [36].Oh GS, Pae HO, Choi BM, Chae SC, Lee HS, Ryu DG, Chung HT. 3-Hydroxyanthranilic acid, one of metabolites of tryptophan via indoleamine 2,3-dioxygenase pathway, suppresses inducible nitric oxide synthase expression by enhancing heme oxygenase-1 expression. Biochemical and Biophysical Research Communications. 2004;320:1156–1162. doi: 10.1016/j.bbrc.2004.06.061. [DOI] [PubMed] [Google Scholar]
- [37].Park EJ, Cheenpracha S, Chang LC, Pezzuto JM. Suppression of cyclooxygenase-2 and inducible nitric oxide synthase expression by epimuqubilin A via IKK/IκB/NF-κB pathways in lipopolysaccharide-stimulated RAW 264.7 cells. Phytochemistry Letters. 2011;4:426–431. doi: 10.1016/j.phytol.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. Journal of Immunological Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- [39].Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- [40].Zhao L, Tao JY, Zhang SL, Jin F, Pang R, Dong JH. N-Butanol extract from Melilotus suaveolens Ledeb affects pro- and anti-inflammatory cytokines and mediators. Evidence-based Complementary and Alternative Medicine. 2010;7:97–106. doi: 10.1093/ecam/nem165. [DOI] [PMC free article] [PubMed] [Google Scholar]


