Abstract
Staphylococcus hominis is a commensal resident of human skin and an opportunistic pathogen. The species is subdivided into two subspecies, S. hominis subsp. hominis and S. hominis subsp. novobiosepticus, which are difficult to distinguish. To investigate the evolution and epidemiology of S. hominis, a total of 108 isolates collected from 10 countries over 40 years were characterized by classical phenotypic methods and genetic methods. One nonsynonymous mutation in gyrB, scored with a novel SNP typing assay, had a perfect association with the novobiocin-resistant phenotype. A multilocus sequence typing (MLST) scheme was developed from six housekeeping gene fragments, and revealed relatively high levels of genetic diversity and a significant impact of recombination on S. hominis population structure. Among the 40 sequence types (STs) identified by MLST, three STs (ST2, ST16 and ST23) were S. hominis subsp. novobiosepticus, and they distinguished between isolates from different outbreaks, whereas 37 other STs were S. hominis subsp. hominis, one of which was widely disseminated (ST1). A modified PCR assay was developed to detect the presence of ccrAB4 from the SCCmec genetic element. S. hominis subsp. novobiosepticus isolates were oxacillin-resistant and carriers of specific components of SCCmec (mecA class A, ccrAB3, ccrAB4, ccrC), whereas S. hominis subsp. hominis included both oxacillin-sensitive and -resistant isolates and a more diverse array of SCCmec components. Surprisingly, phylogenetic analyses indicated that S. hominis subsp. novobiosepticus may be a polyphyletic and, hence, artificial taxon. In summary, these results revealed the genetic diversity of S. hominis, the identities of outbreak-causing clones, and the evolutionary relationships between subspecies and clones. The pathogenic lifestyle attributed to S. hominis subsp. novobiosepticus may have originated on more than one occasion.
Introduction
Distinguishing commensal from pathogenic coagulase-negative staphylococci (CoNS) is a major challenge for clinical microbiology laboratories. The CoNS are part of the normal bacterial flora of human skin, from which they frequently contaminate clinical specimens, yet they have been increasingly isolated over the past 30 years from human infections associated with indwelling medical devices [1], [2]. Staphylococcus epidermidis and Staphylococcus hominis are the most common CoNS from the surfaces of the axillae, arms, and legs [3]. S. hominis appears to be more successful than other CoNS at colonizing drier regions of the skin such as the volar forearm [4], [5]. Larson et al. [6] reported that S. hominis colonization was significantly more frequent on the hands of nurses with damaged skin compared to the hands of nurses with healthy skin. However, S. hominis is not limited to colonization of dry environments. Gunn and Colwell [7] reported S. hominis to be the most common and phenotypically diverse staphylococcal species isolated from unpolluted marine environments. A link to the marine environment is further supported by the isolation of S. hominis from crabs [8] and crabmeat [9].
S. hominis can cause nosocomial bloodstream infections [10]–[12] and other opportunistic infections of humans [13], [14]. An increasing proportion of blood culture isolates of S. hominis that were atypical or misidentified as Staphylococcus equorum, led to the the formal description in 1998 of two S. hominis subspecies, including S. hominis subsp. hominis and S. hominis subsp. novobiosepticus [15]. Novobiocin resistance and failure to produce acid aerobically from D-trehalose and N-acetyl-D-glucosamine are the distinguishing characteristics of S. hominis subsp. novobiosepticus [15]. Outbreaks and other clusters of bloodstream infections among neonates and adults have been attributed to S. hominis subsp. novobiosepticus [16]–[19]. However, various difficulties have been encountered in distinguishing between the two subspecies based on standard panels of phenotypes [20], [21], whole-cell protein profiles [22], and sequences from three housekeeping gene fragments [16]. For example, sequence polymorphisms in 16S rRNA and gyrA genes were able to distinguish between prototype strains of the two subspecies, but many clinical isolates with S. hominis subsp. hominis phenotypes were indistinguishable in sequence from isolates with S. hominis subsp. novobiosepticus phenotypes [16]. Knowledge of the evolutionary relationship of the two subspecies may lead to improved diagnostic tools and an improved understanding of the pathogenic potential of the two subspecies.
Towards this goal, we report the development of a multilocus sequence typing (MLST) scheme based on six housekeeping gene fragments of S. hominis. MLST schemes allow bacterial clones to be precisely defined and their relationships to be elucidated at a coarse level [23]. Among staphylococcal species, MLST schemes have been developed for S. epidermidis, Staphylococcus aureus, Staphylococcus lugdunensis, and the Staphylococcus intermedius group of species [24]–[27]. However, a recent attempt to develop an MLST scheme for Staphylococcus haemolyticus, which may be a close relative of S. hominis, concluded that the scheme was of limited use due to the relatively low sequence diversity detected among the isolates [28].
In addition to its occasional pathogenicity, S. hominis may be a reservoir of specific components of the methicillin resistance genetic element, SCCmec, that may be transferrable to more pathogenic staphylococcal species [29], [30]. Nasal carriage of methicillin-resistant S. hominis can be nearly as common as that of methicillin-resistant S. epidermidis and S. haemolyticus [31]. The relatively high prevalence of methicillin resistance among clinical isolates of S. hominis, and especially among S. hominis subsp. novobiosepticus, has been noted previously [6], [15], [16], [19], [32]. Furthermore, resistance to relatively new antibiotics such as linezolid and quinupristin/dalfopristin has been reported in S. hominis [33], [34]. Thus, the ability to genetically characterize and identify relationships between S. hominis subspecies and clones will provide a framework for future study of this enigmatic pathogen.
Materials and Methods
Bacterial isolates
A total of 108 isolates of S. hominis were included in this study. Sixty-eight isolates were from three hospitals in the USA (Illinois, n = 26; Mississippi, n = 1; New York, n = 41), 17 isolates were from one hospital in Spain, including 15 isolates from an outbreak among neonates and two isolates from adults that were not associated with the outbreak [17], five isolates were from a hospital outbreak among adults in Brazil [18], and 16 isolates were selected to be diverse by geography (Argentina, n = 1; Colombia, n = 1; Italy, n = 1; Japan, n = 1; Portugal, n = 2; Mexico, n = 3; Tunisia, n = 7) and by their different XhoI pulsed-field gel electrophoresis (PFGE) patterns that were characterized according to Bouchami et al. [30]. In addition, American Type Culture Collection (ATCC) strains ATCC 27844T and ATCC 700236T were included as the S. hominis subsp. hominis and S. hominis subsp. novobiosepticus prototype strains, respectively [15]. The isolates were collected from clinical specimens from 1971 to 2011. Ninety-two isolates were from blood cultures, 12 isolates were from other sources such as wound, urine, catheter, and pus specimens, and four isolates were of unknown source. Characteristics of all isolates are listed in Table S1. All isolates were single-colony purified on tryptic soy agar (TSA) plates and stored long-term in tryptic soy broth (TSB) with 15% glycerol at –80°C. Bacterial genomic DNA was extracted with a DNeasy kit (Qiagen, Valencia, USA), according to the manufacturer's instructions.
Phenotypic characterization
TSA plates with 5% defibrinated sheep blood (Cleveland Scientific, Bath, USA) were used to detect hemolytic activity, and mannitol salt agar (Becton Dickinson, Franklin Lakes, USA) plates were used to detect D-mannitol fermentation, after overnight incubation at 37°C. To detect aerobic acid production from D-trehalose and N-acetyl-D-glucosamine, 10 µL loopfuls of bacteria inoculum were streaked 1 cm radially on the surface of purple agar base (Difco, Lawrence, USA) plates that included 1% of either filter-sterilized D-trehalose (Fisher Scientific, Waltham, USA) or N-acetyl-D-glucosamine (Alfa Aesar, Ward Hill, USA). These plates were incubated at 35°C and examined at 24 h and 72 h to score acid production in terms of medium-to-strong (+), weak (±), or none (–), as described by Kloos and Schleifer [35]. Briefly, yellow indicator color extends from the culture streak into the surrounding medium within 72 hours for + isolates, yellow indicator color occurs under the culture streak but does not extend into the surrounding medium within 72 h for ± isolates, and very faint to no yellow indicator color occurs under the culture streak within 72 h for - isolates. Each plate was inoculated with up to six testing isolates, plus the positive (ATCC 27844) and negative (ATCC 700236) control strains.
Susceptibilities to novobiocin and oxacillin were determined with disk diffusion tests [36]. Single colonies were used to inoculate 1 mL of TSB, and bacterial cultures were grown to a turbidity comparable to a 0.5 McFarland standard, 100 µl of which was used to inoculate Mueller Hinton agar (Oxoid, Lenexa, USA) plates. The plates were incubated at 35°C for 18 h in the presence of 5 µg novobiocin (Remel, Lenexa, USA) and 1 µg oxacillin (Oxoid) disks. A zone of inhibition <16 mm was defined as novobiocin-resistant as per the manufacturer’s recommendation. A zone of inhibition <18 mm was defined as oxacillin-resistant [36].
Resistance gene characterization
In staphylococci, resistance to novobiocin can occur by point mutations in gyrB, which encodes the target of novobiocin, DNA gyrase B [37]–[39]. Full-length gyrB sequences were obtained by primer walking from a diverse set of six novobiocin-sensitive and six novobiocin-resistant isolates. The sequences of both DNA strands of gyrB were assembled, edited, and aligned using Lasergene software v7.2.1 (DNAStar, Madison, USA). Nonsynonymous mutations in gyrB were compared with the novobiocin susceptibilities to identify candidate resistance mutations. A fragment of gyrB containing the only identified candidate resistance mutation was subsequently amplified by PCR in all isolates using the same conditions as used for multilocus sequence typing (described below). These gyrB amplicons were subjected to single nucleotide polymorphism (SNP) typing on a Luminex 200 instrument (Millipore, Billerica, USA), according to the protocol outlined in Text S1. PCR primer information is listed in Table S2, and Luminex results for all isolates are listed in Table S3.
Resistance to oxacillin in staphylococci occurs by acquisition of the Staphylococcal Chromosomal Cassette mec (SCCmec) genetic element [40]. The SCCmec element was typed using several PCR assays that score the presence of mecA class A, class B, and class C, and ccrAB1, ccrAB2, ccrAB3, and ccrC gene complexes [41–43; additional information on the classification of these elements is available at: http://www.sccmec.org]. The presence of the mecA gene was scored using a separate PCR assay [43]. Three additional PCR assays, including those of Kondo et al. [43], Oliveira et al. [44], and a modified version of the Oliveira method developed here, were used to score the presence of ccrAB4. The modified Oliveira method consisted of decreasing the annealing temperature to 52°C, increasing the number of PCR cycles to 35, and increasing the final elongation time to 10 min. To confirm the specificity of the modified Oliveira method for amplifying ccrAB4, amplicons from eight isolates were purified and sequenced on both DNA strands. Alignments of the translated ccrAB4 sequences along with a variety of reference sequences were made with MUSCLE v3.7 [45] and curated with Gblocks v0.91b [46], using default settings. A maximum likelihood tree was constructed from the curated alignments using PhyML under a WAG model of amino acid substitution [47].
Multilocus sequence typing (MLST)
A fragment of the tuf gene [48] was amplified by PCR and sequenced on both DNA strands for species identification. The tuf sequences from all isolates were at least 99% identical to the sequences from one or the other ATCC prototype strains of S. hominis used here. The tuf gene fragment and six additional housekeeping gene fragments, selected from loci that had been tested for the S. aureus and S. epidermidis multilocus sequence typing (MLST) schemes [24], [26], were examined for use in a S. hominis MLST scheme. These gene fragments were located on separate contigs (ranging in size from ∼52 kb to ∼396 kb) of the unfinished genome sequence of S. hominis strain SK119 (GenBank accession number: ACLP00000000.1). The same primers were used for PCR and sequencing on both DNA strands. Thermal cycling conditions for PCR were: 95°C for 2 min; 30 cycles of 95°C for 30 sec, 55°C for 30 sec, and 72°C for 1 min; a final elongation step of 72°C for 2 min. Due to its apparently variable presence among the S. hominis isolates, aroE was subsequently dropped from the MLST scheme. The following six gene fragments were included in the final MLST scheme: arcC, glpK, gtr, pta, tpiA, and tuf. PCR primer information is listed in Table S2. For each of the six MLST loci, a unique nucleotide sequence defined an allele. Unique allelic profiles, consisting of the allele numbers at each of the six MLST loci, defined sequence types (STs).
Population genetic and phylogenetic analyses
Nucleotide sequences for each MLST locus were aligned using MUSCLE v3.7 software [45]. The number of polymorphic sites, nucleotide diversity per site (π), and allelic diversity (Hd) were calculated for each locus using DnaSP v5.10 software [49]. ST diversity was measured by Simpson’s index [50], [51]. Multilocus linkage disequilibrium was measured by the standardized index of association (I As), using LIAN v3.5 software [52].
ClonalFrame v1.2 software [53] was used to estimate population-scaled mutation and recombination parameters. The nucleotide sequence alignments of each MLST locus were input as individual blocks, using the sequences from single representatives of each ST. ClonalFrame was run five times, and each run used a Monte Carlo Markov Chain of 500,000 iterations, discarding the first 250,000 iterations as burn-in and saving every 100th iteration thereafter. The mixing and convergence of the runs were judged to be satisfactory based on inspections with the program's built-in tools. Recombination tract length could not be reliably estimated, so it was fixed to 1000 bp to allow comparisons of other parameter estimates with those of S. aureus and S. epidermidis [54]. The other parameter estimates represent the averages of the five runs.
To further test the role of recombination in generating allelic variation, the pairwise homoplasy index (PHI) test [55], implemented in SplitsTree v4.0 software [56], was calculated for each locus and for a concatenate of loci. A genetic algorithm for recombination detection (GARD) was used to identify the number and location of recombination breakpoints within each locus [57]. The concatenated nucleotide sequences were also analyzed for reticulate structure by the neighbor-net algorithm [58] implemented in SplitsTree.
Nucleotide sequences
The S. hominis MLST database will be publicly available at shominis.mlst.net. The ccrAB4 and gyrB sequences were deposited in the GenBank database with accession numbers JQ836536-836543 and JQ836544-JQ836555.
Results and Discussion
Phenotypic and genetic identification of S. hominis subspecies
S. hominis subsp. novobiosepticus is distinguished from S. hominis subsp. hominis by novobiocin resistance and failure to produce acid from D-trehalose and N-acetyl-D-glucosamine under aerobic conditions [15]. In this study, the disk diffusion tests identified 70 novobiocin-sensitive isolates and 38 novobiocin-resistant isolates. Of the 38 novobiocin-resistant isolates, 37 failed to produce acid aerobically from D-trehalose and N-acetyl-D-glucosamine and were classified as S. hominis subsp. novobiosepticus (Table 1). The remaining 71 isolates were classified as S. hominis subsp. hominis. As observed previously by Kloos et al. [15], the S. hominis subsp. hominis isolates were variable in their ability to produce acid aerobically from D-trehalose, N-acetyl-D-glucosamine, and D-mannitol (Table 1). As expected for S. hominis [15], [59], none of the 108 isolates were hemolytic on blood agar plates.
Table 1. Phenotypic and genetic characteristics of S. hominis subspecies.
| Marker category | Marker subcategory | No. (%) positiveSHN isolates(n = 37)a | No. (%) positiveSHH isolates(n = 71)a |
| Subspecies-defining phenotypes | Novobiocin resistance | 37 (100) | 1 (1) |
| D-trehalose | 0 (0) | 61 (86) | |
| N-acetyl-D-glucosamine | 0 (0) | 57 (80) | |
| Other phenotypes | D-mannitol | 0 (0) | 16 (23) |
| β-hemolysis | 0 (0) | 0 (0) | |
| Oxacillin resistance | 37 (100) | 46 (65) | |
| Genetic characteristics | gyrB allele T431 | 37 (100) | 1 (1) |
| mecA | 37 (100) | 46 (65) | |
| mecA class A | 37 (100) | 42 (59) | |
| mecA class B | 0 (0) | 4 (6) | |
| ccrAB1 | 15 (41) | 30 (42) | |
| ccrAB2 | 0 (0) | 4 (6) | |
| ccrAB3 | 22 (59) | 1 (1) | |
| ccrAB4 | 36 (97) | 14 (20) | |
| ccrC | 29 (78) | 21 (30) |
SHN is S. hominis subsp. novobiosepticus, SHH is S. hominis subsp. Hominis.
Examination of full-length gyrB sequences from a diverse set of six novobiocin-sensitive and six novobiocin-resistant isolates revealed one SNP, G431T, resulting in a predicted amino acid polymorphism, R144L, that was perfectly associated with the novobiocin susceptibilities (Table 2). This amino acid position may be a hotspot for novobiocin resistance in staphylococci [37]–[39]. A Luminex SNP typing assay was designed for this SNP and was applied to all 108 isolates (Table S3). All 70 novobiocin-sensitive isolates had the G431 allele, and all 38 novobiocin-resistant isolates had the T431 allele. Comparison of the SNP types with the subspecies identification based on the three defining phenotypes, revealed that the SNP typing assay was 100% (37/37) sensitive and 98.6% (70/71) specific for identifying S. hominis subsp. novobiosepticus (see Table 1). Thus, this SNP typing assay may provide a valid method for subspecies identification after the initial species identification.
Table 2. Identification of a candidate novobiocin resistance mutation in gyrB.
| Nonsynonymous SNPs and corresponding amino acids in gyrB codonsd | ||||||||||||
| 137 | 144 | 165 | 220 | 246 | 299 | 336 | 381 | 567 | ||||
| Strain | Subspeciesa | STb | Novc | 411 | 431 | 493 | 658 | 736 | 896 | 1006 | 1141 | 1700 |
| DAR1263 | SHH | 1 | S | Asp | Arg | Ile | Glu | Ser | Ala | Ile | Ile | Ala |
| GAT | CGT | ATA | GAA | TCT | GCA | ATA | ATC | GCT | ||||
| DAR1286 | SHH | 10 | S | |||||||||
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ||||
| DAR1932 | SHH | 13 | S | Pro | Val | Val | ||||||
| ... | ... | ... | ... | C.. | ... | G.. | G.. | ... | ||||
| DAR2030 | SHH | 1 | S | |||||||||
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ||||
| DAR3114 | SHH | 17 | S | Pro | Val | Val | ||||||
| ... | ... | ... | ... | C.. | .T. | ... | ... | .T. | ||||
| DAR3383 | SHH | 26 | S | Val | Lys | Pro | Val | |||||
| ... | ... | G.. | A.. | C.. | ... | G.. | ... | ... | ||||
| DAR1684 | SHN | 2 | R | Leu | ||||||||
| ... | .T. | ... | ... | ... | ... | ... | ... | ... | ||||
| DAR1919 | SHH | 8 | R | Glu | Leu | Pro | ||||||
| ..G | .T. | ... | ... | C.. | ... | ... | ... | ... | ||||
| DAR3358 | SHN | 2 | R | Leu | ||||||||
| ... | .T. | ... | ... | ... | ... | ... | ... | ... | ||||
| DAR3374 | SHN | 16 | R | Leu | Pro | |||||||
| ... | .T. | ... | ... | C.. | ... | ... | ... | ... | ||||
| DAR3377 | SHN | 2 | R | Leu | ||||||||
| ... | .T. | ... | ... | ... | ... | ... | ... | ... | ||||
| DAR3384 | SHN | 2 | R | Leu | ||||||||
| ... | .T. | ... | ... | ... | ... | ... | ... | ... | ||||
SHN is S. hominis subsp. novobiosepticus, SHH is S. hominis subsp. Hominis.
ST is multilocus sequence type.
Nov is novobiocin susceptibility, S = sensitive, R = resistant.
In column header, top number is amino acid position, bottom number is nucleotide position; dots indicate identity with the sequence from strain DAR1263.
Background genetic variation in S. hominis
An MLST scheme was developed for S. hominis using housekeeping gene fragments from six loci, including arcC, glpK, gtr, pta, tpiA, and tuf. A total of 2,453 bp was sequenced across these six loci for each isolate. One hundred and four SNPs were detected among the isolates, and the number of SNPs per locus ranged from five to 27 (Table 3). Nucleotide diversity, which is the average number of pairwise nucleotide differences per site, ranged from 0.002 to 0.021 for different loci and averaged 0.01 across sequence types (STs) (Table 3). This level of nucleotide diversity is much greater than that recently reported for S. haemolyticus (0.00035) [28], and is greater than that recently reported for S. aureus and S. epidermidis (0.0068 and 0.0064, respectively) [54]. The number of alleles per locus ranged from five to 16, while allelic diversity averaged 0.747 across STs (Table 3). Among all isolates, 40 STs were identified, yielding an ST diversity of 0.887 (0.850, 0.924) by Simpson’s index (95% confidence interval). Among the subset of 16 isolates that were selected to be diverse by geography and their different PFGE patterns (Figure S1), 11 STs were identified, yielding an ST diversity of 0.933 (0.857, 1). These results indicate that S. hominis is a genetically diverse species and that this MLST scheme has a relatively high discriminatory power.
Table 3. Genetic variation detected by the S. hominis MLST scheme.
| All isolates (n = 108) | STs only (n = 40) | ||||||
| Locus | Sequence length (bp) | No. polymorphicsites (SNPs) | No. alleles | Nucleotide diversity (π) | Allelic diversity (Hd) | Nucleotide diversity (π) | Allelic diversity (Hd) |
| arcC | 360 | 27 | 16 | 0.020 | 0.809 | 0.021 | 0.909 |
| glpK | 435 | 24 | 9 | 0.013 | 0.729 | 0.015 | 0.791 |
| gtr | 393 | 20 | 14 | 0.006 | 0.430 | 0.013 | 0.792 |
| pta | 444 | 13 | 11 | 0.004 | 0.594 | 0.004 | 0.731 |
| tpiA | 450 | 15 | 10 | 0.003 | 0.565 | 0.006 | 0.727 |
| tuf | 371 | 5 | 5 | 0.003 | 0.483 | 0.002 | 0.533 |
| Average | 408.8 | 17.3 | 10.8 | 0.008 | 0.602 | 0.010 | 0.747 |
MLST-defined clones of S. hominis, including outbreak clones
No STs were shared between the two subspecies. Three STs were identified among the 37 isolates of S. hominis subsp. novobiosepticus; ST2 (n = 21) was isolated from Argentina, Brazil, Colombia, Spain, and the USA, while ST16 (n = 15) and ST23 (n = 1) were isolated exclusively from Spain. Two sets of S. hominis subsp. novobiosepticus isolates were from hospital outbreaks. One outbreak was among neonates in Spain [17]; all 15 of these isolates from neonates were ST16, whereas two isolates from adults that were not associated with the outbreak were ST2 and ST23. The isolates from neonates were previously found to be indistinguishable based on PFGE [17]. Another outbreak was among adults in Brazil [18]; four of these isolates were S. hominis subsp. novobiosepticus ST2, but the fifth isolate was S. hominis subsp. hominis ST13. The original PFGE data for these Brazilian isolates also showed the fifth isolate to be different from the other four isolates [19]. While the detection of isolate similarities within outbreaks and isolate differences between outbreaks indicates that this MLST scheme may be valid for short-term molecular epidemiological use (i.e. outbreak investigations), some caveats should be noted. First, the outbreaks from Spain and Brazil differed by geography and host age-group. The bacterial clones identified from these two outbreaks may therefore have differed simply because of differences in the geographic or host distributions of these clones. Second, MLST allows for unambiguous, precise identification of clones, but only at a coarse level of detail. In order to distinguish between isolates that have recently diverged from a common ancestor, additional genetic markers may be required. Further studies are needed to determine the extent to which this MLST scheme is appropriate for outbreak investigations.
Thirty-seven STs were identified among the 71 isolates of S. hominis subsp. hominis; ST1 (n = 26) was the most prevalent ST of this subspecies and was isolated from Mexico, Portugal, Tunisia, and the USA. To our knowledge, S. hominis subsp. hominis has not been reported to cause outbreaks; however, the relative abundance and wide geographic distribution of ST1 suggests that this subspecies should be monitored more closely. Of note, 81% (30/37) of the S. hominis subsp. hominis STs were represented by single isolates. Accordingly, ST diversity was significantly higher among S. hominis subsp. hominis, 0.864 (0.785, 0.943), than among the mostly outbreak-causing S. hominis subsp. novobiosepticus, 0.527 (0.455, 0.599).
SCCmec variation in S. hominis
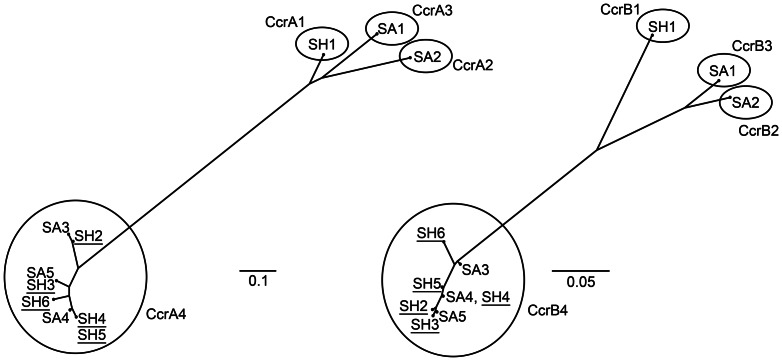
S. hominis has been suggested to be an important source of mecA class A, ccrAB1, and ccrAB4, which are components of some SCCmec genetic elements that may be transferrable to S. aureus [29], [30]. Two PCR assays for detecting ccrAB4 have been published [43], [44], but the relative performance of these assays has not been described. We first tested these two PCR assays and a modified version of the Oliveira method, on the 16 isolates that were selected to be diverse by geography and PFGE. The modified Oliveira method yielded an amplicon for 50% (8/16) of these isolates, whereas each of the other two methods yielded an amplicon in single isolates. Phylogenetic analysis of the translated ccrAB4 sequences from the eight amplicons of the modified Oliveira method revealed these to be genuine ccrAB4 alleles (Figure 1), which are characteristically divergent from other ccrAB alleles [30]. Thus, the modified Oliveira method was determined to be more sensitive than the other two methods and was used to score the presence of ccrAB4 in the remaining isolates.
Figure 1. Phylogeny of CcrA4 (panel A) and CcrB4 (panel B) alleles.
Translated sequences of ccrAB are from the eight S. hominis sequences obtained here (SH, underlined), along with various S. hominis (SH) and S. aureus (SA) reference sequences. Circles outline the major CcrAB alleles. Allelic variant and strain name (and GenBank accession number) are as follows: SH1 = GIFU12263 (AB063171); SH2 = DAR4404 (JQ836542); SH3 = DAR4401 (JQ836541); SH4 = DAR4386 (JQ836536), DAR4388 (JQ836537), DAR4394 (JQ836540), DAR4405 (JQ836543); SH5 = DAR4392 (JQ836539); SH6 = DAR4391 (JQ836538); SA1 = 85-2082 (AB037671); SA2 = N315 (NC_002745); SA3 = HDE288 (AF411935); SA4 = CHE482 (EF126185); SA5 = CHE482 (EF126186).
All 37 isolates of S. hominis subsp. novobiosepticus were oxacillin-resistant based on disk diffusion tests and were positive for the mecA gene based on PCR (Table 1). SCCmec typing of this subspecies revealed three combinations of ccrAB alleles with mecA class A, including those of SCCmec type III (3A, n = 1) and two different nontypeable configurations (3+4A, n = 21; 1+4A, n = 15) (a full listing of SCCmec results is in Table S1). ccrC was carried by 78% (29/37) of the S. hominis subsp. novobiosepticus isolates (Table 1).
In contrast, no perfect association between oxacillin susceptibility and carriage of the mecA gene was observed for S. hominis subsp. hominis. Of the 46 isolates that were positive for the mecA gene, 45 were oxacillin-resistant and, conversely, of the 25 isolates that were negative for the mecA gene, 24 were oxacillin-sensitive; results for these two outlier isolates were confirmed. All isolates that were positive for the mecA gene were also typeable for the mecA gene complex. Nine isolates had a ccrAB complex but no detectable mecA gene or gene complex, whereas 11 isolates had no detectable ccrAB complex but they had a typeable mecA complex. SCCmec type I (1B, n = 2), type II (2A, n = 2), type IV (2B, n = 1), type VI (4B, n = 1), and type VIII (4A, n = 4), and four different nontypeable configurations (1A, n = 21; 1+4A, n = 2; 2+4A, n = 1; 3+4A, n = 1) were identified for this subspecies. ccrC was carried by 30% (21/71) of the S. hominis subsp. hominis isolates (Table 1).
Statistical comparisons of the frequency of SCCmec components in the two subspecies indicated that S. hominis subsp. novobiosepticus was a significant source of mecA class A, ccrAB3, ccrAB4, and ccrC (Table 1); each P<0.0001 in two-tailed Fisher's exact tests. On the other hand, S. hominis subsp. hominis harbored a more diverse array of SCCmec components (Table S1, and described above); Simpson's index of diversity for the different combinations of mecA/ccrAB complex was 0.527 (0.455, 0.599) for S. hominis subsp. novobiosepticus and 0.807 (0.727, 0.888) for S. hominis subsp. hominis, including nontypeable elements but excluding ccrC which can be mobilized to locations other than the integration site of SCCmec [60]. These subspecies differences in SCCmec diversity were somewhat reflected in how strongly STs associated with SCCmec. For example, S. hominis subsp. novobiosepticus ST2 isolates carried the 3A (III) and 3+4A elements and ST16 isolates carried the 1+4A element, whereas the prevalent S. hominis subsp. hominis ST1 isolates carried five different elements, most of which were nontypeable. The nontypeable SCCmec elements identified in this study may represent novel elements, but further work demonstrating that the different components detected by PCR are physically linked is needed to prove this notion. The nontypeable element with mecA class A and ccrAB1 has been reported previously for S. hominis [30], [32]; our data show that this element occurs frequently in S. hominis subsp. hominis, and it occurs together with ccrAB4 in S. hominis subsp. novobiosepticus.
Population genetic signatures of recombination in S. hominis
To gain insight into the population structure of S. hominis, we first measured multilocus linkage disequilibrium between the MLST loci, using the standardized index of association (IAS). An IAS significantly greater than zero indicates a nonrandom association of alleles at different loci. Analysis of all 108 isolates yielded an IAS of 0.339 (Monte Carlo test with 1000 resamplings, P<0.001), and a clone-corrected analysis restricted to single isolates of each of the 40 STs yielded an IAS of 0.246 (P<0.001). Such results are expected for bacterial populations with relatively low recombination rates [61]; however, this index is a crude measure of the impact of recombination on population structure, and linkage disequilibrium can be caused by processes other than rare recombinations [62], [63].
To further examine the role of recombination on S. hominis population structure, the MLST nucleotide sequences were analyzed using the Bayesian model implemented by ClonalFrame. As found previously with S. epidermidis MLST data, the S. hominis MLST data did not allow the recombination tract length parameter to be reliably estimated, so it was fixed to the biologically plausible value of 1000 bp and other parameters were estimated [54]. The per-site mutation rate was based on Watterson's estimator [64], and was the same value as the average nucleotide diversity, 0.01. The per-site recombination rate (95% credibility interval) was estimated to be 0.0016 (0.0009, 0.0025), which does not differ significantly from previous estimates for S. aureus and S. epidermidis assuming 1000 bp tract lengths (0.0011 and 0.0004, respectively) [54]. However, the estimate of the rate at which nucleotides change by recombinations versus point mutations (95% credibility interval) was 1.73 (1.06, 2.69), which was significantly greater than one and indicative of the greater amount of nucleotide diversity transferred by recombinations in S. hominis populations in comparison to those of S. aureus and S. epidermidis (0.68 and 0.72, respectively) [54]. Consistent with this interpretation, the PHI test detected a signature of recombination (P<0.05) within the glpK locus. Followup analysis with the GARD algorithm identified a recombination breakpoint (P<0.01) at 291 bp into the glpK sequence, plus another recombination breakpoint (P<0.01) at 177 bp into the gtr sequence. Thus, recombination has a significant impact on S. hominis population structure.
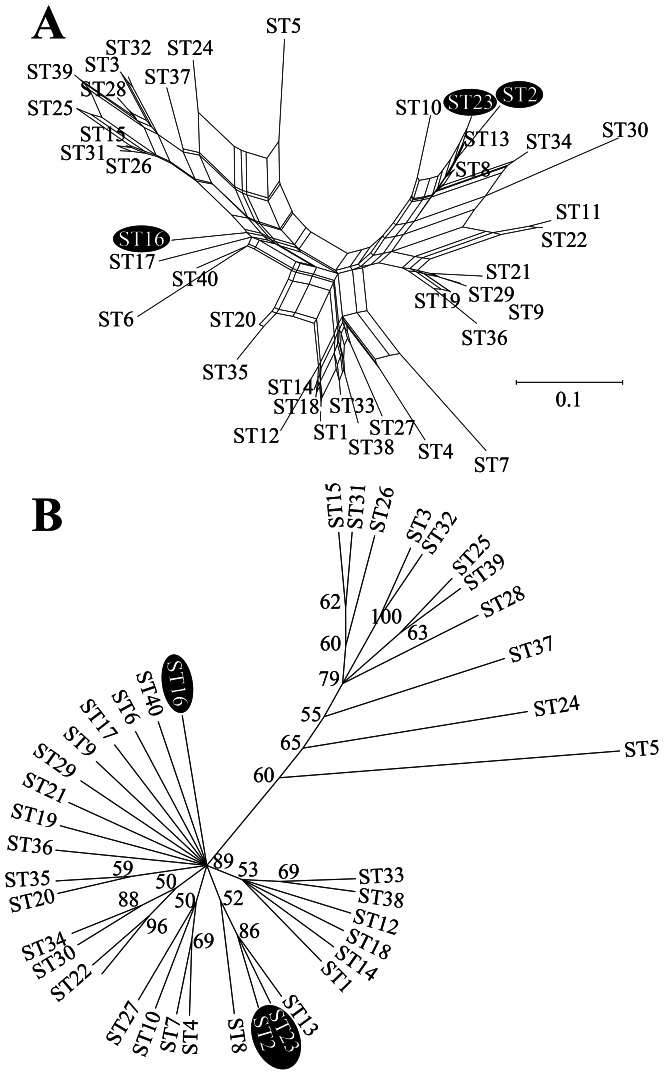
Evidence of S. hominis subsp. novobiosepticus polyphyly
The three S. hominis subsp. novobiosepticus STs differed from each other at three to four MLST loci, suggesting that they were not very closely related to each other, whereas the 37 S. hominis subsp. hominis STs differed from each other at one to six MLST loci. An eBURST analysis of the MLST alleles [65] with default settings showed one major clonal complex of 17 STs, one minor clonal complex of two STs, and 21 divergent STs (Figure S2). A more detailed study of the relationships between STs was made with analyses that accommodate non-treelike patterns, such as those produced by recombination, in the MLST sequences. Analysis with the neighbor-net algorithm revealed a highly reticulate network (Figure 2A). Of note, S. hominis subsp. novobiosepticus ST16 was separated from ST2 and ST23, suggesting that this subspecies may not be a phylogenetically cohesive group. Also, networks for all of the individual MLST loci except gtr separated the S. hominis subsp. novobiosepticus STs in various combinations (Figure S3). The ClonalFrame tree (Figure 2B) accounts for the influence of recombination on tree branch lengths and topology rather than revealing it in a network, and it provided statistical support for the separation of ST16 from ST2 and ST23.
Figure 2. Phylogeny of sequence types (STs) based on neighbor-net (panel A) and ClonalFrame (panel B) algorithms.
Numbers at nodes on the ClonalFrame tree are posterior probabilities >50/100. Highlighting indicates the three S. hominis subsp. novobiosepticus STs.
Interestingly, the single isolate of S. hominis subsp. hominis ST8, which was the only novobiocin-resistant isolate from this subspecies (Tables 1, 2), clustered with S. hominis subsp. novobiosepticus ST2 and ST23 (Figure 2A, 2B). In addition, S. hominis subsp. hominis ST13 was a member of this cluster, and it represented two of the four isolates from this subspecies that failed to produce acid aerobically from D-trehalose and N-acetyl-D-glucosamine (the other two isolates were ST4 and ST21; Table S1). As defined by phenotype, ST8 and ST13 were S. hominis subsp. hominis, but they were more similar to S. hominis subsp. novobiosepticus by MLST. Thus, this cluster appears to include either evolutionary intermediates or recombinants of the two subspecies.
The phylogenetic separation of the S. hominis subsp. novobiosepticus STs indicates that this subspecies may be polyphyletic; hence, it may be an artificial taxon whose members have independently gained novobiocin resistance and lost two metabolic traits. Novobiocin resistance may be caused by a single point mutation in gyrB (Table 2) [37]–[39], and the two metabolic traits were observed to be variable in this species (Table 1) [15]. These three subspecies-defining phenotypes may be labile from an evolutionary perspective. S. hominis subsp. novobiosepticus ST16 and ST2 both caused outbreaks, so their obvious pathogenicity may provide a rationale to retain the subspecies designation despite the evidence for their phylogenetic separation. However, if S. hominis subsp. hominis clones, such as the widespread ST1, were also shown to cause outbreaks, then this rationale for separating the subspecies on the basis of pathogenicity would be less clear.
Conclusions
S. hominis was revealed by MLST to be a genetically diverse species, relative to other staphylococci such as S. aureus and S. epidermidis, and recombination was shown to have a significant role in generating this diversity. Furthermore, PCR typing of SCCmec, using tools adopted from S. aureus, yielded a large number of nontypeable elements in S. hominis, which is suggestive of novel elements in S. hominis. While mecA class A, ccrAB3, ccrAB4, and ccrC are significantly more common in S. hominis subsp. novobiosepticus, a more diverse array of SCCmec components are found in S. hominis subsp. hominis. The two subspecies of S. hominis are distinguished by three phenotypes, but phylogenetic analyses indicate that S. hominis subsp. novobiosepticus STs do not form a single, well-supported cluster to the exclusion of all other STs; that is, the subspecies may be a polyphyletic, artificial taxon. Future genomic investigations of the S. hominis clones identified here should provide more insight into the nature of the subspecies and the role of recombination in generating the apparent polyphyly.
Supporting Information
Pulsed-field gel electrophoresis (PFGE) patterns and sequence types (STs) for 16 isolates selected to be diverse by geography and PFGE patterns.
(PPT)
Relationships between sequence types (STs) as inferred from the eBURST algorithm with default parameters. Each dot represents a different ST. Lines indicate that STs differ at one of the six loci used for multilocus sequence typing.
(TIF)
Neighbor-net networks for each of the six loci used for multilocus sequence typing.
(TIF)
Characteristics of study isolates.
(XLS)
Additional primer sets.
(DOC)
Results for Luminex SNP typing of gyrB .
(XLS)
Supplemental methods.
(DOC)
Acknowledgments
We thank Brande Beddingfield and Xiao Luo for technical assistance.
Funding Statement
This work was supported in part by grant GM080602 from the National Institutes of Health (to DAR) and grant RD06/0008 from the Spanish Network for Research in Infectious Diseases (to FC). The work performed in Portugal was supported by grants PTDC/BIA-EVF/117507/2010 (to MM) and grant #Pest-OE/EQB/LAO004/2011 (to ITQB) from Fundaçao para a Ciência e Tecnologia, Portugal. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. von Eiff C, Peters G, Heilmann C (2002) Pathogenesis of infections due to coagulase-negative staphylococci. Lancet Infect Dis 2: 677–685. [DOI] [PubMed] [Google Scholar]
- 2. Rogers KL, Fey PD, Rupp ME (2009) Coagulase-negative staphylococcal infections. Infect Dis Clin North Am 23: 73–98. [DOI] [PubMed] [Google Scholar]
- 3. Kloos WE, Musselwhite MS (1975) Distribution and persistence of Staphylococcus aureus and Micrococcus species and other aerobic bacteria on human skin. Appl Microbiol 30: 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kloos WE, Bannerman TL (1994) Update on clinical significance of coagulase-negative staphylococci. Clin Microbiol Rev 7: 117–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gao Z, Tseng CH, Pei Z, Blaser MJ (2007) Molecular analysis of human forearm superficial skin bacterial biota. Proc Natl Acad Sci U S A 104: 2927–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Larson EL, Hughes CAN, Pyrek JD, Sparks SM, Cagatay EU, et al. (1998) Changes in bacterial flora associated with skin damage on hands of health care personnel. Am J Infect Control 26: 513–521. [DOI] [PubMed] [Google Scholar]
- 7. Gunn BA, Colwell RR (1983) Numerical taxonomy of staphylococci isolated from the marine environment. Int J Syst Bacteriol 33: 751–759. [Google Scholar]
- 8. Faghri MA, Pennington CL, Cronholm LS, Atlas RM (1984) Bacteria associated with crabs from cold waters with emphasis on the occurrence of potential human pathogens. Appl Environ Microbiol 47: 1054–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ellender RD, Huang L, Sharp SL, Tettleton RP (1995) Isolation, enumeration, and identification of gram-positive cocci from frozen crabmeat. J Food Prot 58: 853–857. [DOI] [PubMed] [Google Scholar]
- 10. Kim S-D, McDonald LC, Jarvis WR, McAllister SK, Jerris R, et al. (2000) Determining the significance of coagulase-negative staphylococci isolated from blood cultures at a community hospital: a role for species and strain identification. Infect Control Hosp Epidemiol 21: 213–217. [DOI] [PubMed] [Google Scholar]
- 11. Ruhe J, Menon A, Mushatt D, Dejace P, Hasbun R (2004) Non-epidermidis coagulase-negative staphylococcal bacteremia: clinical predictors of true bacteremia. Eur J Clin Microbiol Infect Dis 23: 495–498. [DOI] [PubMed] [Google Scholar]
- 12. Al Wohoush I, Rivera J, Cairo J, Hachem R, Raad I (2011) Comparing clinical and microbiological methods for the diagnosis of true bacteraemia among patients with multiple blood cultures positive for coagulase-negative staphylococci. Clin Microbiol Infect 17: 569–571. [DOI] [PubMed] [Google Scholar]
- 13. Iyer MN, Wirostko WJ, Kim SH, Simons KB (2005) Staphylococcus hominis endophthalmitis associated with a capsular hypopyon. Am J Ophthalmol 139: 930–932. [DOI] [PubMed] [Google Scholar]
- 14. Cunha BA, Esrick MD, LaRusso M (2007) Staphylococcus hominis native mitral valve bacterial endocarditis (SBE) in a patient with hypertrophic obstructive cardiomyopathy. Heart Lung 36: 380–382. [DOI] [PubMed] [Google Scholar]
- 15. Kloos WE, George CG, Olgiate JS, Van Pelt L, McKinnon ML, et al. (1998) Staphylococcus hominis subsp. novobiosepticus subsp. nov., a novel trehalose- and N-acetyl-D-glucosamine-negative, novobiocin- and multiple-antibiotic-resistant subspecies isolated from human blood cultures. Int J Syst Bacteriol 48: 799–812. [DOI] [PubMed] [Google Scholar]
- 16. Fitzgibbon JE, Nahvi MD, Dubin DT, John Jr JF (2001) A sequence variant of Staphylococcus hominis with a high prevalence of oxacillin and fluoroquinolone resistance. Res Microbiol 152: 805–810. [DOI] [PubMed] [Google Scholar]
- 17. Chaves F, García-Álvarez M, Sanz F, Alba C, Otero JR (2005) Nosocomial spread of a Staphylococcus hominis subsp. novobiosepticus strain causing sepsis in a neonatal intensive care unit. J Clin Microbiol 43: 4877–4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. d’Azevedo PA, Trancesi R, Sales T, Monteiro J, Gales AC, et al. (2008) Outbreak of Staphylococcus hominis subsp. novobiosepticus bloodstream infections in São Paulo city, Brazil. J Med Microbiol 57: 256–257. [DOI] [PubMed] [Google Scholar]
- 19. Palazzo ICV, d’Azevedo PA, Secchi C, Pignatari ACC, da Costa Darini AL (2008) Staphylococcus hominis subsp. novobiosepticus strains causing nosocomial bloodstream infection in Brazil. J Antimicrob Chemother 62: 1222–1226. [DOI] [PubMed] [Google Scholar]
- 20. Weinstein MP, Mirrett S, van Pelt L, McKinnon M, Zimmer BL, et al. (1998) Clinical importance of identifying coagulase-negative staphylococci isolated from blood cultures: evaluation of microscan rapid and dried overnight gram-positive panels versus a conventional reference method. J Clin Microbiol 36: 2089–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gilad J, Schwartz D (2007) Identification of Staphylococcus species with the VITEK 2 system: the case of Staphylococcus hominis . J Clin Microbiol 45: 685–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. da Silva Santos OC, Laport MS, Teixeira LM, Giambiagi-DeMarval M, Iorio NL, et al. (2009) Reliable identification of clinically prevalent species and subspecies of staphylococci by sodium dodecyl sulfate polyacrylamide gel electrophoresis analysis. Diagn Microbiol Infect Dis 64: 1–5. [DOI] [PubMed] [Google Scholar]
- 23. Maiden MC (2006) Multilocus sequence typing of bacteria. Annu Rev Microbiol 60: 561–588. [DOI] [PubMed] [Google Scholar]
- 24. Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG (2000) Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus . J Clin Microbiol 38: 1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bannoehr J, Ben Zakour NL, Waller AS, Guardabassi L, Thoday KL, et al. (2007) Population genetic structure of the Staphylococcus intermedius group: insights into agr diversification and the emergence of methicillin-resistant strains. J Bacteriol 189: 8685–8692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thomas JC, Vargas MR, Miragaia M, Peacock SJ, Archer GL, et al. (2007) Improved multilocus sequence typing scheme for Staphylococcus epidermidis . J Clin Microbiol 45: 616–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chassain B, Lemee L, Didi J, Thiberge JM, Brisse S, Pons JL, Pestel-Caron M (2012) Multilocus sequence typing analysis of Staphylococcus lugdunensis implies a clonal population structure. J Clin Microbiol 50: 3003–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cavanagh JP, Klingenberg C, Hanssen A-M, Fredheim EA, Francois P, et al. (2012) Core genome conservation of Staphylococcus haemolyticus limits sequence based population structure analysis. J Microbiol Methods 89: 159–166. [DOI] [PubMed] [Google Scholar]
- 29. Katayama Y, Takeuchi F, Ito T, Ma XX, Ui-Mizutani Y, et al. (2003) Identification in methicillin-susceptible Staphylococcus hominis of an active primordial mobile genetic element for the staphylococcal cassette chromosome mec of methicillin-resistant Staphylococcus aureus . J Bacteriol 185: 2711–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bouchami O, Ben Hassen A, de Lencastre H, Miragaia M (2011) Molecular epidemiology of methicillin-resistant Staphylococcus hominis (MRSHo): low clonality and resevoirs of SCCmec structural elements. PLoS One 6: e21940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lebeaux D, Barbier F, Angelbault C, Benmahdi L, Ruppé E, et al. (2012) Evolution of nasal carriage of methicillin-resistant coagulase-negative staphylococci in a remote population. Antimicrob Agents Chemother 56: 315–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Garza-González E, Morfin-Otero R, Martínez-Vázquez MA, Gonzalez-Diaz E, González-Santiago O, et al. (2011) Microbiological and molecular characterization of human clinical isolates of Staphylococcus cohnii, Staphylococcus hominis, and Staphylococcus sciuri . Scand J Infect Dis 43: 930–936. [DOI] [PubMed] [Google Scholar]
- 33. Petinaki E, Spiliopoulou I, Maniati M, Maniatis AN (2005) Emergence of Staphylococcus hominis strains expressing low-level resistance to quinupristin/dalfopristin in Greece. J Antimicrob Chemother 55: 811–812. [DOI] [PubMed] [Google Scholar]
- 34. Ruiz de Gopegui E, Iuliana Marinescu C, Diaz P, Socias A, Garau M, et al. (2011) Nosocomial spread of linezolid-resistant Staphylococcus hominis in two hospitals in Majorca. Enferm Infecc Microbiol Clin 29: 339–344. [DOI] [PubMed] [Google Scholar]
- 35. Kloos WE, Schleifer KH (1975a) Simplified scheme for routine identification of human Staphylococcus species. J Clin Microbiol 1: 82–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.CLSI (2007) Performance standards for antimicrobial disk susceptibility tests. M100-S17. Wayne, PA.
- 37. Fournier B, Hooper DC (1998) Mutations in topoisomerase IV and DNA gyrase of Staphylococcus aureus: novel pleiotropic effects on quinolone and coumarin activity. Antimicrob Agents Chemother 42: 121–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fujimoto-Nakamura M, Ito H, Oyamada Y, Nishino T, Yamagashi J-I (2005) Accumulation of mutations in both gyrB and parE genes is associated with high-level resistance to novobiocin in Staphylococcus aureus . Antimicrob Agens Chemother 49: 3810–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vickers AA, Chopra I, O’Neill AJ (2007) Intrinsic novobiocin resistance in Staphylococcus saprophyticus . Antimicrob Agents Chemother 51: 4484–4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. IWG-SCC (2009) Classification of staphylococcal cassette chromosome mec (SCCmec): . guidelines for reporting novel SCCmec elements. 53: 4961–4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ito T, Katayama Y, Asada K, Mori N, Tsutsumimoto K, et al. (2001) Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus . Antimicrob Agents Chemother 45: 1323–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Robinson DA, Enright MC (2003) Evolutionary models of the emergence of methicillin-resistant Staphylococcus aureus . Antimicrob Agents Chemother 47: 3926–3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN, et al. (2007) Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother 51: 264–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Oliveira DC, Milheiriço C, de Lencastre H (2006) Redefining a structural variant of staphylococcal cassette chromosome mec, SCCmec type VI. Antimicrob Agents Chemother 50: 3457–3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17: 540–552. [DOI] [PubMed] [Google Scholar]
- 47. Dereeper A, Guignon V, Blanc G, Audic S, Buffet S (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36: W465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Heikens E, Fleer A, Paauw A, Florijn A, Fluit AC (2005) Comparison of genotypic and phenotypic methods for species-level identification of clinical isolates of coagulase-negative staphylococci. J Clin Microbiol 43: 2286–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- 50. Hunter PR, Gaston MA (1988) Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol 26: 2465–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Grundmann H, Hori S, Tanner G (2001) Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J Clin Microbiol 39: 4190–4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Haubold B, Hudson RR (2000) Lian 3.0: detecting linkage disequilibrium in multilocus data. Linkage analysis. Bioinformatics 16: 847–848. [DOI] [PubMed] [Google Scholar]
- 53. Didelot X, Falush D (2007) Inference of bacterial microevolution using multilocus sequence data. Genetics 175: 1251–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang L, Thomas JC, Didelot X, Robinson DA (2012) Molecular signatures identify a candidate target of balancing selection in an arcD-like gene of Staphylococcus epidermidis . J Mol Evol 75: 43–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bruen TC, Philippe H, Bryant D (2006) A simple and robust statistical test for detecting the presence of recombination. Genetics 172: 2665–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Huson DH, Bryant D (2006) Application of Phylogenetic Networks in Evolutionary Studies. Mol Biol Evol 23: 254–267. [DOI] [PubMed] [Google Scholar]
- 57. Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SDW (2006) Automated phylogenetic detection of recombination using a genetic algorithm. Mol Biol Evol 23: 1891–1901. [DOI] [PubMed] [Google Scholar]
- 58. Bryant D, Moulton V (2004) Neighbour-Net: An agglomerative method for the construction of phylogenetic networks. Mol Biol Evol 21: 255–265. [DOI] [PubMed] [Google Scholar]
- 59. Kloos WE, Schleifer KH (1975b) Isolation and characterization of staphylococci from human skin. Int J Syst Bacteriol 25: 62–79. [Google Scholar]
- 60. Chen L, Mediavilla JR, Smyth DS, Chavda KD, Ionescu R, Roberts RB, Robinson DA, Kreiswirth BN (2010) Identification of a novel transposon (Tn6072) and a truncated staphylococcal cassette chromosome mec element in methicillin-resistant Staphylococcus aureus ST239. Antimicrob Agents Chemother 54: 3347–3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Smith JM, Smith NH, O’Rourke M, Spratt BG (1993) How clonal are bacteria? Proc Natl Acad Sci U S A 90: 4384–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Smith JM (1994) Estimating the minimum rate of genetic transformation in bacteria. J Evol Biol 7: 525–534. [Google Scholar]
- 63. Slatkin M (2008) Linkage disequilibrium – understanding the evolutionary past and mapping the medical future. Nat Rev Genet 9: 477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Watterson GA (1975) Number of segregating sites in genetic models without recombination. Theor Popul Biol 7: 256–276. [DOI] [PubMed] [Google Scholar]
- 65. Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG (2004) eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol 186: 1518–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pulsed-field gel electrophoresis (PFGE) patterns and sequence types (STs) for 16 isolates selected to be diverse by geography and PFGE patterns.
(PPT)
Relationships between sequence types (STs) as inferred from the eBURST algorithm with default parameters. Each dot represents a different ST. Lines indicate that STs differ at one of the six loci used for multilocus sequence typing.
(TIF)
Neighbor-net networks for each of the six loci used for multilocus sequence typing.
(TIF)
Characteristics of study isolates.
(XLS)
Additional primer sets.
(DOC)
Results for Luminex SNP typing of gyrB .
(XLS)
Supplemental methods.
(DOC)