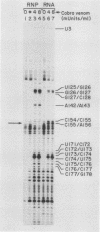
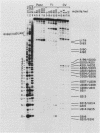
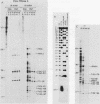
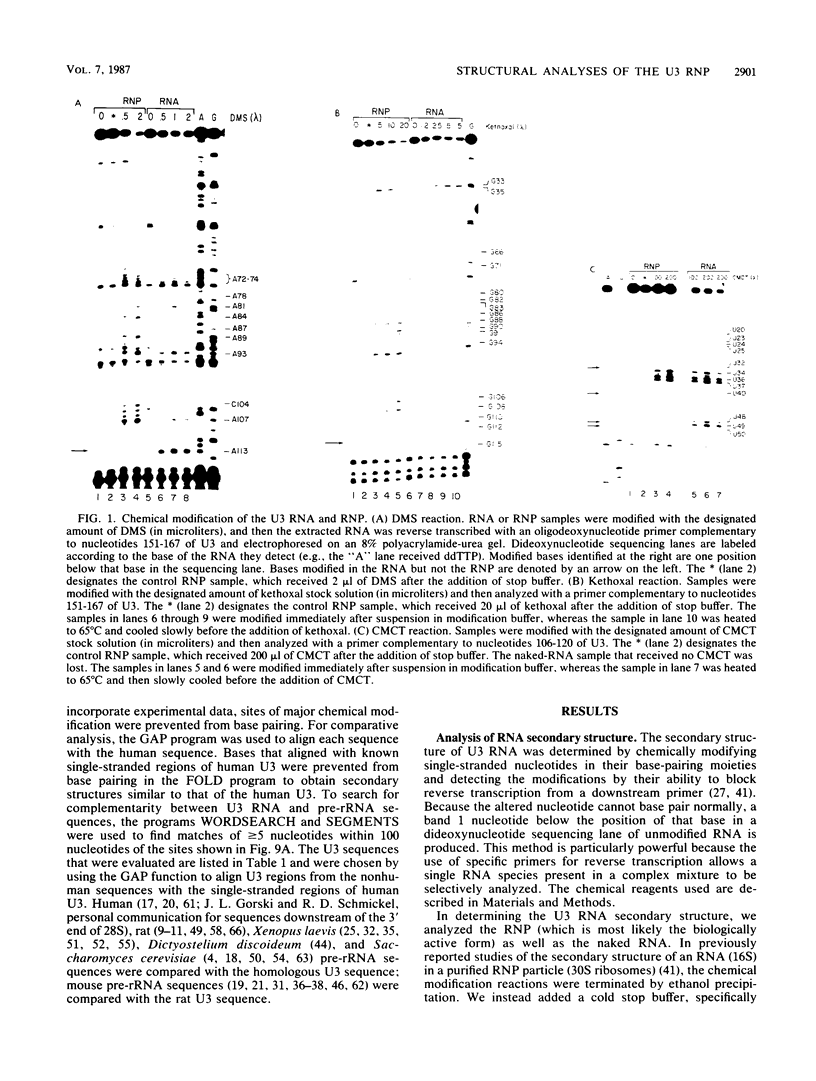
Abstract
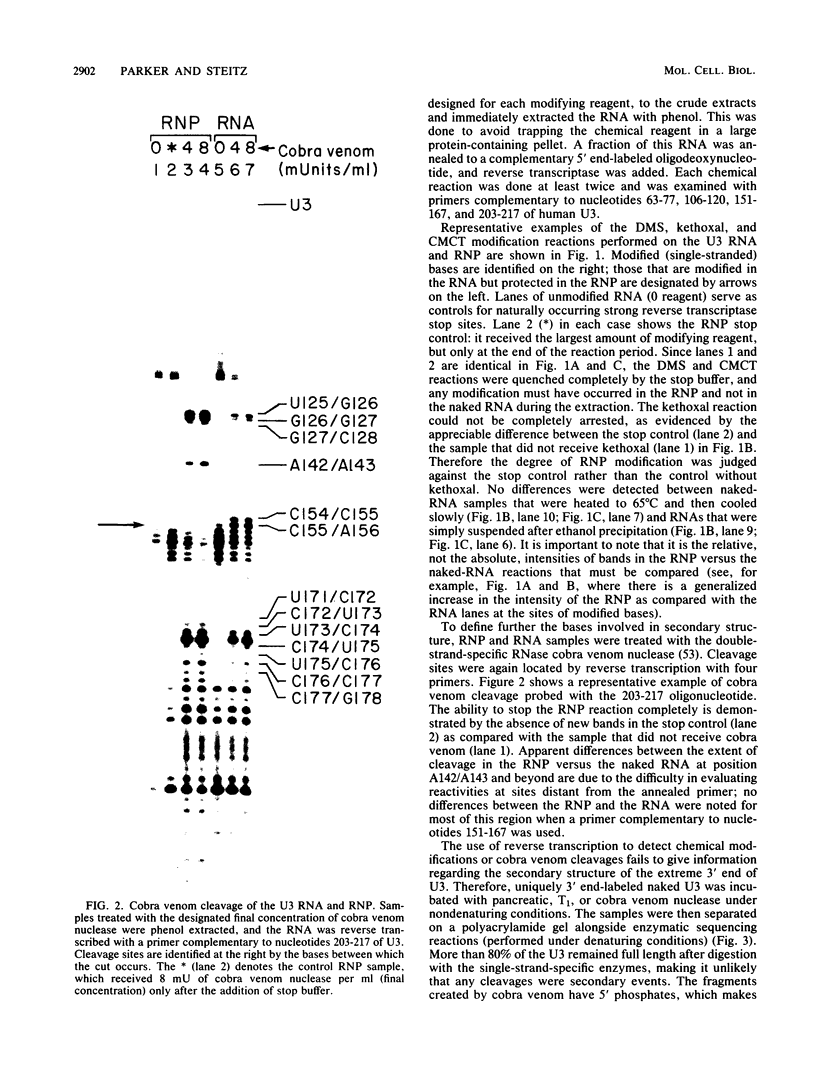
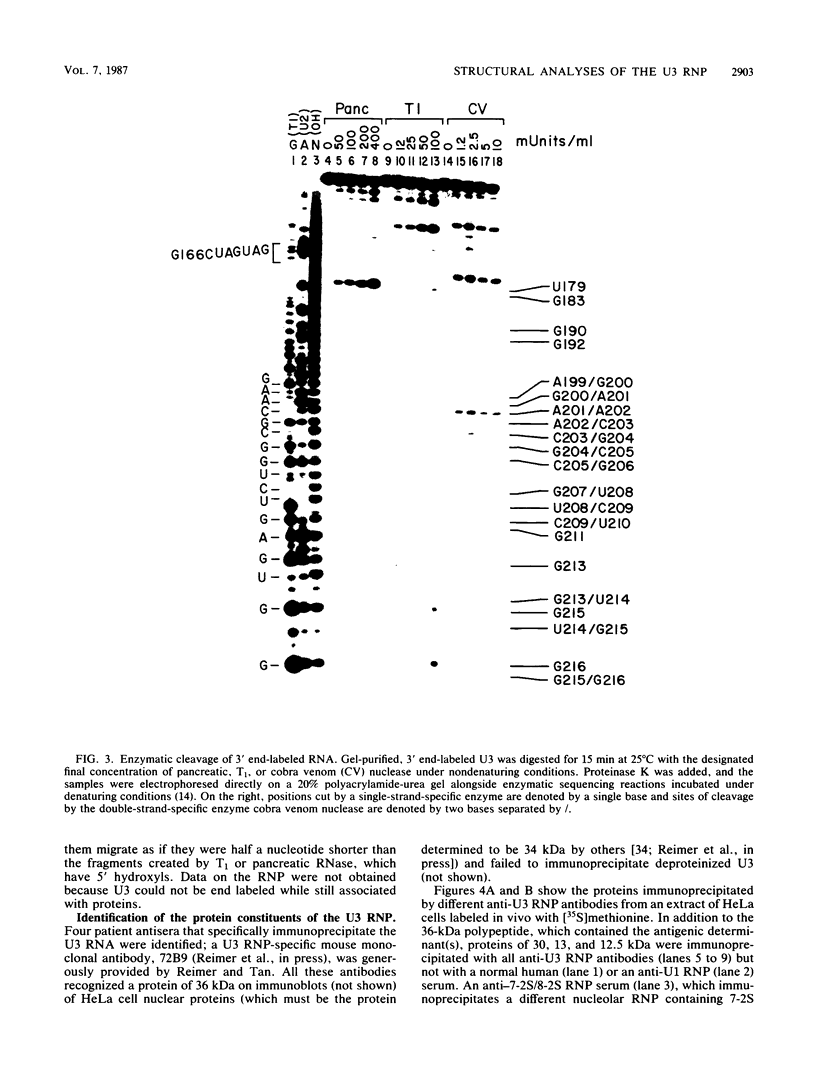
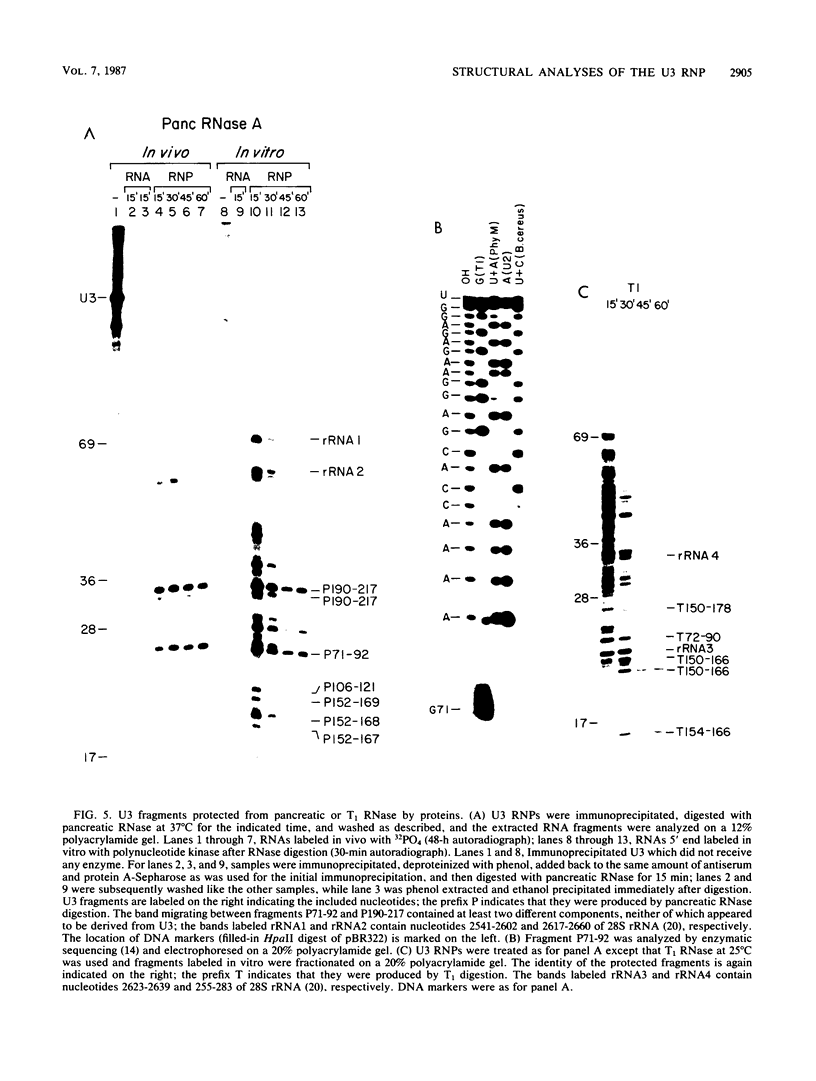
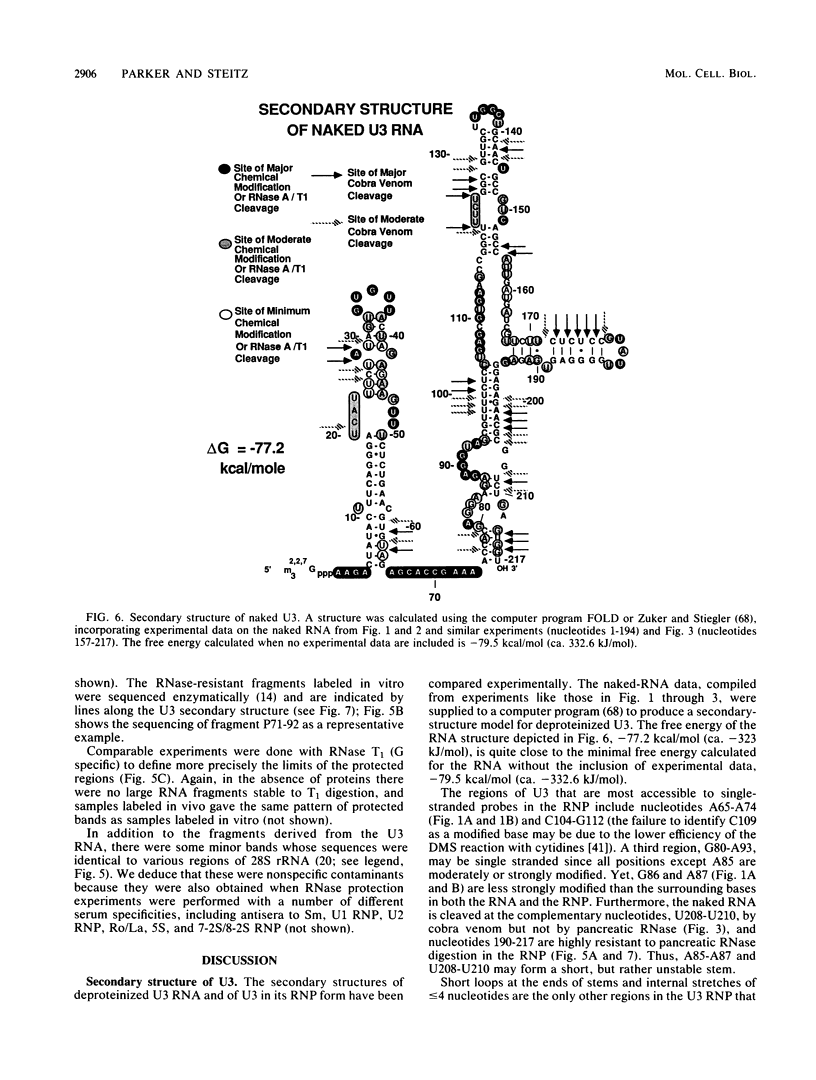
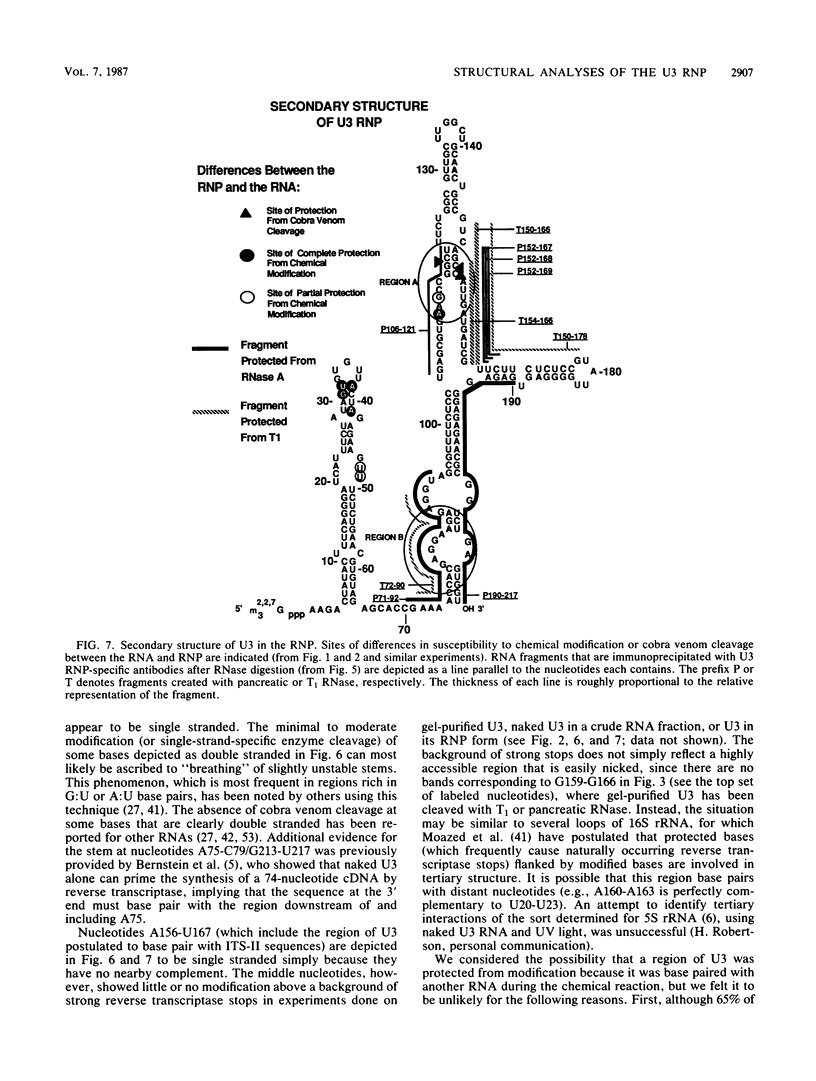
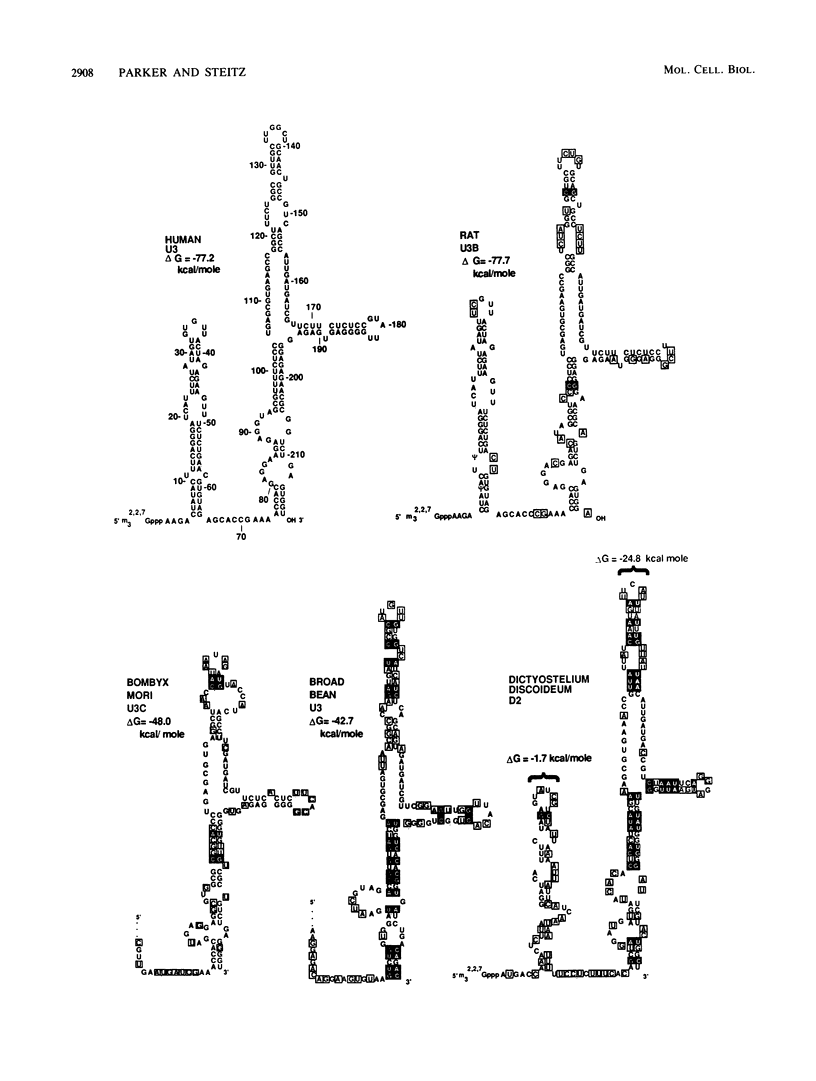
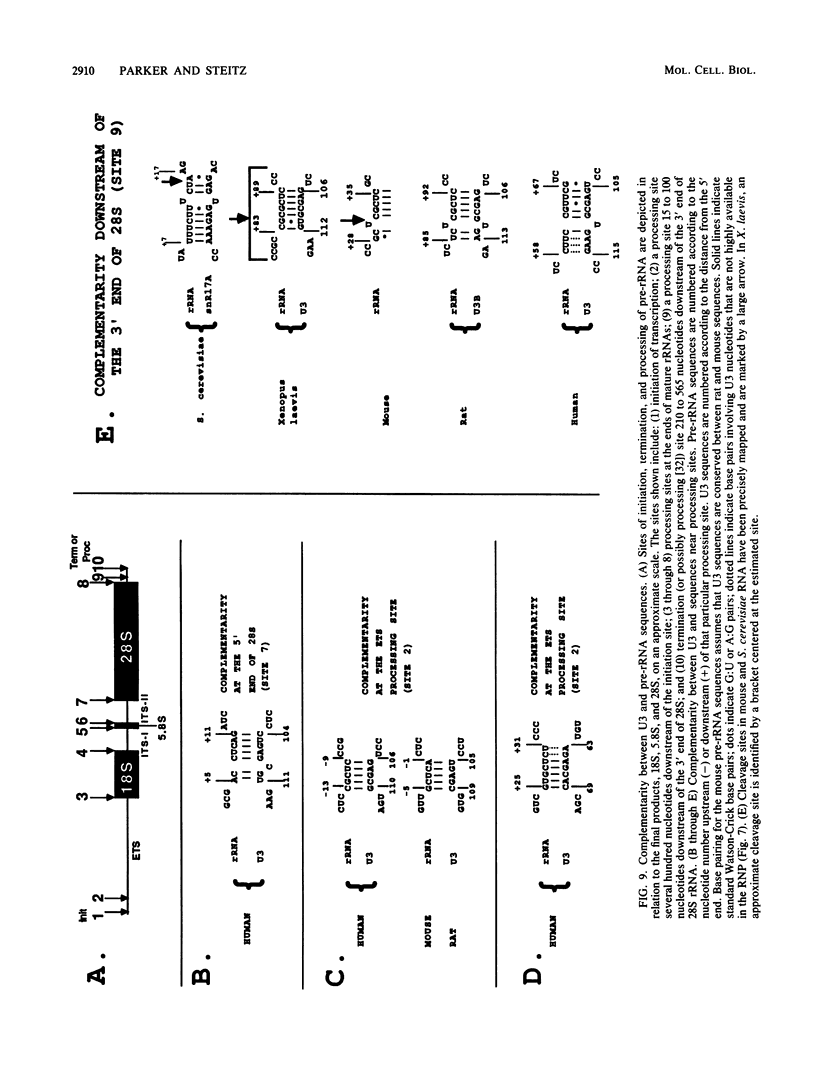
The human U3 ribonucleoprotein (RNP) has been analyzed to determine its protein constituents, sites of protein-RNA interaction, and RNA secondary structure. By using anti-U3 RNP antibodies and extracts prepared from HeLa cells labeled in vivo, the RNP was found to contain four nonphosphorylated proteins of 36, 30, 13, and 12.5 kilodaltons and two phosphorylated proteins of 74 and 59 kilodaltons. U3 nucleotides 72-90, 106-121, 154-166, and 190-217 must contain sites that interact with proteins since these regions are immunoprecipitated after treatment of the RNP with RNase A or T1. The secondary structure was probed with specific nucleases and by chemical modification with single-strand-specific reagents that block subsequent reverse transcription. Regions that are single stranded (and therefore potentially able to interact with a substrate RNA) include an evolutionarily conserved sequence at nucleotides 104-112 and nonconserved sequences at nucleotides 65-74, 80-84, and 88-93. Nucleotides 159-168 do not appear to be highly accessible, thus making it unlikely that this U3 sequence base pairs with sequences near the 5.8S rRNA-internal transcribed spacer II junction, as previously proposed. Alternative functions of the U3 RNP are discussed, including the possibility that U3 may participate in a processing event near the 3' end of 28S rRNA.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams D. S., Herrera R. J., Luhrmann R., Lizardi P. M. Isolation and partial characterization of U1-U6 small RNAs from Bombyx mori. Biochemistry. 1985 Jan 1;24(1):117–125. doi: 10.1021/bi00322a017. [DOI] [PubMed] [Google Scholar]
- Akao M., Reddy R., Busch H. Multiple sequences in the Drosophila melanogaster U3 RNA gene are homologous to vertebrate U3 RNA. Biochem Biophys Res Commun. 1986 Jul 31;138(2):512–518. doi: 10.1016/s0006-291x(86)80526-5. [DOI] [PubMed] [Google Scholar]
- Bachellerie J. P., Michot B., Raynal F. Recognition signals for mouse pre-rRNA processing. A potential role for U3 nucleolar RNA. Mol Biol Rep. 1983 May;9(1-2):79–86. doi: 10.1007/BF00777477. [DOI] [PubMed] [Google Scholar]
- Bayev A., Georgiev O. I., Hadjiolov A. A., Nikolaev N., Skryabin K. G., Zakharyev V. M. The structure of the yeast ribosomal RNA genes. 3. Precise mapping of the 18 S and 25 S rRNA genes and structure of the adjacent regions. Nucleic Acids Res. 1981 Feb 25;9(4):789–799. doi: 10.1093/nar/9.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein L. B., Mount S. M., Weiner A. M. Pseudogenes for human small nuclear RNA U3 appear to arise by integration of self-primed reverse transcripts of the RNA into new chromosomal sites. Cell. 1983 Feb;32(2):461–472. doi: 10.1016/0092-8674(83)90466-x. [DOI] [PubMed] [Google Scholar]
- Branch A. D., Benenfeld B. J., Robertson H. D. Ultraviolet light-induced crosslinking reveals a unique region of local tertiary structure in potato spindle tuber viroid and HeLa 5S RNA. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6590–6594. doi: 10.1073/pnas.82.19.6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch H., Reddy R., Rothblum L., Choi Y. C. SnRNAs, SnRNPs, and RNA processing. Annu Rev Biochem. 1982;51:617–654. doi: 10.1146/annurev.bi.51.070182.003153. [DOI] [PubMed] [Google Scholar]
- Calvet J. P., Pederson T. Base-pairing interactions between small nuclear RNAs and nuclear RNA precursors as revealed by psoralen cross-linking in vivo. Cell. 1981 Nov;26(3 Pt 1):363–370. doi: 10.1016/0092-8674(81)90205-1. [DOI] [PubMed] [Google Scholar]
- Cassidy B. G., Subrahmanyan C. S., Rothblum L. I. The nucleotide sequence of the 5' region of rat 18S rDNA and adjoining spacer. Biochem Biophys Res Commun. 1982 Aug 31;107(4):1571–1576. doi: 10.1016/s0006-291x(82)80179-4. [DOI] [PubMed] [Google Scholar]
- Chan Y. L., Gutell R., Noller H. F., Wool I. G. The nucleotide sequence of a rat 18 S ribosomal ribonucleic acid gene and a proposal for the secondary structure of 18 S ribosomal ribonucleic acid. J Biol Chem. 1984 Jan 10;259(1):224–230. [PubMed] [Google Scholar]
- Chan Y. L., Olvera J., Wool I. G. The structure of rat 28S ribosomal ribonucleic acid inferred from the sequence of nucleotides in a gene. Nucleic Acids Res. 1983 Nov 25;11(22):7819–7831. doi: 10.1093/nar/11.22.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig N., Kass S., Sollner-Webb B. Nucleotide sequence determining the first cleavage site in the processing of mouse precursor rRNA. Proc Natl Acad Sci U S A. 1987 Feb;84(3):629–633. doi: 10.1073/pnas.84.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouch R. J., Kanaya S., Earl P. L. A model for the involvement of the small nucleolar RNA (U3) in processing eukaryotic ribosomal RNA. Mol Biol Rep. 1983 May;9(1-2):75–78. doi: 10.1007/BF00777476. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England T. E., Bruce A. G., Uhlenbeck O. C. Specific labeling of 3' termini of RNA with T4 RNA ligase. Methods Enzymol. 1980;65(1):65–74. doi: 10.1016/s0076-6879(80)65011-3. [DOI] [PubMed] [Google Scholar]
- Epstein P., Reddy R., Busch H. Multiple states of U3 RNA in Novikoff hepatoma nucleoli. Biochemistry. 1984 Nov 6;23(23):5421–5425. doi: 10.1021/bi00318a007. [DOI] [PubMed] [Google Scholar]
- Financsek I., Mizumoto K., Mishima Y., Muramatsu M. Human ribosomal RNA gene: nucleotide sequence of the transcription initiation region and comparison of three mammalian genes. Proc Natl Acad Sci U S A. 1982 May;79(10):3092–3096. doi: 10.1073/pnas.79.10.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev O. I., Nikolaev N., Hadjiolov A. A., Skryabin K. G., Zakharyev V. M., Bayev A. A. The structure of the yeast ribosomal RNA genes. 4. Complete sequence of the 25 S rRNA gene from Saccharomyces cerevisae. Nucleic Acids Res. 1981 Dec 21;9(24):6953–6958. doi: 10.1093/nar/9.24.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman W. E., Goldberg G., Bowman L. H., Steinmetz D., Schlessinger D. Mouse rDNA: sequences and evolutionary analysis of spacer and mature RNA regions. Mol Cell Biol. 1983 Aug;3(8):1488–1500. doi: 10.1128/mcb.3.8.1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez I. L., Gorski J. L., Campen T. J., Dorney D. J., Erickson J. M., Sylvester J. E., Schmickel R. D. Variation among human 28S ribosomal RNA genes. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7666–7670. doi: 10.1073/pnas.82.22.7666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I., Maier U., Ohrlein A., Hassouna N., Bachellerie J. P. Transcription of mouse rDNA terminates downstream of the 3' end of 28S RNA and involves interaction of factors with repeated sequences in the 3' spacer. Cell. 1985 Dec;43(3 Pt 2):801–810. doi: 10.1016/0092-8674(85)90253-3. [DOI] [PubMed] [Google Scholar]
- Grummt I., Sorbaz H., Hofmann A., Roth E. Spacer sequences downstream of the 28S RNA coding region are part of the mouse rDNA transcription unit. Nucleic Acids Res. 1985 Apr 11;13(7):2293–2304. doi: 10.1093/nar/13.7.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurney T., Jr Characterization of mouse 45S ribosomal RNA subspecies suggests that the first processing cleavage occurs 600 +/- 100 nucleotides from the 5' end and the second 500 +/- 100 nucleotides from the 3' end of a 13.9 kb precursor. Nucleic Acids Res. 1985 Jul 11;13(13):4905–4919. doi: 10.1093/nar/13.13.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall L. M., Maden B. E. Nucleotide sequence through the 18S-28S intergene region of a vertebrate ribosomal transcription unit. Nucleic Acids Res. 1980 Dec 20;8(24):5993–6005. doi: 10.1093/nar/8.24.5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto C., Steitz J. A. Sequential association of nucleolar 7-2 RNA with two different autoantigens. J Biol Chem. 1983 Feb 10;258(3):1379–1382. [PubMed] [Google Scholar]
- Inoue T., Cech T. R. Secondary structure of the circular form of the Tetrahymena rRNA intervening sequence: a technique for RNA structure analysis using chemical probes and reverse transcriptase. Proc Natl Acad Sci U S A. 1985 Feb;82(3):648–652. doi: 10.1073/pnas.82.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan L. S., Chandrasegaran S., Pulford S. M., Miller P. S. Detection of a guanine X adenine base pair in a decadeoxyribonucleotide by proton magnetic resonance spectroscopy. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4263–4265. doi: 10.1073/pnas.80.14.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempers-Veenstra A. E., Oliemans J., Offenberg H., Dekker A. F., Piper P. W., Planta R. J., Klootwijk J. 3'-End formation of transcripts from the yeast rRNA operon. EMBO J. 1986 Oct;5(10):2703–2710. doi: 10.1002/j.1460-2075.1986.tb04554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T., Tóth M., Solymosy F. Plant small nuclear RNAs. Nucleolar U3 snRNA is present in plants: partial characterization. Eur J Biochem. 1985 Oct 15;152(2):259–266. doi: 10.1111/j.1432-1033.1985.tb09192.x. [DOI] [PubMed] [Google Scholar]
- Kominami R., Mishima Y., Urano Y., Sakai M., Muramatsu M. Cloning and determination of the transcription termination site of ribosomal RNA gene of the mouse. Nucleic Acids Res. 1982 Mar 25;10(6):1963–1979. doi: 10.1093/nar/10.6.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. Characterization of three sites of RNA 3' end formation in the Xenopus ribosomal gene spacer. Cell. 1986 May 9;45(3):431–443. doi: 10.1016/0092-8674(86)90329-6. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lischwe M. A., Ochs R. L., Reddy R., Cook R. G., Yeoman L. C., Tan E. M., Reichlin M., Busch H. Purification and partial characterization of a nucleolar scleroderma antigen (Mr = 34,000; pI, 8.5) rich in NG,NG-dimethylarginine. J Biol Chem. 1985 Nov 15;260(26):14304–14310. [PubMed] [Google Scholar]
- Maden B. E., Moss M., Salim M. Nucleotide sequence of an external transcribed spacer in Xenopus laevis rDNA: sequences flanking the 5' and 3' ends of 18S rRNA are non-complementary. Nucleic Acids Res. 1982 Apr 10;10(7):2387–2398. doi: 10.1093/nar/10.7.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michot B., Bachellerie J. P., Raynal F., Renalier M. H. Sequence of the 3'-terminal domain of mouse 18 S rRNA. Conservation of structural features with other pro- and eukaryotic homologs. FEBS Lett. 1982 Jun 7;142(2):260–266. doi: 10.1016/0014-5793(82)80148-8. [DOI] [PubMed] [Google Scholar]
- Michot B., Bachellerie J. P., Raynal F. Sequence and secondary structure of mouse 28S rRNA 5'terminal domain. Organisation of the 5.8S-28S rRNA complex. Nucleic Acids Res. 1982 Sep 11;10(17):5273–5283. doi: 10.1093/nar/10.17.5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michot B., Bachellerie J. P., Raynal F. Structure of mouse rRNA precursors. Complete sequence and potential folding of the spacer regions between 18S and 28S rRNA. Nucleic Acids Res. 1983 May 25;11(10):3375–3391. doi: 10.1093/nar/11.10.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K. G., Sollner-Webb B. Transcription of mouse rRNA genes by RNA polymerase I: in vitro and in vivo initiation and processing sites. Cell. 1981 Nov;27(1 Pt 2):165–174. doi: 10.1016/0092-8674(81)90370-6. [DOI] [PubMed] [Google Scholar]
- Mimori T., Hinterberger M., Pettersson I., Steitz J. A. Autoantibodies to the U2 small nuclear ribonucleoprotein in a patient with scleroderma-polymyositis overlap syndrome. J Biol Chem. 1984 Jan 10;259(1):560–565. [PubMed] [Google Scholar]
- Moazed D., Stern S., Noller H. F. Rapid chemical probing of conformation in 16 S ribosomal RNA and 30 S ribosomal subunits using primer extension. J Mol Biol. 1986 Feb 5;187(3):399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- Noller H. F. Structure of ribosomal RNA. Annu Rev Biochem. 1984;53:119–162. doi: 10.1146/annurev.bi.53.070184.001003. [DOI] [PubMed] [Google Scholar]
- Ochs R. L., Lischwe M. A., Spohn W. H., Busch H. Fibrillarin: a new protein of the nucleolus identified by autoimmune sera. Biol Cell. 1985;54(2):123–133. doi: 10.1111/j.1768-322x.1985.tb00387.x. [DOI] [PubMed] [Google Scholar]
- Ozaki T., Hoshikawa Y., Iida Y., Iwabuchi M. Sequence analysis of the transcribed and 5' non-transcribed regions of the ribosomal RNA gene in Dictyostelium discoideum. Nucleic Acids Res. 1984 May 25;12(10):4171–4184. doi: 10.1093/nar/12.10.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestayko A. W., Tonato M., Busch H. Low molecular weight RNA associated with 28 s nucleolar RNA. J Mol Biol. 1970 Feb 14;47(3):505–515. doi: 10.1016/0022-2836(70)90318-9. [DOI] [PubMed] [Google Scholar]
- Raynal F., Michot B., Bachellerie J. P. Complete nucleotide sequence of mouse 18 S rRNA gene: comparison with other available homologs. FEBS Lett. 1984 Feb 27;167(2):263–268. doi: 10.1016/0014-5793(84)80139-8. [DOI] [PubMed] [Google Scholar]
- Reddy R., Henning D., Busch H. Nucleotide sequence of nucleolar U3B RNA. J Biol Chem. 1979 Nov 10;254(21):11097–11105. [PubMed] [Google Scholar]
- Reddy R., Tan E. M., Henning D., Nohga K., Busch H. Detection of a nucleolar 7-2 ribonucleoprotein and a cytoplasmic 8-2 ribonucleoprotein with autoantibodies from patients with scleroderma. J Biol Chem. 1983 Feb 10;258(3):1383–1386. [PubMed] [Google Scholar]
- Rothblum L. I., Reddy R., Cassidy B. Transcription initiation site of rat ribosomal DNA. Nucleic Acids Res. 1982 Nov 25;10(22):7345–7362. doi: 10.1093/nar/10.22.7345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsov P. M., Musakhanov M. M., Zakharyev V. M., Krayev A. S., Skryabin K. G., Bayev A. A. The structure of the yeast ribosomal RNA genes. I. The complete nucleotide sequence of the 18S ribosomal RNA gene from Saccharomyces cerevisiae. Nucleic Acids Res. 1980 Dec 11;8(23):5779–5794. doi: 10.1093/nar/8.23.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim M., Maden B. E. Nucleotide sequence encoding the 5' end of Xenopus laevis 18S rRNA. Nucleic Acids Res. 1980 Jul 11;8(13):2871–2884. doi: 10.1093/nar/8.13.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim M., Maden B. E. Nucleotide sequence of Xenopus laevis 18S ribosomal RNA inferred from gene sequence. Nature. 1981 May 21;291(5812):205–208. doi: 10.1038/291205a0. [DOI] [PubMed] [Google Scholar]
- Shelness G. S., Williams D. L. Secondary structure analysis of apolipoprotein II mRNA using enzymatic probes and reverse transcriptase. Evaluation of primer extension for high resolution structure mapping of mRNA. J Biol Chem. 1985 Jul 15;260(14):8637–8646. [PubMed] [Google Scholar]
- Skryabin K. G., Eldarov M. A., Larionov V. L., Bayev A. A., Klootwijk J., de Regt V. C., Veldman G. M., Planta R. J., Georgiev O. I., Hadjiolov A. A. Structure and function of the nontranscribed spacer regions of yeast rDNA. Nucleic Acids Res. 1984 Mar 26;12(6):2955–2968. doi: 10.1093/nar/12.6.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Reeder R. H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell. 1979 Oct;18(2):485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]
- Solnick D. Alternative splicing caused by RNA secondary structure. Cell. 1985 Dec;43(3 Pt 2):667–676. doi: 10.1016/0092-8674(85)90239-9. [DOI] [PubMed] [Google Scholar]
- Stroke I. L., Weiner A. M. Genes and pseudogenes for rat U3A and U3B small nuclear RNA. J Mol Biol. 1985 Jul 20;184(2):183–193. doi: 10.1016/0022-2836(85)90372-9. [DOI] [PubMed] [Google Scholar]
- Subrahmanyam C. S., Cassidy B., Busch H., Rothblum L. I. Nucleotide sequence of the region between the 18S rRNA sequence and the 28S rRNA sequence of rat ribosomal DNA. Nucleic Acids Res. 1982 Jun 25;10(12):3667–3680. doi: 10.1093/nar/10.12.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh D., Busch H., Reddy R. Isolation and characterization of a human U3 small nucleolar RNA gene. Biochem Biophys Res Commun. 1986 Jun 30;137(3):1133–1140. doi: 10.1016/0006-291x(86)90343-8. [DOI] [PubMed] [Google Scholar]
- Tague B. W., Gerbi S. A. Processing of the large rRNA precursor: two proposed categories of RNA-RNA interactions in eukaryotes. J Mol Evol. 1984;20(3-4):362–367. doi: 10.1007/BF02104742. [DOI] [PubMed] [Google Scholar]
- Torczynski R. M., Fuke M., Bollon A. P. Cloning and sequencing of a human 18S ribosomal RNA gene. DNA. 1985 Aug;4(4):283–291. doi: 10.1089/dna.1985.4.283. [DOI] [PubMed] [Google Scholar]
- Urano Y., Kominami R., Mishima Y., Muramatsu M. The nucleotide sequence of the putative transcription initiation site of a cloned ribosomal RNA gene of the mouse. Nucleic Acids Res. 1980 Dec 20;8(24):6043–6058. doi: 10.1093/nar/8.24.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman G. M., Klootwijk J., van Heerikhuizen H., Planta R. J. The nucleotide sequence of the intergenic region between the 5.8S and 26S rRNA genes of the yeast ribosomal RNA operon. Possible implications for the interaction between 5.8S and 26S rRNA and the processing of the primary transcript. Nucleic Acids Res. 1981 Oct 10;9(19):4847–4862. doi: 10.1093/nar/9.19.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg R. A., Penman S. Small molecular weight monodisperse nuclear RNA. J Mol Biol. 1968 Dec;38(3):289–304. doi: 10.1016/0022-2836(68)90387-2. [DOI] [PubMed] [Google Scholar]
- Wise J. A., Weiner A. M. Dictyostelium small nuclear RNA D2 is homologous to rat nucleolar RNA U3 and is encoded by a dispersed multigene family. Cell. 1980 Nov;22(1 Pt 1):109–118. doi: 10.1016/0092-8674(80)90159-2. [DOI] [PubMed] [Google Scholar]
- Yavachev L. P., Georgiev O. I., Braga E. A., Avdonina T. A., Bogomolova A. E., Zhurkin V. B., Nosikov V. V., Hadjiolov A. A. Nucleotide sequence analysis of the spacer regions flanking the rat rRNA transcription unit and identification of repetitive elements. Nucleic Acids Res. 1986 Mar 25;14(6):2799–2810. doi: 10.1093/nar/14.6.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieve G., Penman S. Small RNA species of the HeLa cell: metabolism and subcellular localization. Cell. 1976 May;8(1):19–31. doi: 10.1016/0092-8674(76)90181-1. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]