Abstract
Cd2+ causes damages to several human tissues. Although the toxicological and carcinogenetic mechanisms of Cd2+ have been previously established, some basic questions on this toxicant remain unclear. In this study, we constructed Met-cad 1.57, a new fluorescent resonance energy transfer (FRET)-based Cd2+ indicator, which contains a portion of a Cd2+-binding protein (CadR) obtained from Pseudomonas putida as the Cd2+ sensing key. We produced a human embryonic kidney cell line HEK-MCD157 which stably expresses the Met-cad 1.57 for further investigations. Both fluorescence spectroscopy and FRET microscopic ratio imaging were used to monitor the Cd2+ concentration within the living HEK-MCD157 cells. The dissociation constant of Met-cad 1.57 was approximately 271 nM. The function of Ca2+ channels as a potential Cd2+ entry gateway was further confirmed in the HEK-MCD157 cells. The organelle-targeted property of the protein-based Cd2+ indicator directly reveals the nucleus accumulation phenomena. In summary, a human kidney cell line that stably expresses the FRET-based Cd2+ indicator Met-cad 1.57 was constructed for reliable and convenient investigations to determine the Cd2+ concentration within living cells, including the identification of the entry pathway of Cd2+ and sub-cellular sequestration.
Introduction
In contrast to ions that are important to life, heavy metal/metalloid ions such as Pb2+, Cd2+, Hg2+, and As3+ are extremely toxic and not beneficial to living organisms [1]. Molecular mimicry is one of the most accepted concepts that explain the toxic mechanism of these ions [2], [3], including Cd2+ [4]. In particular, Cd2+ impairs certain biochemical reactions involved in normal physiological functions, thereby causing severe damages in certain targeted cells [1], [4]. Clinically, toxicants are monitored by evaluating the concentrations in the blood or urine through atomic absorption spectroscopy. For Cd2+, the acceptable blood Cd2+ concentration (BCdC) is approximately 5 µg/L to 10 µg/L [1], [5].
Considering the concepts in molecular and cellular toxicology, some basic questions on Cd2+ remain unclear. For example, whether BCdC can fully represent the whole intoxication status in certain human tissues is unknown. Various membrane proteins that have been proposed as transport molecules for Cd2+ entry is yet to be confirmed [6], [7], [8]. For metal ion trafficking, the sub-cellular sequestration and accumulation of Cd2+ have been observed in several organelles such as the mitochondria or nucleus [9], but have not been completely elucidated. Conclusions for these issues cannot be easily obtained partially because of the lack of suitable live-cell tools that are specific for cytosolic and sub-cellular Cd2+ sensing [10], [11]. Ideally, the patch-clamp electrophysiological method can precisely determine the specific ionic current flux into or out of living cells, and thus has been used as the first choice for cytotoxic Cd2+ investigations [6]. However, the ion channel recording can neither reveal the Cd2+ currents chronically (e.g., at least 1 h to 2 h long-term recording), not monitor the sub-cellular dynamics of Cd2+. To obtain accurate information on the succeeding sequestration and the bio-magnification of Cd2+ after cytotoxic entry, appropriate live-cell sensing approaches for intra/sub-cellular Cd2+ monitoring have been attempted. Reliable method that can directly observe the Cd2+ toxicity status within intact cells should be developed because this method can potentially help to identify the agents that can efficiently remove Cd2+ from the human body.
In this study, we established a fluorescent protein-based Cd2+ indicator, Met-cad 1.57, which is applicable for both intracellular and sub-cellular Cd2+ detections. The sensing element is CadR, a dimer protein responsible for the Cd2+ resistance of Pseudomonas putida [12]. The sensing ability of Met-cad was characterized for the content monitoring of Cd2+. The human embryonic kidney (HEK) cells that stably express Met-cad 1.57 (HEK-MCD157) were produced for further investigations.
Results
Development of Met-cads
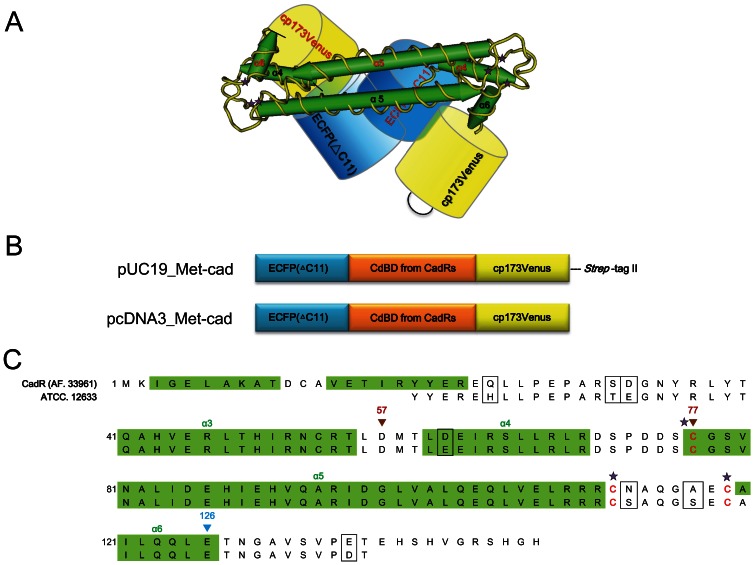
From our recent report, we proposed that a certain part of MerR-like protein is a possible metal ion sensing toolbox that produces an identical metal ion indicator, such as Met-lead with PbrR [13]. In this study, we selected CadR, another MerR family member, to design Met-cad, a Cd2+ indicator (Figure 1). The proposed dimer structure of a FRET-based Cd2+ indicator is shown in Figure 1A according to previous report about the structure property of CadR [12]. The FRET event can occur if ECFP (ΔC11) and cpVenus (as an effective FRET pair) can be driven close together upon the CadR-Cd2+ binding. Figure 1B reveals the genetic maps of Met-cads designed for bacteria and mammalian cell expressions by using pUC19 and pcDNA3 vectors, respectively. Two versions of Met-cads were constructed: Met-cad 1.57, with the protein sequence from residue 57 to 126; and Met-cad 1.77, with the protein sequence from residue 77 to 126 (Figure 1C). The following sub-cellular targeted constructs for mammalian cell were produced with specific organelle-localized sequences.
Figure 1. Design of fluorescent resonance energy transfer (FRET)-based Cd2+ indicators.
(A) Proposed protein structure of Met-cad, a FRET-based Cd2+ indicator, which contains the FRET pair ECFP (ΔC11) (blue) and cpVenus (yellow) as well as CadR (green) with the Cd2+ sensing site (yellow with 3 stars). The ZntR structure (PDB ID code 1Q08) was used as a model for CadR [39]. (B) Genetic map for Met-cad constructs. For bacterial expression, pUC19 is the major vector that contains the FRET pair gene and the Cd2+-binding domain, CdBD. For the large-scale preparation of Met-cad, Strep-tag was ligated at the end of the constructs. For the mammalian cell expression, pcDNA3 vector was used to carry the Met-cad construct into the cells or sub-cellular compartments. (C) Detailed protein sequence of CadR designed as the Cd2+-sensing center to construct Met-cads. The P. putida (ATCC, 12633) was so far the only available source for CadR in our laboratory. The proposed CadR protein fragment from ATCC 12633 was therefore compared with the known CadR sequence from AF 33961 (P. putida 06909) as previous reported [12]. These two are found to be highly conserved. For Met-cad 1.57, the protein sequence begins at the 57th residue (red inverted triangle), whereas the protein sequence for Met-cad 1.77 begins at the 77th residue (red inverted triangle). Both constructs ended at the 126th residue (blue inverted triangle). The three stars labeled cysteine residues of each CadR unit are the proposed Cd2+ binding sites under the conformation of intermolecular dimer [12].
FRET Spectra of Met-cads for Cd2+ Sensing
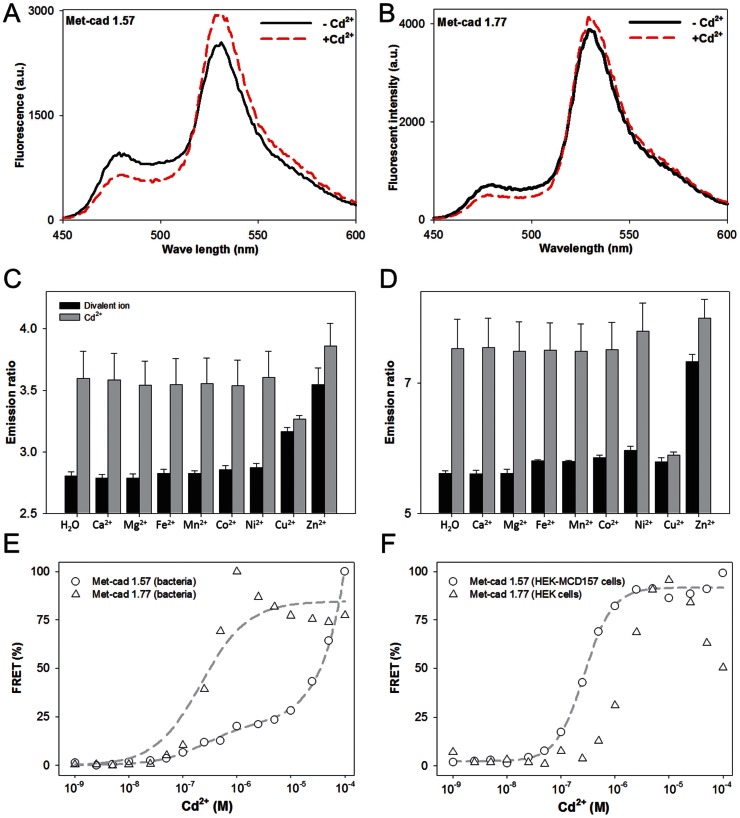
The spectral patterns of ECFP (ΔC11) [fluorescent intensity (FI) at 475 nm] and cp173Venus (FI at 535 nm) from bacterial and mammalian Met-cad proteins between the control solution (without Cd2+; solid line in Figures 2A, 2B, and S1) and the Cd2+ solution (100 µM, dashed line in Figures 2A, 2B, and S1) are shown. The YFP/CFP FRET emission ratio of Met-cads was used to present the Cd2+-sensing capacity qualitatively. Such emission ratios can be obtained from the original spectral data (e.g., Figure 2C and 2D). The increase in FI of YFP and the decrease in FI of CFP indicate the occurrence of a successful Cd2+-dependent FRET event and correspond to the Cd2+-sensing ability of Met-cads (Figures 2A, 2B, and S2).
Figure 2. Spectral analysis of Met-cads.
(A and B) Pattern changes in fluorescence spectra from bacterial Met-cads 1.57 (A) and 1.77 (B) between the control group [(–)Cd, solid line] and Cd [(+)Cd, 100 µM of Cd2+; dashed line]. The YFP/CFP (emission ratio) values of Met-cads range from 520 nm to 550 nm for YFP and 460 nm to 490 nm for CFP, which correspond to the fluorescent resonance energy transfer (FRET) status. (C and D) Ion selectivity of Met-cads 1.57 (C) and 1.77 (D) to Cd2+ (1 µM) was tested (compared with the emission ratios) on various ions (1 µM): Ca2+, Mg2+, Fe2+, Mn2+, Co2+, Ni2+, Cu2+, and Zn2+ without (black bars) or with (gray bars) Cd2+. (E and F) Titration curves of bacterial (E) and mammalian (F) Met-cads, 1.57 (circle) and 1.77 (triangle). Normalized emission ratio data presented as FRET percentage (%) of Met-cads were recorded under different Cd2+ concentrations (10–9 M to 10–4 M). The fitting curves (dash lines) of bacterial Met-cads 1.57 and 1.77 as well as the mammalian Met-cad 1.57 are also shown.
For sensing specificity, the Met-cads were then challenged with metal ions other than Cd2+. The emission ratio changes in Met-cads (1 µM) of the control group (H2O) or selected divalent ions (1 µM), such as Ca2+, Mg2+, Fe2+, Mn2+, Co2+, Ni2+, Cu2+, and Zn2+ (black bars in Figures 2C to 2D) are shown to present the ion selectivity of Met-cads on Cd2+ sensing. To demonstrate the competitive ability of Met-cads (gray bars in Figure 2C to 2D), Met-cads were also treated with the selected ions and Cd2+. The increase was approximately 0.8 (2.8 to 3.6, about 1.29 fold increase) for Met-cad 1.57 (Figure 2C) and 2.6 (5.2 to 7.8, 1.5 fold increase) for Met-cad 1.77 (Figure 2D). There are some adverse effects of Zn2+ and Cu2+ on the Cd2+ sensing (Figure S2). Even though Zn2+ alone can induce increase of FRET ratio (Figure S2A), Cd2+ still can cause additional significant FRET ratio increase under the existence of equal content of Zn2+ (Figure 2C for Met-cad 1.57 and 2D for Met-cad 1.77). Therefore the results here indicated that Cd2+ can induce a specific increase in FRET ratio without the influence of other divalent ions, except for Cu2+ (Figure S2B).
To determine the detailed sensing capability of Met-cads, the ratios under the spectral platform were further obtained and analyzed under various Cd2+ concentrations (10–9 M to 10–4 M). The titration curves (Cd2+ concentrations versus emission ratios; curve fitted as described in the Materials and Methods section) of bacterial (Figure 2E) as well as mammalian (Figure 2F) Met-cad Met-cads 1.57 (circle) and 1.77 (triangle) are shown in Figures 2E and 2F. K ds of the bacterial Met-cad 1.57 and Met-cad 1.77 were 250 (calculated by one-site saturation with a nonspecific factor Ns = 796460) and 221 nM (calculated by one-site saturation without Ns), and that of mammalian Met-cad 1.57 was 271 nM (calculated by Hill equation).
The length of Met-cad 1.57 is longer than that of Met-cad 1.77 containing additional α4 helix. This causes spatial difference between ECFP (ΔC11) and cpVenus among these two sensors. Various basal FRET value and different FRET response can be acquired in the presence of Cd2+. The fact that the pattern of FRET ratio values in Met-cad 1.77 dropped when the concentration of Cd2+ is rising higher than 10−6 M (triangle in Figures 2E and 2F) is still hard to be explained. We finally chose Met-cad 1.57 for the rest of the experiments.
As described above in Introduction that both MerR including CadR (as well as PbrR [13]) and MerR-based FRET sensors essentially form dimer to construct functional Cd2+ binding (similar for Pb2+ binding in PbrR). It might raise a concern that such structure could disturb the FRET event for real sensing. The result that we still can observe the FRET event after adding Cd2+ (in Met-cads, Figure 2) or identical metal ion (Pb2+ in Met-leads in [13]) answered this question.
From experimental data in Figure 2E (dashed line alone with circle), there did exist a novel nonspecific factor (as described above) correlated to the dramatic increase of FRET ratio under high concentration of Cd2+ (from 5×10−5 M) in bacterial Met-cad 1.57. Alternatively, a stable-expressed cell line HEK-MCD157 was adapted to express large amount of Met-cad protein without the tag to perform the curve fitting (dashed line alone with circle in Figure 2F). The FRET ratio of mammalian 1.57 was saturated (around 5×10−6 M), significantly different from the increase pattern shown in bacterial Met-cad 1.57. Base on genetic sequence, the Strep-tag could be one of the main differences between bacterial and mammalian Met-cad 1.57, in addition to the two different expression systems (perhaps more complicated modifications like glycosylation make them different). More experiments need to be done in the future.
Live-Cell Spectral Sensing and Visualization of Cd2+
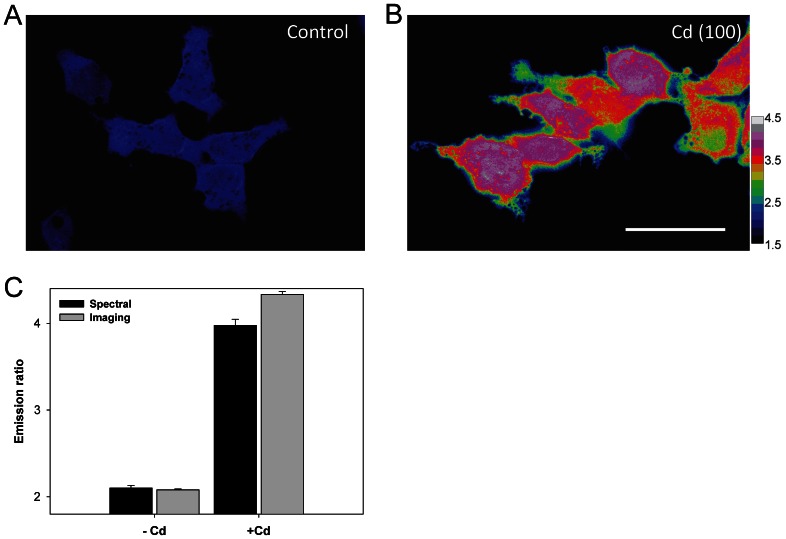
For the easy handling of live-cell Cd2+ sensing with an acceptable throughput scale, HEK-MCD157, a Met-cad 1.57 that is stably expressed in the HEK293 cell line, was obtained (see Material and Methods section). Considering Met-cad 1.57 under a visualization aspect (from blue to pink and gray under the ratio color bar defined from 1.5 to 4.5), the homogeneous blue (ratio ≈ 2) cytosolic space in the control cells (Figure 3A) and the pink areas (ratio ≈ 4.35) in the cells that were exposed to 100 µM of Cd2+ (Figure 3B) clearly and directly reveal the real status of cytosolic Cd2+ contents. Figure 3 shows that the ratio from both spectral and imaging systems may be comparable for further usage. We used ionomycin to test the sensing range of Met-cad 1.57 in vivo since this chemical agent can be used as ionic carrier into living cells [13], with a ratio that ranges from 1.86 (basal without Cd2+) to 4.29 (100 µM of Cd2+), thereby yielding approximately 2.3-fold (data not shown).
Figure 3. Live-cell fluorescent resonance energy transfer (FRET)-based Cd2+ sensing within human embryonic (HEK)-MCD157 cells.
(A to B) Representative FRET ratio images of Met-cad 1.57 stably expressed in cells (HEK-MCD157) without (A, Control) or with 100 µM of Cd2+ incubated for 2 h [B, Cd (100)]. The ratio color bar ranges from 1.5 to 4.5 (color change sequence: black, blue, green, yellow, orange, red, pink, gray, and white). Scale bar = 10 µm. (C) YFP/CFP emission ratios of living HEK-MCD157 cells. The original data were obtained from fluorescent spectroscopy (Spectral, black bars) or FRET ratio imaging (Imaging, gray bars). These HEK-MCD157 cells were treated without [(–) Cd] or with 100 µM of Cd2+ [(+) Cd] for 2 h.
Monitoring the Cytosolic Entry of Cd2+
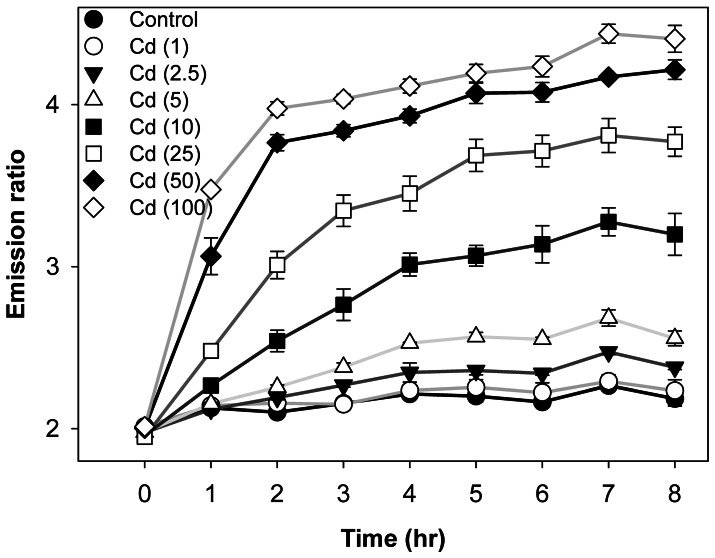
HEK-MCD 157 cells were used in the following live-cell experiments to monitor the intracellular Cd2+ content with the passage of time (recorded every 1 h to 8 h of exposure) in various concentrations of extracellular/environmental Cd2+ [from low (1, 2.5, and 5 µM) to high concentrations (10, 25, 50, and 100 µM)]. Figure 4 shows that 1 h to 2 h of relatively high dosage treatments [10 µM (closed square); 25 µM (open square); and 50 µM (closed diamond)] can significantly increase the emission ratio compared with the basal level. By contrast, low concentration (e.g., 2.5 µM) exposure with sufficient incubation time, such as 7 h, also caused a significant increase in ratio (inverted triangle in Figure 4). Although the Cd2+ level did not change throughout the 8 h incubation period at 1 µM of Cd2+ (empty circle in Figure 4), such trace amounts of Cd2+ for a longer exposure time (at least 24 h) can be sensed by Met-cad 1.57 (imaging data shown in Figure S3). The ratios reached 2.401±0.075 and 2.687±0.075 after 24 and 48 h of treatments, respectively, which are significantly higher than those of the basal level (2.013±0.001).
Figure 4. Content monitoring of intracellular Cd2+.
Human embryonic kidney (HEK)-MCD157 cells cultured in a 96-well plate treated with Cd2+ from 1 h to 8 h under various concentrations: control (closed circle), 1 µM (open circle), 2.5 µM (closed inverted triangle), 5 µM (open triangle), 10 µM (closed rectangle), 25 µM (open rectangle), 50 µM (closed diamond), and 100 µM (open diamond). The emission ratio data were obtained by fluorescent spectroscopy.
Ca2+ Channels are Involved in the Cytosolic Entry of Cd2+
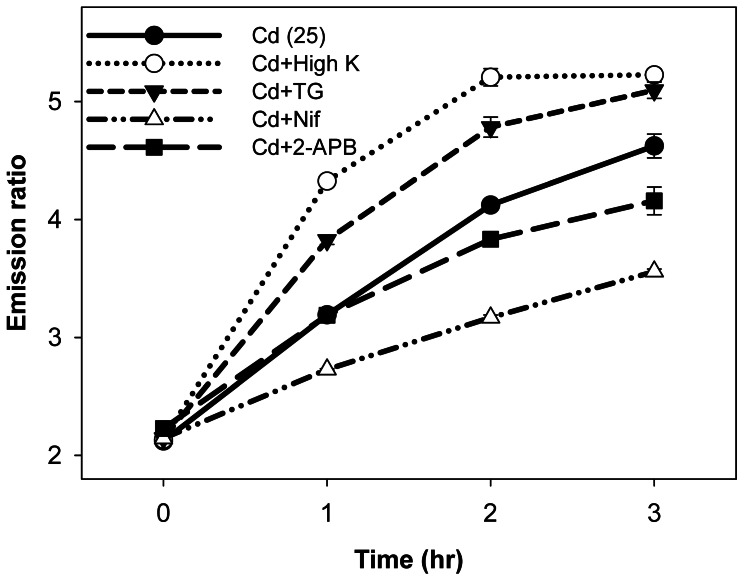
We investigated the entry routes of Cd2+ into the HEK-MCD157 cells. The data show that the ratios reached 3 after 2 h of incubation at 25 µM of Cd2+ (Figure 4). This condition was used as the experimental background to test the possible transport mechanisms for Cd2+ entry (Figure 5). Either the agonists (stimulants of Ca2+ channels) or the antagonists (blockers of Ca2+ channels) were applied (30 min prior to the Cd2+ recording, Table S1) as the pharmacological strategy to challenge the potential gateways for Cd2+ entry. Figure 5 shows that the activation of different types of Ca2+ channels [high K+ buffer for voltage-gated channels (open circle with dotted line); thapsigargin (TG) for storage-operated channels (inverted triangle with dashed line)] significantly enhanced Cd2+ entry. These channels can be blocked using the corresponding antagonists (nifedipin for voltage-gated Ca2+ channels, open triangle with dashed-dotted line; 2-APB for storage-operated Ca2+ channels, closed rectangle with long dashed line), which significantly affected Cd2+ entry (Figure 5) in agreement with the finding from previous report [6]. The results suggested that various types of Ca2+ channels are involved in Cd2+ entry.
Figure 5. Cd2+ Uptake Mechanism.
Human embryonic kidney (HEK)-MCD 157 cells seeded in a 96-well culture plate without [Cd (25), closed circle with solid line] or with different additional agents: high K+ buffer (Cd+High K, 150 mM; open circle with dotted line), thapsigargin (Cd+TG, 1 µM; closed inverted triangle with dashed line), nifedipine (Cd+Nif, 50 µM; open triangle with dashed-dotted line), and 2-APB (Cd+2-APB, 25 µM, closed rectangle with long dashed line).
Sub-Cellular Sequestration of Cd2+
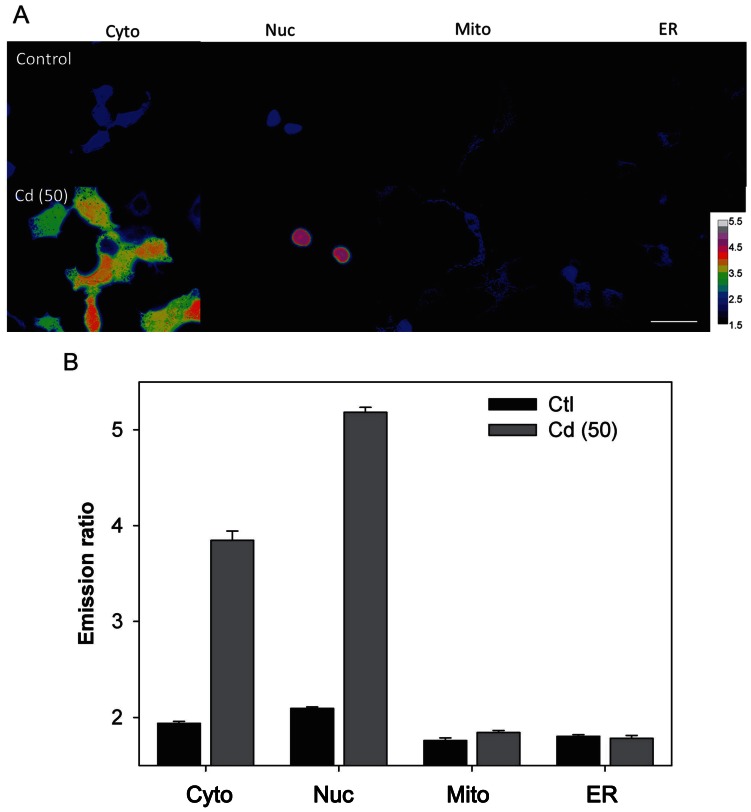
The organelle-targeted versions of Met-cad 1.57 were used to visualize the sub-cellular localization of Cd2+ that successfully entered the cell (Figure S4). Representative ratio color images of cytosolic (Cyto) and organelle-targeted Met-cad 1.57 within living cells (nucleus, Nuc; mitochondria, Mito; endoplasmic reticulum, ER) are shown in Figure 6A without (control) or with 2 h of exposure to 50 µM of Cd2+ [Cd (50)]. The bar figures in Figure 6B indicate the different sub-cellular Cd2+ levels. A significant increase in Cd2+ contents in the nucleus after 2 h of incubation was observed (higher than the cytosolic Cd2+ level). In contrast to the cytosolic and nuclear spaces, no further increase in ratio was found within the mitochondria and ER under the same treatment condition.
Figure 6. Sub-cellular Cd2+ accumulation.
(A) Representative images of Met-cads with various organelle-targeted versions (Cyto for cytosol; Nuc for nucleus; Mito for mitochondria; ER for endoplasmic reticulum) without (Control) and with 2 h incubation period at 50 µM of Cd2+ [Cd (50)]. The color bars show the ratio that ranges from 1.5 to 5.5. Scale bar = 10 µm. (B) The emission ratios from different sub-cellular compartments are shown without (Control in black bars) and with 50 µM of Cd2+ [Cd (50) in gray bars].
Discussion
The selection of a suitable sensing key is considered one of the difficult processes in manufacturing protein-based metal ion indicators [13], and only three have been successfully manufactured, specifically Ca2+ [14], [15], Zn2+ [16], and Cu+ [17]. In this study, we constructed additional ones for Pb2+ [13] and Cd2+. Bacteria such as Cupriavidus metallidurans CH34 and P. putida survive in extremely harsh environments by evolving protective anti-metal systems [12], [18]. MerR is one of the specific metal-binding systems that comprise five major members: PbrR, CadR, MerR, CueR, and ZntR. We initially produced the MerR-based indicator (Met-lead 1.59) for live-cell Pb2+ sensing with PbrR used as the metal-binding domain [13]. In this study, part of the CadRs was designed as the Cd2+-binding domain to create Met-cads, the second MerR-based indicator for Cd2+ (Figure 1). Such serial MerRs can be used as a novel sensing toolbox for individual metal ion sensing (the comparisons on sensing part between Met-lead 1.59 and Met-cad 1.57 are shown in Figure S5). Although we preliminarily established some of the other MerR-related metal ion biosensors, such as those for Cu+ (CueR, unpublished data) and Zn2+ (ZntR, unpublished data), but those for Hg2+ (MerR) have not been developed yet. Further studies should be conducted to prove this concept.
Compared with the currently available fluorescent indicators for live-cell Cd2+ sensing, such as the commercial fura-5F [19], fluo-3 [20], and even indo-1, which are originally designed for Ca2+ sensing, the sensors can only detect the presence of Cd2+ that is incubated at 30 µM for 24 h, and Cd2+ cannot be distinguished from Ca2+. New developed chemical indicators own much better sensing ability especially the ionic selectivity to distinguish Cd2+ from Zn2+ [21]–[23]. These indicators tend to be ratiometric within suitable emission wavelength [22], [23] for popular usage on conventional fluorescent imaging. Among these three, CadQM developed by Taki et al., might be the most convincing one for live-cell sensing among these developed chemical-based Cd2+ indicators [21]–[23]. The basal level of a Cd2+ ratio dye CadMQ is about 0.75 increased to 0.84 under 5 µM Cd2+ exposure for 3 hrs (about 1.12 fold increases). (Met-cad: 2.1±0.02; reached to 2.5±0.06, about 1.19 fold increase, shown in Figure 4), and decreased its ratio to 0.74 upon TPEN chelation [23] (we also did this chelation test to confirm, ratio changed back to 2.2±0.03). However, the sensing ability of CadMQ was also affected by Zn2+ and Cu2+ (Zn2+ and Cu2+ did have certain influence on Met-cad sensing as shown in Figure S2) and even Hg2+. The rest limitations of these chemical dyes are probably the inability to fully organelle targeting specifically (Figures 6, S4) as well as within whole animal sensing (whole animal genetically expressed protein-based biosensor, e.g. Met-cad, can be archived through transgenic way in the near future).
In this study, the newly developed Met-cad 1.57 exhibits a sub-µM level of K d (250 nM), relatively high sensitivity, and good selectivity without the influence of Ca2+ as well as organelle-targetable characteristic. Met-cad 1.57 can sense the accumulation of Cd2+ at very low concentrations (1 µM) for 24 and 48 h of incubation periods. The non-cytotoxicity at 1 µM [19] requires further investigations.
Although the gating mechanisms of Cd2+ entry are considered as different types of Ca2+ channels [24], [25], [26], divalent metal-ion transporter (DMT1) [8], [27], [28], and Zn2+ transporter (ZIP8/14) [29], [30], the conclusions on the Cd2+ entry pathway remain controversial. We used the HEK-MCD157 cells to confirm that Ca2+ channels are one of the corresponding gates for Cd2+ entry (Figure 5). The other candidates such as DMT1 and ZIP8/14 would be tested in future investigations.
Among the available live-cell sensing methods, including chemicals and proteins, the protein-based indicators are probably one of the most promising indicators because of the organelle-targeting and genetic-encoding ability. These indicators can monitor the target biomolecules in the intra/sub-cellular space within living cells and in organisms as a whole [10], [14], [15]. In the current study, we produced three kinds of organelle-targeted versions of Met-cads for the nucleus, mitochondria, and ER. Cd2+ accumulation was found inside the nucleus within 2 h of exposure, but not in other sub-cellular compartments (Figure 6). Such Cd2+ accumulation within the nucleus was higher than that in the cytosolic Cd2+ contents. Cd2+ has been considered as a human carcinogen for years [31], [32], [33]. Thus, the nuclear transport mechanism of Cd2+ should be investigated in the future because this phenomenon may be correlated with the carcinogenic property of Cd2+. The data from sub-cellular localization of heavy Cd2+ metal provide valuable insight on heavy metal ion toxicology.
In addition, we established a live-cell system that stably expresses Met-cad 1.57 for numerous benefits. For example, large amounts of Met-cad proteins were provided to help reveal the hidden problem of Strep-tag in Met-cads. The live-cell system can be seeded in multi-well cell culture plates to perform easy median-scale experiments such as titration (Figure 2F), time-dose response (Figure 4), and Ca2+ channel tests (Figure 5). Furthermore, the live-cell system can be applied in real high-throughput platforms in either spectral or imaging modality with a 96−/386-well plate for more advanced investigations on the molecular and cellular toxicology of Cd2+. The possible effective agent(s) for therapeutic usage on Cd2+ poisoning would be possibly established through a large-scale screening.
In summary, we produced Met-cad 1.57, another MerR-based indicator for Cd2+, according to the FRET strategy. This live-cell Cd2+ indicator was further incorporated into a stable HEK cell line or to become organelle-targetable for advanced research on sub-cellular Cd2+ dynamics. These constructs and cell line can be further applied in several investigations on the molecular and cellular toxicology of Cd2+.
Materials and Methods
Chemicals
All reagents were purchased from Sigma Chemical Co. (St. Louis, MO, USA) unless indicated otherwise. Most of the reagents for molecular cloning, such as restriction enzymes, ligases, and others, were from New England BioLabs. The polymerase chain reaction kit and the primers used were purchased from Finnzymes Oy (Finland) and Mission Biotech (Taiwan), respectively. The DNA extraction kit was purchased from Viogene.
Gene Construction and Sub-Cellular Targeting of Met-cad
According to the gene alignment program Kalign (EMBL-EBI) and the structure prediction PredictProtein [34], the gene fragments of CadR from P. putida [American Type Culture Collection (ATCC, Rockville, MD, USA) 12633] were compared with AF 33961 [12] and were cloned as the Cd2+-binding domain (CdBD) for the Cd2+-sensing. The protein sequence map (Figure 1A) shows that the CdBDs, ECFP (ΔC11), and cp173Venus were ligated together [13], [15] as the Met-cads, which were further cloned into pcDNA3 vector (Invitrogen) to form pcDNA3_Met-cads (Figure 1B). Organelle-targeted versions of Met-cad 1.57 were constructed by adding specific targeted sequences as previously reported, such as the sequences in the nucleus (two tandem repeats of DPKKKRKV derived from the simian virus large T-antigen) [35], mitochondria (four tandem repeats of SVLTPLLLRGLTGSARRLPVPRAKIHSL derived from the precursor of subunit VIII of human cytochrome C oxidase) [36], and endoplasmic reticulum (calrericulin signal sequence LLSVPLLLGLLGLAVA with the retention sequence KDEL) [37], [38].
Cell Culture, Transfection, and Stable Expression of Met-cad in HEK-MCD157 Cell Line
HEK293 cells from the ATCC were cultured in 22 mm coverslip and transfected [13] with pcDNA3_Met-cad 1.57 or other organelle-targeted versions of Met-cad 1.57 for live-cell fluorescent resonance energy transfer (FRET)-based microscopic ratio imaging.
The HEK-MCD157 cells, which stably express Met-cad 1.57, were produced through antibiotic (G418) selection. These cells were seeded in 96-well culture plates (COSTAR 3599, Corning, USA) for fluorescent spectroscopy or cultured as described for FRET ratio imaging.
Fluorescent Spectroscopy
The fluorescent spectrum of Met-cad 1.57 and its YFP/CFP emission ratios were obtained using a spectrophotometer (Infinite M1000, TECAN, Switzerland) as previously reported [13]. The band settings for CFP and YFP are 460 nm to 490 nm and 520 nm to 550 nm, respectively.
Live-cell FRET-based Microscopic Ratio Imaging
The live cell Cd2+ imaging was performed by acquiring the emission ratios under the FRET imaging platform. The microscope (Axio D1, Zeiss, German) was equipped with a 40× objective lens (NA = 0.75). A metal fluorescent light (X-Cite, Lumen Dynamics Group Inc., Canada) filtered with a band-pass filter (426 nm to 446 nm) and a dichroic mirror (442 nm) was used as the excitation light source. Ratio images of the Met-cads within living cells that contain the fluorescent signals of CFP (483/32 nm) and YFP (542/27 nm) were captured using a dual charge-coupled device camera system (ORCA-D2, Hamamatsu, Japan) with HCImage software (Hamamatsu). Ratio imaging was further displayed in a ratio color plate using the ImageJ analysis software (NIH).
For the experiment using ionomycin as ionophore to transport Cd2+ across cell membrane into cytosol, a Ca2+ free buffer containing 150 mM NaCl, 5 mM KCl, 1 mM MgCl2, 10 mM HEPES, and 5 mM D-glucose (pH 7.4) with 5 µM of ionomycin was used to be without or with a defined Cd2+ solution (100 µM) to cells for 10 min before taking ratio imaging [13].
Extraction of Met-cad Protein, Titration, and Selectivity Tests
The expression and purification of bacterial Met-cad proteins were performed using the same strategy as previous report, the Strep-tag [13]. In this study, the Strep-tag was used for the following protein purification instead of applying His-tag to avoid non-selective metal ion interaction, i.e. the binding of Ni2+ to His-tag. Actually Ni2+, as predicted, did not influence the sensing ability of Met-cad with Strep-tag (Figures 2C and 2D on Ni2+). The only concern about Strep-tag would be its possible involvement on the nonspecific factor found in bacterial Met-cad 1.57 (Figure 2E). The mammalian Met-cad 1.57 without the Strep-tag stably expressing in the HEK cell line was therefore alternatively prepared by partial purification to test this issue. Briefly, the supernatant with a large amount of Met-cad protein expressed from HEK-MCD157 cells after sonication were further centrifuged to remove the cell debris from the lysate. The data from mammalian source was found to be well fitted on Hill equation.
Simple quantification of these protein indicators was performed prior to titration and selectivity experiments (from 10–9 M to 10–4 M of Cd2+) by fluorescent spectroscopy. To estimate the dissociation constant (K d) of Met-cads expressed from bacterial sources or HEK-MCD157 cells, the ratio data from the titration experiments were normalized as percentages of FRET relative to the maximal difference of the emission values (R). The fitting curve was further analyzed using the following equations: (1) One-site saturation with a nonspecific factor, [Cd2+] = [(R max × R)/(K d+R)]+n × R (bacterial version); (2) Hill equation [14], [Cd2+] = K d [(R – R min)/(R max – R)(1/n) (mammalian version), where n is the nonspecific binding coefficient. For the selectivity tests (specificity and competition), 1 µM of divalent ions (such as CaCl2, MgCl2, FeSO4, MnCl2, CoCl2, NiCl2, CuCl2, and ZnCl2) were used with or without equal CdCl2 concentrations (1 µM).
Data Analyses and Statistics
All experiments were conducted thrice with at least three different sets of cell samples. Data gathered from the different batches were integrated to calculate the emission ratio. Significant changes were considered significant at the 0.05 level by ANOVA (SPSS statistics 19, IBM) described previously [13].
Supporting Information
Spectral Pattern changes of mammalian Met-cad 1.57 between the control group [(–)Cd, solid line] and Cd [(+)Cd, 100 µM of Cd2+; red dashed line].
(TIFF)
Effects of Zn2+ and Cu2+ on the spectral pattern of Met-cad 1.57. The spectral patterns of Met- cad 1.57 to Zn 2+ (A, 1 µM in dotted line) and to Cu2+ (B, 1 µM in dashed line; 10 µM in dashed-dotted line; 100 µM in dotted line) compared with the control without specific ions (Apo in solid line).
(TIF)
Long-term monitoring of intracellular Cd2+ content. The representative images of cells expressing Met-cad for monitoring intracellular content of Cd2+ are shown in YFP (top), CFP (middle), and ratio (bottom) with 8, 12, 24, and 36 hours of incubations (1 µM). The color bars show the ratio that ranges from 1 to 5.
(TIF)
Sub-cellular targeting of Met-cads within cells. On top, the representative confocal images (LSM 5 Pascal, Zeiss, Germany, with a 63× oil objective, NA = 1.4, on an inverted microscope, Axiovert 200M) [1], [2] of Met-cads (EX: 458 nm; EM: LP475 nm) of cells transfected with various organelle-targeted versions (Met-cad-nuc for nucleus; Met-cad-mito for mitochondria; Met-cad-ER for endoplasmic reticulum, ER) are displayed in green color. In the middle, the red images (EX: 543 nm; EM: LP560 nm) of cells were stained with certain organelle dyes, e.g. propidium iodine (Vector Lab.) for nucleus; MitoTracker Orange CMTMRos (Invitrogen) for mitochondria; ER-Tracker Red (Invitrogen) for ER. The merged images are show in the bottom part. The white arrows indicate that certain targeted Met-cads co-localize with specific organelles within the same compartments of the cells (yellow color). Scale bar = 20 µm.
(TIF)
The comparisons between Met-lead 1.59 and Met-cad 1.57. Sensing key of Met-lead 1.59 was from part of PbrR (starting from the residue D between α3 and α4 helix in bold) [1]. And sensing key of Met-cad 1.57 was from part of CadR, in this study. The most different parts in sequences are after the 2nd cysteine (denoted as star, after the end of the α5 helix) through the whole α6 helix (in red color).
(TIF)
The effects of reagents used (high K, TG, Nif, and 2-APB) on ratio value in Figure 5 . There’s no significant changes between the two experimental sets, i.e. before and 30 min. after reagent treatments).
(TIFF)
Supplemental Bibliography
(pdf)
Acknowledgments
The authors would like to thank Professor Takeharu Nagai for the generous sharing of YC 3.6. This work is part of the master thesis of Po-Hsun Chen in Institute of Biophotonics, National Yang-Ming University. We appreciate Profs. Te-Chang Lee and Yueh-Hsin Ping, and Dr. Yu-Fen Chang providing valuable suggestions to this work.
Funding Statement
This work is supported by grants from the National Science Council of the Republic of China, Taiwan (Grants NSC-100-2320-B-075-004), Taipei Veterans General Hospital (V100C1-032, V101C1-072, V102C-163), the Joint Projects of UTVGH (University System of Taiwan together with Taipei Veterans General Hospital: VGHUST100-G1-1-2, VGHUST102-G7-1-2E), and National Yang-Ming University (Ministry of Education, Aim for the Top University Plan) and Biophotonics and Molecular Imaging Research Center. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Goyer RA, Waalkes MP (2008) Toxic effects of metals. In Casarett and Doull’s Toxicology: The Basic Science of Poisons (seventh edition), Edit by Klaassen, C. D. The McGraw-Hill Companies, Inc. [Google Scholar]
- 2. Clarkson TW (1993) Molecular and ionic mimicry of toxic metals. Ann Rev Pharma Tox 33: 545–571. [DOI] [PubMed] [Google Scholar]
- 3. Bridges CC, Zalups RK (2005) Molecular and ionic mimicry and the transport of toxic metals. Toxicol Appl Pharmacol 204: 274–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Moulis JM (2010) Cellular mechanisms of cadmium toxicity related to the homeostasis of essential metals. Biometals 23: 877–896. [DOI] [PubMed] [Google Scholar]
- 5.ATSDR. (2005) CERCLA Priority List of Hazzardous Substances Available: http://www.atsdr.cdc.gov.cercla.
- 6. Fukuda J, Kawa K (1997) Permeation of manganese, cadmium, zinc, and beryllium through calcium channels of an insect muscle membrane. Science 196: 309–311. [DOI] [PubMed] [Google Scholar]
- 7. Thevenod B (2010) Catch me if you can! Novel aspects of cadmium transport in mammalian cells. Biometals 23: 857–875. [DOI] [PubMed] [Google Scholar]
- 8. Vesey DA (2010) Transport pathways for cadmium in the intestine and kidney proximal tubule: focus on the interaction with essential metals. Toxicol Lett 198: 13–19. [DOI] [PubMed] [Google Scholar]
- 9. Lin CJ, Wu KH, Yew FH, Lee TC (1995) Differential cytotoxicity of cadmium to rat embryonic fibroblasts and human skin fibroblasts. Toxicol Appl Pharmacol 133: 20–26. [DOI] [PubMed] [Google Scholar]
- 10. Li IT, Pham E, Truong K (2006) Protein biosensors based on the principle of fluorescence resonance energy transfer for monitoring cellular dynamics. Biotechnol Lett 28: 1971–1982. [DOI] [PubMed] [Google Scholar]
- 11. Dean KM, Qin Y, Palmer AE (2012) Visualizing metal ions in cells: An overview of analytical techniques, approaches, and probes. Bioch Biophys Acta 1823: 1406–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee SW, Glickmann E, Cooksey DA (2001) Chromosomal locus for cadmium resistance in Pseudomonas putida consisting of a cadmium-transporting ATPase and a MerR family response regulator. Appl Environ Microbiol 67: 1437–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chiu TY, Yang DM (2012) Intracellular Pb2+ content monitoring using a protein-based Pb2+ indicator. Toxicol Sci 126: 436–445. [DOI] [PubMed] [Google Scholar]
- 14. Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, et al. (1997) Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 388: 882–887. [DOI] [PubMed] [Google Scholar]
- 15. Nagai T, Yamada S, Tominaga T, Ichikawa M, Miyawaki A (2004) Expanded dynamic range of fluorescent indicators for Ca(2+) by circularly permuted yellow fluorescent proteins. Proc Natl Acad Sci USA 101: 10554–10559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vinkenborg JL, Koay MS, Merkx M (2010) Fluorescent imaging of transition metal homeostasis using genetically encoded sensors. Curr Opin Chem Biol 14: 231–237. [DOI] [PubMed] [Google Scholar]
- 17. Wegner SV, Arslan H, Sunbul M, Yin J, He C (2010) Dynamic copper(I) imaging in mammalian cells with a genetically encoded fluorescent copper(I) sensor. J Am Chem Soc 132: 2567–2569. [DOI] [PubMed] [Google Scholar]
- 18. Brown NL, Stoyanov JV, Kidd SP, Hobman JL (2003) The MerR family of transcriptional regulators. FEMS Microbiol Rev 27: 145–163. [DOI] [PubMed] [Google Scholar]
- 19. Tvermoes BE, Bird GS, Freedman JH (2011) Cadmium induces transcription independently of intracellular calcium mobilization. PLoS One 6: e20542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nishimura Y, Yamaguchi JY, Kanada A, Horimoto K, Kanemaru K, et al. (2006) Increase in intracellular Cd(2+) concentration of rat cerebellar granule neurons incubated with cadmium chloride: cadmium cytotoxicity under external Ca(2+)-free condition. Toxicol In Vitro 20: 211–216. [DOI] [PubMed] [Google Scholar]
- 21. Peng X, Du J, Fan J, Wang J, Wu Y (2007) A selective fluorescent sensor for imaging Cd2+ in living cells. J Am Chem Soc 129: 1500–1501. [DOI] [PubMed] [Google Scholar]
- 22. Cheng T, Xu Y, Zhang S, Zhu W, Qian X, et al. (2008) A highly sensitive and selective OFF-ON fluorescent sensor for cadmium in aqueous solution and living cell. J Am Chem Soc 130: 16160–16161. [DOI] [PubMed] [Google Scholar]
- 23. Taki M, Desaki M, Ojida A, Iyoshi S, Hirayama T, et al. (2008) Fluorescence imaging of intracellular cadmium using a dual-excitation ratiometric chemosensor. J Am Chem Soc 130: 12564–12565. [DOI] [PubMed] [Google Scholar]
- 24. Weidner WJ, Sillman AJ (1997) Low levels of cadmium chloride damage the corneal endothelium. Arch Toxicol 71: 455–460. [DOI] [PubMed] [Google Scholar]
- 25. Leslie EM, Liu J, Klaassen CD, Waalkes MP (2006) Acquired cadmium resistance in metallothionein-I/II(−/−) knockout cells: role of the T-type calcium channel Cacnalpha1G in cadmium uptake. Mol Pharmaco 69: 629–639. [DOI] [PubMed] [Google Scholar]
- 26. Martineau C, Abed E, Médina G, Jomphe LA, Mantha M, et al. (2010) Involvement of transient receptor potential melastatin-related 7 (TRPM7) channels in cadmium uptake and cytotoxicity in MC3T3-E1 osteoblasts. Toxicol Lett 199: 3573–63. [DOI] [PubMed] [Google Scholar]
- 27. Bannon DI, Portnoy ME, Olivi L, Lees PS, Culotta VC, et al. (2002) Uptake of lead and iron by divalent metal transporter 1 in yeast and mammalian cells. Biochem Biophys Res Commun 295: 978–984. [DOI] [PubMed] [Google Scholar]
- 28. Bressler JP, Olivi L, Cheong JH, Kim Y, Bannona D (2004) Divalent metal transporter 1 in lead and cadmium transport. Ann NY Acad Sci 1012: 142–152. [DOI] [PubMed] [Google Scholar]
- 29. Dalton TP, He L, Wang B, Miller ML, Jin L, et al. (2005) Identification of mouse SLC39A8 as the transporter responsible for cadmium-induced toxicity in the testis. Proc Natl Acad Sci USA 102: 3401–3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. He L, Wang B, Hay EB, Nebert DW (2009) Discovery of ZIP transporters that participate in cadmium damage to testis and kidney. Toxicol Appl Pharmacol 238: 250–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Waisberg M, Joseph P, Hale B, Beyersmann D (2003) Molecular and cellular mechanisms of cadmium carcinogenesis. Toxicol 192: 95–117. [DOI] [PubMed] [Google Scholar]
- 32. Bertin G, Averbeck D (2006) Cadmium: cellular effects, modifications of biomolecules, modulation of DNA repair and genotoxic consequences (a review). Biochimie 88: 1549–1559. [DOI] [PubMed] [Google Scholar]
- 33. Joseph P (2009) Mechanisms of cadmium carcinogenesis. Toxicol Appl Pharmacol 238: 272–279. [DOI] [PubMed] [Google Scholar]
- 34. Rost B, Yachdav G, Liu J (2004) The PredictProtein server. Nucleic Acids Res 32: W321–W326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kalderon D, Roberts BL, Richardson WD, Smith AE (1984) A short amino acid sequence able to specify nuclear location. Cell 39: 499–509. [DOI] [PubMed] [Google Scholar]
- 36. Rizzuto R, Nakase H, Darras B, Francke U, Fabrizi GM, et al. (1989) A gene specifying subunit VIII of human cytochrome c oxidase is localized to chromosome 11 and is expressed in both muscle and non-muscle tissues. J Biol Chem 264: 10595–10600. [PubMed] [Google Scholar]
- 37. Munro S, Pelham HR (1987) A C-terminal signal prevents secretion of luminal ER proteins. Cell 48: 899–907. [DOI] [PubMed] [Google Scholar]
- 38. Fliegel L, Burns K, MacLennan DH, Reithmeier RA, Michalak M (1989) Molecular cloning of the high affinity calcium-binding protein (calreticulin) of skeletal muscle sarcoplasmic reticulum. J Biol Chem 264: 21522–21528. [PubMed] [Google Scholar]
- 39. Changela A, Chen K, Xue Y, Holschen J, Outten CE, et al. (2003) Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science 301: 1383–1387. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Spectral Pattern changes of mammalian Met-cad 1.57 between the control group [(–)Cd, solid line] and Cd [(+)Cd, 100 µM of Cd2+; red dashed line].
(TIFF)
Effects of Zn2+ and Cu2+ on the spectral pattern of Met-cad 1.57. The spectral patterns of Met- cad 1.57 to Zn 2+ (A, 1 µM in dotted line) and to Cu2+ (B, 1 µM in dashed line; 10 µM in dashed-dotted line; 100 µM in dotted line) compared with the control without specific ions (Apo in solid line).
(TIF)
Long-term monitoring of intracellular Cd2+ content. The representative images of cells expressing Met-cad for monitoring intracellular content of Cd2+ are shown in YFP (top), CFP (middle), and ratio (bottom) with 8, 12, 24, and 36 hours of incubations (1 µM). The color bars show the ratio that ranges from 1 to 5.
(TIF)
Sub-cellular targeting of Met-cads within cells. On top, the representative confocal images (LSM 5 Pascal, Zeiss, Germany, with a 63× oil objective, NA = 1.4, on an inverted microscope, Axiovert 200M) [1], [2] of Met-cads (EX: 458 nm; EM: LP475 nm) of cells transfected with various organelle-targeted versions (Met-cad-nuc for nucleus; Met-cad-mito for mitochondria; Met-cad-ER for endoplasmic reticulum, ER) are displayed in green color. In the middle, the red images (EX: 543 nm; EM: LP560 nm) of cells were stained with certain organelle dyes, e.g. propidium iodine (Vector Lab.) for nucleus; MitoTracker Orange CMTMRos (Invitrogen) for mitochondria; ER-Tracker Red (Invitrogen) for ER. The merged images are show in the bottom part. The white arrows indicate that certain targeted Met-cads co-localize with specific organelles within the same compartments of the cells (yellow color). Scale bar = 20 µm.
(TIF)
The comparisons between Met-lead 1.59 and Met-cad 1.57. Sensing key of Met-lead 1.59 was from part of PbrR (starting from the residue D between α3 and α4 helix in bold) [1]. And sensing key of Met-cad 1.57 was from part of CadR, in this study. The most different parts in sequences are after the 2nd cysteine (denoted as star, after the end of the α5 helix) through the whole α6 helix (in red color).
(TIF)
The effects of reagents used (high K, TG, Nif, and 2-APB) on ratio value in Figure 5 . There’s no significant changes between the two experimental sets, i.e. before and 30 min. after reagent treatments).
(TIFF)
Supplemental Bibliography
(pdf)