Abstract
FoxD4/5, a forkhead transcription factor, plays a critical role in establishing and maintaining the embryonic neural ectoderm. It both up-regulates genes that maintain a proliferative, immature neural ectoderm and down-regulates genes that promote the transition to a differentiating neural plate. We constructed deletion and mutant versions of FoxD4/5 to determine which domains are functionally responsible for these opposite activities, which regulate the critical developmental transition of neural precursors to neural progenitors to differentiating neural plate cells. Our results show that up-regulation of genes that maintain immature neural precursors (gem, zic2) requires the Acidic blob (AB) region in the N-terminal portion of the protein, indicating that the AB is the transactivating domain. Additionally, down-regulation of those genes that promote the transition to neural progenitors (sox) and those that lead to neural differentiation (zic, irx) involves: 1) an interaction with the Groucho co-repressor at the Eh-1 motif in the C-terminus; and 2) sequence downstream of this motif. Finally, the ability of FoxD4/5 to induce the ectopic expression of neural precursor genes in the ventral ectoderm also involves both the AB region and the Eh-1 motif; FoxD4/5 accomplishes ectopic neural induction by both activating neural precursor genes and repressing BMP signaling and epidermal genes. This study identifies the specific, conserved domains of the FoxD4/5 protein that allow this single transcription factor to regulate a network of genes that controls the transition of a proliferative neural ectodermal population to a committed neural plate population poised to begin differentiation.
Keywords: Neural induction, sox2, sox3, sox11, soxD, zic2, zic1, zic3, irx1, irx2, irx3, Transcription, foxD4, foxD4L1
Introduction
The vertebrate neural ectoderm is induced by antagonists of the BMP and Wnt pathways that are secreted by cells in the Organizer region of the dorsal mesoderm (reviewed in De Robertis and Kuroda, 2004; Itoh and Sokol, 2007; Rogers et al., 2009a; Stern, 2005). These antagonists enable the expression of a large number of transcription factors in the dorsal ectoderm that in turn promote its conversion to a neural ectodermal fate and prevent its reversion to a non-neural fate. One of these transcription factors, FoxD4/5, acts very early in the nascent neural ectoderm to promote the formation of the immature neural ectoderm, expand the neural plate and delay the onset of neural differentiation (Fetka et al., 2000; Sölter et al., 1999; Sullivan et al., 2001). It both up-regulates genes that maintain an immature, proliferative neural ectoderm and down-regulates genes that promote the transition to neural progenitors and lead to neural differentiation (Sullivan et al., 2001; Yan et al., 2009, 2010). Determining how FoxD4/5 both up-regulates and down-regulates its various target genes is key to understanding the transcriptional network that regulates the critical developmental transition of an immature, proliferative neural ectoderm to a definitive neural plate comprised of neurally-committed, differentiating cells.
Forkhead/Fox genes constitute a large family of transcription factors that play key roles in numerous developmental processes in nearly every tissue (Carlsson and Mahlapuu, 2002; Pohl and Knochel, 2005; Wijchers et al., 2006). They all contain a highly conserved winged-helix DNA-binding domain that defines the family. However, sequences flanking this domain are so divergent, that the family has been classified into 18 sub-families in vertebrates (http://biology.pomona.edu/fox/). Some Fox proteins regulate transcription by activation, some by repression, and a few by both, depending upon the cell type, the developmental state or the availability of interacting proteins. In addition, some Fox proteins act as “pioneer” transcription factors during development (Zaret, 2002; Zaret et al., 2008). They stably bind to their recognition sites in chromatin domains of nuclear DNA that other factors cannot access, and their binding then causes a conformational change to allow other factors to engage the DNA (Cirillo et al., 2002). Because the DNA-binding domain is so similar across the Fox family members, the sequences flanking it must account for their divergent activities.
It is important to establish which flanking regions of Fox proteins account for these different kinds of transcriptional activities because these proteins play critical roles in numerous developmental and differentiation processes, and mutations or fusions of protein domains underlie certain cancers (Carlsson and Mahlapuu, 2002; Pohl and Knochel, 2005; Wijchers et al., 2006). Some Fox proteins contain acidic domains, in either the amino (N)- or carboxy (C)-terminal regions, that are thought to be involved in target gene activation (Ptashne, 1988; Schuddekopf et al., 1996); in Xenopus, only members of the FoxD class contain an Acidic blob region (AB), which has yet to be functionally characterized (Pohl and Knochel, 2005). The C-terminal regions of some Fox proteins contain domains implicated in transcriptional repression. These include a P/A/Q-rich region, a highly charged Region II (R-II) and an Engrailed homology region-1 [Eh-1] that can bind the well-known co-repressor protein, Groucho [Gro; Grg in vertebrates; TLE in humans] (reviewed in Pohl and Knochel, 2005; Sullivan et al., 2001; Yaklichkin et al., 2007b). The Eh-1 motif is conserved in about 50% of metazoan Fox proteins and in all FoxD proteins (Yaklichkin et al., 2007b). Recently, the functional significance of this motif in FoxD3, FoxA1 and FoxA2 was revealed: Gro/ Grg proteins bind to the Eh-1 motif and this interaction is required for repression of downstream targets (Sekiya and Zaret, 2007; Yaklichkin et al., 2007a).
Despite the key role demonstrated for FoxD4/5 in early neural development in Xenopus (Fetka et al., 2000; Sölter et al., 1999; Sullivan et al., 2001; Yan et al., 2009), little is known of its function in other vertebrates. Homologues of FoxD4/5 have been identified across vertebrates and they are expressed in the early nervous systems of zebrafish, mouse and human (Freyaldenhoven et al., 2004; Kaestner et al., 1995; Katoh and Katoh, 2004; Odenthal and Nusslein-Volhard, 1998; Pohl and Knochel, 2005; Suda et al., 1999; Tuteja and Kaestner, 2007; Yaklichkin et al., 2007b). Interestingly, this gene has been duplicated in primates; one gene (FoxD4) is most similar to mouse foxd4 and one gene (FoxD4L1) is most similar to fish and frog foxD5. Due to this homology, Xenopus foxD5 recently was reassigned the name foxD4L1.1; since prior publications refer to it as foxD5, herein we use the name foxD4/5.
We found that several vertebrate FoxD4/5 proteins contain many of the aforementioned domains (Fig. 1). To identify which regions are responsible for up-regulating and/or down-regulating several known downstream targets, we made several deleted and mutated versions of Xenopus FoxD4/5. We show that the ability of FoxD4/5 to up-regulate two genes that maintain an immature neural precursor state (gem, zic2) requires the AB region, indicating that the AB is the transactivating domain. Additionally, down-regulation of genes that promote the transition to neural progenitors (sox) and of those that lead to neural differentiation (zic, irx) involves: 1) an interaction with the Gro/Grg4 co-repressor at the Eh-1 motif; and 2) sequence C-terminal to this motif. Thus, FoxD4/5 contains both activating and repressing domains, and can regulate the transition of an immature neural ectoderm to a differentiating neural plate by up-regulating some targets and down-regulating others. In addition, we show that the previously demonstrated ability of FoxD4/5 to induce the ectopic expression of neural genes in the ventral ectoderm (Yan et al., 2009) involves both the AB region and the Eh-1 motif. We show that FoxD4/ 5 accomplishes ectopic neural induction by both activating neural genes and repressing BMP signaling and epidermal genes.
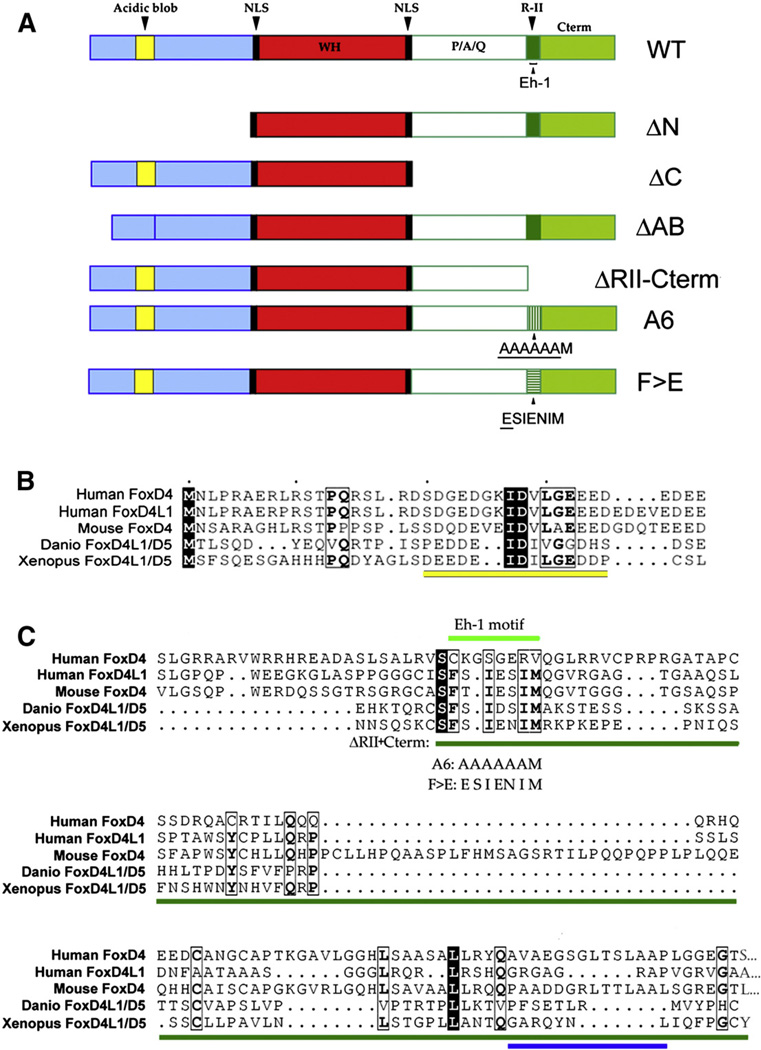
Fig. 1.
Conserved domains in the FoxD4/5 proteins. (A) Wild type (WT) and mutant constructs of Xenopus FoxD4/5. In ΔN, all of the amino acids upstream of a nuclear localization signal (NLS, black) and winged helix (WH, red) DNA binding domain were removed. In ΔC, all of the amino acids downstream of a second NLS and WH were removed. In ΔAB, only the 14 amino acids constituting the Acidic blob were deleted. In ΔRII-Cterm, the entire R-II domain (dark green) as well as all of the amino acids downstream of it (light green) were deleted. In A6, the amino acids constituting the Eh-1 motif (FSIENEM) within the R-II domain were mutated to AAAAAAM. In F>E, FSIENEM was mutated to ESIENIM. (B) CLUSTALW alignment, viewed in ESPript (Gouet et al., 1999), of the N-terminal region of several vertebrate proteins in the FoxD4/5 family shows that within the AB domain (underlined in yellow) there are several highly conserved residues. The black boxes highlight identical amino acids, the light boxes highlight conserved amino acids and the bold letters indicate identical amino acids within a region of conserved amino acids. (UniProtKB/Swiss Prot Accession numbers are: human FoxD4, Q12950; human FoxD4L1, Q9NU39; mouse FoxD4, Q60688; Danio FoxD4L1, O73784; Xenopus FoxD4L1.1, Q9PRJ8). (C) CLUSTALW alignment of the C-terminal regions of several vertebrate FoxD4/5 proteins shows that the Eh-1 motif (light green line) is highly conserved. The positions of the A6 and F>E mutations are indicated. The dark green line indicates the sequence that was deleted in the ΔRII-Cterm construct. The C-terminal amino acids of the Danio (C) and Xenopus (Y) proteins are shown, whereas the mammalian proteins contain 3 (mouse), 9 (human FoxD4L1) or 19 (human FoxD4) more amino acids that are not shown (indicated by: …). Note several highly conserved residues (boxes and bold as in Fig. 1B) downstream of the Eh-1 motif, and the location of a predicted α-helical region near the C-terminus in the Xenopus protein (blue line).
These results identify the specific domain that enables FoxD4/5 to activate immature neural genes, and identify two domains that enable it to down-regulate neural progenitor and differentiation-promoting genes, as well as epidermal genes. These findings illustrate how this single transcription factor can regulate the transition of the immature neural ectoderm, composed of proliferative precursor cells, to neurally-committed progenitor cells, and then to definitive neural plate cells that are beginning to differentiate.
Materials and methods
Creation of mutant FoxD4/D5 plasmids
The ΔN- and ΔC-FoxD4/5 plasmids were previously described (Sullivan et al., 2001; Fig. 1A). We deleted and mutated additional sites in Myc-tagged-foxD4/5, (Fig. 1A) in the pCS2+ vector using the Quik-change mutagenesis kit (Stratagene). To delete the Acidic blob sequence, the primer 5′-GAT TAT GCA GGA CTT TCT CCC TGC AGC CTA AAG TCA C-3′ and its gene complement were used with an annealing temperature of 55.0 °C for 1 min and extension was performed at 68.0 °C for 10 min for 18 cycles. The ΔRII/Cterm-FoxD5 mutant was similarly constructed using the primer 5′-CCA TCC CAA TTC ACA GAG CAA ATG TTG ATC TAG AAC TAT AGT GAG TCG-3′ and its gene complement. The FSNIEI to AAAAAA mutant (A6-FoxD5) was similarly constructed using the primer 5′-CCA TCC CAA TAA TTC ACA GAG CAA ATG TTC AGC CGC TGC TGC GGC CGC CAT GAG GAA ACC CAA GGA GCC-3′ and its gene complement. The FSNIEI to ESNIEI point mutant (F>E-FoxD5) was similarly generated using the primer 5′-CAG AGC AAA TGT TCA GAGA AGT ATT GAG AAC ATC ATG AGG AAA CCC-3′ and its gene complement.
mRNA synthesis and injection
mRNAs encoding foxD4/5 wild-type and mutant proteins were synthesized in vitro (Ambion, mMessage mMachine kit). These mRNAs (100 pg/nl each) were mixed with nuclear-localized βgal mRNA (100 pg/nl) as a lineage tracer. Embryos were obtained, cultured and microinjected as previously described (Moody, 1999, 2000). One nanoliter of each mRNA mixture was microinjected into a defined precursor of the neural ectoderm (blastomere D1.1; Moody, 1987) on one side of the 16-cell embryo. This results in FoxD4/5 protein expression in about 50% of the neural plate only on the experimental side of the embryo, ensuring that the mutant proteins do not disrupt earlier morphogenesis and avoiding nonspecific effects or embryonic lethal phenotypes. The uninjected side of the embryo was used as an internal control.
Whole embryo in situ hybridization
Embryos were cultured to stage 10.5 (nascent neural ectoderm), stage 12 (transition to neural plate) and stage 14/15 (differentiating neural plate), and processed for in situ hybridization (ISH) as previously described (Sive et al., 2000). Anti-sense Dig-labeled RNA probes were synthesized as previously described (Yan et al., 2009). The expression patterns of gem, sox2, sox3, sox11, soxD, zic1–3, and irx1–3 were compared on the experimental and control sides of embryos derived from at least three different clutches of eggs from different sets of adult parents. The frequency at which embryos showed altered expression was compared to the frequency from wt-FoxD5-injected samples using the Chi-squared statistic (p<0.001).
Decreasing Gro/Grg4
Anti-sense morpholino oligonucleotides directed against Xenopus Gro/Grg4 (GroMO; GGTACATCTTGCTCAAGTCTCGAAT, Gene Tools, LLC) were used to decrease the levels of endogenous Gro/Grg4. Based on sequence analysis, GroMO will not bind to any other member of the Xenopus Gro/TLE family. The effectiveness of GroMO to block translation of an HA-tagged Gro/Grg4 protein was demonstrated by injecting Xenopus oocytes with 5 ng of in vitro transcribed mRNA encoding either wild-type Gro/Grg4 or a mutant harboring 5 point mutations generated by PCR in the wobble codons of amino acids 2–6 of Gro/Grg4 (rescue mRNA). These mRNAs were injected alone or in combination with 5 ng of GroMO, and the oocytes cultured overnight at 21 °C. Lysates were prepared and Western analysis using HA antibody was performed (Supplemental Fig. 1). In addition, the reversal of the GroMO phenotype in whole embryos was demonstrated by co-injecting 60 pg of the rescue Gro/Grg4 mRNA.
In some experiments GroMO was injected into a single dorsal-animal 8-cell blastomere (20 ng or 40 ng) and subsequently one of the 16-cell daughters was injected with 1 nl of wt-foxD4/5 mRNA (100 pg/nl or 50 pg/nl) plus nuclear-localized βgal mRNA (100 pg/ nl) as a lineage tracer. In other experiments GroMO (20 ng) was injected into a single ventral-animal 8-cell blastomere and subsequently the equatorial 16-cell daughter was injected with 1 nl of ΔAB-foxD4/5 mRNA (100 pg/nl) plus βgal mRNA or with ΔAB-foxD4/5 mRNA (100 pg/nl) plus rescue Gro/Grg4 mRNA (60 pg) plus βgal mRNA.
Western blots and Co-IPs
Oocytes were injected with mRNAs coding for wt-FoxD4/5, mutant FoxD4/5 constructs and/or Gro/Grg4 and cultured, as above. For each immunoprecipitation reaction, 150 µl of lysate (15 oocyte equivalents) was mixed with 650 µl ice-cold TNSG lysis buffer and 1 µg of antibody (raised against HA or Flag; Applied Biological Materials) and incubated at 4 °C for 1–2 h, after which 25 µl protein A/G agarose beads (Santa Cruz Biotechnology) were added to the reaction and rotated in an orbital mixer overnight at 4 °C. Beads were briefly pelleted at 4 °C and rinsed 3 times with ice-cold TNSG lysis buffer. All residual buffer was removed with a flat pipette tip and beads were resuspended in 45 µl 1× RIPA sample buffer (RIPA Buffer: 150 mM NaCl, 1% NP40, 0.5% Na Deoxycholate, 0.1% SDS, 50 mM Tris (8.0); 4× sample buffer: 4 mL 10%SDS, 2 mL glycerol, 0.3086 g DTT, 0.00001 g Brom-phenol Blue; 4× sample buffer was diluted to 1× in RIPA buffer). Samples were boiled at 100 °C for 10 min prior to loading on Tris-glycine SDS-Polyacrylamide 10% gels. For expression checks, 15 µl (1.5 oocyte equivalents) lysate was prepared with 4× sample buffer and loaded on Tris-glycine SDS-Polyacrylamide 10% gels. Proteins were resolved by SDS/PAGE, transferred to Immobilon-P transfer membranes (Millipore) using standard methods, and blocked in Tris-buffered saline (25 mM Tris)+0.2% Tween-20 (TBST)+5% nonfat dry milk for at least 2 h to overnight at 4 °C. Whenever possible, IP-Western blots were incubated with the following HRP-conjugated primary antibodies to reduce background: anti-HA-HRP-conjugated (Roche), and anti-Myc-HRP (Thermo Scientific). Following antibody incubation, blots were rinsed with TBST, blotted with HyGLO Chemilu-minescent HRP antibody detection reagent (Denville Scientific Inc.) and exposed to film.
Immunostaining
To demonstrate that mutant FoxD4/5 proteins had access to the nucleus, dorsal blastomeres were injected with myc-tagged mRNAs and embryos fixed in 4% paraformaldehyde at stage 11. Frozen sections were cut with a cryostat and subjected to standard immunofluorescence staining protocols using an anti-Myc-tag primary antibody (#9B11, Cell Signaling Tech., 1:2000), a goat anti-mouse IgG Alexa Fluor 488 conjugated secondary antibody (#4408, Cell Signaling Tech., 1:1000) followed by counterstaining of the nuclei with DAPI. Images were collected using a Zeiss LSM 710 confocal system equipped with 32-channel spectral photomultiplier. Thirty-two channel spectral stacks were collected at spectral resolution of 9.6 nm within the range of 418–726 nm. To obtain the signature spectral curves of autofluorescence, DAPI and Alexa Fluor 488 emissions, spectral confocal images were taken with excitation of either the 405 nm diode laser (DAPI and autofluorescence) or the argon 488 laser line (Alexa Fluor 488); these spectral curves were then used to unmix the DAPI, autofluorescence and Alexa Fluor 488 emissions registered upon simultaneous excitation of the samples with 405 and 488 laser lines (Supplemental Fig. 2).
To test whether C-terminal mutant FoxD4/5 proteins blocked BMP signaling, ventral blastomeres were co-injected with non-tagged mRNAs plus cytoplasm-localized βgal mRNA as a lineage tracer (100 pg/nl each). Embryos were fixed in MEMPHA at stage 11 and processed for whole mount immunostaining as previously described (Yan et al., 2009) using an anti-Phospho-SMAD1,5,8 primary antibody (#9511, Cell Signaling Tech., 1:100) and a goat anti-rabbit IgG HRP-conjugated secondary antibody (#7074, Cell Signaling Tech., 1:250).
Results
FoxD4/5 proteins contain highly conserved Acidic blob and Eh-1 domains
We previously demonstrated that FoxD4/5 can both up-regulate and down-regulate downstream targets (Sullivan et al., 2001; Yan et al., 2009, 2010). To determine whether FoxD4/5 contains domains indicative of both activating and repressing transcriptional activity, we performed a CLUSTALW alignment (Thompson et al., 1994) of several vertebrate FoxD4/5 proteins (Fig. 1). Within the region N-terminal to the winged-helix DNA-binding domain (WH) in Xenopus FoxD4/5 there is a 14 amino acid Acidic blob (AB) region (Fig. 1A). Within the AB there are several residues that are highly conserved across vertebrates (Fig. 1B). Within the region C-terminal to the WH in Xenopus FoxD4/5 there is a P/A/Q-rich region, an R-II domain, and within the R-II there is an Eh-1 motif (Fig. 1A). The Eh-1 motif and several downstream amino acids are highly conserved across vertebrates (Fig. 1C). To determine whether any of these regions are specifically required for up-regulating or down-regulating FoxD4/5 targets, we made several deletion constructs (Fig. 1A).We deleted: 1) the entire region (ΔN) upstream of a nuclear localization signal (NLS); 2) just the Acidic Blob (ΔAB); 3) the entire region (ΔC) downstream of another NLS; or 4) the R-II domain plus all the sequence C-terminal to it (ΔRII-Cterm). Each deletion construct produces abundant protein that can access the nucleus (Sullivan et al., 2001; Supplemental Fig. 3).
FoxD4/5 activates two neural precursor genes via an Acidic blob domain in the N-terminus
We previously showed that wild-type (wt) FoxD4/5 up-regulates the expression of gem and zic2 in both the nascent neural ectoderm of the gastrulating embryo and in the neural plate, and that this occurs via transcriptional activation (Yan et al., 2009). To identify the region(s) of FoxD4/5 that are responsible for target gene activation, we injected mRNA encoding each FoxD4/5 deletion construct (Fig. 1A) into a dorsal blastomere that gives rise to clones in the medial neural plate, and analyzed gem and zic2 expression within the lineage-labeled clone by in situ hybridization (ISH). As previously reported, β-Gal-tagged wt-FoxD4/5-positive cells express higher levels of gem and zic2 compared to neighboring cells (Fig. 2A). For both genes, the ΔN-FoxD4/5 construct caused a significant reduction in the percentage of embryos showing up-regulated gene expression within the labeled clone, whereas the ΔC-FoxD4/5 construct did not (Fig. 2A). This result demonstrates that an activation domain likely resides in the N-terminal part of the protein. Due to the acidity and high level of sequence conservation in the AB, we tested this region for activation activity. The percentage of embryos showing upregulated gem or zic2 expression was significantly reduced in the ΔAB-expressing labeled clone, whereas deleting the ΔRII-Cterm region had no significant effect on gem or zic2 up-regulation (Fig. 3A). Thus, the activation of gem and zic2 by FoxD4/5 requires the AB domain and is independent of the RII+Cterm region.
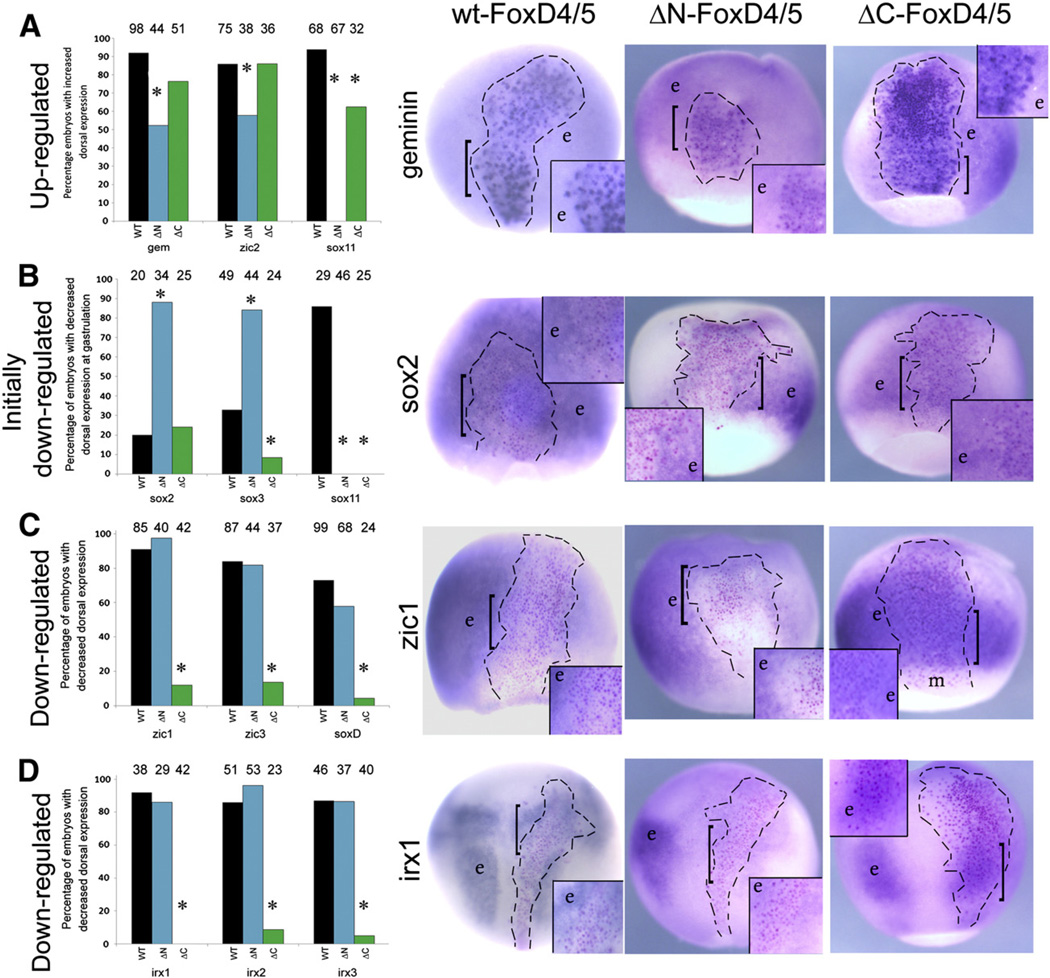
Fig. 2.
N-terminal sequences are required for up-regulation, and C-terminal sequences are required for down-regulation of FoxD4/5 targets. (A) The graph presents the percentage of embryos in which WT-, ΔN- or ΔC-FoxD4/5 caused up-regulation of gem, zic2 or sox11 (the latter at neural plate stages). Numbers above each bar indicates sample size; * indicates significant difference from WT at the p<0.001 level. The images for gem expression are representative for zic2; examples of sox11 are presented in Fig. 4. Examples of endogenous expression patterns can be found in Supplemental Fig. 2. The FoxD4/5-expressing clones, marked by nuclear β-Gal (red or purple dots), are located in the neural ectoderm and indicated by hatched lines. Boxed insets are higher magnifications of the clone, the position of which is indicated on the whole embryo by a bracket. In the insets for wt-FoxD4/5 and ΔC-FoxD4/5, the β-Gal labeled cells are more intensely stained than neighboring cells (e) that show the endogenous level of gem expression. The intense blue ISH label often obscures the red-labeled nuclei in these cases. In the inset for ΔN-FoxD4/5, the β-Gal labeled cells stain for gem expression only slightly higher than the endogenous level in neighboring cells (e). This example would be scored as a positive up-regulation in the graph, even though the level of up-regulation is much lower compared to wt-FoxD4/5 and ΔCFoxD4/ 5. (B) The graph presents the percentage of embryos in which WT-, ΔN- or ΔC-FoxD4/5 caused down-regulation of sox2, sox3 or sox11 at gastrulation stages. The images for sox2 expression are representative for sox3; examples of sox11 are presented in Fig. 4. In the insets for wt-FoxD4/5 and ΔN-FoxD4/5, the β-Gal labeled cells are less intensely stained than neighboring cells (e) that show the endogenous level of sox2 expression. Often with wt-FoxD4/5 the effect is not uniform throughout the clone. The extent of down-regulation of sox2 is greater for ΔN-FoxD4/5. In the inset for ΔC-FoxD4/5, the β-Gal labeled cells stain for sox2 expression about the same as the endogenous level in neighboring cells (e), which is the most frequent phenotype. (C) The graph presents the percentage of embryos in which WT-, ΔN- or ΔC-FoxD4/5 caused down-regulation of zic1, zic3 or soxD. The images for zic1 expression are representative for the other two genes. In the insets for wt-FoxD4/5 and ΔN-FoxD4/5, nearly all of the β-Gal labeled cells are less intensely stained than neighboring cells (e) that show the endogenous level of zic1 expression. In the inset for ΔC-FoxD4/5, the β-Gal labeled cells stain with the same intensity as neighboring cells (e), indicating a lack of down-regulation. m, non-involuted mesoderm that does not normally express zic1. (D) The graph presents the percentage of embryos in which WT-, ΔN- or ΔC-FoxD4/5 caused down-regulation of irx1, irx2 or irx3. The images for irx1 expression are representative for the other two genes. In the insets for wt- FoxD4/5 and ΔN-FoxD4/5, nearly all of the β-Gal labeled cells are less intensely stained than neighboring cells (e) that show the endogenous expression levels of irx1. In the inset for ΔC-FoxD4/5, the β-Gal labeled cells stain with the same intensity as neighboring cells (e), indicating a lack of down-regulation. All images are dorsal views with vegetal pole to the bottom.
N-terminal sequences are required for up-regulation, and C-terminal sequences are required for down-regulation of FoxD4/5 targets. (A) The graph presents the percentage of embryos in which WT-, ΔN- or ΔC-FoxD4/5 caused up-regulation of gem, zic2 or sox11 (the latter at neural plate stages). Numbers above each bar indicates sample size; * indicates significant difference from WT at the p<0.001 level. The images for gem expression are representative for zic2; examples of sox11 are presented in Figure 4. Examples of endogenous expression patterns can be found in Supplemental Figure 2. The FoxD4/5-expressing clones, marked by nuclear β-Gal (red or purple dots), are located in the neural ectoderm and indicated by hatched lines. Boxed insets are higher magnifications of the clone, the position of which is indicated on the whole embryo by a bracket. In the insets for wt-FoxD4/5 and ΔC-FoxD4/5, the β-Gal labeled cells are more intensely stained than neighboring cells (e) that show the endogenous level of gem expression. The intense blue ISH label often obscures the red-labeled nuclei in these cases. In the inset for ΔN-FoxD4/5, the β-Gal labeled cells stain for gem expression only slightly higher than the endogenous level in neighboring cells (e). This example would be scored as a positive up-regulation in the graph, even though the level of up-regulation is much lower compared to wt-FoxD4/5 and ΔC-FoxD4/5. (B) The graph presents the percentage of embryos in which WT-, ΔN- or ΔC-FoxD4/5 caused down-regulation of sox2, sox3 or sox11 at gastrulation stages. The images for sox2 expression are representative for sox3; examples of sox11 are presented in Figure 4. In the insets for wt-FoxD4/5 and ΔN-FoxD4/5, the β-Gal labeled cells are less intensely stained than neighboring cells (e) that show the endogenous level of sox2 expression. Often with wt-FoxD4/5 the effect is not uniform throughout the clone. The extent of down-regulation of sox2 is greater for ΔN-FoxD4/5. In the inset for ΔC-FoxD4/5, the β-Gal labeled cells stain for sox2 expression about the same as the endogenous level in neighboring cells (e), which is the most frequent phenotype. (C) The graph presents the percentage of embryos in which WT-, ΔN- or ΔC-FoxD4/5 caused down-regulation of zic1, zic3 or soxD. The images for zic1 expression are representative for the other two genes. In the insets for wt-FoxD4/5 and ΔN-FoxD4/5, nearly all of the β-Gal labeled cells are less intensely stained than neighboring cells (e) that show the endogenous level of zic1 expression. In the inset for ΔC-FoxD4/5, the β-Gal labeled cells stain with the same intensity as neighboring cells (e), indicating a lack of down-regulation. m, non-involuted mesoderm that does not normally express zic1. (D) The graph presents the percentage of embryos in which WT-, ΔN- or ΔC-FoxD4/5 caused downregulation of irx1, irx2 or irx3. The images for irx1 expression are representative for the other two genes. In the insets for wt-FoxD4/5 and ΔN-FoxD4/5, nearly all of the β-Gal labeled cells are less intensely stained than neighboring cells (e) that show the endogenous expression levels of irx1. In the inset for ΔC-FoxD4/5, the β-Gal labeled cells stain with the same intensity as neighboring cells (e), indicating a lack of down-regulation. All images are dorsal views with vegetal pole to the bottom.
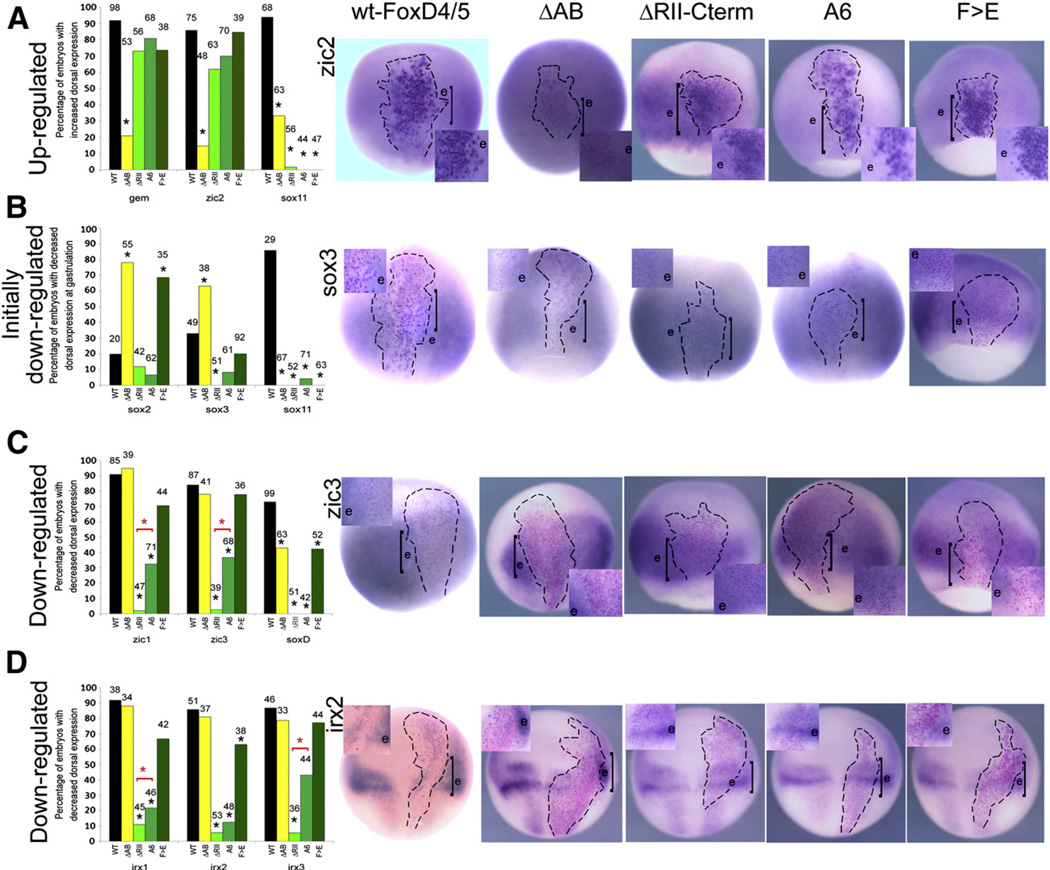
Fig. 3.
The Acidic blob domain is required for up-regulation, and the Eh-1 domain and the C-terminal region downstream of it are required for down-regulation of FoxD4/5 targets. (A) The graph presents the percentage of embryos in which WT- and mutant FoxD4/5 caused up-regulation of gem, zic2 or sox11 (the latter at neural plate stages). Numbers above each bar indicates sample size; * indicates significant difference from WT at the p<0.001 level. The images for zic2 expression are representative for gem; examples of sox11 are presented in Fig. 4. The FoxD4/5-expressing clones, marked by nuclear β-Gal (red or purple dots), are located in the neural ectoderm and indicated by hatched lines. Boxed insets are higher magnifications of the clone, the position of which is indicated on the whole embryo by a bracket. For WT-FoxD4/5, ΔRII-Cterm, A6 and F>E, the β-Gal labeled cells are more intensely stained than neighboring cells (e), that show the endogenous level of gem expression. The intense blue ISH label often obscures the red-labeled nuclei in these cases. For ΔAB-FoxD4/5, the β-Gal labeled cells stain at the same level as endogenous (e), and thus do not show up-regulation. (B) The graph presents the percentage of embryos in which WT- and mutant FoxD4/5 caused an initial down-regulation of sox2, sox3 or sox11 at gastrulation stages. The images for sox3 expression are representative for sox2; examples of sox11 are presented in Fig. 4. For WT- and ΔAB-FoxD4/5, the β-Gal labeled cells are more weakly stained than neighboring cells (e) that show the endogenous level of sox3 expression. For the other mutants, the β-Gal labeled cells stain at levels similar to the endogenous expression in neighboring cells (e). (C) The graph presents the percentage of embryos in which WT- and mutant FoxD4/5 caused down-regulation of zic1, zic3 or soxD. The images for zic3 expression are representative for the other two genes. For WT-, ΔAB and F>E-FoxD4/5, the β-Gal labeled cells are less intensely stained than neighboring cells (e) that show the endogenous level of zic3 expression. For ΔRII-Cterm and A6-FoxD4/5 the β-Gal labeled cells stain at levels similar to the endogenous expression in neighboring cells (e). *, indicates that the ΔRII-Cterm construct represses significantly less frequently than the A6 construct (p<0.001). (D) The graph presents the percentage of embryos in which WT- and mutant FoxD4/5 caused down-regulation of irx1, irx2 or irx3. The images for irx2 expression are representative for the other two genes. For WT-, ΔAB and F>E-FoxD4/5, the β-Gal labeled cells are less intensely stained than neighboring cells (e) that show the endogenous level of irx2 expression. For ΔRII-Cterm and A6-FoxD4/5, the β-Gal labeled cells stain at levels similar to the endogenous expression in neighboring cells (e). *, indicates that the ΔRII-Cterm construct represses significantly less frequently than the A6 construct (p<0.001). All images are dorsal views with vegetal pole to the bottom.
Down-regulation of sox neural progenitor genes is affected by both N-terminal and C-terminal domains
Over-expression of FoxD4/5 down-regulates the expression of three neural progenitor genes (sox2, sox3, sox11) in the gastrula neural ectoderm (Yan et al., 2009). We found that the down-regulation of each sox gene was altered by both N-terminal and C-terminal sequences. First, while deletion of the entire C-terminus did not alter the percentage of embryos showing down-regulation of sox2, it caused a significant reduction in down-regulation of sox3 and sox11 (Fig. 2B). Injection of the ΔRII-Cterm construct identified this region as required for the down-regulation of sox3 and sox11 (Fig. 3B). Second, the N-terminal region also affected the expression levels of all three sox genes. For both sox2 and sox3, deleting the entire N-terminus or just the AB domain significantly increased the percentage of embryos showing down-regulation in the gastrula neural ectoderm and increased the extent of the down-regulation within the clone (Figs. 2B, 3B). This result indicates that the AB domain normally ameliorates the repression of sox2 and sox3 by wt-FoxD4/5. Either wt-FoxD4/5 directly activates sox2 and sox3,which are secondarily repressed by other genes, or it activates genes that repress sox gene repressors; distinguishing between these possibilities requires further investigation. In contrast, deleting either the entire N-terminus or just the AB domain caused sox11 to be up-regulated in the gastrula neural ectoderm (ΔN: 95.7%, n=46; ΔAB: 74.6%, n=67; Fig. 4), suggesting that wt-FoxD4/5 normally activates a gene that represses sox11. In fact, we previously showed that sox11 expression is down-regulated by Zic2 (Yan et al., 2009). Consequently, we propose that deletion of the AB domain, which causes a loss of zic2 up-regulation, also leads to a de-repression of sox11.
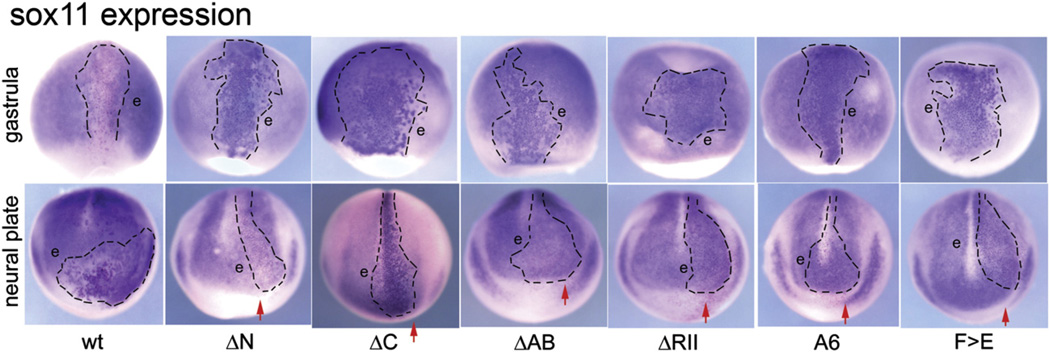
Fig. 4.
Effects of FoxD4/5 constructs on the expression of sox11 at gastrula and neural plate stages. Top panel: At gastrulation stages, wt-FoxD4/5 causes β-Gal labeled cells (clone is outlined) to express sox11 at lower levels than neighboring cells (e) that show the endogenous level of sox11 expression. In contrast, each mutant construct caused β-Gal labeled cells to express sox11 at higher levels, indicated by darker staining compared to neighboring cells (e). All images are dorsal views with vegetal pole to the bottom. Bottom panel: At neural plate stages, wt-FoxD4/5 causes β-Gal labeled cells (outlined within the normal expression domain in the neural plate) to express sox11 at higher levels than neighboring cells (e). The same effect is observed with the ΔC-FoxD4/5 mutant, but in significantly fewer embryos compared to wtr-FoxD4/5 (see Fig. 2A). However, for all other mutant clones (outlined only within the neural plate) the sox11 expression levels are similar to those of the neighboring cells (e) that show endogenous levels. Note that the neural plate is broader on the injected side (red arrow) in embryos expressing N-terminal mutants (ΔN, ΔAB) but not in embryos expressing C-terminal mutants (ΔC, ΔRII, A6, F>E), consistent with a previous report that the C-terminal domain is required for neural plate expansion (Sullivan et al., 2001). The ΔC image is a dorsal view with anterior to the bottom; all other images are anterior views with dorsal to the top.
In contrast to gastrula stage embryos, sox11 is up-regulated by wt- FoxD4/5 at neural plate stages (Fig. 4), and this appears to be by direct activation (Yan et al., 2009). Consistent with the results obtained for gem and zic2, the neural plate stage up-regulation of sox11 requires the N-terminus, and specifically the AB domain (Figs. 2A, 3A, 4). However, the up-regulation of sox11 in the neural plate also requires the RII-Cterm domain, and specifically the Eh-1 motif in the C-terminus (Figs. 3A, 4). These results indicate that the up- regulation of sox11 by FoxD4/5 can be achieved directly by activation mediated by the AB domain and indirectly by repressing other target genes mediated by C-terminal sequences. It should be noted that while the deletion of the entire C-terminus significantly reduces up-regulation of sox11 in the neural plate (Fig. 2A) the phenotype is less frequent than for the more discreet deletions/mutations made within the C-terminus (Fig. 3A). This requires further experimental investigation, but may indicate involvement of the P/A/Q domain (Fig. 1A) or reflect a conformational change that affects function when such a drastic alteration to the protein is imposed.
SoxD is a member of the SoxG family that is unique to amphibians and appears to act downstream of Sox2 to expand neural progenitors (Rogers et al., 2009a). Its expression in the neural plate is down-regulated by wt-FoxD4/5 (Yan et al., 2009). We found that the percentage of embryos in which soxD is down-regulated in the neural plate is moderately, but significantly reduced by deletion of the AB domain and eliminated by deletion of the RII-Cterm domain (Fig. 3C). As for the other sox genes, these results indicate that wt-FoxD4/5 affects soxD transcription by both activation and repression most likely by involving intermediate genes.
FoxD4/5 represses genes that promote neural differentiation via the C-terminal region
Wt-FoxD4/5 strongly down-regulates five other neural transcription factors that promote the expression of the bHLH neural differentiation genes (zic1, zic3, irx1–3), and this appears to be mediated by transcriptional repression (Yan et al., 2009). We found that both the ΔN and ΔAB constructs caused repression of all five genes at frequencies equivalent to wt-FoxD4/5 (Figs. 2C, D, 3C, D). In contrast, both the ΔC and ΔRII-Cterm constructs failed to repress their expression, indicating that the RII-Cterm domain is required for transcriptional repression. Unlike the sox genes, the N-terminal portion of the FoxD4/ 5 protein does not have a role in the repression of these neural differentiation-promoting genes.
The role of Groucho binding in FoxD4/5-mediated transcriptional repression
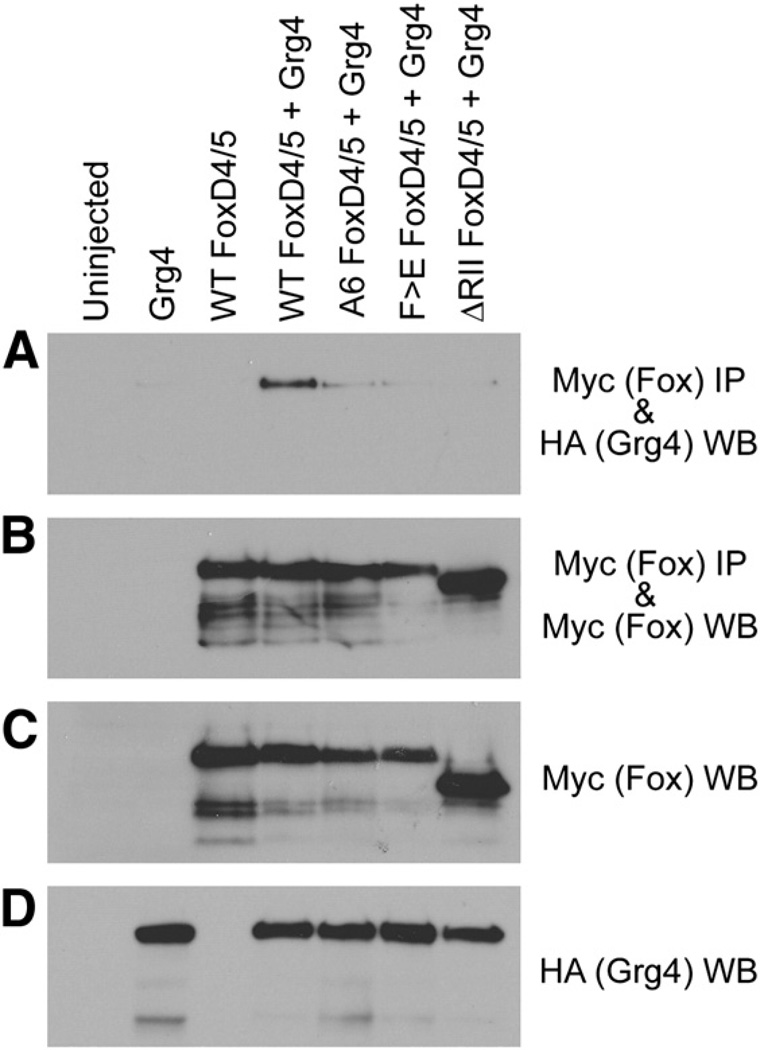
Groucho (Gro) is a well-studied transcriptional co-repressor that can bind to the Eh-1 motif and thereby mediate the repressive effects of some Fox proteins (Sekiya and Zaret, 2007; Yaklichkin et al., 2007a, 2007b). Because the FoxD4/5 protein in a number of vertebrates also contains an Eh-1 motif within the R-II domain (Fig. 1C), we tested whether Gro/Grg4 is responsible for the ability of FoxD4/5 to repress downstream genes. In Xenopus, Gro/Grg4 is widely expressed throughout the neural ectoderm from the earliest stages (Molenaar et al., 1997; Neilson et al., 2010). To assess whether FoxD4/5 and Gro/Grg4 can interact, we conducted an immunoprecipitation (IP) analysis of lysates from Xenopus oocytes co-expressing either wt- FoxD4/5 or C-terminal deletion or amino acid substitution constructs of FoxD4/5 along with Gro/Grg4. Co-IP analysis demonstrates that Gro/Grg4 is found in the FoxD4/5 immunoprecipitates (Fig. 5A), indicating that these two proteins interact in an in vivo Xenopus expression system.
Fig. 5.
Gro/Grg4 binds to the Eh-1 motif of FoxD4/5. (A–D) Myc-tagged versions of wild-type (WT), as well asmutants harboring amino acid substitutions in the Eh-1 domain (A6, F>E) or deleted for the Eh-1 domain (ΔRII) in FoxD4/5 were expressed in Xenopus oocytes along with HA-tagged wild-type Gro/Grg4 (Grg4). Co-immunoprecipitation (IP) and Western blot (WB) analyses of Xenopus oocyte lysates expressing HA- and Myc-tagged constructs are indicated. (A) Although all constructs are equivalently expressed, only full-length FoxD4/5 effectively binds with Gro/Grg4. The control panels (B–D) show that the IPs each contain similar levels of FoxD4/5 wild-type and mutant proteins (B), as do the direct lysates (C). Gro/Grg4 expressing lysates also show similar levels of this protein (D).
To test whether this interaction has a role in repressing downstream targets, we made the same mutations in the Eh-1 motif thatYaklichkin et al. (2007a), showed prevents Gro/Grg4 binding to Xenopus FoxD3. In one construct (A6), the first six amino acids of the Eh-1 motif (FSIENIM) were changed to alanine (AAAAAAM), and in the second construct (F>E), the first amino acid was changed to glutamic acid (ESIENIM) (Fig. 1C). Both Eh-1 mutant constructs are abundantly expressed in oocytes in the presence of Gro/Grg4, but they do not interact with Gro/Grg4 in a co-IP assay (Fig. 5A). It should be noted that the RII-Cterm deletion construct also does not interact with Gro/Grg4 (Fig. 5A), which is expected because the entire Eh-1 motif plus downstream sequence is deleted. The controls show that the IPs contain similar levels of FoxD4/5 wild-type and mutant proteins and that expression of the proteins was similar among oocytes lysates (Fig. 5B–D).
We next tested whether the A6 or F>E mutants would fail to down-regulate the sox, zic or irx genes, and thus implicate the requirement for Gro/Grg4 binding to the Eh-1 motif. The down-regulated target genes fell into two groups. Some target genes were repressed by the A6 mutant at a frequency indistinguishable from the ΔRII-Cterm deletion, suggesting that the binding of Gro/Grg4 to the Eh-1 motif is responsible for repression (sox3, sox11, soxD, irx2) (Figs. 3B, C, 4). There also was no significant difference between the ΔRII-Cterm and A6 constructs for sox2 (Fig. 3B), but since neither these nor the ΔC-FoxD4/ 5 construct altered the frequency of sox2 repression compared to wt-FoxD4/5, we cannot with confidence conclude that binding of Gro/Grg4 to the Eh-1 motif is involved. Other target genes were repressed by the A6 mutant at a significantly lower frequency than the ΔRII-Cterm construct (zic1, zic3, irx1, irx3) (Figs. 3C, D). This latter result indicates that there are additional regions in the RII-Cterm domain that are needed for full repression of the zic and irx genes. This is supported by the observation that the point mutation, F>E, which does not bind Gro/Grg4 in a co-IP oocyte assay (Fig. 5A), nonetheless represses sox2, sox3, zic1, zic3, and irx1–3 at a frequency indistinguishable from that of wt-FoxD4/5 (Figs. 3B–D).
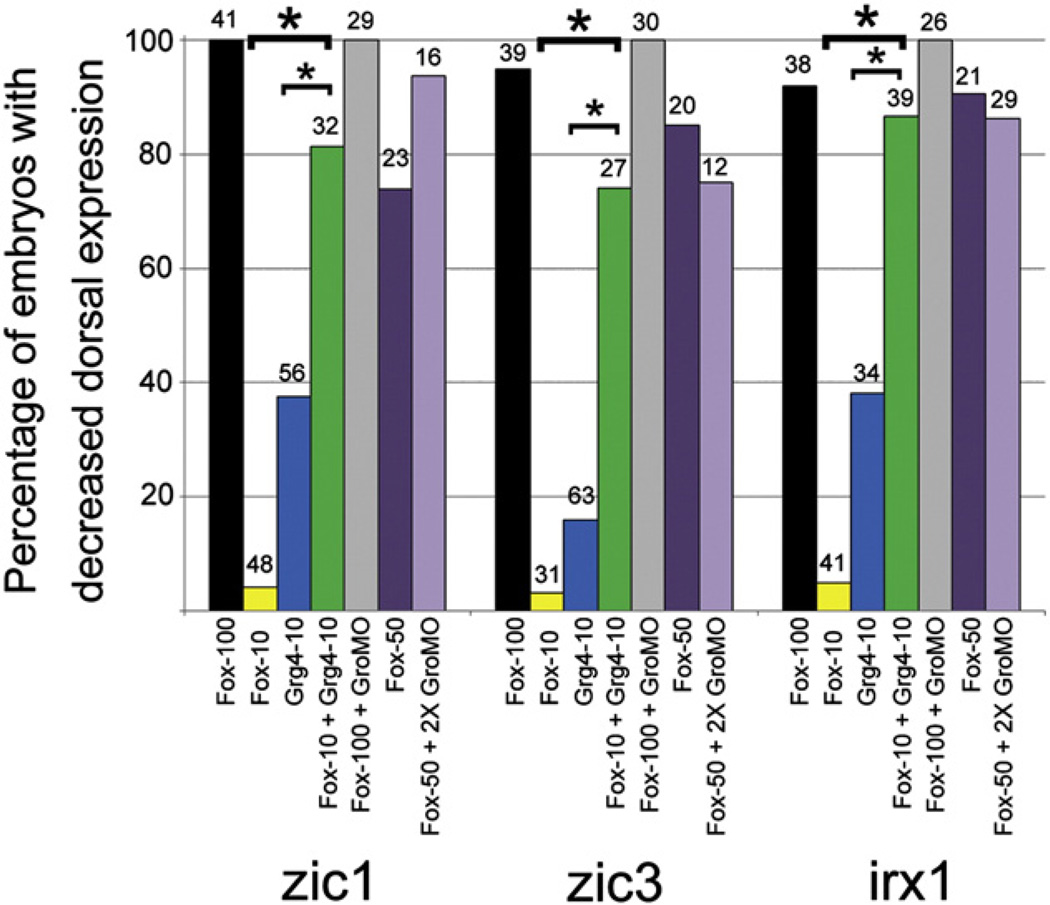
We next tested whether an interaction between FoxD4/5 and Gro/Grg4 contributes to repressing those genes (zic1, zic3, irx1) for which down-regulation requires additional C-terminal sequence. First, we injected either foxD4/5 or gro/grg4 mRNAs at several concentrations to find a dose of each that is not effective at repressing downstream genes. For zic1, zic3, and irx1, 100 pg of foxD4/5 mRNA per 16-cell blastomere caused repression in the majority of embryos (Yan et al., 2009), whereas 10 pg caused repression in only a few embryos (Fig. 6). Likewise, 10 pg of gro/grg4 mRNA per blastomere caused repression of these genes in less than half of the embryos (Fig. 6). Next, we co-injected these sub-optimal doses (10 pg foxD4/5+10 pg gro/ grg4 mRNAs); repression occurred at significantly higher frequencies than either mRNA alone, nearly equivalent to the optimal 100 pg dose of wt-foxD4/5 mRNA alone (Fig. 6). Thus, FoxD4/5 and Gro/Grg4 can cooperatively repress these three neural genes in a dose dependent manner. However, depleting endogenous Grg4 with a specific MO (GroMO) did not reduce the ability of exogenous FoxD4/5 to efficiently repress zic1, zic3, or irx1. Co-injecting embryos with a combination of 100 pg wt-foxD4/5 mRNA+20 ng GroMO or with 50 pg wt-foxD4/5 mRNA+40 ng GroMO did not significantly reduce the repression of these genes compared to mRNA injection alone. These results indicate that Gro/Grg4 is not required for FoxD4/5 to repress zic1, zic3 or irx1. It is possible that endogenous levels of other Groucho family members that are expressed in the neural ectoderm (Molenaar et al., 1997; Neilson et al., 2010) and are not targeted by the GroMO may have substituted for Gro/Grg4 under these experimental conditions. Alternatively, since binding site affinity can affect transcription factor occupancy and activation versus repressive function (Essien et al., 2009), Gro/Grg4 may facilitate FoxD4/5 repression of these genes when the concentration/occupancy of FoxD4/5 is low, and not be required when the concentration/occupancy of FoxD4/5 is higher.
Fig. 6.
Gro/Grg4 and FoxD4/5 co-operate to cause transcriptional repression. The percentages of embryos showing decreased expression of zic1, zic3, or irx1 after injection of: 100 pg wt-FoxD4/5 mRNA (Fox-100), 10 pg wt-FoxD4/5 mRNA (Fox-10), 10 pg Gro/Grg4 mRNA (Grg4-10), 10 pg FoxD4/5 mRNA plus 10 pg Gro/Grg4 mRNA (Fox-10+Grg4-10), 100 pg wt-FoxD4/5 mRNA plus 20 ng GroMO (Fox-100+GroMO), 50 pg wt-FoxD4/5 mRNA (Fox-50), or 50 pg wt-FoxD4/5 mRNA plus 40 ng GroMO (Fox-50+2X GroMO). A low dose (10 pg) of either FoxD4/5 or Gro/Grg4 down-regulates the expression of these three genes in many fewer embryos than a higher dose of FoxD4/5 alone (100 pg). However, low doses of FoxD4/5 plus Gro/Grgr4 act synergistically to restore down-regulation at a level significantly higher than either mRNA alone (*, p<0.001), and at a level approaching 100 pg FoxD4/5 alone. However, MO knock-down of endogenous Gro/Grg4 expression does not reduce the ability of FoxD4/5 to cause down-regulation. At either a high (100 pg, black bar) or lower (50 pg, dark purple bar) dose of FoxD4/5, addition of GroMO did not significantly change the frequency of down-regulation of these genes (cf. to black bar to grey bar and dark purple bar to light purple bar). Numbers above bars indicate sample sizes.
Ectopic induction of neural genes in the ventral ectoderm requires both the AB and Eh-1 domains
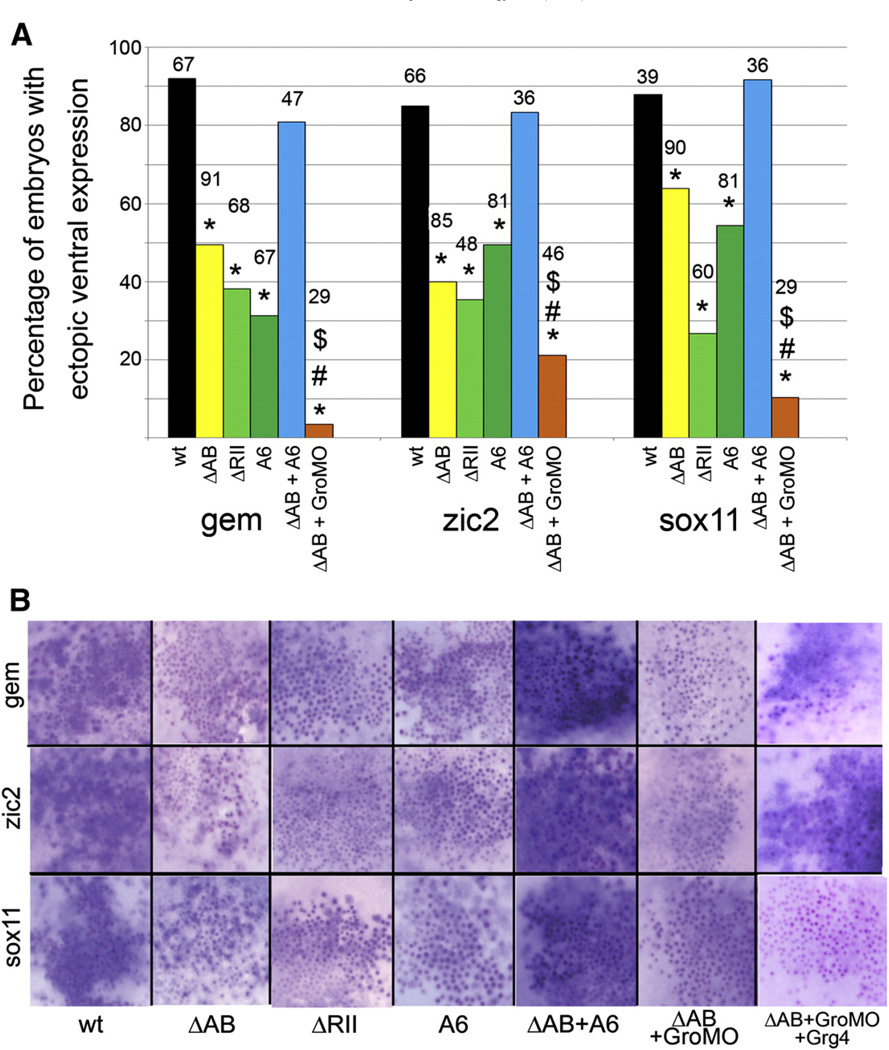
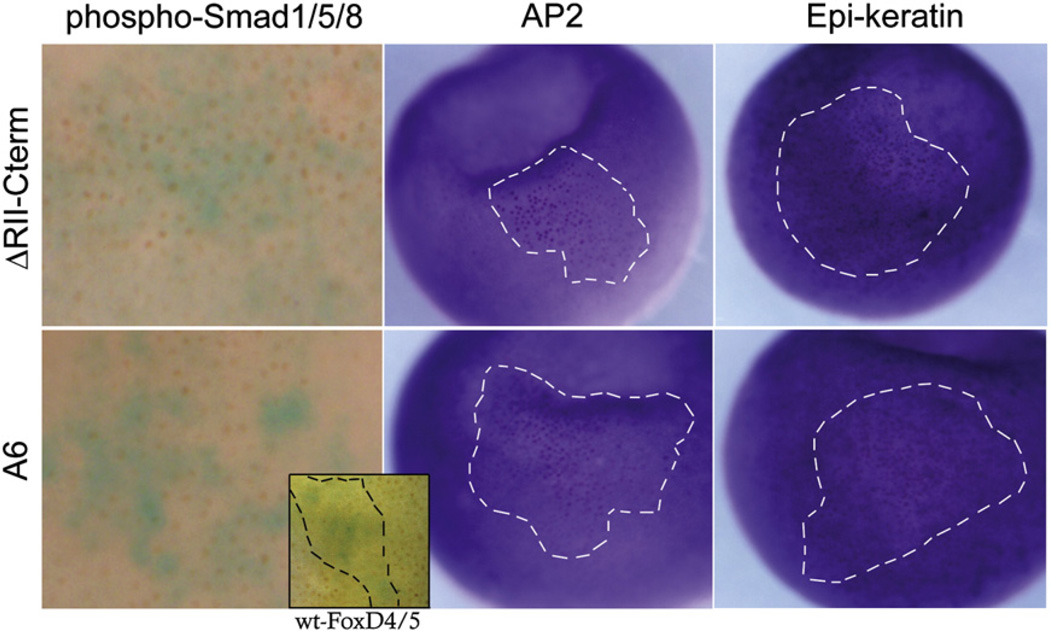
Injection of FoxD4/5 mRNA into the ventral epidermal lineage induces the ectopic expression of gem, zic2 and sox11, and down-regulates BMP signaling and subsequent expression of epidermal genes (Yan et al., 2009, 2010). We hypothesized that both gene activation and gene repression would be involved since ectodermal cells must switch from an epidermal to a neural fate. In fact, deleting either the N-terminus or the C-terminus reduced, but did not eliminate ectopic ventral induction of gem, zic2 and sox11 (data not shown), suggesting that both domains, and thereby transcriptional activities, are involved. This was confirmed by injecting the ΔAB or ΔRII-Cterm constructs; each significantly reduced, but did not eliminate, the frequency of ectopic expression (Fig. 7A). Also, the number of ventral ectodermal cells expressing neural genes and the intensity of that expression was reduced compared to wt-FoxD4/5 (Fig. 7B). A role for Gro/Grg4 is implicated because the A6 mutant caused a similar reduction in ectopic ventral induction of each gene. Because both activation and repression are involved, we hypothesized that for optimal ectopic induction the neural genes need to be up-regulated via an intact AB domain and a second set of genes needs to be down-regulated via binding of Gro/Grg4 to the Eh-1 motif. Two experiments support this idea. First, co-injection of the ΔAB and A6 constructs completely restored the frequency and intensity of ectopic ventral induction (Figs. 7A, B). Since we assume that these constructs bind to DNA independently, because only FoxP proteins have been shown to form dimers (Carlsson and Mahlapuu, 2002; Li et al., 2004; Wijchers et al., 2006), they are likely to affect at least two different targets, activating some (the A6 mutant contains an intact AB domain) and repressing others (the ΔAB construct has an intact Eh-1 motif). Second, simultaneously preventing either activation or repression by co-injecting the ΔAB construct with GroMO significantly reduced ventral ectopic induction; this effect was rescued for gem (13/15) and zic2 (12/15; Fig. 7B) by co-injecting a morpholino-insensitive version of Gro/ Grg4 (rescue; Supplemental Fig. 1). Together, these data demonstrate that the ability of FoxD4/5 to ectopically induce neural genes in the ventral epidermis requires both activation of the neural genes, mediated via the AB domain, and repression, mediated via the Eh-1 motif binding to Gro/Grg4. We predicted that the likely targets of repression are BMP signaling and epidermal genes because both are significantly repressed by wt-FoxD4/5 (Yan et al., 2009, 2010). This was confirmed by showing: 1) neither the ΔRII-Cterm nor A6 construct prevented the nuclear localization of phosphorylated Smad1/5/8, which indicates intact BMP signaling; and 2) neither repressed the expression of two epidermis specific genes (Fig. 8).
Fig. 7.
Both the Acidic blob and the Eh-1 motif are required for ectopic expression of gem, zic2 and sox11 in the ventral epidermis. (A) The percentages of embryos showing ectopic ventral expression of gem, zic2, and sox11 after injection of wt- and mutant FoxD4/5 mRNAs. Although the AB and C-terminal mutants significantly reduced the frequency of ectopic ventral expression compared to wt-FoxD4/5 (*, p<0.001), none eliminated it, indicating that both activating and repressing activities are required. Providing both activating and repressing activities, by co-expressing both ΔAB and A6 mutants (ΔAB+A6), restored the frequency of ectopic ventral expression to wt levels. Conversely, eliminating both activating and repressing activities, by co-expressing both ΔAB and GroMOs (ΔAB+GroMO), significantly (p<0.001) reduced the frequency of ectopic ventral expression of all three genes, compared to wt (*), ΔAB (#) or ΔAB+A6 ($). Numbers above bars indicate sample sizes. (B) Examples of the ventral ectopic expression of gem, zic2 and sox11 after injection of each mutant mRNA (plus βgal, indicated by red or purple dots) into an epidermal precursor blastomere. In wt and ΔAB+A6 clones, most cells exhibit a high level of expression (blue stain). In ΔRII, A6 and ΔAB+GroMO clones, fewer cells express the gene and expression is only faintly detectable. Co-expressing a morpholino-insensitive Grg4 mRNA with ΔAB+GroMO rescued the high level of gem and zic2 ectopic ventral expression; surprisingly, this was not observed for sox11 (n=21).
Fig. 8.
C-terminal domains are required to prevent BMP signaling and repress epidermal gene expression in ventral ectodermal lineages. Wt-FoxD4/5 prevents nuclear accumulation of phospho-Smad1/5/8 (see inset) and expression of epidermal genes (Yan et al., 2009), but C-terminal mutants do not. Left column: ventral ectodermal cells expressing either the ΔRII-Cterm or A6-FoxD4/5 mutant protein (blue cytoplasm) are positive for nuclear-localized phospho-Smad1/5/8 (brown nuclei), indicating a response to BMP signaling (ΔRII-Cterm: 100%, n=19; A6: 85.7%, n=21). For comparison, inset shows a wt-FoxD4/5 clone within the hatched lines (blue cytoplasm) in which phospho-Smad 1/5/8 staining is not detected in most nuclei, whereas all nuclei outside the clone are stained. Middle and right columns: ventral ectodermal cells expressing either the ΔRII-Cterm or A6-FoxD4/5 mutant protein (red or purple nuclei within the hatched lines) express normal levels of epidermal genes (ΔRII-Cterm: AP2, 100%, n=45; Epi-ker, 100%, n=60; A6: AP2, 100%, n=34; Epi-ker, 100%, n=62).
Discussion
FoxD4/5 contains both activating and repressing domains
FoxD4/5 has been shown to play a key role in early neural development in Xenopus (Fetka et al., 2000; Sölter et al., 1999; Sullivan et al., 2001; Yan et al., 2009). FoxD4/5 both up-regulates genes that are involved in maintaining an immature neural ectoderm and down-regulates genes that promote neural differentiation in the neural plate. To understand how this transcription factor regulates its numerous targets, it is important to establish which regions of a Fox protein outside the DNA-binding domain account for these different kinds of transcriptional activities. Therefore, we sought to identify the specific domains that are responsible for transcriptional activation and transcription repression.
Our results demonstrate that within the N-terminus the AB domain is required for the up-regulation of gem and zic2 in both the neural ectoderm( Fig. 2) and the ventral epidermis (Fig. 7). In contrast, deletions and mutations in the C-terminal part of the protein have a minimal effect on their expression in the neural ectoderm. Since the up-regulation of gem and zic2 occurs in the absence of protein synthesis, indicating that FoxD4/5 directly activates these two genes, and is mimicked by a VP16-FoxD4/5 activating construct (Yan et al., 2009), we conclude that the AB comprises the transactivation domain of FoxD4/5. This is consistentwith an activating role for highly acidic regions in other transcription factors (Ptashne, 1988; Schuddekopf et al., 1996).
Our studies also show that the Eh-1 motif is responsible for the down-regulation of the sox genes and irx2 (Fig. 3). The Eh-1 motif has been shown in Xenopus FoxD3 (Yaklichkin et al., 2007a), and now in FoxD4/5, to bind the Gro/Grg4 co-repressor. In Xenopus, gro/grg4 is expressed at low levels ubiquitously (Molenaar et al., 1997), and is enhanced in the neural ectoderm and neural plate regions (Neilson et al., 2010). Thus, it is endogenously available to interact with FoxD4/5. Our experiments demonstrate that FoxD4/5 and Gro/ Grg4 can interact via the Eh-1 motif, and that Gro/Grg4 binding enhances the ability of low concentrations of FoxD4/5 to repress zic and irx genes.
However, while the A6 mutant showed that the Eh-1 motif was sufficient to account for the repression of some downstream neural genes, this motif does not account for all of the repressive activity of FoxD4/5. Repression of zic1, zic3, irx1 and irx3 requires additional sequence downstream of the Eh-1 motif (Figs. 3, 6). Analysis of the FoxD4/5 amino acid sequence using a variety of bioinformatics servers to predict secondary structure and identify additional functional domains (PSIPRED: Jones, 1999; McGuffin et al., 2000; GlobPlot: Linding et al., 2003; Porter: Pollastri and McLysaght, 2005) showed that outside the WH domain, FoxD4/5 is predicted to be random coiled and disordered (data not shown). Disordered proteins are predicted to be flexible and dynamic, forming multiple meta-stable conformations that enable the protein to bind multiple targets (protein and/or DNA), causing the protein to undergo transitions to more structured states (Dyson and Wright, 2005). While no known functional domains or protein interaction domains were identified, our sequence analyses predicted a short α-helical region close to the C-terminus of the protein (Fig. 1C, blue bar) adjacent to a region that is highly conserved across species. Interestingly, our analyses showed that mouse and human FoxD4 proteins are also predicted to have a short α-helical region in this area. We propose that this conserved C-terminal region with predicted secondary structure may influence the efficacy of transcriptional repression by FoxD4/5, either by strengthening the interaction with Gro/Grg at the Eh-1 motif or by interacting with other proteins. The fact that this region is intact in the F>E point mutant may account for this construct's near wild-type ability to repress target genes (Fig. 3). We found that mouse FoxA1 and FoxA2 also are predicted to a have short α-helical regions, but they are located in close proximity to the Eh-1 motif rather than near the C-terminus. Because the ability of these two proteins to repress target genes relies on an interaction with Gro/Grg that subsequently binds to acetylated histone to compact nucleosomes (Sekiya and Zaret, 2007), we speculate that the secondary structure of FoxD4/5 may participate in interactions of Gro/Grg4 with other proteins.
FoxD4/5 regulates the transition of an immature neural ectoderm to a differentiating neural plate by both gene activation and repression
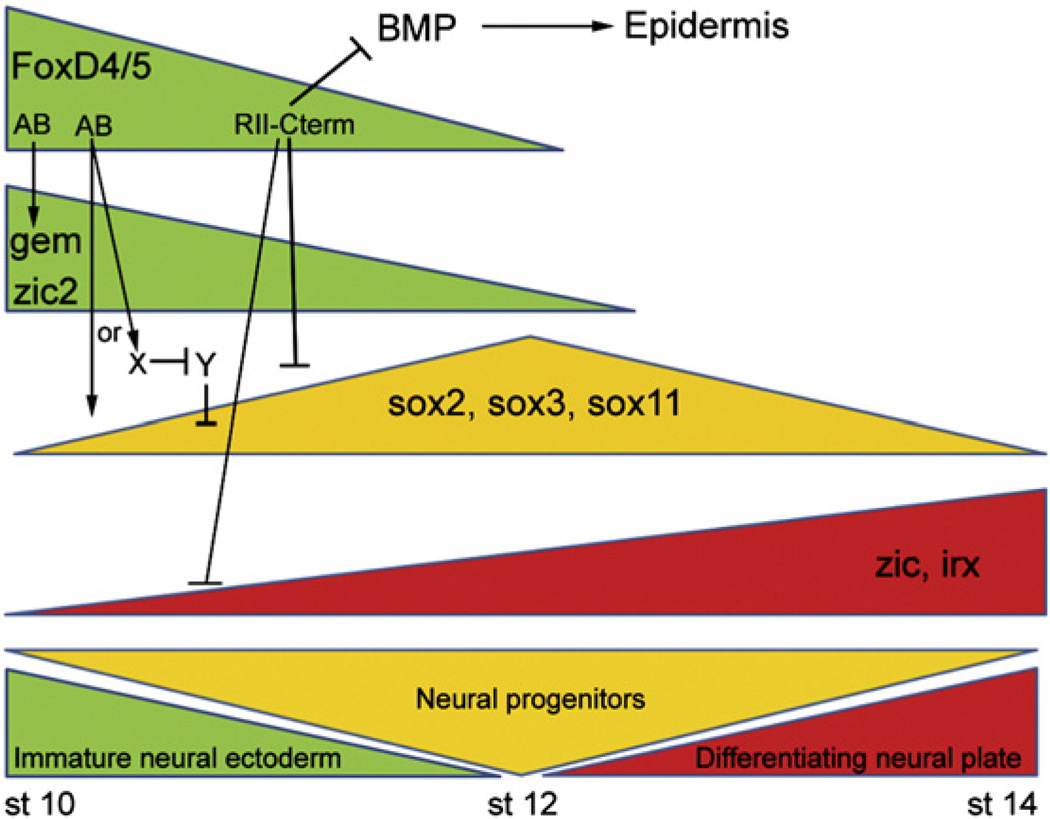
Gene regulatory networks define the transcription factors involved in a developmental process, the hierarchy of their functional interactions, and the regulatory loops that maintain a particular cellular state, and thereby elucidate the molecular regulation of a developmental process (Levine and Davidson, 2005). We experimentally defined the general epistatic relations between several early neural transcription factor genes by gain-of-function and loss-of-function studies in whole embryos, and showed that FoxD4/5 is a critical upstream component of this network (Yan et al., 2009). Increasing the level of FoxD4/5 in the neural ectoderm differentially affected the expression levels of 11 other early neural genes (Fig. 9): two that are known to promote a proliferative, immature neural ectoderm were up-regulated by direct transcriptional activation; three that are known to promote a neural progenitor state were down-regulated transiently in the neural ectoderm of the gastrula (but not neural plate); and six that are known to promote the expression of the bHLH neural differentiation genes were down-regulated via transcriptional repression. Because these results indicated that FoxD4/5 can act as both a transcriptional activator and repressor, we sought to identify those protein domains that influence these various genes that together regulate the transition of an immature, proliferative neural ectoderm to a neurally-committed neural plate whose cells are beginning to differentiate (Fig. 9). Three functional regions of the protein were revealed: the AB, Eh-1 and RII+C-terminal domains.
Fig. 9.
Fox D4/5 plays a critical role in regulating a gene network that controls the transition of an immature neural ectoderm to neural progenitors, and then to a differentiating neural plate. Wild-type FoxD4/5 affects the expression of three classes of neural transcription factors affiliated with each of these phases of neural development, and does so via different functional regions of the protein. First, the ability of FoxD4/5 to up-regulate two genes that maintain a proliferative, immature neural ectodermal state (gem, zic2) requires the AB activation domain. Second, the ability of FoxD4/5 to down-regulate during gastrulation three sox genes that promote the transition to neural progenitors involves both repression via the RII-Cterm region and activation via the AB domain. It is possible that the AB domain directly activates sox genes, or activates an unknown factor (X), which in turn represses another gene (Y) that represses sox expression. Third, the ability of FoxD4/5 to down-regulate genes that promote neural differentiation (zic, irx) requires the RII-Cterm region. FoxD4/5 additionally inhibits BMP signaling, dependent upon the Eh-1 domain within the RII-Cterm region that leads to repression of epidermal fate. Approximate timeline in Nieuwkoop and Faber (1967) stages is given below.
When FoxD4/5 levels are high in the immature neural ectoderm, gem and zic2 are up-regulated by transcriptional activation dependent upon the AB domain. Like FoxD4/5, Gem and Zic2 both promote a proliferative neural ectoderm and suppress neural differentiation (Rogers et al., 2009a). gem blocks bHLH neural differentiation gene transcription by regulating SWI/SNF chromatin-remodeling proteins and keeping cells in the cell cycle (Kroll, 2007; Kroll et al., 1998; Seo and Kroll, 2006; Seo et al., 2005). Zic2 represses bHLH neural differentiation genes and counteracts the formation of ectopic neurons produced by over-expression of Ngnr1 (Brewster et al., 1998). Our previous work showed that downstream of FoxD4/5, Gem and Zic2 up-regulate each other and down-regulate zic1, zic3 and irx1–3 (Yan et al., 2009). Therefore, we propose that at the initiation of neural ectoderm formation, FoxD4/5 is expressed at high levels to coordinately activate these two genes via the AB (Fig. 9), and together they subsequently maintain an immature neural ectoderm and prevent premature neural differentiation.
When FoxD4/5 levels are high in the immature neural ectoderm, three sox genes are down-regulated by activities in both the AB and Eh-1 domains (Fig. 9). sox2 and sox3 are transcriptional activators thought to counteract the ability of SoxB2 repressor proteins to induce neuronal differentiation (Bylund et al., 2003; Graham et al., 2003; Uchikawa et al., 1999). Sox2 and Sox3 are required for the expression of bHLH neural differentiation genes, but when their levels are experimentally increased in the embryo, there is a delay in bHLH gene expression (Dee et al., 2008; Graham et al., 2003; Kishi et al., 2000; Rogers et al., 2009b; Schlosser et al., 2008). This indicates that when levels of Sox2/Sox3 are high they hold cells in an intermediate neural progenitor state. Fewer functional studies are available for sox11, but it also appears to be involved in the transition of an immature neural progenitor to an early differentiating state (Bergsland et al., 2006; Hyodo-Miura et al., 2002; Uwanogho et al., 1995). Because FoxD4/5 delays the onset of bHLH neural differentiation gene expression (Sullivan et al., 2001), we hypothesized that it delays the transition to a neural progenitor state by down-regulating the expression of the sox genes during gastrulation (Rogers et al., 2009a; Yan et al., 2009). Herein, we show that the early down-regulation of sox2, sox3 and sox11 by FoxD4/5 in the neural ectoderm is affected by both the AB and the Eh-1 domains (Fig. 9). These results suggest several possible regulatory mechanisms including: 1) FoxD4/5 may both activate and repress sox genes, dependent upon the concentration of the protein, the affinities of the Fox binding sites in their enhancers, or interactions with other factors at adjacent binding sites; 2) FoxD4/5 may indirectly cause the down-regulation of sox genes by activating and/or repressing intermediate genes; or 3) FoxD4/5 may activate both sox genes and repressors of sox genes. For sox11, we favor the last explanation based on two previous observations: 1) in the absence of protein synthesis, FoxD4/5 can directly activate sox11; and 2) when Zic2 levels are increased, sox11 expression is down-regulated (Yan et al., 2009). In this model FoxD4/5 likely activates both zic2 and sox11, but in those cells in which Zic2 achieves high levels, sox11 would be repressed. However, FoxD4/5 repression of another gene at neural plate stages also must be involved in sox11 up-regulation, since deletions and mutations in the C-terminal region prevent this phenotype. Distinguishing between these possibilities requires further investigation.
High levels of FoxD4/5 down-regulate the expression of zic and irx genes via the R-II plus downstream C-terminal sequence (Fig. 3). These genes are effectors of initiating the gene program that activates the bHLH neural differentiation genes (Rogers et al., 2009a); they expand the expression of bHLH neural differentiation genes in whole embryo and explant assays, and the irx genes are required for the expression of the bHLH genes (Bellefroid et al., 1998; Gomez-Skarmeta et al., 1998; Mizuseki et al., 1998; Nakata et al., 1997). Together with the observation that the EnR-FoxD4/5 construct down-regulates the zic and irx genes as effectively as wt-FoxD4/5 (Yan et al., 2009), we conclude that these genes are directly repressed by FoxD4/5 when levels of this protein are high. As endogenous levels of FoxD4/5 drop during the formation of the neural plate, these differentiation-promoting genes would be released from repression from both FoxD4/5 and the transitional sox genes (Fig. 9).
We also provide evidence that the Eh-1 domain has an additional involvement in repressing BMP signaling, which in turn prevents expression of epidermal fate (Figs. 8, 9). In this manner, FoxD4/5 provides a signaling environment that allows the formation of neural ectoderm, even in ectopic locations. It will be critical to determine exactly how the C-terminal regions interact with Gro/Grg4 to repress the BMP pathway and to detail which specific parts of the BMP pathways are affected.
The identified functional domains are highly conserved across vertebrates
These studies define for the first time several functional domains in the FoxD4/5 protein that enable it to both activate and repress transcription of downstream targets that play critical roles in expanding the nascent neural ectoderm and regulating the onset of neural differentiation. While it is notable that most of the target neural genes have been studied in many other animals, only in Xenopus have they been studied in relation to FoxD4/5. In fact, to our knowledge there are no functional studies of the activity of FoxD4/5 in the nervous system of any other vertebrate. Since these important functional domains are highly conserved across vertebrates (Fig. 1), we predict that the results reported herein are likely to apply to the function of the FoxD4/5 protein in many other animals, including humans.
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.ydbio.2012.03.004.
Supplementary Material
Acknowledgements
This work was supported in part by NSF grant IOS-0817902, funds from the George Washington University Medical Center, and the Intramural Research Program of the NIH, National Cancer Institute. Confocal microscopy was performed at the GWU Center for Microscopy and Image Analysis with support from the NIH grants S10 RR025565 and P30 HD040677 (IDDRC at CNMC). We thank Dr. Anastas Popratiloff (GWUMC) for confocal microscopy, Dr. John Orban (Univ. Maryland) for advice on protein structure analyses and Ms. Rebecca He for immunostaining. Dr. Klein’s efforts were supported by the National Science Foundation while working at the Foundation. Any opinion, finding, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation.
References
- Bellefroid EJ, Kobbe A, Gruss P, Pieler T, Gurdon JB, Papalopulu N. Xiro3 encodes a Xenopus homolog of the Drosophila Iroquois genes and functions in neural specification. EMBO. J. 1998;17:191–203. doi: 10.1093/emboj/17.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsland M, Werme M, Malewicz M, Perlmann T, Muhr J. The establishment of neuronal properties is controlled by Sox4 and Sox11. Genes Dev. 2006;20:3475–3486. doi: 10.1101/gad.403406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewster R, Lee J, Ruiz i Altaba A. Gli/Zic factors pattern the neural plate by defining domains of cell differentiation. Nature. 1998;393:579–583. doi: 10.1038/31242. [DOI] [PubMed] [Google Scholar]
- Bylund M, Andersson E, Novitch BG, Muhr J. Vertebrate neurogenesis is counteracted by Sox1–3 activity. Nature Neurosci. 2003;6:1162–1168. doi: 10.1038/nn1131. [DOI] [PubMed] [Google Scholar]
- Carlsson P, Mahlapuu M. Forkhead transcription factors: key players in development and metabolism. Dev. Biol. 2002;250:1–23. doi: 10.1006/dbio.2002.0780. [DOI] [PubMed] [Google Scholar]
- Cirillo L, Lin FR, Cuesta I, Jarnik M, Friedman D, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FOXA) and GATA-4. Mol. Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- De Robertis EM, Kuroda H. Dorsal-ventral patterning and neural induction in Xenopus embryos. Annu. Rev. Cell Dev. Biol. 2004;20:285–308. doi: 10.1146/annurev.cellbio.20.011403.154124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dee CT, Hirst CS, Shih YH, Tripathi VB, Patient RK, Scotting PJ. Sox3 regulates both neural fate and differentiation in the zebrafish ectoderm. Dev. Biol. 2008;320:289–301. doi: 10.1016/j.ydbio.2008.05.542. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- Essien K, Vigneau S, Apreleva S, Singh LN, Bartolomei MS, Hannenhalli S. CTCF binding site classes exhibit distinct evolutionary, genomic, epigenomic and transcriptomic features. Genome Biol. 2009;10:R131. doi: 10.1186/gb-2009-10-11-r131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fetka I, Doederlein G, Bouwmeester T. Neuroectodermal specification and regionalization of the Spemann organizer in Xenopus. Mech. Dev. 2000;93:49–58. doi: 10.1016/s0925-4773(00)00265-3. [DOI] [PubMed] [Google Scholar]
- Freyaldenhoven B, Fried C, Wielckens K. FOXD4a and FOXD4b, two new winged helix transcription factors, are expressed in human leukemia cell lines. Gene. 2004;294:131–140. doi: 10.1016/s0378-1119(02)00702-3. [DOI] [PubMed] [Google Scholar]
- Gomez-Skarmeta JL, Glavic A, de La Calle-Mustienes E, Modolell J, Mayor R. Xiro, a Xenopus homolog of the Drosophila Iroquois complex genes, controls development at the neural plate. EMBO. J. 1998;17:181–190. doi: 10.1093/emboj/17.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- Hyodo-Miura J, Urushiyama S, Nagai S, Nishita M, Ueno N, Shibuya H. Involvement of NLK and Sox11 in neural induction in Xenopus development. Genes Cells. 2002;7:487–496. doi: 10.1046/j.1365-2443.2002.00536.x. [DOI] [PubMed] [Google Scholar]
- Itoh K, Sokol SY. Early Development of epidermis and neural tissue. In: Moody SA, editor. Principles of Developmental Genetics. NY: Elsevier; 2007. pp. 241–257. [Google Scholar]
- Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- Kaestner KH, Monaghan AP, Kern H, Ang SL, Weitz S, Lichter P, Schütz G. The mouse fkh-2 gene. Implications for notochord, foregut, and midbrain regionalization. J. Biol. Chem. 1995;270:30029–30035. doi: 10.1074/jbc.270.50.30029. [DOI] [PubMed] [Google Scholar]
- Katoh M, Katoh M. Human FOX gene family. Int. J. Oncol. 2004;25:1495–1500. [PubMed] [Google Scholar]
- Kishi M, Mizuseki K, Sasai N, Yamazaki H, Shiota K, Nakanishi S, Sasai Y. Requirement of Sox 2-mediated signaling for differentiation of early Xenopus neuroectoderm. Development. 2000;127:791–800. doi: 10.1242/dev.127.4.791. [DOI] [PubMed] [Google Scholar]
- Kroll KL. Geminin in embryonic development: coordinating transcription and the cell cycle during differentiation. Front. Biosci. 2007;12:1395–1409. doi: 10.2741/2156. [DOI] [PubMed] [Google Scholar]
- Kroll KL, Salic AN, Evans LM, Kirschner MW. Geminin, a neuralizing molecule that demarcates the future neural plate at the onset of gastrulation. Development. 1998;125:3247–3258. doi: 10.1242/dev.125.16.3247. [DOI] [PubMed] [Google Scholar]
- Levine M, Davidson EH. Gene regulatory networks in development. Proc. Natl. Acad. Sci. U.S.A. 2005;102:4936–4942. doi: 10.1073/pnas.0408031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Weidenfeld J, Morrisey EE. Transcriptional and DNA binding activity of the Foxp1/2/4 family is modulated by heterotypic and homotypic protein interactions. Mol. Cell Biol. 2004;24:809–822. doi: 10.1128/MCB.24.2.809-822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linding R, Russell RB, Neduva V, Gibson TJ. GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003;31:3701–3708. doi: 10.1093/nar/gkg519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- Mizuseki K, Kishi M, Shiota K, Nakanishi S, Sasai Y. SoxD: An essential mediator of induction of anterior neural tissues in Xenopus embryos. Neuron. 1998;21:77–85. doi: 10.1016/s0896-6273(00)80516-4. [DOI] [PubMed] [Google Scholar]
- Molenaar M, Brian E, Roose J, Clevers H, Destree O. Differential expression of the Groucho-related genes 4 and 5 during early development of Xenopus laevis. Mech. Dev. 1997;91:311–315. doi: 10.1016/s0925-4773(99)00259-2. [DOI] [PubMed] [Google Scholar]
- Moody SA. Fates of the blastomeres of the 16-cell stage Xenopus embryo. Dev. Biol. 1987;119:560–578. doi: 10.1016/0012-1606(87)90059-5. [DOI] [PubMed] [Google Scholar]
- Moody SA. Cell Lineage Analysis in Xenopus Embryos. In: Tuan RS, Lo CW, editors. Methods in Molecular Biology.: Developmental Biology Protocols. vol. 135. Totowa, NJ: Humana Press Inc.; 1999. [Google Scholar]
- Moody SA. Testing the cell fate commitment of Single blastomeres in Xenopus laevis. In: Richter J, editor. Advances in Molecular Biology. Oxford University Press; 2000. pp. 355–381. [Google Scholar]
- Nakata K, Nagai T, Aruga J, Mikoshiba K. Xenopus Zic3, a primary regulator both in neural and neural crest development. Proc. Natl. Acad. Sci. U.S.A. 1997;94:11980–11985. doi: 10.1073/pnas.94.22.11980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilson KM, Pignoni F, Yan B, Moody SA. Developmental expression patterns of candidate co-factors for vertebrate Six family transcription factors. Dev. Dyn. 2010;239:3446–3466. doi: 10.1002/dvdy.22484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal table of Xenopus laevis (Daudin) North-Holland, Amsterdam: 1967. [Google Scholar]
- Odenthal J, Nusslein-Volhard C. Fork head domain genes in zebrafish. Dev. Genes Evol. 1998;208:245–258. doi: 10.1007/s004270050179. [DOI] [PubMed] [Google Scholar]
- Pohl BS, Knochel W. Of Fox and Frogs: Fox (fork head/winged helix) transcription factors in Xenopus development. Gene. 2005;344:21–32. doi: 10.1016/j.gene.2004.09.037. [DOI] [PubMed] [Google Scholar]
- Pollastri G, McLysaght A. Porter: a new accurate server for protein secondary structure prediction. Bioinformatics. 2005;21:1719–1720. doi: 10.1093/bioinformatics/bti203. [DOI] [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988;335:683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- Rogers CD, Moody SA, Casey ES. Neural induction and factors that stabilize a neural fate. Birth Defects Res. C Embryo Today. 2009a;87:249–262. doi: 10.1002/bdrc.20157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers CD, Harafuji N, Archer T, Cunningham DD, Casey ES. Xenopus Sox3 activates sox2 and geminin and indirectly represses Xvent2 expression to induce neural progenitor formation at the expense of non-neural ectodermal derivatives. Mech. Dev. 2009b;126:42–55. doi: 10.1016/j.mod.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlosser G, Awtry T, Brugmann SA, Jensen ED, Neilson KM, Ruan G, Stammler A, Voelker D, Yan B, Zhang C, Klymkowsky MW, Moody SA. Eya1 and Six1 promote neurogenesis in the cranial placodes in a SoxB1-dependent fashion. Dev. Biol. 2008;320:199–214. doi: 10.1016/j.ydbio.2008.05.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuddekopf K, Schorpp M, Boehm T. The whn transcription factor encoded by the nude locus contains an evolutionarily conserved and functionally indispensable activation domain. Proc. Natl. Acad. Sci. U.S.A. 1996;93:9661–9664. doi: 10.1073/pnas.93.18.9661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya T, Zaret KS. Repression by Groucho/TLE/Grg proteins: genomic site recruitment generates a compacted chromatin structure in vitro and impairs activator binding in vivo. Mol. Cell. 2007;28:291–303. doi: 10.1016/j.molcel.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S, Kroll KL. Geminin's double life: chromatin connections that regulate transcription at the transition from proliferation to differentiation. Cell Cycle. 2006;5:374–379. doi: 10.4161/cc.5.4.2438. [DOI] [PubMed] [Google Scholar]
- Seo S, Herr A, Lim JW, Richardson GA, Kroll KL. Geminin regulates neuronal differentiation by antagonizing Brg1 activity. Genes Dev. 2005;19:1723–1734. doi: 10.1101/gad.1319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, Harland RM. A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. Early Development of Xenopus laevis. [Google Scholar]
- Sölter M, Köster M, Holleman T, Brey A, Pieler T, Knöchel W. Characterization of a subfamily of related winged helix genes, XFD-12/12′/12″ (XFLIP), during Xenopus embryogenesis. Mech. Dev. 1999;89:161–165. doi: 10.1016/s0925-4773(99)00195-1. [DOI] [PubMed] [Google Scholar]
- Stern CD. Neural induction: old problem, new findings, yet more questions. Development. 2005;132:2007–2021. doi: 10.1242/dev.01794. [DOI] [PubMed] [Google Scholar]
- Suda Y, Nakabayashi J, Matsuo I, Aizawa S. Functional equivalency between Otx2 and Otx1 in development of rostral head. Development. 1999;126:743–757. doi: 10.1242/dev.126.4.743. [DOI] [PubMed] [Google Scholar]
- Sullivan SA, Akers L, Moody SA. foxD5a, a Xenopus winged helix gene, maintains an immature neural ectoderm via transcriptional repression that is dependent on the C-terminal domain. Dev. Biol. 2001;232:439–457. doi: 10.1006/dbio.2001.0191. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja G, Kaestner KH. SnapShot: forkhead transcription factors I. Cell. 2007;130:1160. doi: 10.1016/j.cell.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Uchikawa M, Kamachi Y, Kondoh H. Two distinct subgroups of Group B Sox genes for transcriptional activators and repressors: their expression during embryonic organogenesis of the chicken. Mech. Dev. 1999;84:103–120. doi: 10.1016/s0925-4773(99)00083-0. [DOI] [PubMed] [Google Scholar]
- Uwanogho D, Rex M, Cartwright EJ, Pearl G, Healy C, Scotting PJ, Sharpe PT. Embryonic expression of the chicken Sox2, Sox3 and Sox11 genes suggests an interactive role in neuronal development. Mech. Dev. 1995;49:23–36. doi: 10.1016/0925-4773(94)00299-3. [DOI] [PubMed] [Google Scholar]
- Wijchers PJ, Burbach JP, Smidt MP. In control of biology: of mice, men and Foxes. Biochem. J. 2006;397:233–246. doi: 10.1042/BJ20060387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaklichkin S, Steiner AB, Lu Q, Kessler DS. FoxD3 and Grg4 physically interact to repress transcription and induce mesoderm in Xenopus. J. Biol. Chem. 2007a;282:2548–2557. doi: 10.1074/jbc.M607412200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaklichkin S, Vekker A, Stayrook S, Lewis M, Kessler DS. Prevalence of the EHI Groucho interaction motif in the metazoan Fox family of transcriptional regulators. BMC Genomics. 2007b;8:201–218. doi: 10.1186/1471-2164-8-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B, Neilson KM, Moody SA. FoxD5 plays a critical upstream role in regulating neural fate and onset of differentiation. Dev. Biol. 2009;329:80–95. doi: 10.1016/j.ydbio.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B, Neilson KM, Moody SA. Microarray identification of novel downstream targets of FoxD5, a critical component of the neural ectodermal transcriptional network. Dev. Dyn. 2010;239:3467–3480. doi: 10.1002/dvdy.22485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS. Regulatory phases of early liver development: Paradigms of organogenesis. Nat. Rev. Genet. 2002;3:499–512. doi: 10.1038/nrg837. [DOI] [PubMed] [Google Scholar]
- Zaret KS, Watts J, Xu J, Wandzioch E, Smale ST, Sekiya T. Pioneer factors, genetic competence, and inductive signaling: programming liver and pancreas progenitors from the endoderm. Cold Spring Harb. Symp. Quant. Biol. 2008;73:119–126. doi: 10.1101/sqb.2008.73.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.