Abstract
Macrophages play a key role in host defense against microbes, in part, through phagocytosis. MARCO (macrophage receptor with collagenous structure) is a scavenger receptor on the cell surface of macrophages that mediates opsonin-independent phagocytosis. The goal of our study is to investigate the role of MARCO in lipopolysaccharide (LPS) or lipotechoic acid (LTA)-induced macrophage tolerance. While it has been established that expression of MARCO and phagocytosis is increased in tolerant macrophages, the transcriptional regulation and biological role of MARCO in tolerant macrophages has not been investigated. Here, we confirm that tolerized mouse bone marrow derived macrophages (BMDM) selectively increase expression of MARCO (both transcript and cell surface receptor) and increase phagocytosis. We found that H3K4me3 dynamic modification of a promoter site of MARCO was increased in tolerized BMDM. Blocking methylation by treatment with 5-Aza-2′-deoxycytidine (5-AZA) resulted in reduced H3K4me3 binding in the promoter of MARCO, decreased expression of MARCO, and impaired phagocytosis in tolerized BMDM. However, 5-AZA had no effect on the inflammatory component of innate immune tolerance. In aggregate, we found that histone methylation was critical to MARCO expression and phagocytosis in tolerized macrophages but did not affect the inflammatory component of innate immune tolerance.
Keywords: Phagocytosis, MARCO, Macrophage tolerance, Chromatin modification
Introduction
Innate immune tolerance plays a key role in protecting the host from potential damage caused by excessive inflammation (1). With repetitive pathogen stimulation, tolerant macrophages are characterized by suppressed secretion of proinflammatory cytokines, upregulation of anti-inflammatory genes, and increased phagocytosis (2). The increased phagocytosis helps clear microorganisms, is mediated by the CD64 (Fc receptor) (3), and limits further inflammation. There are two phagocytic processes: opsonin-dependent phagocytosis via Fc receptor and complement receptors (4, 5), and opsonin-independent through scavenger receptors.
Scavenger receptors on the surface of macrophages, such as class A scavenger receptor MARCO, mediate binding of unopsonized particles and phagocytosis (6). Other scavenger receptors on macrophages include LOX1 and class B scavenger receptors (SR-A, SR-B, and CD36) , which mediate phagocytosis of modified LDL (low-density lipoprotein), bacteria, and apoptotic cells (6, 7). It has been reported that MARCO is the major binding receptor for oxidized lipids, unopsonized particles, and bacteria in human alveolar macrophages (8–10). In addition, MARCO serves as a signaling receptor involved in responses to CpG oligodeoxynucleotides and mycobacterial trehalose dimycolate (11, 12).
While previous studies have shown that the expression of MARCO was significantly upregulated in LPS-induced tolerant macrophages (13), the mechanisms controlling this process and the role of MARCO in innate immune tolerance has not been comprehensively investigated. Macrophage tolerance can be induced by different toxic signals including tissue damage and components of microbial pathogens through various receptors and signaling pathways (14–16). Here, we investigated the expression of MARCO in both LPS- and LTA-induced tolerant cells. While our results confirmed the increased expression of MARCO (transcript and cell surface expression) in tolerant macrophages, we also detected tolerance induced epigenetic marks (trimethylation of the H3K4 residue in the promoter of MARCO) that regulate the expression of MARCO. In aggregate, we found that histone methylation was critical to MARCO expression and phagocytosis in tolerized macrophages but did not affect the inflammatory component of innate immune tolerance.
Materials and Methods
Mice
Male C57BL/6 mice, 8–12 weeks old were purchased from Jackson Laboratories. The mice were maintained at the animal facilities at National Jewish Health and University of Colorado Denver. The femurs from MARCO−/− mice on C57BL/6 genetic background were provided by Dr. Lester Kobzik (10).
Reagents
LPS (cat. no.421) was purchased from List Biological Laboratories, INC (Campbell, CA). Lipoteichoic acid (LTA), cytochalasin D, 5-Aza-2′-deoxycytidine (5-AZA) and Fluorescein diacetate were purchased from Sigma. ELISA kits and M-CSF were purchased from R&D Systems, Inc. Vybrant Phagocytosis Assay Kit (cat. No. V-6694) and Deep Red plasma membrane stains (cat. no.c10046) were purchased from Invitrogen. cDNA Reverse Transcription Kit and SYBR Green PCR Master Mix were purchased from AB applied biosystem. RNeasy Mini Kit was purchased from QIAGEN. CHIP kits were purchased from Active Motif and Covaris Inc. Rat anti mouse MARCO-FITC (cat. No. MCA1849FA) and rat IgG1 negative control-FITC (cat. No. MCA1211F) were purchased from AbD Serotec. Rabbit anti-Goat IgG Antibody, HRP conjugate (cat. No. AP106P) was purchased from EMD Millipore Corporation (Chicago, IL). Mouse MARCO Affinity Purified Polyclonal Ab (cat. No. AF2956) was purchased from R&D Systems Inc. Anti-NFκB p65 antibody (cat. No. ab7970) was purchased from Abcam Ltd (Cambridge, GB). CellMask™ Deep Red plasma membrane stain was purchased from Invitrogen. Anti-trimethyl-Histone H3 (Lys4) (cat. No. 07-473) and Anti-dimethyl-Histone H3 (Lys9) (cat. No. 07-521) were purchased from Millipore.
Cell Culture, Endotoxin Assay, Fluorescein diacetate assay, and Cytokine ELISA
BMDM were harvested from femurs of male C57BL/6. Bone marrow progenitors were cultured for 6 days in DMEM/high glucose supplemented with 10% (v/v) heat-inactivated fetal bovine serum, 5% penicillin/streptomycin and M-CSF (final concentration 20–25 ng/ml). Cells were lifted with warm PBS and replated on tissue-culture treated plates. On day 7, macrophages were left untreated (nontolerant, NT) or stimulated with 5 EU (endotoxin unit)/ml LPS or 20ug/ml LTA for 24 h (tolerant, T). At 24h, cell cultures were washed twice with warm PBS and given fresh media or conditioned media with 5 EU/ml LPS (LPS/LPS-incubation time or LTA/LPS-incubation time). After various times of incubation, media were tested for cytokines content. To ensure the same activity of LPS from different lots, we measured the concentration of LPS by Endosofe-PTS kinetic reader (Charles River Laboratories).
For Fluorescein diacetate assay, live cells were stained by fluorescein diacetate (final concentration 100ug/ml) for 5 min at room temperature (RT). Lysed cells were obtained after incubation with the same amount of triton-100 (1%) at RT for 5min. The plate was read by GEN5 (BioTek, VT) using ~480 nm excitation, ~ 520 nm emissions.
IL-6 and TNF-α concentrations in macrophage culture supernatants were measured by ELISA, as specified by manufacturer's instruction (R&D Systems). The data were collected from at least 3 independent experiments and normalized with cell numbers measured by Fluorescein diacetate assay.
Phagocytosis assays and Immunofluorescence Microscopy
BMDM were cultured and treated as described above. After 24h incubation at 37°C, cells were lifted with warm PBS and replated on 96-well tissue culture plates with the density of 106/ml for phagocytosis assays. Cells were seeded in coverslip coated with collagen on 6-well tissue culture plates with the density of 5×105/ml for microscopic imaging. Cells were treated with LPS (5EU) and incubated at 37°C for 4 h. For phagocytosis assays, the detail protocol follows the manufacturer's instructions of Vybrant Phagocytosis Assay. In brief, cells were fed with prepared fluorescein-labeled BioParticles and incubated for 2h. Cells were quenched with trypan blue (250μg/ml) for 1 min. For microscopy imaging, cells were incubated with Deep Red plasma membrane stain (10μg/ml) for 30 min and fixed with 4% paraformaldehyde for 10 min at various time points (5min, 15min, 35min, 70min and 2h). Cells were then extensively washed with PBS. Coverslips were mounted in VectaShield (Vector laboratories). Cells were imaged with an inverted Zeiss Axiovert 200M deconvolution microscope. Image processing was performed using Intelligent Imaging Innovations three-dimensional rendering and exploration software. NFkB p65 colocalization was analyzed by Olympus 6×63 Upright Microscope and CellSens software.
Reverse transcription and Quantitative real time PCR (RT-qPCR)
Total RNA from tolerant (T) and nontolerant (NT) cells was isolated with RNeasy Kit. Total RNA was reverse transcribed with random primers using MultiScribe reverse transcriptase (Applied Biosystems). Complementary DNA was analyzed in triplicate by qPCR amplification using SYBR Green QPCR Master Mix (MARCO and other scavenger receptors) on the Applied Biosystems ViiA™7 Real-Time PCR System. For SYBR Green, the qPCR amplification conditions were: 50 °C (2 min), 95 °C (10 min), 40 cycles of 95 °C (15 s), 60 °C (1min) and melt curve stage of 95 °C (15 s), 60 °C (1 min), 95 °C (15 s). Primer pairs were designed to amplify mRNA-specific fragments and unique products were tested by melt-curve analysis.
MARCO (F 5'-gaagacttcttgggcagcac-3', R 5'-cttcttgggcactggatcat-3')
SR AI (F 5'-tcaaactcaaaagccgacct-3', R 5'-ccagtgaattcccatgttcc-3')
SR B (F 5'-aagtggtcaacccaaacgag-3', R- 5'-acggtgtcgttgtcattgaa-3')
CD36 (F 5'-gcttgcaactgtcagcacat-3', R-5'-gccttgctgtagccaagaac-3')
LOX1 (F 5'-tggtggatccagatgtttga-3', R 5'-gttggttgggagactttgga-3')
B-actin (F 5'-agggctatgctctccctcac-3', R 5'-ctctcagctgtggtggtgaa-3')
TLR2 (F 5'-ctggagcatccgaattgca-3', R 5'-catcctctgagatttgacgct-3')
TLR4 (F 5'-ggcaacttggacctgaggag-3', R 5'-catgggctctcggtccatag-3')
Relative quantification was calculated by the ΔΔCT method ((17)). Data are represented as the fold induction over NT (nontolerant) cells.
Chromatin immunoprecipitation assay (CHIP)
1 × 107 BMDM were stimulated, washed with PBS, and fixed with 1% formaldehyde (16% methanol free, Thermo Scientific) for 5 min at room temperature. Formaldehyde fixation was stopped with the addition of quench buffer (375 mg glycine in 50 ml). Fixed cells were sonicated for 8–10 min in S220 Covaris according to manufacturer's instruction (Covaris Inc.). Chromatin shearing efficiency was analyzed on a 1% agarose gel to ensure that sheared fragments range from 200 to 700 bp in size. 25 μl of sonicated material was saved for qPCR analysis (input). The rest of the sonicated material was incubated with antibody (Anti- H3K4me3, Anti- H3K9me2 and IgG) and protein G magnetic beads while rotating overnight according to manufacturer's instruction (Active Motif). After the elution of protein-DNA complexes, DNA was desalted and concentrated by QIAEX II Gel Extraction Kits (QIAGEN). Immunoprecipitated DNA and input DNA were amplified with MARCO (415F 5'-atttgcacctgaggctcact-3', 415R 5'-gtgaaaaccaggtgtcgttg-3'; 255F 5'-gttatgcttgcctgatgtgc-3', 255R 5'-ccacacagaccaaat ccaaa-3') CD4 (F 5'- tgtaagccttgcctcacgtt-3', R 5'- atgcctgagcagaatcaagc-3') and β-actin (F 5'-gagacattgaatggggcagt -3', R 5'-aagcgagagagaaagcgaga -3') specific primers by qPCR, using input DNA to generate a standard curve. ChIP data is represented as fold changes of tolerant (T) and NT which were calculated by 2-[(ΔT target gene-ΔT input)- (ΔT IgG- ΔT input)].
Flow cytometry
BMDMs were treated with or without LPS (5 EU) at 37°C for 24 hours followed by a second LPS (5 EU) stimulation. Cells were then harvested and fixed with 4% paraformaldehyde for 10 min, followed by quenching with quench buffer (375 mg glycine in 50 ml) 2× for 5 min. Cells were washed with 1×PBS, blocked with incubation buffer (0.5 g BSA, 2ml serum in 50ml PBS) for 30 min at RT, followed by incubation with anti-MARCO-FITC or anti-IgG-FITC (10 μg/ml) in 200μl incubation buffer for 2h at RT. After extensive washing with incubation buffer, cells were resuspended in 500 μl PBS. Cell-associated FITC-conjugated antibodies were analyzed by flow cytometry. Flow cytometry analysis was performed using a FACS Scan flow cytometry equipped with 488nm lasers in the National Jewish Health Flow Cytometry Core Facility.
Western Blot
BMDMs were treated with or without LPS (5 EU) at 37°C for 24 hours followed by a second LPS (5 EU) stimulation. Cells were harvested at 4h, 6h, 12h and 24h after second stimulation. Cell lysates were extracted by Nuclear and Cytoplasmic Extraction Reagent (Thermo Scientific, IL) and loaded with 7ug protein per lane. Cell extracts were resolved by SDS-PAGE and were analyzed by immunoblot. The membranes were probed with antibodies to MARCO at a dilution of 1:2000 and RCP/FIP1. Blots were developed with SuperSignal West Dura (Thermo Scientific, Rockford, IL).
Statistics
The phagocytic ability of macrophages was defined as the average number of fluorescein-labeled BioParticles in random cells of multiple × 63 high-power fields. Data were presented as the mean ± SEM. Statistical comparisons or fold changes were performed by t-test as indicated using GraphPad Prism 5 software.
Results
MARCO is significantly upregulated in LPS- and LTA-induced tolerant and cross tolerant macrophages
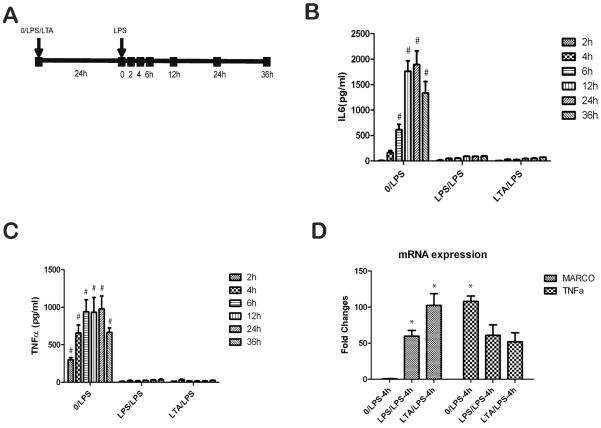
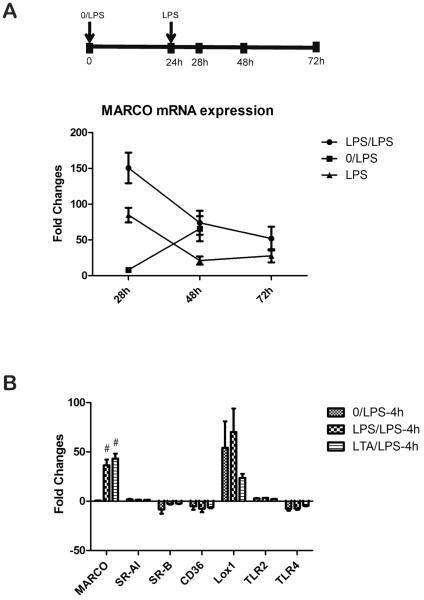
Previous studies have established that MARCO expression is significantly upregulated in LPS stimulated tolerant cells (13). Here, we asked whether the expression of MARCO is generally upregulated in tolerant cells or is specific for LPS-induced tolerance. To address this question, we measured MARCO expression in LPS and LTA induced tolerant and cross tolerant macrophages derived from mouse bone marrow. As shown in Supplemental Figures 1A and 1B, IL-6 and TNF-α concentration were highest when stimulated with 5 EU LPS at several time points (6h, 12h, 24h). LTA at a concentration of 20μg/ml also produced a high concentration of TNF-α (Supplemental Figure 1C). Tolerized BMDM resulted in significantly reduced concentrations of IL-6 and TNF-α levels when restimulated with either LPS or LTA (Figures 1B, 1C). Consistent with previous studies, BMDM were highly tolerant to LPS secondary stimulation in both LPS- and LTA-induced tolerance at several time points. The extent of decreased TNF-α mRNA in tolerant macrophages is less obvious than the decreased protein level in the media, suggesting the important role of post-transcriptional regulation (Figure 1D). We next examined the expression of MARCO in tolerant and cross tolerant cells (Figure 1D). After secondary LPS stimulation, the expression of MARCO was significantly increased in both tolerant and cross tolerant macrophages. To understand the kinetic response of MARCO expression, we measured MARCO mRNA level at various time-points with single LPS or double LPS stimulation (Figure 2A). The results showed that MARCO mRNA was increased after LPS stimulation in all three groups and all three time-points. At the 28h time-point, LPS/LPS group had maximum fold changes compared with median fold changes of the LPS group and minimum fold changes of the 0/LPS group. The increased mRNA levels peaked after 24h stimulation of LPS and slowed down in the following 24–48h. The increased MARCO mRNA with single dose of LPS stimulation can last for at least 72 hours. These results were consistent with previous studies that showed increased expression depends on protein synthesis at early stage. The rapid responses to the second dose of LPS and the maximum fold changes at 28h in the LPS/LPS group indicate the MARCO gene expression loses its dependency on protein synthesis showing the gene converts to a rapid response (primary response) gene following the second LPS stimulation The presence of LPS continues to induce the persistent expression of MARCO (13).
Figure 1.
LPS and LTA induced tolerant in MBMM. (A) The schematic used to induce tolerance in MBMM. Cells were treated with LPS (5EU), LTA (20ug/ml), or no treatment control. Following 24h incubation, media were removed and second stimulation with LPS (5EU) or no treatment control was used to establish non-tolerant (NT) and tolerant (T) status. Media were collected at various time points and IL-6 (B) and TNF-α (C) were measured by ELISA. (D) Expression of MARCO and TNF-α were examined by qPCR in cells that harvested 4h after second LPS stimulation. Data are representative of 3 independent experiments; shown are mean ± s.e.m from triplicate values (#, p<0.01; *, p<0.05, compared with LPS/LPS or 0/LPS groups).
Figure 2.
The mRNA expression level of MARCO was significant upregulated in LPS or LTA induced tolerant cells. (A) Dynamic changes of MARCO mRNA expression level in the process of LPS stimulation. BMDMs were exposed to LPS with single stimulation at 0 h (LPS) and 24 h (0/LPS) or double stimulations (LPS/LPS). At timepoints of 28h, 48h and 72h, total mRNA of BMDMs were purified and analyzed by qPCR. (B) In addition to MARCO, Lox1 is upregulated in all three conditions while expression of other scavenger receptors is unchanged. Data are representative of 3 independent experiments; shown are mean ± s.e.m from triplicate values (#, p<0.01, compared with 0/LPS group).
Cell surface MARCO is specifically increased in tolerant macrophages
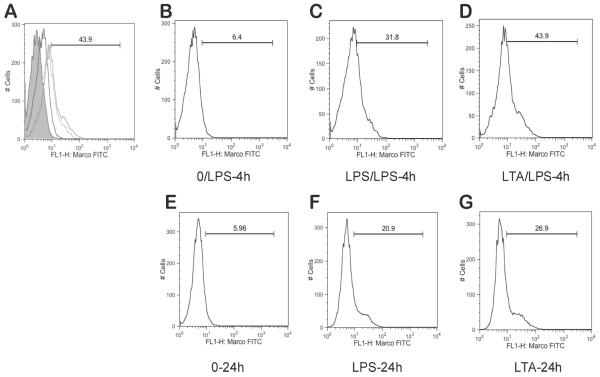
We next wanted to examine whether the expression of other scavenger receptors are also increased in tolerant and cross tolerant cells (Figure 2B). Consistent with previous studies, TLR4 was down-regulated after stimulation with LPS or LTA (18). In contrast, expression of MARCO was again significantly increased in tolerant and cross tolerant cells, but the expression of other scavenger receptor was not increased indicating a specific role of MARCO in tolerant cells. To explore the protein level of MARCO on the cell surface, we measured cell surface MARCO in both nontolerant and tolerant macrophages by flow cytometry (Figure 3). After BMDMs were incubated with LPS (Figure 3F) or LTA (Figure 3G), cells were further stimulated with LPS for 4h (Figure 3C–D). Figure 3A clearly demonstrates the increase of MARCO on cell surfaces of tolerant and cross tolerant cells.
Figure 3.
MARCO expression on cell surface is increased with persistent stimulations. (A) Merge of (B-D). Experimental conditions are as follows: 4 hour stimulation with LPS following 24h incubation with no treatment (B), 4 hour stimulation with LPS following 24h incubation with LPS (C) or LTA (D). Additional control conditions: cells incubated for 24h with no treatment (E), LPS (F), or LTA (G).
LPS and LTA induced tolerant macrophages have increased phagocytosis
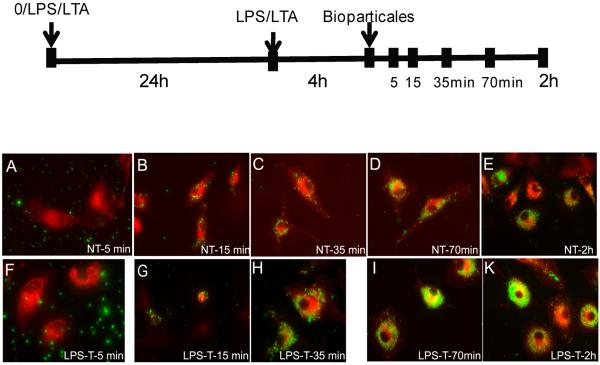
MARCO plays an important role in mediating non-opsonin dependent phagocytosis, which is a key defensive mechanism of macrophages for clearing foreign pathogens. Here, we determined whether LPS- or LTA-induced tolerant or cross tolerant macrophages have increased phagocytosis. More fluorescein-labeled BioParticles were internalized into nontolerant and LPS- or LTA-induced tolerant macrophages with prolonged incubation with fluorescence-labeled BioParticles (Figure 4). Macrophages were fed BioParticles after 4h of LPS stimulation and fixed at different time points as shown in the plot in Figure 4. At each time point, nontolerant macrophages have significantly less internalized Escherichia coli BioParticles than corresponding tolerant macrophages (Figure 4, panels A–K). These data demonstrate that phagocytosis is increased in both tolerant and cross tolerant cells.
Figure 4.
Phagocytosis ability is increased in LPS or LTA induced tolerant macrophage. The schematic on the top represents the various time points for cell fixation after adding fluorescent Bioparticles in nontolerant and tolerant cells. Nontolerant (A-E) and tolerant (F-K) macrophages from different time points were fed with fluorescent E. Coli Bioparticles (green) and incubated with cell membrane marker.
The role of epigenetic regulation of MARCO in tolerant macrophages
TLR4 and TLR2 signaling pathways both lead to activation of the transcription factor NF-κB. We evaluated the localization of active NF-κB p65 in macrophages and quantified NF-κB localized in the nucleus in 200–300 cells in each condition, including no treatment, LPS stimulation, and LPS induced tolerance (Supplemental Figure 2). Interestingly, NF-κB p65 accumulated in nuclei of both nontolerant and tolerant macrophages, suggesting that loci-specific targeting of p65 rather than nuclear localization was altered in tolerant macrophages. Previous studies also demonstrated the role of histone modification in LPS induced tolerant cells (13, 19–21). Here, we ask whether epigenetic mechanisms also play a role in regulation of the expression of MARCO in tolerant and cross tolerant cells.
According to previous studies, H3K4me3 (permissive) and H3K9me2 (repressive) modifications play important roles in expression of proinflammatory cytokines (13, 20, 21). We performed a ChIP assay to test whether there is differential methylation of histones in the MARCO promoter between nontolerant and tolerant cells. As shown in Figure 5A, the promoter region (−255) of MARCO was specifically immunoprecipitated by H3K4me3 in tolerant and cross tolerant cells compared to nontolerant cells. In contrast, there were no differences observed in the H3K9me2 mark between tolerant and nontolerant cells (Supplemental Figure 3A).
Figure 5.
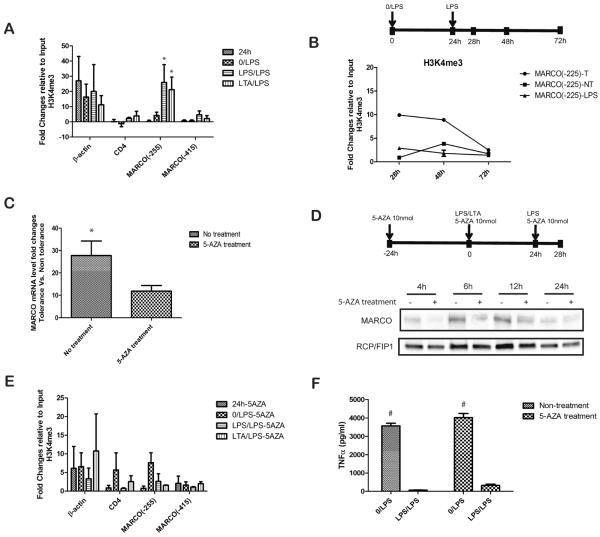
The histone modification (H3K4me3) in the promoter of MARCO is differentially regulated between T and NT cells. The schematic on top shows the treatment regimen for 5-AZA treatment experiments. Cells treated with 5-AZA (10 nmol; or no treatment control) 24h prior to first stimulation with LPS as well at first and second LPS stimulations. Cells were harvested 4h after second LPS (5EU) stimulation. (A) Sheared nuclear chromatins from T and NT macrophages were immunoprecipitated with anti-H3K4me3. Immunoprecipitated DNA and input DNA were amplified with specific primers for the promotor region of β-actin, CD4 and MARCO. qPCR data show that promoter of MARCO (−255) is differentially regulated between T and NT cells. (B) H3K4me3 histone modifications in the MARCO promoter dynamically change over time for MARCO (−255)-T, MARCO (−255)-NT, and MARCO (−255)-LPS macrophages. (C) The expression of MARCO is decreased after the demethylation treatment by 5-AZA. (D) The protein level of MARCO is decreased in 5-AZA treatment tolerant macrophages compared with no 5-AZA treatment tolerant macrophages. (E) Histone modification (H3K4me3) in the promoter of MARCO is not differentially regulated in tolerant and non-tolerant 5-AZA treated cells. (F) The tolerance was measured by ELISA testing of TNF-α in 5-AZA treatment and non-treatment cells. Data are representative of 3 or more independent experiments; shown are mean ± s.e.m from triplicate and quadruplicate values (*, p<0.05 A. compared with 0/LPS group; C. compared with 5-AZA treatment. F. # p<0.01 compared with LPS/LPS group).
To understand the dynamic process of H3K4me3 histone modification and whether this is the only specific permissive modification at MARCO promotor region, we observed H3K4me3 and H3K9Ac histone modifications at the MARCO gene promoter (−255) at different time-points in MARCO(−255)-T, MARCO(−255)-NT, and MARCO(−255)-LPS macrophages. MARCO(−255)-T macrophages were exposed to LPS at 0h and 24h, MARCO(−255)-NT macrophages were exposed to LPS at 24h only, and MARCO(−255)-LPS macrophages were exposed to LPS at 0h only (see diagram). At time points, 28h, 48h, and 72h, these three groups were analyzed by ChIP (H3K4me3 and H3K9Ac). MARCO(−255)-T macrophages had increased H3K4me3 histone modifications at 28h and 48h compared with MARCO(−255)-NT macrophages, but this modification had a decreased trend at 72h. MARCO(−255)-NT macrophages had fewer H3K4me3 histone modifications at 28h and an increased trend at 48h and 72h. MARCO(−255)-LPS macrophages had intermediate H3K4me3 histone modifications at 28h compared to NT and T macrophages and more steady at 48h and 72h (Figure 5B). H3K4me3 histone modification levels at the beta actin promoter are steady over time for all three groups of macrophages (Supplemental Figure 3B). Interestingly, H3K9Ac modification had mild changes and a similar trend compared to H3K4me3 modifications (Supplemental Figure 3B). These data suggest the dynamic nature of histone modifications during the tolerant process, which involves multiple permissive histone modifications.
We next considered DNA methylation in regulation of MARCO expression. We used 5-Aza-2′-deoxycytidine (5-AZA) treatment as shown in Figure 5. Interestingly, the extent of increased MARCO expression between tolerance and nontolerance analyzed by qPCR was significantly reduced in tolerant cells treated with 5-AZA (Figure 5C). The protein level of MARCO were also decreased in 5-AZA treatment tolerant macrophages compared with no 5-AZA treatment tolerant macrophages at the time point of 4h, 6h and 12h after second LPS stimulation. There are 12 CpG motifs located 2000 bp upstream of the MARCO transcription start site. Recent studies have shown 5-AZA play a role in reorganizing genomic histone modification patterns (22). It is possible that DNA methylation in combination with histone modification play a role in regulation of MARCO expression. Consistent with this, H3K4me3 modification at the promoter region of MARCO was significantly decreased after 5-AZA treatment (Figure 5E). These data suggest that the expression of MARCO in tolerized cells can be reduced by treatment with a demethylating agent. This is likely caused by inhibition of H3K4me3 which specifically regulates MARCO expression in tolerant cells. However, 5-AZA treatment did not change LPS-induced tolerance as measured by the expression of TNFα (Figure 5F). Thus, MARCO appears to be regulated by epigenetic mechanisms in innate immune tolerance, and the reduced expression of TNFα in innate immune tolerance appears to be controlled by other mechanisms. This is consistent with differentially expressed MARCO and TNFα following tolerance (Figure 1D).
MARCO contributes to increased phagocytosis in tolerant macrophages
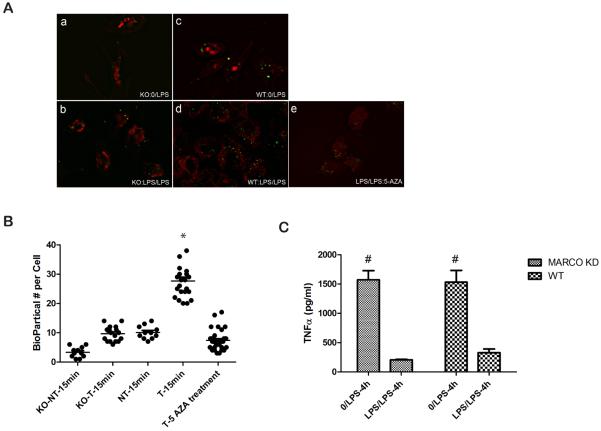
To determine whether elevated MARCO expression is responsible for increased phagocytosis in tolerant cells and whether MARCO is necessary for the establishment of tolerance, wild type C57BL6 (WT) and MARCO-deficient macrophages were analyzed for tolerance (Figure 6A a–e). To quantify internalized Escherichia coli BioParticles in the phagocytosis assay, the images were captured from top to bottom in 0.2μm distance and the BioParticles were counted in random cells in 3D images (Supplemental Figure 4). The average fluorescent signal of internalized BioParticles in WT macrophages were increased 3.4 times while it was increased 1.8 in MARCO-deficient macrophages (Fig 6B). Although the internalized BioParticles were increased in both WT and MARCO-deficient tolerant cells compared to nontolerant macrophages, WT cells were capable of far more phagocytosis than MARCO-deficient cells. However, MARCO-deficient macrophages are capable of inducing innate immune tolerance. TNFα (Figure 6C) were decreased in both WT and MARCO-deficient tolerant macrophages, and the extent of decrease was comparable. In addition, 5-AZA treatment reduced the phagocytosis in WT tolerant macrophages further suggesting that DNA methylation is involved in the regulation of phagocytosis (Figure 6B).
Figure 6.
MARCO contributes to the increased Phagocytosis in tolerance cells and does not affect the extent of tolerance. (Panel A a-d) NT and T cells derived from wild and MARCO−/−C57BL/6, (Panel A e) WT tolerant cell with 5-AZA treatment are fed with fluorescent E.Coli Bioparticles for 15 min and phagocytotic Bioparticles were counted in random cells (B). The tolerance was measured by ELISA testing of TNF-α in MARCO KO (C), Data are representative for 3 or more independent experiments; shown are mean ± s.e.m from triplicate values (B, * p<0.05 WT T-15 min compared with NT-15min and 5-AZA treated cells; C, # p<0.01compared with LPS/LPS group).
Discussion
Our findings demonstrate that expression of MARCO in both LPS- and LTA-induced tolerant macrophages is significantly increased, is regulated by H3K4me3, and partially contributes to increased phagocytosis. However, MARCO is not essential for inducing the inflammatory component of innate immune tolerance. Thus, we conclude that phagocytosis and inflammation associated with innate immune tolerance are differentially regulated.
The expression of MARCO is TLR4-dependent and mediated by MyD88 (23). It has been shown to be associated with the p38 mitogen-activated protein kinase (MAPK) pathway in dendritic cells (24). However, the regulatory mechanism of MARCO in tolerant macrophages remains unknown. Interestingly, both LPS and LTA signaling pathways include MAPK pathway, and MKP-1 has been proven to be the negative regulator of MAPK signaling pathway in tolerant cells (25, 26).
Our findings demonstrate that innate immune tolerance increases MARCO associated phagocytosis. The regulation of phagocytosis is involved in multiple levels including the detection of target by cell surface receptor, recruitment of associated cytoskeleton componentsand clearance in lysosomes. Cell surface receptors involved in phagocytosis include opsonin-dependent receptors (Fc receptors and complement receptors) and opsonin-independent receptors (scavenger receptors). In our study, we focused on MARCO in tolerant macrophages because of its significant upregulation in both LPS and LTA induced tolerance (Figure 1B). Many studies have reported the role of MARCO in clearance of bacteria in macrophages (27, 28), but not crucial for the survival of a LPS challenging model (23). This is suggested by increased anti-LPS antibodies and activation of marginal zone B cells in class A scavenger receptor deficient mice. Phagocytosis is also initiated by Toll-like receptors (29). The stimulation of macrophages by LPS is associated with increased phagocytosis and followed by activation of proinflammatory pathways (30–32). Recent studies showed that a blockade of class B scavenger receptors SR-A, SR-B, and CD36 was associated with reduced inflammation and increased phagocytosis(33). Here, we have shown the phagocytosis and proinflammatory activation in tolerant macrophages represent distinct biological processes (Figure 6). The increased phagocytosis may be due to the increased expression of MARCO. This is consistent with our finding (Figure 2) that TLR4 expression is decreased and MARCO is specifically increased in tolerant macrophage and with previous studies about the negative regulation of TLR4 signaling pathways (34, 35). Interestingly, MARCO mainly contributes to host defense by capture and clearance of invading microorganisms. These findings lead us to hypothesize that different mechanisms regulate the expression of TLR4 and MARCO in immune tolerance and account for the divergent responses in inflammation and phagocytosis.
Tolerance induced phagocytosis involving MARCO is regulated by epigenetic factors. Epigenetics play an important role in the regulation of immune cell functions in LPS induced tolerance (36). Consistent with this, our findings demonstrated that NF-κB is not functional and accumulates in the nucleus of both nontolerant and tolerant macrophages (Supplemental Figure 2). Many studies have shown that chromatin modification regulate specific genes in their promoter regions during this process (13, 20, 21, 37). Among these chromatin modifications, repressive histone modification H3K9me2 was increased in the promoter region of TNFα (21) and permissive histone modification H3K4me3 was lost at the promoter of IL6 in tolerant cells (13). Here, we demonstrated that H3K4me3 was involved in controlling the expression of MARCO (Figure 5A). This modification dynamically changes within the first 48 hours of stimulation with LPS (Figure 5B). H3k4me3 is not the only permissive modification at the promotor region of MARCO. Another modification (H3K9Ac) may be involved in this process (Supplemental Figure 3C), but this needs to be further explored. The dynamic histone modification changes is similar to MARCO mRNA level dynamic changes (Figure 2A) but not identical (gradually diminished histone modification between 48 and 72h instead of persistent mRNA level in this period) indicating multiple level regulation of MARCO expression. Beside histone modification, DNA methylation also contributes to the epigenetic regulation in tolerant cells (21). Effector proteins that are recruited by methylated DNA and histones can orchestrate the complexes that include chromatin structure and regulate specific gene expression (38, 39). Our experiments showed that 5-AZA treatment did change the extent of increased MARCO expression in tolerance macrophages (Figure 5C) and the H3K4me3 modification was also reversed in promoter region of MARCO (Figure 5D). Although this could be affected indirectly by other genes involved in MARCO regulation due to the global effect of 5-AZA, it is possible that changes in epigenetic patterns involved both DNA methylation and histone modification. It is also possible that gene expression can be regulated by DNA methylation in other regulatory elements (40). Although our findings indicate that H3K4me3 modifications and methylation are involved in controlling the expression of MARCO, further work is needed to understand the precise mechanisms.
In summary, the establishment of macrophage tolerance is associated with massive gene expression changes that modulate the effects of persistent pathogen stimulation. This process includes suppression of proinflammatory cytokines and upregulation of genes responsible for phagocytosis such as scavenger receptor MARCO. We show that MARCO expression is significantly elevated through histone modification in LPS and LTA induced tolerant macrophages and that increased expression of MARCO on the surface of macrophages is partially responsible for the enhanced phagocytosis in tolerant cells.
Supplementary Material
Acknowledgements
We thank Dr. Steven R. Kleeberger, Dr. Jacqui Marzec and Dr. S Jeyaseelan-Jey for providing materials. This work was supported in part by grant from R01-HL095393, P01-ES18181, and RC2-HL101715.
Footnotes
Disclosures The authors have no financial conflicts of interest.
References
- 1.Cavaillon JM, Adib-Conquy M. Bench-to-bedside review: endotoxin tolerance as a model of leukocyte reprogramming in sepsis. Crit Care. 2006;10:233. doi: 10.1186/cc5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biswas SK, Lopez-Collazo E. Endotoxin tolerance: new mechanisms, molecules and clinical significance. Trends in immunology. 2009;30:475–487. doi: 10.1016/j.it.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 3.del Fresno C, Garcia-Rio F, Gomez-Pina V, Soares-Schanoski A, Fernandez-Ruiz I, Jurado T, Kajiji T, Shu C, Marin E, Gutierrez del Arroyo A, Prados C, Arnalich F, Fuentes-Prior P, Biswas SK, Lopez-Collazo E. Potent phagocytic activity with impaired antigen presentation identifying lipopolysaccharide-tolerant human monocytes: demonstration in isolated monocytes from cystic fibrosis patients. J Immunol. 2009;182:6494–6507. doi: 10.4049/jimmunol.0803350. [DOI] [PubMed] [Google Scholar]
- 4.Ravetch JV, Bolland S. IgG Fc receptors. Annual review of immunology. 2001;19:275–290. doi: 10.1146/annurev.immunol.19.1.275. [DOI] [PubMed] [Google Scholar]
- 5.van Lookeren Campagne M, Wiesmann C, Brown EJ. Macrophage complement receptors and pathogen clearance. Cellular microbiology. 2007;9:2095–2102. doi: 10.1111/j.1462-5822.2007.00981.x. [DOI] [PubMed] [Google Scholar]
- 6.Palecanda A, Kobzik L. Receptors for unopsonized particles: the role of alveolar macrophage scavenger receptors. Current molecular medicine. 2001;1:589–595. doi: 10.2174/1566524013363384. [DOI] [PubMed] [Google Scholar]
- 7.Taylor PR, Martinez-Pomares L, Stacey M, Lin HH, Brown GD, Gordon S. Macrophage receptors and immune recognition. Annual review of immunology. 2005;23:901–944. doi: 10.1146/annurev.immunol.23.021704.115816. [DOI] [PubMed] [Google Scholar]
- 8.Arredouani MS, Palecanda A, Koziel H, Huang YC, Imrich A, Sulahian TH, Ning YY, Yang Z, Pikkarainen T, Sankala M, Vargas SO, Takeya M, Tryggvason K, Kobzik L. MARCO is the major binding receptor for unopsonized particles and bacteria on human alveolar macrophages. J Immunol. 2005;175:6058–6064. doi: 10.4049/jimmunol.175.9.6058. [DOI] [PubMed] [Google Scholar]
- 9.Hamilton RF, Jr., Thakur SA, Mayfair JK, Holian A. MARCO mediates silica uptake and toxicity in alveolar macrophages from C57BL/6 mice. The Journal of biological chemistry. 2006;281:34218–34226. doi: 10.1074/jbc.M605229200. [DOI] [PubMed] [Google Scholar]
- 10.Dahl M, Bauer AK, Arredouani M, Soininen R, Tryggvason K, Kleeberger SR, Kobzik L. Protection against inhaled oxidants through scavenging of oxidized lipids by macrophage receptors MARCO and SR-AI/II. The Journal of clinical investigation. 2007;117:757–764. doi: 10.1172/JCI29968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bowdish DM, Sakamoto K, Kim MJ, Kroos M, Mukhopadhyay S, Leifer CA, Tryggvason K, Gordon S, Russell DG. MARCO, TLR2, and CD14 are required for macrophage cytokine responses to mycobacterial trehalose dimycolate and Mycobacterium tuberculosis. PLoS pathogens. 2009;5:e1000474. doi: 10.1371/journal.ppat.1000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jozefowski S, Sulahian TH, Arredouani M, Kobzik L. Role of scavenger receptor MARCO in macrophage responses to CpG oligodeoxynucleotides. Journal of leukocyte biology. 2006;80:870–879. doi: 10.1189/jlb.0705357. [DOI] [PubMed] [Google Scholar]
- 13.Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- 14.Sato S, Nomura F, Kawai T, Takeuchi O, Muhlradt PF, Takeda K, Akira S. Synergy and cross-tolerance between toll-like receptor (TLR) 2- and TLR4-mediated signaling pathways. J Immunol. 2000;165:7096–7101. doi: 10.4049/jimmunol.165.12.7096. [DOI] [PubMed] [Google Scholar]
- 15.Buckley JM, Wang JH, Redmond HP. Cellular reprogramming by gram-positive bacterial components: a review. Journal of leukocyte biology. 2006;80:731–741. doi: 10.1189/jlb.0506312. [DOI] [PubMed] [Google Scholar]
- 16.Jacinto R, Hartung T, McCall C, Li L. Lipopolysaccharide- and lipoteichoic acid-induced tolerance and cross-tolerance: distinct alterations in IL-1 receptor-associated kinase. J Immunol. 2002;168:6136–6141. doi: 10.4049/jimmunol.168.12.6136. [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Zhong B, Ma HY, Yang Q, Gu FR, Yin GQ, Xia CM. Decrease in toll-like receptors 2 and 4 in the spleen of mouse with endotoxic tolerance. Inflammation research : official journal of the European Histamine Research Society … [et al.] 2008;57:252–259. doi: 10.1007/s00011-007-7104-4. [DOI] [PubMed] [Google Scholar]
- 19.Chen X, El Gazzar M, Yoza BK, McCall CE. The NF-kappaB factor RelB and histone H3 lysine methyltransferase G9a directly interact to generate epigenetic silencing in endotoxin tolerance. The Journal of biological chemistry. 2009;284:27857–27865. doi: 10.1074/jbc.M109.000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan C, Li L, McCall CE, Yoza BK. Endotoxin tolerance disrupts chromatin remodeling and NF-kappaB transactivation at the IL-1beta promoter. J Immunol. 2005;175:461–468. doi: 10.4049/jimmunol.175.1.461. [DOI] [PubMed] [Google Scholar]
- 21.El Gazzar M, Yoza BK, Chen X, Hu J, Hawkins GA, McCall CE. G9a and HP1 couple histone and DNA methylation to TNFalpha transcription silencing during endotoxin tolerance. The Journal of biological chemistry. 2008;283:32198–32208. doi: 10.1074/jbc.M803446200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komashko VM, Farnham PJ. 5-azacytidine treatment reorganizes genomic histone modification patterns. Epigenetics : official journal of the DNA Methylation Society. 2010;5 doi: 10.4161/epi.5.3.11409. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y, Wermeling F, Sundqvist J, Jonsson AB, Tryggvason K, Pikkarainen T, Karlsson MC. A regulatory role for macrophage class A scavenger receptors in TLR4-mediated LPS responses. European journal of immunology. 2010;40:1451–1460. doi: 10.1002/eji.200939891. [DOI] [PubMed] [Google Scholar]
- 24.Matsushita N, Komine H, Grolleau-Julius A, Pilon-Thomas S, Mule JJ. Targeting MARCO can lead to enhanced dendritic cell motility and anti-melanoma activity. Cancer immunology, immunotherapy : CII. 2010;59:875–884. doi: 10.1007/s00262-009-0813-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nimah M, Zhao B, Denenberg AG, Bueno O, Molkentin J, Wong HR, Shanley TP. Contribution of MKP-1 regulation of p38 to endotoxin tolerance. Shock. 2005;23:80–87. doi: 10.1097/01.shk.0000145206.28812.60. [DOI] [PubMed] [Google Scholar]
- 26.West AP, Koblansky AA, Ghosh S. Recognition and signaling by toll-like receptors. Annual review of cell and developmental biology. 2006;22:409–437. doi: 10.1146/annurev.cellbio.21.122303.115827. [DOI] [PubMed] [Google Scholar]
- 27.Elomaa O, Kangas M, Sahlberg C, Tuukkanen J, Sormunen R, Liakka A, Thesleff I, Kraal G, Tryggvason K. Cloning of a novel bacteria-binding receptor structurally related to scavenger receptors and expressed in a subset of macrophages. Cell. 1995;80:603–609. doi: 10.1016/0092-8674(95)90514-6. [DOI] [PubMed] [Google Scholar]
- 28.Arredouani M, Yang Z, Ning Y, Qin G, Soininen R, Tryggvason K, Kobzik L. The scavenger receptor MARCO is required for lung defense against pneumococcal pneumonia and inhaled particles. The Journal of experimental medicine. 2004;200:267–272. doi: 10.1084/jem.20040731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.West MA, Wallin RP, Matthews SP, Svensson HG, Zaru R, Ljunggren HG, Prescott AR, Watts C. Enhanced dendritic cell antigen capture via toll-like receptor-induced actin remodeling. Science. 2004;305:1153–1157. doi: 10.1126/science.1099153. [DOI] [PubMed] [Google Scholar]
- 30.Underhill DM, Goodridge HS. Information processing during phagocytosis. Nature reviews. Immunology. 2012;12:492–502. doi: 10.1038/nri3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swanson JA. Shaping cups into phagosomes and macropinosomes. Nature reviews. Molecular cell biology. 2008;9:639–649. doi: 10.1038/nrm2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alpuche-Aranda CM, Racoosin EL, Swanson JA, Miller SI. Salmonella stimulate macrophage macropinocytosis and persist within spacious phagosomes. The Journal of experimental medicine. 1994;179:601–608. doi: 10.1084/jem.179.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leelahavanichkul A, Bocharov AV, Kurlander R, Baranova IN, Vishnyakova TG, Souza AC, Hu X, Doi K, Vaisman B, Amar M, Sviridov D, Chen Z, Remaley AT, Csako G, Patterson AP, Yuen PS, Star RA, Eggerman TL. Class B scavenger receptor types I and II and CD36 targeting improves sepsis survival and acute outcomes in mice. J Immunol. 2012;188:2749–2758. doi: 10.4049/jimmunol.1003445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scott MJ, Liu S, Shapiro RA, Vodovotz Y, Billiar TR. Endotoxin uptake in mouse liver is blocked by endotoxin pretreatment through a suppressor of cytokine signaling-1-dependent mechanism. Hepatology. 2009;49:1695–1708. doi: 10.1002/hep.22839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quinn EM, Wang J, Redmond HP. The emerging role of microRNA in regulation of endotoxin tolerance. Journal of leukocyte biology. 2012;91:721–727. doi: 10.1189/jlb.1111571. [DOI] [PubMed] [Google Scholar]
- 36.Carson WF, Cavassani KA, Dou Y, Kunkel SL. Epigenetic regulation of immune cell functions during post-septic immunosuppression. Epigenetics : official journal of the DNA Methylation Society. 2011;6:273–283. doi: 10.4161/epi.6.3.14017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.El Gazzar M, Yoza BK, Chen X, Garcia BA, Young NL, McCall CE. Chromatin-specific remodeling by HMGB1 and linker histone H1 silences proinflammatory genes during endotoxin tolerance. Molecular and cellular biology. 2009;29:1959–1971. doi: 10.1128/MCB.01862-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010;143:470–484. doi: 10.1016/j.cell.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sasai N, Defossez PA. Many paths to one goal? The proteins that recognize methylated DNA in eukaryotes. The International journal of developmental biology. 2009;53:323–334. doi: 10.1387/ijdb.082652ns. [DOI] [PubMed] [Google Scholar]
- 40.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nature reviews. Genetics. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.