Summary
Revolutionary new technologies, namely in the areas of DNA sequencing and molecular imaging, continue to impact new discoveries in plant science and beyond. For decades we have been able to determine properties of enzymes, receptors and transporters in vitro or in heterologous systems, and more recently been able to analyze their regulation at the transcriptional level, use GFP reporters to obtain insights into cellular and subcellular localization, and measure ion and metabolite levels with unprecedented precision using mass spectrometry. However, we lack key information on location and dynamics of the substrates of enzymes, receptors and transporters, and on the regulation of these proteins in their cellular environment. Such information can now be obtained by transitioning from in vitro to in vivo biochemistry using biosensors. Genetically encoded fluorescent protein-based sensors for ion and metabolite dynamics provide highly resolved spatial and temporal information, and are complemented by sensors for pH, redox, voltage, and tension. They serve as powerful tools for identifying missing processes (e.g. glucose transport across ER membranes), components (e.g. SWEET sugar transporters for cellular sugar efflux), and signaling networks (e.g. from systematic screening of mutants that affect sugar transport or cytosolic and vacuolar pH). Combined with the knowledge of properties of enzymes and transporters and their interactions with the regulatory machinery, biosensors promise to be key diagnostic tools for systems and synthetic biology.
Keywords: metabolic signaling, metabolic pathways, sugar signaling, imaging
Introduction into genetically encoded small molecule biosensors
To understand a biological process or system, we need to know the components of that process, the role of each component, the physical properties and chemical conditions, and the dynamic features such as fluxes and enzymatic activities. Biochemical and biophysical methods have brought us close to complete resolution of components and conditions, but unveiling their dynamics in living cells still poses a great challenge. To measure a dynamic process under physiological conditions we need means to visualize the actual process, e.g. the translocation of molecules, the conversion of one metabolite into another, or the triggering of signaling events. This visualization can be achieved by sensors that respond dynamically, e.g. to changing concentrations of a particular molecule or to modifications or structural changes of involved enzymes and complexes. As some of the first sensors in biology, pH indicators were used to visualize the activities of proton pumps [1]. Later, fluorescent sensor molecules were designed that could be injected into cells to measure levels and periodicity of calcium [2,3]. However, the way of applying such sensors by injection or other forms of membrane penetration posed a major risk of artifacts. Being minimally invasive is a critical requirement for sensors to ensure that the measured process is mostly unperturbed by the measurement itself.
The discovery, characterization and engineering of fluorescent proteins (i.e. green fluorescent protein, GFP) has played a key role in allowing many minimally invasive measurements, particularly the analysis of the localization and dynamics of proteins in live cells as well as for the development of genetically encoded small molecule biosensors [4,5]. Roger Tsien’s lab developed the first prototype protein sensors, i.e. to measure caspase activity and to monitor calcium in live cells [for review see 5]. These sensors made use of Förster resonance energy transfer (FRET) between two spectral variants of GFP; shortly afterwards single fluorophore sensors based on circular permutated GFPs (cpGFPs) were developed, e.g. for calcium [6,7]. The biosensor tool set has been vastly expanded to detect sugars, amino acids, phosphate, as well as many ions and signaling intermediates (http://dpb.carnegiescience.edu/labs/frommer-lab/biosensor_database). Key features of genetically encoded biosensors are: (i) universal utility, i.e. transient or stable introduction into most organisms by transfection, transformation or protein transduction [for detailed protocols in yeast, plants and human cells see 8,9–11]; their ability to report (ii) steady state levels of ions and metabolites, (iii) dynamics with high temporal resolution, and (iv) with cellular and even subcellular information. They can be used in complex solutions and cell cultures as well as in tissues and intact organisms. Despite recent progress, many key compounds await sensors for high-resolution monitoring in vivo. In plant biology, the dynamics of phytohormones represent a highly attractive target for such monitoring, although some progress has been made using lower-resolution sensors, e.g. for indirect imaging of plant hormones. Reporters of either hormone responsive gene expression, such as DR5-GFP, or hormone dependent protein degradation, such as IAA28 DII-Venus have proven highly valuable tools [12,13]. Nevertheless, spatial and temporal dynamics of the full spectrum of phytohormones would ideally be monitored by FRET hormone sensors.
Sensor design
Small molecule fluorescent biosensors make use of conformational changes in a recognition element induced by the analytic target (analyte) as proxies for analyte concentrations. There are several different types of sensor designs and several classes of analytes with available sensors [5]. In designing a sensor for a new analyte, the selection or creation of a recognition element is the primary parameter. Recognition elements are typically chosen from proteins or protein domains that are known to directly bind or be modified by a specific analyte. The ligand recognition domains of receptor proteins are good sources for new recognition elements, with the periplasmic binding proteins of bacterial ABC transport systems proving an especially rich source for metabolite recognition elements. A variant of this approach makes use of the interaction of two domains that come together when an analyte is present. Other variants include use of a molecular spring recognition element to measure tension and co-opting intrinsic fluorophore properties to detect changes in environmental conditions such as pH or voltage.
In the simplest case, two fluorophores are fused to the recognition element at either terminus and/or internal sites to generate a FRET sensor. Because the design of new sensors is a largely empirical process, structural information for recognition elements is desirable but not an absolute requirement. These sensors are ratiometric in that they report on cognate analytes through changes in the relative intensities of the two fluorophore emissions, and thus require appropriate fluorescence detection/imaging equipment [for details see 11,14*]. The list of fluorescent proteins that form FRET pairs is ever growing, leading to a wealth of options for designing new sensors with appropriate biophysical properties (e.g. pH optimum, emission wavelength) for the intended application (http://flowcyt.salk.edu/fluo.html). Ratiometric sensors can also be generated from a single fluorophore with multiple excitation or emission maxima (e.g. the HyPer sensor has two excitation maxima, the relative strengths of which are altered by hydrogen peroxide [15]). Alternatively, single fluorophores whose fluorescence has been engineered to vary in proportion to changes in an analyte can be used to generate intensity-based sensors. Although easier to image, non-ratiometric single fluorophore intensity changes are more susceptible to artifacts resulting from variation in sensor protein level or optical parameters of the sample (background fluorescence, light scattering). Additional important parameters for any sensor include specificity, brightness, signal-to-noise ratio and a dynamic range that matches the concentrations of analyte to be examined. Typically, the dynamic range of a sensor is ~2 orders of magnitude, which means multiple sensors with different affinities are necessary if large changes in analyte concentration are to be tracked. However, the dynamic range of a recently developed lactate sensor exceeds four orders of magnitude owing to the kinetics of the recognition element [16]. Testing of such parameters is required to evaluate the effectiveness of a new sensor and to optimize existing sensors. Many of the requisite tests are best carried out on purified sensor in vitro (e.g. after expression in and purification from an E. coli host system), but in vivo evaluation and optimization can also be necessary to minimize interaction with off-target endogenous components and detrimental buffering of the analyte caused by the presence of the sensor, and to improve expression and/or stability of the sensor protein and signal-to-noise ratio in the cellular environment and imaging platform. For example, silencing of biosensor transgenes has been a recurring setback for in planta applications, but recent evidence suggests that the use of the UBQ10 promoter can greatly improve biosensor expression levels [17*].
Biosensors are complementary to both surveying ions and metabolites using mass spectrometry (ionomics and metabolomics, [18,19]) and fluxomics (an approach for estimating metabolic flux using stable isotope labeling [20]). Furthermore, biosensors can also be used in the context of other investigations, e.g. in genetic screens (see below). Yet there are still many areas in which biosensor potential has not fully been developed, and the continuing optimization of fluorophores and sensors is expected to lead to further improvements in detection range and sensitivity, as well as more options for multiplexing. Exploiting the full potential of biosensors will require accelerated methods of protein engineering (e.g. using Gateway, Gibson assembly or Golden Gate cloning; see also [21](In-Fusion® cloning, Clontech) and expression because all parts of a sensor protein (fluorophore type and site of fusion, recognition elements, linkers, etc.) are potential targets for rational or empirical modification and even small changes in any of these parts can have large impacts on sensor behavior [22,23*]. The development of plant hormone sensors is in principle feasible using recent progress in identification of hormone receptors [24]. Development of such sensors is expected to provide a significant step forward.
Quantifying and visualizing molecules over time at the cellular and subcellular level
Advancing technologies have long driven major progress in determining – at cellular and even subcellular resolution - gene expression levels [25], ion and metal distributions [26,27], membrane composition [e.g. using nanoSIMS, 28], or metabolites [29]. High spatial resolution measurements can reveal patterns that lead to the discovery of novel physiological processes [30]. Yet subcellular compartments are often not accessible to current methods. For example, organelles such as the endoplasmic reticulum can be biochemically enriched using differential centrifugation gradients, but the isolated organelle is then out of its native cellular context. Thus, we require tools for measuring with high resolution in native environments under physiological conditions.
Genetically encoded fluorescent protein based sensors can be targeted to subcellular compartments to reveal subcellular patterns (Figure 1). For example, glucose produced from G6P in the ER lumen was measured by targeting glucose sensors to the ER [22]. The data from these sensors indicate that the residence time of GLUT glucose uniporters during their transit via the ER to the plasma membrane is sufficient to provide high capacity ER import and export of glucose [31]. Alternatively, biosensors can be targeted to the cell surface, e.g. to monitor glutamate release triggered by depolarization of hippocampal neurons [32]. Subcellular analysis of calcium dynamics in plant cells has been facilitated by Cameleon biosensors [33*], and an array of Cameleons targeted to different subcellular compartments is now available [17*]. In addition to subcellular spatial resolution, biosensors combined with appropriate imaging platforms permit time-course analysis with near real-time temporal resolution. Rapid dynamics of analytes are only revealed at such higher resolutions. Plant growth techniques that allow for continuous imaging under controllable environmental conditions also facilitate advanced study of analyte dynamics over time (Figure 2).
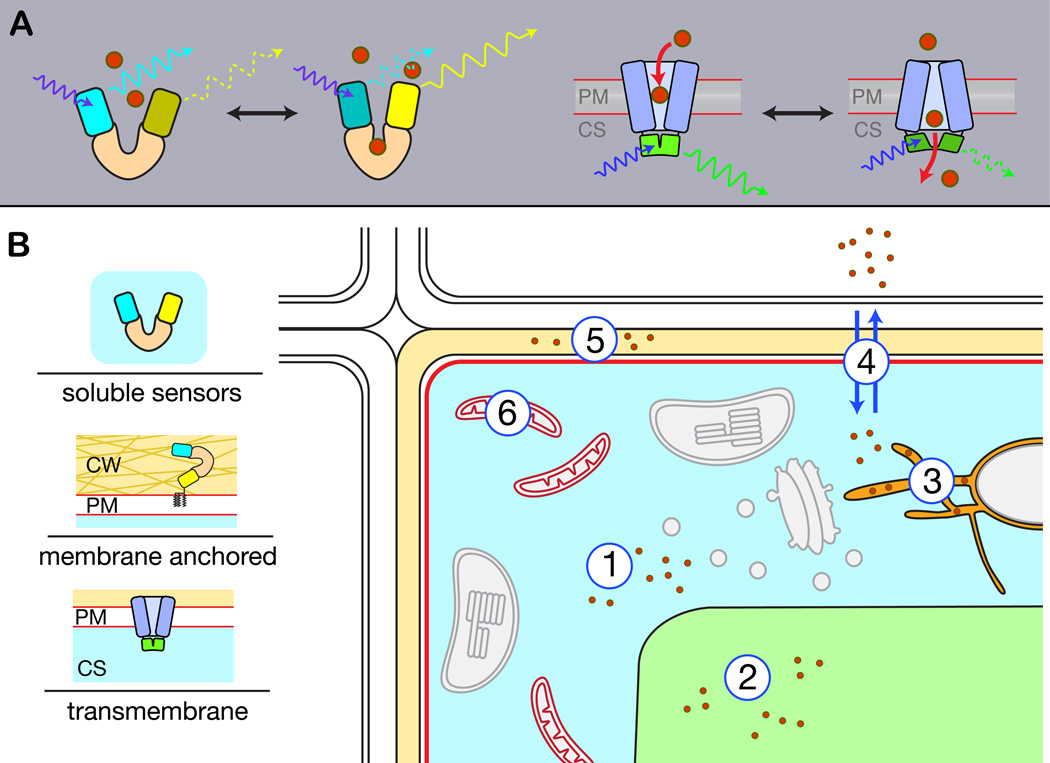
Figure 1. Biosensor design principles and their targeting to subcellular compartments.
A. Left: The recognition element (orange) is fused to two spectrally overlapping fluorophores (SWL short wavelength fluorophore, e.g. cyan version of GFP (cyan barrel); LWL long wavelength fluorophore, yellow version of GFP (yellow barrel). Binding of ligand (red) results in a conformational change of the recognition element and a change in orientation and/or distance of the two fluorophores, altering FRET efficiency. Right: cpGFP is inserted into the recognition element (here: transmembrane protein, blue). Conformational changes due to transmembrane protein activity are translated into signal intensity changes of cpGFP. B. Left: Example diagrams of soluble, membrane anchored and transmembrane sensors. Right: Genetically encoded fluorescent protein based sensors can be targeted to subcellular compartments to reveal local substrate level changes or enzyme activity. Cytosol (1): untargeted, nucleoplasm with nuclear localization signal (NLS), or extranuclear cytoplasm with nuclear export signal (NES). ER (2): ER targeting sequence, retain with ER retention signal (KDEL). Vacuole (3): lumen with vacuolar signal sequence, vacuole with targeting peptide. Apoplasm (4): secrete with export sequence. PM (5): display on surface using membrane anchor, e.g. platelet-derived growth factor (PDGF) receptor transmembrane (TM) domain; membrane integral with sensing domain cytosolic. Mitochondria (6): targeting with mitochondrial signal sequence.
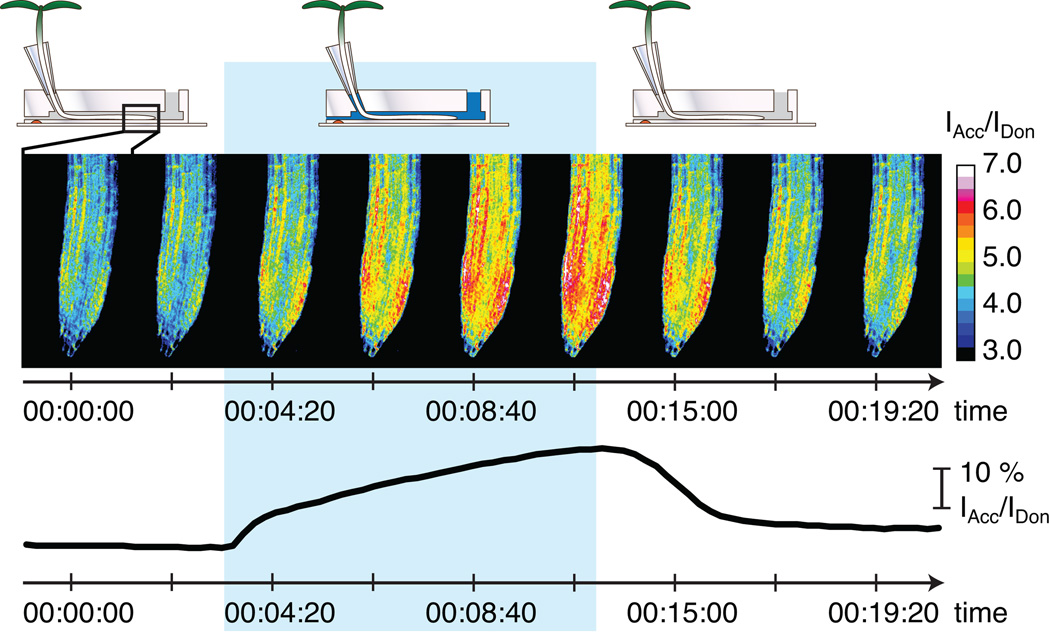
Figure 2. Time course analysis of the response of a fluorescent biosensor for glucose (FLII12P) in a RootChip.
Upon perfusion of a root with ligand solution (blue background) the cytosolic sensor reports intracellular increase in ligand concentration by a changed ratio of donor and acceptor intensities (here: increase in ratio I(Acc)/I(Don); for details see [10, 46**].
In vivo application of biosensors
High resolution measurements can open up previously undetectable patterns to primary observation, which can lead to new understanding of plant physiological processes. For example, the early discovery of differential calcium accumulation in different leaf cell types [26] led to functional models of calcium flux and regulation through the leaf [30]. The spatial (sub-cellular) and temporal (real-time) resolution afforded by biosensors has similarly led to deeper understanding of processes like the calcium oscillations required for symbiosis signaling in root hair cells. Initial observations that calcium oscillations in root hair cells were concentrated in the perinuclear region [34] were reproduced using Cameleon calcium sensors [33*]. However, the downstream signal transducer calmodulin-dependent protein kinase (CCaMK) is nuclear-localized [35,36], suggesting that nucleo-cytoplasmic calcium oscillations, rather than perinuclear calcium oscillations, are involved in the symbiotic signaling pathway of legumes. The presence on the inner nuclear membrane of two proteins required for symbiotic calcium oscillations - the cation channel DMI1 and the sarco/endoplasmic reticulum calcium ATPase SERCA – provides further evidence that nucleo-cytoplasmic calcium oscillations are important in legume symbiosis signaling [37**]. Observation of calcium oscillations in the nucleus was made possible through the use of nuclear localized Cameleon [38], and biosensor imaging combined with flux modeling indicates that calcium is released from the inner nuclear membrane [37**]. Thus, sub-cellular monitoring of a regulator (calcium oscillations) provided key support for a more detailed model of how that regulator controls a process (symbiosis signaling). High temporal resolution imaging of calcium oscillations using Cameleon sensors also led to important insights. For example, the finding that number of calcium spikes is more important than duration of spiking for symbiosis signaling was arrived at using time-course analysis of plants expressing Cameleon [33*]. Cameleon biosensors have been widely adopted for in vivo analysis and have led to insights into the role of calcium in pollen tube growth [39], root mechano-sensing [40], guard cell regulation by ABA [41,42] and JA [43], and root response to treatment with aluminum [44]. Furthermore, metabolite biosensors have revealed in vivo metabolite dynamics and transport in intact roots [45,46**]. Additional sensors have been applied to measuring redox status, reactive oxygen species and pH in planta [for review, see 47**]. However, the majority of available biosensors (http://dpb.carnegiescience.edu/labs/frommer-lab/biosensor_database) have yet to be applied to in planta studies.
Biosensors for gene discovery
Biosensors can be used to identify missing components related to the metabolism, regulation, or transport of the analyte, either in a heterologous system or in the native environment. For example, a FRET sensor for sucrose led to the identification of proteins that carry out a previously uncharacterized transport step in phloem loading – sucrose efflux from the mesophyll. Sucrose is made in mesophyll cytosol, but imported into the cytosol of phloem companion cells by a proton sucrose cotransporter of the SUT family. Thus, efflux from the mesophyll is required. A human cell line that expresses a sucrose sensor but does not import sucrose was used to screen heterologously expressed Arabidopsis proteins for sucrose transport activity. An analogous approach identified AtSWEET1, a glucose uniporter and the first member of the SWEET family of transporters [48*]. The screen for sucrose transporters led to the characterization of AtSWEET11 and AtSWEET12 as sucrose uniporters involved in net sucrose efflux from the mesophyll [49*]. When expressed in a native cell environment, a biosensor can be used to systematically identify components and processes that affect the analyte. Fluorimeter-based assays with FRET sugar sensors were successfully used to identify sugar transporters that can function immediately after exposure of starved yeast cells to glucose [50,51*]. Similar assays were successfully deployed to identify genes that affect cytosolic or vacuolar pH in yeast [52,53]. These results prove that biosensors can be widely applied in genetic screens provided imaging technologies of suitable throughput are available.
Future applications of biosensors
Today it is possible to track gene expression at the cellular level using cell-type specific sorting of protoplasts [54] or from RNA in affinity purified nuclei or ribosomes [25,55], protein abundance and localization with subcellular resolution using GFP fusions, and small molecule/metabolite dynamics with subcellular resolution using biosensors. Some biosensors can also track other protein dynamics such as phosphorylation events or receptor activation [for review see 5]. In the future, we anticipate biosensors to track these dynamics with even greater spatial and temporal resolution, and even to report directly on critical biophysical mechanisms such as enzyme or transporter activities. A biosensor approaching such direct reporting of transport activity tracks pH changes at the site of Cl−/HCO3− exchangers [56], but an ideal tool would detect conformational changes during the transport cycle. In addition to reporting on fine scale activities, current and future biosensors will be able to readily report with high resolution on dynamic molecules and processes that regulate fundamental check points in cell control. Biosensor-facilitated measurement of such dynamics (e.g. calcium oscillations) will permit the dissection of their regulation and co-regulation by multiple signals and inputs. The development of integrated imaging platforms like the RootChip presents a first step towards enabling technologies for integrated and high-resolution analysis of various inputs on Arabidopsis lines that express biosensors [46**], possibly even in medium to high throughput. Future directions include new applications of biosensors already expressed in plants (e.g. metabolite tracking at sites of nutrient exchange), new applications of biosensors that have not yet been expressed in plants (e.g. cell-cycle control measurement with FUCCI [57], and new applications of sensors yet to be developed (e.g. mapping phytohormone dynamics with biosensors). We would like to note that in addition to protein-based sensors described here, there are many other approaches, e.g. RNA-based sensors like Spinach and aptamer sensors as well as a suite of non-genetically encoded chemical dyes that can be used as complementary tools for monitoring ions and metabolites in vivo [58**].
Highlights.
Biosensors provide information on location and dynamics of substrates of enzymes and transporters
Biosensors provide information on regulation of enzymes/transporters in the native environment
Biosensors provide highly resolved spatial and temporal information on ions and metabolites
Biosensors help identifying unknown processes and molecular components, and signaling networks
Acknowledgements
This work was made possible by an EAGER grant from the National Science Foundation (NSF IOS-1045181) and an EMBO long-term fellowship (GG).
Abbrev
- FLIP
fluorescent indicator protein
- FRET
Förster resonance energy transfer
- GFP
green fluorescent protein
- eCFP
enhanced cyan fluorescent protein
- eYFP
enhanced yellow fluorescent protein
- LWL
long wavelength fluorophore
- SWL
short wavelength fluorophore
- cpGFP
circular permutated GFP
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lucas WJ, Smith FA. The formation of alkaline and acid regions at the surface of Chara corallina cells. J Exp Bot. 1973;24:1–14. [Google Scholar]
- 2.Blinks JR, Wier WG, Hess P, Prendergast FG. Measurement of Ca2+ concentrations in living cells. Prog Biophys Mol Biol. 1982;40:1–114. doi: 10.1016/0079-6107(82)90011-6. [DOI] [PubMed] [Google Scholar]
- 3.Grynkiewicz G, Poenie M, Tsien RY. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J Biol Chem. 1985;260:3440–3450. [PubMed] [Google Scholar]
- 4.Miyawaki A. Development of probes for cellular functions using fluorescent proteins and fluorescence resonance energy transfer. Annu Rev Biochem. 2011;80:357–373. doi: 10.1146/annurev-biochem-072909-094736. [DOI] [PubMed] [Google Scholar]
- 5.Okumoto S. Quantitative imaging using genetically encoded sensors for small molecules in plants. Plant J. 2012;70:108–117. doi: 10.1111/j.1365-313X.2012.04910.x. [DOI] [PubMed] [Google Scholar]
- 6.Topell S, Glockshuber R. Circular permutation of the green fluorescent protein. Methods Mol Biol. 2002;183:31–48. doi: 10.1385/1-59259-280-5:031. [DOI] [PubMed] [Google Scholar]
- 7.Tian L, Hires SA, Looger LL. Imaging neuronal activity with genetically encoded calcium indicators. Cold Spring Harb Protoc. 2012;2012:647–656. doi: 10.1101/pdb.top069609. [DOI] [PubMed] [Google Scholar]
- 8.Hou BH, Takanaga H, Grossmann G, Chen LQ, Qu XQ, Jones AM, Lalonde S, Schweissgut O, Wiechert W, Frommer WB. Optical sensors for monitoring dynamic changes of intracellular metabolite levels in mammalian cells. Nat Protoc. 2011;6:1818–1833. doi: 10.1038/nprot.2011.392. [DOI] [PubMed] [Google Scholar]
- 9.Bermejo C, Haerizadeh F, Takanaga H, Chermak D, Frommer WB. Optical sensors for measuring dynamic changes of cytosolic metabolite levels in yeast. Nat Protoc. 2011;6:1806–1817. doi: 10.1038/nprot.2011.391. [DOI] [PubMed] [Google Scholar]
- 10.Grossmann G, Meier M, Cartwright HN, Sosso D, Quake SR, Ehrhardt DW, Frommer WB. Time-lapse fluorescence imaging of Arabidopsis root growth with rapid manipulation of the root environment using the RootChip. J Vis Exp. 2012;7:65. doi: 10.3791/4290. e4290 10.3791/4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okumoto S. Imaging approach for monitoring cellular metabolites and ions using genetically encoded biosensors. Curr Opin Biotechnol. 2010;21:45–54. doi: 10.1016/j.copbio.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pagnussat GC, Alandete-Saez M, Bowman JL, Sundaresan V. Auxin-dependent patterning and gamete specification in the Arabidopsis female gametophyte. Science. 2009;324:1684–1689. doi: 10.1126/science.1167324. [DOI] [PubMed] [Google Scholar]
- 13.Brunoud G, Wells DM, Oliva M, Larrieu A, Mirabet V, Burrow AH, Beeckman T, Kepinski S, Traas J, Bennett MJ, et al. A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature. 2012;482:103–106. doi: 10.1038/nature10791. [DOI] [PubMed] [Google Scholar]
- 14. Okumoto S, Jones A, Frommer WB. Quantitative imaging with fluorescent biosensors. Annu Rev Plant Biol. 2012;63:663–706. doi: 10.1146/annurev-arplant-042110-103745. Comprehensive review of biosensor design and application.
- 15.Belousov VV, Fradkov AF, Lukyanov KA, Staroverov DB, Shakhbazov KS, Terskikh AV, Lukyanov S. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat Methods. 2006;3:281–286. doi: 10.1038/nmeth866. [DOI] [PubMed] [Google Scholar]
- 16.San Marten A, Gutierrez R, Ceballo S, Ruminot I, Lerchundi R, Baeza-Lehnert F, Frommer WB, Barros FL. A genetically encoded FRET lactate sensor and its use to detect the Warburg effect in single cancer cells. PLoS ONE. 2013 doi: 10.1371/journal.pone.0057712. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Krebs M, Held K, Binder A, Hashimoto K, Den Herder G, Parniske M, Kudla J, Schumacher K. FRET-based genetically encoded sensors allow high-resolution live cell imaging of Ca2+ dynamics. Plant J. 2012;69:181–192. doi: 10.1111/j.1365-313X.2011.04780.x. Development of an improved set of calcium sensor plant lines.
- 18.Yu D, Danku JM, Baxter I, Kim S, Vatamaniuk OK, Vitek O, Ouzzani M, Salt DE. High-resolution genome-wide scan of genes, gene-networks and cellular systems impacting the yeast ionome. BMC Genom. 2012;13:623. doi: 10.1186/1471-2164-13-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caldana C, Degenkolbe T, Cuadros-Inostroza A, Klie S, Sulpice R, Leisse A, Steinhauser D, Fernie AR, Willmitzer L, Hannah MA. High-density kinetic analysis of the metabolomic and transcriptomic response of Arabidopsis to eight environmental conditions. Plant J. 2011;67:869–884. doi: 10.1111/j.1365-313X.2011.04640.x. [DOI] [PubMed] [Google Scholar]
- 20.O'Grady J, Schwender J, Shachar-Hill Y, Morgan JA. Metabolic cartography: experimental quantification of metabolic fluxes from isotopic labelling studies. J Exp Bot. 2012;63:2293–2308. doi: 10.1093/jxb/ers032. [DOI] [PubMed] [Google Scholar]
- 21.Bourdes A, Rudder S, East AK, Poole PS. Mining the Sinorhizobium meliloti transportome to develop FRET biosensors for sugars, dicarboxylates and cyclic polyols. PLoS ONE. 2012;7:e43578. doi: 10.1371/journal.pone.0043578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takanaga H, Chaudhuri B, Frommer WB. GLUT1 and GLUT9 as major contributors to glucose influx in HepG2 cells identified by a high sensitivity intramolecular FRET glucose sensor. Biochim Biophys Acta. 2008;1778:1091–1099. doi: 10.1016/j.bbamem.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hires SA, Zhu Y, Tsien RY. Optical measurement of synaptic glutamate spillover and reuptake by linker optimized glutamate-sensitive fluorescent reporters. Proc Natl Acad Sci USA. 2008;105:4411–4416. doi: 10.1073/pnas.0712008105. Example illustrating the empirical nature of biosensor development and optimization.
- 24.Shan X, Yan J, Xie D. Comparison of phytohormone signaling mechanisms. Curr Opin Plant Biol. 2012;15:84–91. doi: 10.1016/j.pbi.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 25.Mustroph A, Zanetti ME, Jang CJ, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey-Serres J. Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:18843–18848. doi: 10.1073/pnas.0906131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leigh R, Tomos A. Ion distribution in cereal leaves - pathways and mechanisms. Phil. Transact Royal Soc Biol Sci. 1993;341:75–86. [Google Scholar]
- 27.Donner E, Punshon T, Guerinot ML, Lombi E. Functional characterisation of metal(loid) processes in planta through the integration of synchrotron techniques and plant molecular biology. Anal Bioanal Chem. 2012;402:3287–3298. doi: 10.1007/s00216-011-5624-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore KL, Lombi E, Zhao FJ, Grovenor CR. Elemental imaging at the nanoscale: NanoSIMS and complementary techniques for element localisation in plants. Anal Bioanal Chem. 2012;402:3263–3273. doi: 10.1007/s00216-011-5484-3. [DOI] [PubMed] [Google Scholar]
- 29.Lee YJ, Perdian DC, Song Z, Yeung ES, Nikolau BJ. Use of mass spectrometry for imaging metabolites in plants. Plant J. 2012;70:81–95. doi: 10.1111/j.1365-313X.2012.04899.x. [DOI] [PubMed] [Google Scholar]
- 30.Conn SJ, Gilliham M, Athman A, Schreiber AW, Baumann U, Moller I, Cheng NH, Stancombe MA, Hirschi KD, Webb AA, et al. Cell-specific vacuolar calcium storage mediated by CAX1 regulates apoplastic calcium concentration, gas exchange, and plant productivity in Arabidopsis. Plant Cell. 2011;23:240–257. doi: 10.1105/tpc.109.072769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takanaga H, Frommer WB. Facilitative plasma membrane transporters function during ER transit. FASEB J. 2010;24:2849–2858. doi: 10.1096/fj.09-146472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okumoto S, Looger LL, Micheva KD, Reimer RJ, Smith SJ, Frommer WB. Detection of glutamate release from neurons by genetically encoded surface-displayed FRET nanosensors. Proc. Natl Acad Sci USA. 2005;102:8740–8745. doi: 10.1073/pnas.0503274102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Miwa H, Sun J, Oldroyd GE, Downie JA. Analysis of calcium spiking using a cameleon calcium sensor reveals that nodulation gene expression is regulated by calcium spike number and the developmental status of the cell. Plant J. 2006;48:883–894. doi: 10.1111/j.1365-313X.2006.02926.x. Use of a calcium biosensor to reveal insights into the role of calcium oscillations in legume symbiosis signaling.
- 34.Ehrhardt DW, Wais R, Long SR. Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell. 1996;85:673–681. doi: 10.1016/s0092-8674(00)81234-9. [DOI] [PubMed] [Google Scholar]
- 35.Kalo P, Gleason C, Edwards A, Marsh J, Mitra RM, Hirsch S, Jakab J, Sims S, Long SR, Rogers J, et al. Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science. 2005;308:1786–1789. doi: 10.1126/science.1110951. [DOI] [PubMed] [Google Scholar]
- 36.Smit P, Raedts J, Portyanko V, Debelle F, Gough C, Bisseling T, Geurts R. NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science. 2005;308:1789–1791. doi: 10.1126/science.1111025. [DOI] [PubMed] [Google Scholar]
- 37. Capoen W, Sun J, Wysham D, Otegui MS, Venkateshwaran M, Hirsch S, Miwa H, Downie JA, Morris RJ, Ane JM, et al. Nuclear membranes control symbiotic calcium signaling of legumes. Proc Natl Acad Sci USA. 2011;108:14348–14353. doi: 10.1073/pnas.1107912108. Combined use of biosensor imaging data, flux modeling, and electron microscopy to propose a more precise model of calcium oscillations in legume symbiosis signaling.
- 38.Sieberer BJ, Chabaud M, Timmers AC, Monin A, Fournier J, Barker DG. A nuclear-targeted cameleon demonstrates intranuclear Ca2+ spiking in Medicago truncatula root hairs in response to rhizobial nodulation factors. Plant Physiol. 2009;151:1197–1206. doi: 10.1104/pp.109.142851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iwano M, Entani T, Shiba H, Kakita M, Nagai T, Mizuno H, Miyawaki A, Shoji T, Kubo K, Isogai A, et al. Fine-tuning of the cytoplasmic Ca2+ concentration is essential for pollen tube growth. Plant Physiol. 2009;150:1322–1334. doi: 10.1104/pp.109.139329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monshausen GB, Bibikova TN, Weisenseel MH, Gilroy S. Ca2+ regulates reactive oxygen species production and pH during mechanosensing in Arabidopsis roots. Plant Cell. 2009;21:2341–2356. doi: 10.1105/tpc.109.068395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Allen GJ, Chu SP, Harrington CL, Schumacher K, Hoffmann T, Tang YY, Grill E, Schroeder JI. A defined range of guard cell calcium oscillation parameters encodes stomatal movements. Nature. 2001;411:1053–1057. doi: 10.1038/35082575. [DOI] [PubMed] [Google Scholar]
- 42.Allen GJ, Chu SP, Schumacher K, Shimazaki CT, Vafeados D, Kemper A, Hawke SD, Tallman G, Tsien RY, Harper JF, et al. Alteration of stimulus-specific guard cell calcium oscillations and stomatal closing in Arabidopsis det3 mutant. Science. 2000;289:2338–2342. doi: 10.1126/science.289.5488.2338. [DOI] [PubMed] [Google Scholar]
- 43.Munemasa S, Hossain MA, Nakamura Y, Mori IC, Murata Y. The Arabidopsis calcium-dependent protein kinase, CPK6, functions as a positive regulator of methyl jasmonate signaling in guard cells. Plant Physiol. 2011;155:553–561. doi: 10.1104/pp.110.162750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rincon-Zachary M, Teaster ND, Sparks JA, Valster AH, Motes CM, Blancaflor EB. Fluorescence resonance energy transfer-sensitized emission of yellow cameleon 3.60 reveals root zone-specific calcium signatures in Arabidopsis in response to aluminum and other trivalent cations. Plant Physiol. 2010;152:1442–1458. doi: 10.1104/pp.109.147256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chaudhuri B, Hörmann F, Lalonde S, Brady SM, Orlando DA, Benfey P, Frommer WB. Protonophore- and pH-insensitive glucose and sucrose accumulation detected by FRET nanosensors in Arabidopsis root tips. Plant J. 2008;56:948–962. doi: 10.1111/j.1365-313X.2008.03652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Grossmann G, Guo WJ, Ehrhardt DW, Frommer WB, Sit RV, Quake SR, Meier M. The RootChip: an integrated microfluidic chip for plant science. Plant Cell. 2011;23:4234–4240. doi: 10.1105/tpc.111.092577. Development of a plant growth system that permits time course imaging of biosensors in plant roots with precise and minimally invasive control over growth conditions.
- 47. Choi WG, Swanson SJ, Gilroy S. High-resolution imaging of Ca2+, redox status, ROS and pH using GFP biosensors. Plant J. 2012;70:118–128. doi: 10.1111/j.1365-313X.2012.04917.x. Recent review of in planta applications of calcium, reactive oxygen species, pH and redox.
- 48. Chen LQ, Hou BH, Lalonde S, Takanaga H, Hartung ML, Qu XQ, Guo WJ, Kim JG, Underwood W, Chaudhuri B, et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature. 2010;468:527–532. doi: 10.1038/nature09606. Application of FRET sensors in human cells to identify novel glucose transporters
- 49. Chen LQ, Qu XQ, Hou BH, Sosso D, Osorio S, Fernie AR, Frommer WB. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science. 2012;335:207–211. doi: 10.1126/science.1213351. Application of FRET sensors in human cells to identify novel sucrose transporters
- 50.Bermejo C, Ewald JC, Lanquar V, Jones AM, Frommer WB. In vivo biochemistry: quantifying ion and metabolite levels in individual cells or cultures of yeast. Biochem. J. 2011;438:1–10. doi: 10.1042/BJ20110428. [DOI] [PubMed] [Google Scholar]
- 51. Bermejo C, Haerizadeh F, Takanaga H, Chermak D, Frommer WB. Dynamic analysis of cytosolic glucose and ATP levels in yeast using optical sensors. Biochem. J. 2010;432:399–406. doi: 10.1042/BJ20100946. Simple assay formonitoring glucose accumulation in yeast cells and cell cultures
- 52.Brett CL, Kallay L, Hua Z, Green R, Chyou A, Zhang Y, Graham TR, Donowitz M, Rao R. Genome-wide analysis reveals the vacuolar pH-stat of Saccharomyces cerevisiae. PLoS ONE. 2011;6:e17619. doi: 10.1371/journal.pone.0017619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Orij R, Urbanus ML, Vizeacoumar FJ, Giaever G, Boone C, Nislow C, Brul S, Smits GJ. Genome-wide analysis of intracellular pH reveals quantitative control of cell division rate by pHc in Saccharomyces cerevisiae. Genome Biol. 2012;13:R80. doi: 10.1186/gb-2012-13-9-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN. A gene expression map of the Arabidopsis root. Science. 2003;302:1956–1960. doi: 10.1126/science.1090022. [DOI] [PubMed] [Google Scholar]
- 55.Deal RB, Henikoff S. The INTACT method for cell type-specific gene expression and chromatin profiling in Arabidopsis thaliana. Nat Protoc. 2011;6:56–68. doi: 10.1038/nprot.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson DE, Casey JR. Cytosolic H+ microdomain developed around AE1 during AE1-mediated Cl−/HCO3− exchange. J Physiol. 2011;589:1551–1569. doi: 10.1113/jphysiol.2010.201483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 58. Paige JS, Nguyen-Duc T, Song W, Jaffrey SR. Fluorescence imaging of cellular metabolites with RNA. Science. 2012;335:1194. doi: 10.1126/science.1218298. An innovative approach using RNA in combination with fluorescent dyes to monitor ions and metabolites in live cells.