SUMMARY
Alternative splicing contributes to diverse aspects of cancer pathogenesis including altered cellular metabolism, but the specificity of the process or its consequences are not well understood. We characterized genome-wide alternative splicing induced by the activating EGFRvIII mutation in glioblastoma (GBM). EGFRvIII upregulates the heterogeneous nuclear ribonucleoprotein (hnRNP) A1 splicing factor, promoting glycolytic gene expression and conferring significantly shorter survival in patients. HnRNPA1 promotes splicing of a transcript encoding the Myc-interacting partner Max, generating Delta Max, an enhancer of Myc-dependent transformation. Delta Max, but not full length Max, rescues Myc-dependent glycolytic gene expression upon induced EGFRvIII loss, and correlates with hnRNPA1 expression and downstream Myc-dependent gene transcription in patients. Finally, Delta Max is shown to promote glioma cell proliferation in vitro and augment EGFRvIII expressing GBM growth in vivo. These results demonstrate an important role for alternative splicing in GBM and identify Delta Max as a mediator of Myc-dependent tumor cell metabolism.
INTRODUCTION
Through alternative splicing, cancer cells preferentially express transcripts whose protein products suppress apoptosis, activate proliferative signaling pathways and reprogram cellular metabolism (David et al., 2009; Clower et al., 2010). The effect of specific oncogenes on genome-wide alternative splicing, how it is be regulated through splicing factors, and the functional consequences of splice variants for human cancer are not well understood.
Here, we explore the role of alternative splicing in glioblastoma (GBM), the most common and highly lethal form of adult brain cancer. GBMs are highly glycolytic and contain mutations in growth factor-receptor-initiated signaling pathways, most commonly the epidermal growth factor receptor (EGFR). EGFR is amplified in nearly 50% of patients, often in association with the constitutively activated mutant, EGFRvIII (Cancer Genome Atlas Research Network, 2008; Parsons et al., 2008). EGFRvIII induces major shifts in GBM cell metabolism (Guo et al., 2009a), however the contribution of alternative splicing to these metabolic shifts is not currently known. We performed an unbiased analysis of EGFRvIII-dependent genome wide-splicing coupled to mechanistic studies in cell lines, xenotransplants and clinical samples and we report here identification of a novel mechanism of metabolic reprogramming and tumor growth through alternative splicing of the Myc-interacting protein Max.
RESULTS AND DISCUSSION
EGFRvIII induces genome-wide changes in alternative pre-mRNA splicing and promotes altered metabolism
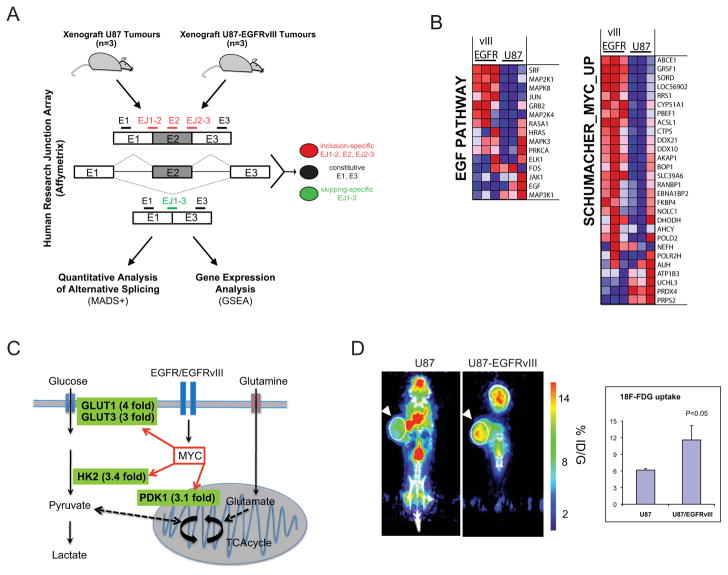
To identify EGFRvIII-dependent alternative splicing we overexpressed the EGFRvIII mutant in the U87 glioma cell line. We isolated and analyzed mRNA from U87 and U87-EGFRvIII expressing xenografts using Affymetrix Human Research Junction Arrays (Figure 1A). MADS+ bioinformatic analysis (Shen et al., 2010) revealed alternative splicing changes for 298 cassette exons (Table S1). Gene ontology (GO) enrichment analysis of differentially spliced transcripts revealed significantly enhanced representation of transcripts encoding proteins involved in transcription/translation (Figure S1). In addition, gene expression analysis of the arrays identified not only upregulation of EGF-pathway target genes as expected, but also showed upregulation of multiple Myc-target genes, including glucose transporters GLUT1 and 3, hexokinase2 (HK2), and pyruvate dehydrogenase kinase (PDK1) (Figure 1B and 1C). These data suggested altered cellular metabolism in EGFRvIII expressing tumors. In fact, EGFRvIII-expressing tumors of the same size as their isogenic counterparts, took-up nearly twice as much 18F — FDG-labeled glucose, confirming an enhanced glycolytic phenotype consistent with their growth advantage as xenografts (Guo et al., 2009a; Furnari et al., 2007; Guo, et al., 2009b, Guo et al., 2011) (Figure 1D).
Figure 1. EGFRvIII enhances alternative splicing, induces expression of Myc-target genes, and promotes glucose uptake in vivo.
(A) Schematic illustration of experimental design used to coordinately analyze gene expression and alternative splicing promoted by EGFRvIII in vivo.
(B) Heat map of expression for genes in the indicated gene sets (EGF-Pathway and Myc). Gene expression in U87 and U87-EGFRvIII tumors from three mice is shown.
(C) Schematic showing increased expression of Myc-regulated genes in EGFRvIII-expressing tumors. The fold upregulation of these transcripts in EGFRvIII expressing tumors is shown in green.
(D) MicroPET/CT imaging comparing U87 with U87-EGFRvIII xenograft tumors (arrow). The graph represents FDG uptake averaged for three tumors (white-dotted circles) presented as percentage injected dose per gram (%ID/G) (p value by t test).
HnRNPA1 splicing factor downstream of EGFRvIII regulates glycolytic gene expression
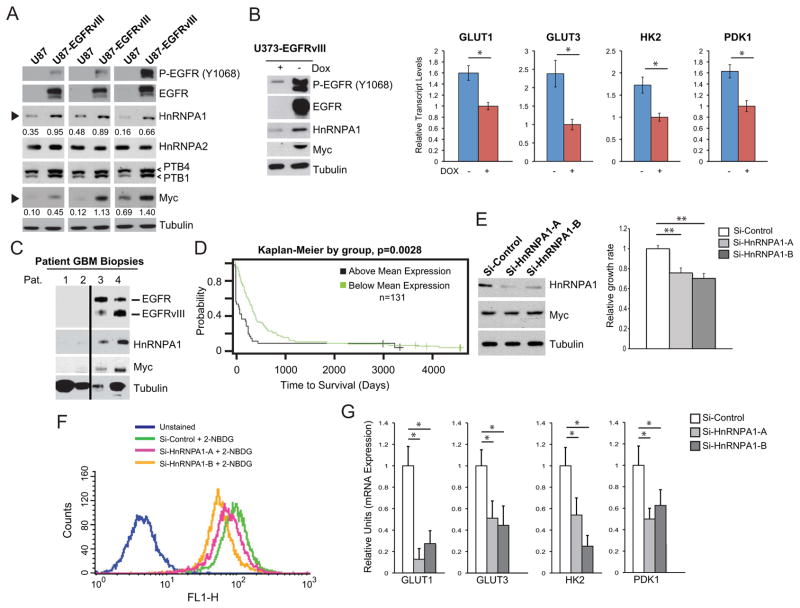
Alternative splicing of transcripts encoding glycolytic enzymes, including the pyruvate kinase isoform PKM2 (Christofk et al., 2008a; Christofk et al., 2008b) are mediated at least in part by the heterogeneous nuclear ribonuclear proteins (hnRNPs) A1 and A2, as well as polypyrimidine tract binding protein (PTB) (David et al., 2009; Clower et al., 2010; Sun et al., 2011). HnRNPA1 expression was increased in EGFRvIII expressing xenografts, whereas no consistent differences in hnRNPA2 or PTB isoform 4 were detected, although there was a small change in PTB isoform 1 expression (Figure 2A). Therefore, we focused on the potential role of hnRNPA1 in mediating the EGFRvIII-splicing associated changes in glycolysis.
Figure 2. EGFRvIII regulates expression of Myc and hnRNPA1 splicing factor promoting glycolysis, proliferation and shorter patient survival.
(A) Immunoblot analysis of Myc and downstream splicing factors from three U87 xenograft tumor lysates and three U87-EGFRvIII xenograft tumor lysates. Numerical values below HnRNPA1 and Myc blots (black arrows) is densitometric quantification of bands normalized to tubulin.
(B) (Left) Immunoblot analysis of lysates from U373 with doxycycline-regulated EGFRvIII expression, and (right) relative mRNA transcript levels of GLUT1, GLUT3, HK2, and PDK1 upon acute loss of EGFRvIII (n=4; shown is the mean±SD; * p<0.05).
(C) Immunoblot analysis of patient GBM biopsy samples having low EGFR expression or EGFR amplification and EGFRvIII expression.
(D) Kaplan-Meier overall survival plots of 131 primary GBM patients stratified by survival plots for hnRNPA1 expression. The single p-value is based on the difference of the two curves, and was calculated using the log-rank test.
(E) Cell proliferation of U87-EGFRvIII cells with hnRNPA1 knockdown (n=3; shown is mean±SD; ** p<0.01).
(F) Uptake of fluorescein-conjugated glucose (2NBDG) after 1 hour in U87-EGFRvIII cells transfected with the indicated siRNA. Unstained are cells without 2NBDG.
(G) RT-qPCR analysis for glycolytic gene expression in U87-EGFRvIII cells with hnRNPA1 knockdowns (n=3; shown is mean±SD; * p<0.05).
To examine the effect of EGFRvIII on hnRNPA1 expression in the acute setting, we analyzed U373 GBM cells expressing EGFRvIII under a doxycycline-regulated promoter (tet off) (Mukasa et al., 2010). Myc and hnRNPA1 levels were tightly linked to those of EGFRvIII (Figure 2A and 2B). Induced loss of EGFRvIII in an acute setting resulted in reduced hnRNPA1 and decreased expression of glycolytic genes GLUT1, GLUT3, HK2, and PDK1 (Figure 2B). Importantly, in tumor lysates obtained from GBM patients, hnRNPA1 expression levels correlated with those of EGFRvIII expression (Figure 2C). To assess clinical relevance for these findings, we determined the correlation between hnRNPA1 expression and GBM patient survival. Stratification of 131 primary GBM patients from the UCLA brain tumor gene expression database (http://www.probesetanalyzer.com/) by hnRNPA1 gene expression levels, revealed a highly significant correlation between elevated hnRNPA1 expression and short overall survival (p=0.0028) (Figure 2D). Taken together, these results place the splicing factor hnRNPA1 downstream of EGFRvIII in GBM, and demonstrate that high levels of hnRNPA1 expression are associated with aggressive tumors in brain cancer patients.
Pharmacological inhibition of mTOR downstream of EGFRvIII, or combined inhibition of EGFRvIII and the MAPK pathway, inhibited hnRNPA1 expression in glioma cells (Figure S2A-S2C). Genetic inhibition of mTOR complex 1 and mTOR complex 2 by siRNA knockdown of raptor and rictor alone, or in combination, further confirmed the mTOR-dependence on the induction of hnRNPA1, consistent with previous findings (Sun et al., 2011) (Figure S2B). We then examined the functional impact of hnRNPA1 on cell proliferation, glucose uptake, and glycolytic gene expression. Two independent hnRNPA1 knockdown constructs significantly inhibited cell proliferation (Figure 2E). Surprisingly, loss of hnRNPA1 inhibited glucose uptake (Figure 2F), which was accompanied by decreased expression of glycolytic genes GLUT1, GLUT3, HK2, and PDK1 (Figure 2G).
HnRNPA1 promotes splicing of the transcript encoding Myc-binding partner Max
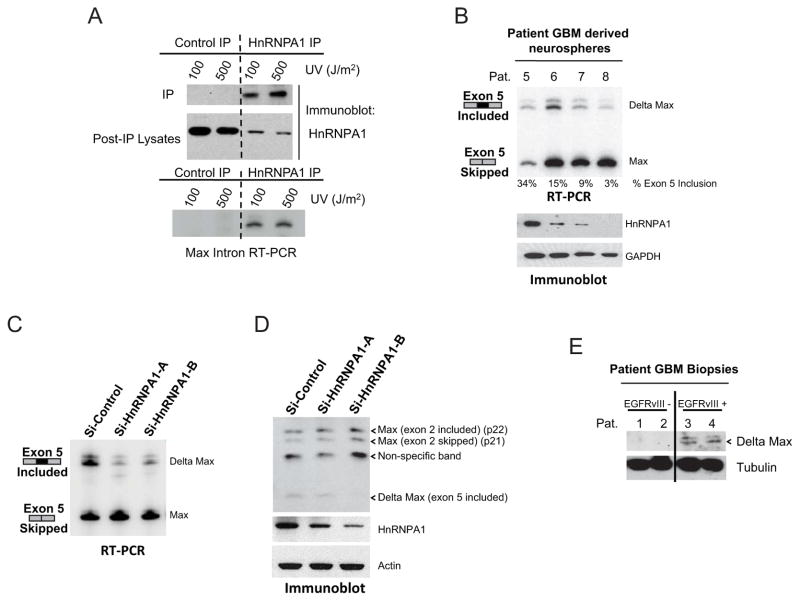
We then searched for potential hnRNPA1-dependent alternatively spliced transcripts involved in EGFRvIII pathogenesis. Bioinformatic analysis identified 40 differentially spliced exons with optimal hnRNPA1 binding sites (UAGGGA/U) within 200bp upstream or downstream of the exon. Among the top “hits”, we identified an alternatively spliced form of the Myc interacting protein Max, an obligate heterodimeric c-Myc binding partner that regulates Myc function (Blackwood et al., 1991; Hurlin et al., 2006). The intronic region upstream of exon 5 of Max contains a potential hnRNPA1-binding site that is highly conserved, suggesting evolutionary significance (Figure S3A). Inclusion of exon 5 generates a truncated Max protein referred to as Delta Max (Makela et al., 1992; Vastrik et al., 1993; Hirvonen et al., 1994) (Figure S3B and S3C). Although Delta Max was previously shown to augment Myc and Ras-dependent transformation, its function in human cancer has not been established (Makela et al., 1992). To obtain direct evidence for a physical association between hnRNPA1 and the intronic region upstream of exon 5 of Max, we performed crosslinking and immunoprecipitation (CLIP) assay. The intronic region harboring the potential hnRNPA1 binding site was immunoprecipitated with hnRNPA1 (Figure 3A). In patient-derived GBM neurospheres, the extent of exon 5 inclusion was tightly linked to the level of hnRNPA1 expression (Figure 3B). We then determined whether hnRNPA1 could mediate alternative splicing of Max to yield the Delta Max transcript and protein. HnRNPA1 knockdown significantly decreased Delta Max mRNA and protein expression in GBM cells (Figure 3C and 3D). We confirmed Delta Max expression in a panel of GBM tumor lysates, in association with EGFRvIII (Figure 3E). Taken together, these results suggest that EGFRvIII promotes Delta Max splicing through hnRNPA1.
Figure 3. HnRNPA1-dependent splicing of the Myc-heterodimerization partner Max results in the truncated variant Delta Max.
(A) RT-PCR for Max intron upstream of exon 5 using RNA extracted from hnRNPA1 immunoprecipitated from U87-EGFRvIII cells after UV crosslinking (CLIP) as described in Supplemental Experimental Procedures.
(B) RT-PCR splicing analysis for Max exon 5 and immunoblot analysis for hnRNPA1 in neurospheres established from patient GBM biopsies.
(C) RT-PCR splicing analysis for Max exon 5 in hnRNPA1 knockdown cells from Figure 2E.
(D) Max immunoblot of lysates from U87-EGFRvIII cells transfected with siRNAs to hnRNPA1.
(E) Immunoblot for Max in Patient GBM biopsies from Figure 2C.
Delta Max promotes glycolytic gene expression and contributes to EGFRvIII- mediated GBM tumor growth
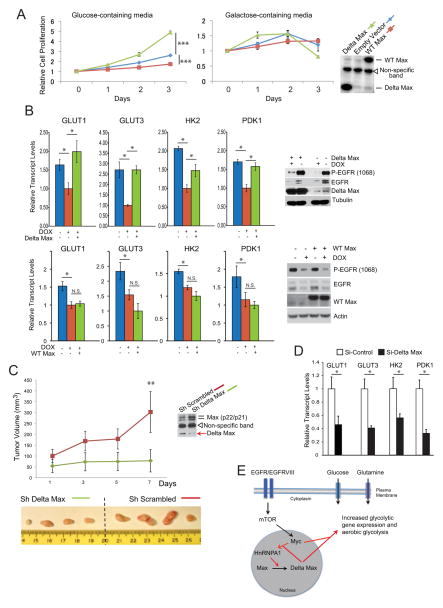
Delta Max is predicted to mediate its effects through Myc (Makela et al., 1992), which regulates the growth and proliferation of cancer cells at least in part by increasing the uptake and metabolism of glucose (Dang, 2012; Dang, 2011). Therefore, we sought to determine whether Delta Max could promote GBM cell proliferation; whether it required the presence of glucose; whether it was Myc-dependent, and whether it was specific to this alternatively spliced form of Max. Overexpression of Delta Max, but not wild-type Max, increased GBM cell proliferation in glucose-containing media but not galactose-containing media, suggesting that the effect on proliferation is glucose dependent and that it is specific for the alternatively spliced form of Max (Figure 4A). Concomitantly, Delta Max, but not wild-type Max, increased the expression of the glucose transporter GLUT3 and HK2 in U87 glioma cells, which was abrogated by Myc knockdown (Figure S4A and S4B). Taken together, these results indicated that Delta Max promotes glycolytic gene expression in a Myc-dependent fashion.
Figure 4. Delta Max promotes glycolytic gene expression and tumor growth in vitro and in vivo.
(A) Proliferation of U87 cells on glucose- or galactose-containing media after transfection with empty vector (EV), wild-type (WT) Max, or Delta Max (n=4; shown is the mean±SD; ***p<0.001).
(B) Relative mRNA transcript levels of GLUT1, GLUT3, HK2, and PDK1 upon acute loss of EGFRvIII with or without Delta Max or wild-type (WT) Max overexpression (n=3; shown is the mean±SD; * p<0.05; N.S. is non-significant).
(C) Xenograft tumors from U87-EGFRvIII cells with stable knockdown of Delta Max grew slower that control (Sh Scrambled) cells as measured by tumor volume (n=4; shown is the mean±SD; **p<0.01).
(D) RT-qPCR analysis of glycolytic gene expression in Delta Max knockdown U87-EGFRvIII cells (n=3; shown is the mean±SD; * p<0.05).
(E) Diagram illustrating EGFRvIII activation of mTOR upregulates Myc which stimulates hnRNPA1 expression and promotes splicing of Max generating Delta Max which augments Myc activity and increases aerobic glycolysis.
To determine whether EGFRvIII mediates GBM cell proliferation through Delta Max-dependent effects on glycolytic metabolism, we examined the impact of Delta Max on glycolytic gene expression in U373 GBM cells in which EGFRvIII is under the control of a doxycycline-regulated promoter (tet-off) (Mukasa et al., 2010). Silencing of EGFRvIII suppressed the expression of glycolytic genes GLUT1, GLUT3, HK2 and PDK1, consistent with its known effect on promoting glycolytic metabolism (Guo et al., 2009). Importantly, over-expression of Delta Max, but not wild-type Max, maintained glycolytic gene expression in the absence of EGFRvIII signaling (Figure 4B). These results suggest an epistatic relationship by which EGFRvIII regulates GBM cell glycolysis through Delta Max. The inability of wild-type Max to rescue glycolytic gene expression (Figure 4B) and to promote GBM growth in glucose-containing media (Figure 4A), demonstrates that alternative splicing of Max to Delta Max is required to promote glucose-dependent GBM cell proliferation.
To determine whether Delta Max was required for the growth of EGFRvIII-expressing GBMs we examined the effect of Delta Max depletion on tumor growth in vivo. Genetic depletion of Delta Max resulted in a nearly five-fold reduction in tumor size relative to control, p<0.01(Figure 4C). Consistent with the demonstrated effect of Delta Max on glycolytic gene expression (Figure 4B), siRNA-mediated depletion of Delta Max specifically inhibited GLUT1, GLUT3, HK2 and PDK1 gene expression (Figure S4C), as well as reducing glucose consumption and lactate production, two readouts of aerobic glycolysis (Figure 4D and S4D). Overexpression of the si-resistant Delta Max construct fully rescued the effect of the knockdown, confirming the specificity of the siRNA targeting Delta Max (Figure S4D).
Myc has previously been shown to transcriptionally upregulate hnRNPA1 levels (David et al., 2010), raising the possibility of a “feed forward” loop. Indeed, overexpression of Delta Max increased hnRNPA1 transcript levels, which was abrogated by Myc siRNA knockdown (Figure S4E). Taken together, these results suggest a feed forward regulatory loop in which EGFRvIII, through mTOR-dependent upregulation of Myc and splicing of Delta Max, feeds back to further elevate hnRNAP1 through Myc-dependent transcriptional regulation (Figure 4E).
Glycolytic gene expression in brain cancers correlates with EGFRvIII and hnRNPA1
Finally, to assess clinical relevance of our findings, we examined the correlation between EGFRvIII expression and hnRNPA1 levels, and between hnRNPA1 and Myc-regulated glycolytic target gene expression in the 120 classical, mesenchymal and neural GBMs analyzed in the Cancer Genome Atlas (TCGA) data from the Unified Scaled dataset (Verhaak et al., 2010). HnRNPA1 expression was significantly elevated in EGFRvIII positive GBMs (p=0.00881) (Table S2). Further, downstream glycolytic genes were significantly correlated with hnRNPA1 in these clinical samples (Table S3). Most importantly, like hnRNAP1, high-level expression of the glycolytic genes GLUT1, GLUT3, HK2, and PDK1 correlated with short overall survival of GBM patients (Figure S4F). Taken together, these results provide clinical evidence that that the hnRNPA1-dependent glycolytic pathway confers more biologically aggressive behavior in patients.
Delta Max was identified nearly 20 years ago when it was reported to augment Myc-dependent transformation (Makela et al., 1992; Vastrik et al., 1993; Hirvonen et al., 1994). However, it was lost from scientific discourse relatively soon thereafter, for lack of apparent relevance in human cancer. We now demonstrate a novel and clinically important role for Delta Max in mediating the intersection of a key oncogenic signaling pathway with a metabolic transformation that promotes glioblastoma progression (Figure 4E). Myc integrates cellular proliferation with metabolism in many cancers (Vander Heiden et al., 2009; Koppenol et al., 2011; Levine and Puzio-Kuter, 2010; Deberardinis et al., 2008), but is amplified or mutated in only a subset. These results provide an alternative splicing-dependent mechanism, in addition to regulating Myc levels (Gan et al., 2010), by which GBMs lacking Myc amplification or mutation engage Myc signaling to regulate tumor metabolism.
EXPERIMENTAL PROCEDURES
Cell culture and transfections
U87-EGFRvIII, U87, and U373-EGFRvIII glioma cell lines were maintained in Dulbecco’s Modified Eagle Media (DMEM; Cellgro) supplemented with 10% fetal bovine serum (FBS; Omega Scientific), 1% penicillin/streptomycin/glutamine (Invitrogen), and cells incubated at 5% CO2 at 37°C. Transfection for overexpression was using FuGene6 (Roche) according to manufacturer’s instructions. Delta Max, wild-type Max, and Flag-tagged Delta Max constructs were in vector pDream 2.1 and synthesized by GenScript USA Inc. Doxycycline induction of U373-EGFRvIII cells was also as described previously (Mukasa et al., 2010).
Xenograft tumor studies
Isogenic human glioma cells U87 and U87-EGFRvIII were implanted subcutaneous (s.c.) into immunodeficient SCID/Beige mice. SCID/Beige mice were bred and kept under defined-flora pathogen-free conditions at the AALAC-approved Animal Facility of the Division of Experimental Radiation Oncology, UCLA. U87 and U87-EGFRvIII cell lines were resuspended at 1 × 106 cells/ml in PBS with Matrigel (BD) and were implanted s.c. on opposite sides of the mouse abdomen. For MicroPET/CT imaging U87 and U87-EGFRvIII cells were implanted s.c. in the abdomen of separate mice. MicroPET/CT scanning and analysis was performed as previously described (Guo et al., 2009).
Microarray and survival data analysis
Human Research Junction Arrays (Affymetrix) were used to identify differentially expressed and spliced transcripts between U87 and U87-EGFRvIII xenograft tumors from NOD/SCID/gamma (NSG) mice. RNA was isolated from xenograft tumors with Trizol (Invitrogen). For microarray analysis RNA was DNAse (Ambion) treated and amplified using the WT Expression Kit (Ambion) and WT Terminal Labeling kit (Affymetrix). Human Research Junction Arrays were obtained from Affymetrix, and labeled samples were hybridized and scanned in the UCLA DNA Microarray Facility. Gene expression and alternative splicing were analyzed with MADS+ software (Shen et al., 2010). Heat maps were generated using Gene Set Enrichment Analysis (GSEA) software (Mootha et al., 2003; Subramanian et al., 2005). Complete microarray profiles are available at GEO at GSE#####.
Survival data and gene correlation data in clinical samples: These microarray data are from the TCGA, and were downloaded from the Cancer Cell publication: “An integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR and NF1”. https://tcga-data.nci.nih.gov/docs/publications/gbm_exp/unifiedScaled.txt Survival curves for the UCLA data are publicly available through the following website: http://probesetanalyzer.com/
Alternative splicing analysis
Alternative splicing was assayed using semi-quantitative reverse-transcriptase PCR. Primers were designed to constitutive exons flanking alternative exons of interest. Primers were: MAX_(85)_F 5′-tcagtcccatcactccaagg-3′; MAX_(85)_R 5′-gcacttgacctcgccttct-3′. Reverse primers were 32P end-labeled and 22 cycle PCR reactions were resolved by denaturing PAGE. Radioactive signal was detected using a Typhoon phosphorimager (GE) and quantified with ImageQuant TL software (Amersham Biosciences).
Real-Time (RT) quantitative PCR
Total RNA was extracted with Trizol (Invitrogen) followed by DNase treatment (Ambion) and Proteinase K treatment (Roche) according to manufacturer’s instructions. For cDNA synthesis 500ng of RNA was used with SuperScript VILO cDNA Synthesis kit (Invitrogen), and 1ul of this used with iQ SYBR Green Supermix (Bio-Rad) for quantitative real-time PCR. Primers were synthesized by IDT and were designed (using Primer3) to amplify approximately 200bp of the 3′UTR region of targets. Sequence of the primers is provided in the supplemental information. RT-PCR was performed in triplicate using Bio-Rad iCycler iQ Real-Time PCR Detection System. Analysis was using the iCycler iQ Optical System Software (Version 3.0a) and data normalized to vinculin internal control.
Glucose uptake assay
Cells were grown in a 6 well plate in DMEM with 0.5%FBS without antibiotics for 24 hr, media was replaced with DMEM containing 10%FBS (no antibiotics) and 100uM 2-NBDG (Invitrogen). Cells were incubated for 1 hr at 37°C 5% CO2 incubator, washed 3 times with PBS, followed by lifting with trypsin, washed and subsequent flow cytometric analysis using FACSCalibur Analyzer (BD).
Cell proliferation assays
Cells were transfected with siRNA and 24 hr later seeded in a 96 well plate with DMEM containing 0.5% FBS. Relative growth rate was determined after 72 hr using Cell Proliferation Assay Kit (Millipore). Cells were incubated 30 min at 37°C 5%CO2 incubator after adding tetrazolium salt WST-1 (Millipore), and the absorbance measured using a microplate reader (Bio-Rad) at 420 to 480 nm. For determining cell number, cells were transfected with siRNA in a 6 well plate and 24 hr later cells were washed and replaced with DMEM supplemented with 0.5% FBS. After 48 hr and 72 hr cells were counted with a haemocytometer. For proliferation on glucose- and galactose-containing media, cells were placed in 96-well plates at 2.5 × 103 cells/well in 100 μl of growth medium and incubated in each condition of treatment. For the measurement of glucose-dependent proliferation, DMEM containing glucose (Cellgro) or no-glucose DMEM (Gibco) supplemented with 4.5 g/l Galactose (Sigma) was used as previously reported (Finley et al., 2011). Cell proliferation was examined with Cell Proliferation Assay Kit (Millpore) according to the manufacturer’s instructions. The absorbance of the treated and untreated cells was measured with a microplate reader (Bio-Rad) at 420 to 480nm. Data represent the mean ± SD of four independent experiments.
Antibodies, cell lysate preparation, and immunoblotting
Specific antibodies were purchased from the following commercial sources: anti-Max antibody was rabbit polyclonal from Abcam (ab68578). Anti-HnRNPA1 was mouse monoclonal from Sigma (clone 9H10). Anti-EGFR/EGFRvIII cocktail antibody was from Upstate. Anti-Actin was from Novus Biologicals (AC-15). Anti-Tubulin was mouse monoclonal (T4026) from Sigma. Anti-Myc (D84C12), anti- Anti-HnRNPA2 (2A2), anti-phosphoEGFR (Y1068), anti-HA (6E2), anti-phosphoAkt (S473), anti-Akt, anti-rS6, anti-phospho-rS6 (S235/S236), anti-rictor, and anti-raptor were from Cell Signaling. Anti-PTB was provided by D.L. Black. For cell lysate preparation, cells were washed with PBS and lysed with RIPA lysis buffer (50mM Tris-HCL pH 7.4, 150mM NaCl, 0.5%deoxycholate, 1% Nonidet P-40, and 0.1% SDS) (Boston BioProducts) containing half protease inhibitor cocktail (Thermo Scientific) and half phosphatase inhibitor cocktail (Thermo Scientific). Protein concentration of lysates was determined using the Bradford assay kit (Bio-Rad). Protein electrophoresis was with 12% NuPAGE Bis-Tris pre-cast Mini gels (Invitrogen) with MES running buffer (Invitrogen), immunoblotting was according to antibody supplier instructions, and Western blot detection was using SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific). For detection of both Myc and Max SuperSignal West Femto Chemiluminescent Substrate (Thermo Scientific) was used. Image J analysis software (NIH) was used for densitometric quantification of imunoblots.
Patient GBM biopsy samples and neurosphere culture
Patient GBM samples were collected following surgical resection at UCLA. GBM samples were provided, with patient consent, by the Brain Tumor Translational Resource (BTTR) at UCLA. Neurospheres were isolated and cultured as previously described (Nakano and Kornblum, 2009).
RNAi experiments and inhibitor studies
Transfections of siRNA was using Lipofectamine RNAiMAX (Invitrogen) according to manufacturer’s instructions. The siRNA targeting exon 5 of Delta Max (target sequence: CAUGGAAGAUGCAAGUAAA) was purchased from Sigma. The siRNAs targeting hnRNPA1 (target sequence (A): GAAUGGUUAUAAAAGUGAU; target sequence (B): GGUUCUAUUUGGAAUUUAU) was purchased from Ambion (Applied Biosystems). The siRNA targeting Max (target sequence: CAAAGACAGCUUUCACAGU) was purchased from Ambion (Applied Biosystems). The control siRNA (Silencer Select Negative Control #2) was purchased from Ambion (Applied Biosystems). The siRNAs targeting Rictor (catalog #L-016984-00-0005) and Raptor (catalog #L-004107-00-0005) were on-TARGETplus SMARTpool siRNAs (Thermo Scientific, Dharmacon Division).
Lentiviral production and xenograft tumor measure
Si RNA sequence targeting Delta Max exon 5 was cloned into pLKO.1-puro vector (Sigma). A scrambled shRNA (Sigma MISSION shRNA non-mammalian control (SHC002)) was used as a control. Lentivirus was made using a three-plasmid packaging system. In brief, shRNAs in the pLKO.1-puro vector were co-transfected into 293T cells along with expression vectors containing the vsvg, gag/pol, and rev genes. 48 h after transfection, lentivirus was harvested and 8 ug/ml of polybrene was added. Subconfluent U87-EGFRvIII cells were infected with harvested virus and selected in 0.7 ug/ml of puromycin for 1 week. Stable cell lines were then used for xenograft tumor growth study as described above. When tumors were palatable external caliper measures were taken over a span of 1 week. For tumor volume the greatest longitudinal diameter (length) and the greatest transverse diameter (width) were determined, and tumor volume calculated by the ellipsoidal formula (tumor volume = ½ (length x width2).
Statistical methods
Tumor volume in the xenograft experiment and cell proliferation on glucose- and galactose-containing media, was compared by two-way ANOVA followed by Tukey’s multiple comparison’s test using GraphPad Prism 6. For relative cell growth with hnRNPA1 siRNA, one-way ANOVA followed by Tukey’s multiple comparison’s test was performed in GraphPad Prism 6. For Real-Time (RT) quantitative PCR, significance was determined by nonparametric Mann-Whitney test (one-tailed) (GraphPad Prism 6) and p values <0.05 were considered significant.
Supplementary Material
RESEARCH HIGHLIGHTS.
EGFRvIII regulates hnRNPA1 splicing factor expression in glioblastoma (GBM)
HnRNPA1 promotes splicing of the Myc interacting protein Max to generate Delta Max
Delta Max promotes Myc-dependent glycolysis and tumor growth in vivo
HnRNPA1 and glycolytic gene expression correlate with poor patient survival
Acknowledgments
We thank Dr. George Thomas and Dr. Dan Geschwind for careful reading of the manuscript and helpful comments. This work was supported by: National Institutes of Health (NIH) grants CA119347 and NS73831 (P.S.M.), The Ben and Catherine Ivy Foundation (P.S.M, W.H.Y. and T.F.C.), Art of the Brain Fund (W.H.Y. and T.F.C.), Accelerate Brain Cancer Cure (P.S.M.), STOP Cancer (P.S.M.), The Fred Miller Family (P.S.M.), The John W. Carson Foundation (P.S.M.), Ziering Family Foundation in memory of Sigi Ziering (T.F.C. and P.S.M.), Henry Singleton Brain Cancer Fund (W.H.Y. and T.F.C.), NIH grant R01 GM084317 (Manuel Ares, Xiang-Dong Fu, Eugene Yeo, and D.L.B.), NIH grant P01-CA95616 and Fellow Award from the National Foundation for Cancer Research (W.K.C.). D.L.B. is an Investigator of the Howard Hughes Medical Institute. This work was also supported by the Moores Cancer Center Core Grant NCI P30CA23100.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Blackwood EM, Eisenman RN. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251(4998):1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research, N. . Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452(7184):230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, et al. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452(7184):181–186. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- Clower CV, Chatterjee D, et al. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc Natl Acad Sci U S A. 2010;107(5):1894–1899. doi: 10.1073/pnas.0914845107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV. Therapeutic targeting of Myc-programmed cancer cell metabolism. Cold Spring Harb Symp Quant Biol. 2011;76:369–374. doi: 10.1101/sqb.2011.76.011296. [DOI] [PubMed] [Google Scholar]
- Dang CV. MYC on the path to cancer. Cell. 2012;149(1):22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David CJ, Chen M, et al. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463(7279):364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, et al. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7(1):11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Finley LW, Carracedo A, et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1α destabilization. Cancer Cell. 2011;19(3):416–428. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnari FB, Fenton T, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21(21):2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- Gan B, Lim C, et al. FoxOs enforce a progression checkpoint to constrain mTORC1-activated renal tumorigenesis. Cancer Cell. 2010;18(5):472–484. doi: 10.1016/j.ccr.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D, I, Hildebrandt J, et al. The AMPK agonist AICAR inhibits the growth of EGFRvIII-expressing glioblastomas by inhibiting lipogenesis. Proc Natl Acad Sci U S A. 2009;106(31):12932–12937. doi: 10.1073/pnas.0906606106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D, Prins RM, et al. EGFR signaling through an Akt-SREBP-1-dependent, rapamycin-resistant pathway sensitizes glioblastomas to antilipogenic therapy. Sci Signal. 2009;2(101):ra82. doi: 10.1126/scisignal.2000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D, Reinitz F, et al. An LXR agonist promotes glioblastoma cell death through inhibition of an EGFR/AKT/SREBP-1/LDLR-dependent pathway. Cancer Discov. 2011;1(5):442–456. doi: 10.1158/2159-8290.CD-11-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirvonen HE, Salonen R, et al. Differential expression of myc, max and RB1 genes in human gliomas and glioma cell lines. Br J Cancer. 1994;69(1):16–25. doi: 10.1038/bjc.1994.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlin PJ, Huang J. The MAX-interacting transcription factor network. Semin Cancer Biol. 2006;16(4):265–274. doi: 10.1016/j.semcancer.2006.07.009. [DOI] [PubMed] [Google Scholar]
- Koppenol WH, Bounds PL, et al. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11(5):325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- Levine AJ, Puzio-Kuter AM. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science. 2010;330(6009):1340–1344. doi: 10.1126/science.1193494. [DOI] [PubMed] [Google Scholar]
- Makela TP, Koskinen PJ, et al. Alternative forms of Max as enhancers or suppressors of Myc-ras cotransformation. Science. 1992;256(5055):373–377. doi: 10.1126/science.256.5055.373. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Mukasa A, Wykosky J, et al. Mutant EGFR is required for maintenance of glioma growth in vivo, and its ablation leads to escape from receptor dependence. Proc Natl Acad Sci U S A. 2010;107(6):2616–2621. doi: 10.1073/pnas.0914356107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Jones S, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Warzecha CC, et al. MADS+: discovery of differential splicing events from Affymetrix exon junction array data. Bioinformatics. 2010;26(2):268–269. doi: 10.1093/bioinformatics/btp643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Chen X, et al. Mammalian target of rapamycin up-regulation of pyruvate kinase isoenzyme type M2 is critical for aerobic glycolysis and tumor growth. Proc Natl Acad Sci U S A. 2011;108(10):4129–4134. doi: 10.1073/pnas.1014769108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, et al. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastrik I, Koskinen PJ, et al. Alternative mRNA forms and open reading frames of the max gene. Oncogene. 1993;8(2):503–507. [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.