Abstract
Phototropins are one of several classes of photoreceptors used by plants and algae to respond to light. These proteins contain flavin-binding LOV (Light-Oxygen-Voltage) domains that form covalent cysteine-flavin adducts upon exposure to blue light, leading to the enhancement of phototropin kinase activity. Several lines of evidence suggest that adduct formation in the phototropin LOV2 domains leads to the dissociation of an alpha helix (Jα) from these domains as part of the light-induced activation process. However, crystal structures of LOV domains both in the presence and absence of the Jα helix show very few differences between dark and illuminated states, and thus the precise mechanism through which adduct formation triggers helical dissociation remains poorly understood. Using Avena sativa phototropin 1 LOV2 as a model system, we have studied the interactions of the LOV domain core with the Jα helix through a series of equilibrium molecular dynamics simulations. Here we show that conformational transitions of a conserved glutamine residue in the flavin binding pocket are coupled to altered dynamics of the Jα helix both through a shift in dynamics of the main β-sheet of the LOV domain core and through a secondary pathway involving the N-terminal A′α helix.
Introduction
The ability to sense and respond to light is critical to plants, as it allows them to make optimal use of available light and avoid potentially harmful conditions such as drying or overexposure. Phototropism, chloroplast movement, and stomatal opening in plants are all regulated in part by blue light photoreceptors known as phototropins 3,5,6,8,9,29,32,34,49. These proteins each contain two LOV (Light-Oxygen-Voltage) domains, LOV1 and LOV2, and a kinase domain that is activated in response to blue light 4,10. The LOV domains, in turn, are members of the Per-ARNT-Sim family 57,60 which noncovalently bind flavin chromophores, either flavin mononucleotide (FMN) or flavin adenine dinucleotide (FAD) 8.
The two phototropin LOV domains are known to undergo broadly similar photoreactions upon exposure to blue light: the isoalloxazine ring of the chromophore becomes covalently bound to a conserved cysteine residue in the protein 14,50,51,56, giving rise to a signaling state which recovers spontaneously on a time scale of seconds to minutes 51. The LOV domains differ, however, both in the quantum yield of their photoreaction 53 and in their importance to phototropin activation. LOV2 is known to be both necessary and sufficient for phototropin signaling 11; the function of LOV1 remains less clear, but it is implicated in phototropin dimerization 52.
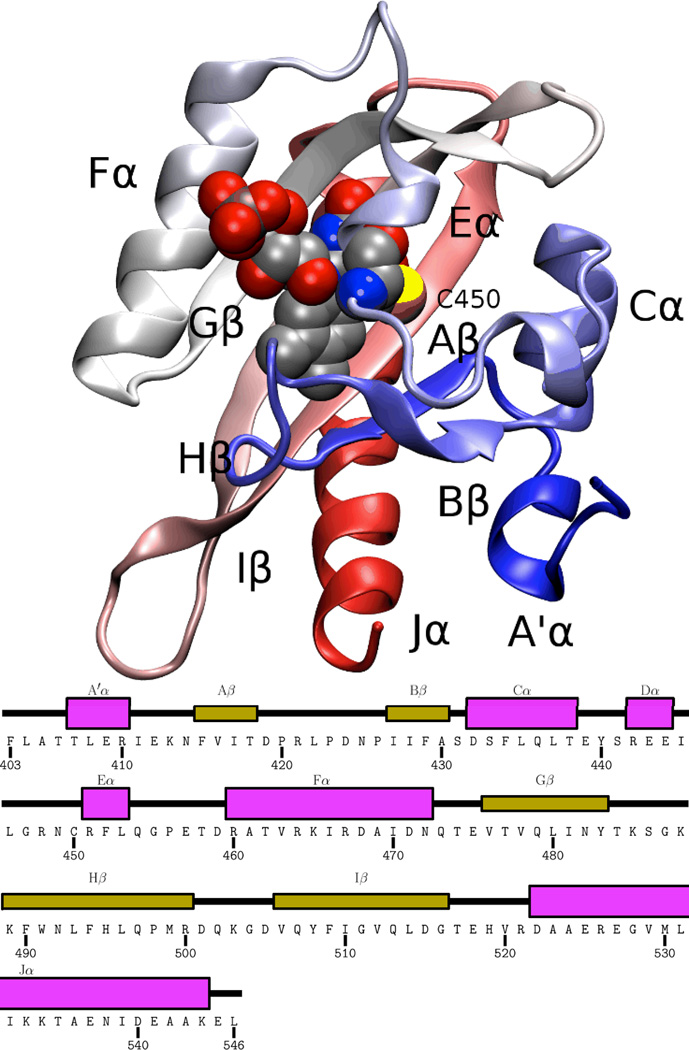
While the precise structural mechanism linking the LOV2 photoreaction with kinase activation remains open, biophysical and biochemical studies have identified several residues which appear to be important at various points along this pathway 36. Early crystal structures of a phototropin LOV1 domain from C. reinhardtii 17 and a phytochrome LOV2 domain from A. capillus-veneris 13,14 in both dark and light states confirmed the chemical change occurring in the LOV domain photoreaction and hinted at rearrangements of the FMN binding site associated with the light state. More recently, crystal structures of the LOV2 domain from A. sativa phototropin 1 (Phot1) 23 and the related Vivid LOV domain 63 demonstrated small light-induced changes both at the chromophore binding site and relayed across the adjacent beta sheet to affect protein-specific elements involved in signaling. An overview of the structure of the LOV2 domain is shown in Fig. 1.
Figure 1.
Overview of the AsPhot1-LOV2 structure 23. The backbone of the homology model is shown in cartoon representation, with the FMN and photoreactive cysteine (C450) shown in vdW representation, and all secondary structure elements labeled. Coloring of the protein backbone runs blue to red from N terminus to C terminus.
Several lines of biophysical data suggest a major loss of α-helical character upon LOV2 photoactivation 12,26. These data include those from a series of solution NMR and biochemical experiments on A. sativa Phot1 LOV2 (AsPhot1-LOV2) by Gardner and colleagues that correlate phototropin kinase activation with the light-induced unfolding of a helix (named the Jα helix) which is normally located on the β-sheet surface of the LOV domain core in the dark state 24,25,26. More recently, time-resolved measurements of the diffusion coefficient of Arabidopsis Phot1-LOV2 have indicated the presence of a two-stage dissociation pathway. It was proposed that after illumination, Jα dissociates from the LOV2 core to form a “precursor state” with a time constant of 300 µs; subsequently, the Jα helix of this precursor unfolds to yield the signaling state with a time constant of 1 ms 40. Time-resolved optical rotary dispersion spectroscopy measurements of the same process using A. sativa Phot1-LOV2 indicated the presence of an intermediate with a partially unfolded Jα helix which appears after photoactivation with a time constant of 90 µs 7. More recently, a combination of NMR and circular dichroism measurements on a variety of AsPhot1-LOV2 domain mutants implicated a short N-terminal A′α helix in the light-activated dissociation of the C-terminal Jα, providing further insight into the structural transition occuring during LOV domain activation 62.
Despite extensive characterization of the LOV domain photocycle itself 1,16,33,42, as well as the presence of crystal structures of LOV2 in both dark and light states 13,14,23 and high-resolution solution NMR data from both states 24,25,26,61, the precise mechanism through which the photoreaction leads to Jα dissociation and unfolding is still unclear. Due to the involvement of a strong allosteric change which links chemical bond formation at a chromophore binding site to a distant secondary structure rearrangement, the mechanism of LOV domain photoactivation is of interest both for understanding phototropin signaling and as a general representative for conformational changes in signaling proteins.
Molecular dynamics (MD) simulations are an ideal tool for the study of allosteric interactions such as those occuring in the LOV domain, as they provide atomic-resolution trajectories of structural transitions while at the same time allowing easy manipulation of the system (e.g., switching between the dark and photoadduct states in the LOV domain). While MD simulations cannot directly reach the relevant timescales for Jα dissociation, thorough analysis of shorter simulations can nevertheless provide significant insight into such (relatively) slow events. Several recent MD studies on LOV domains have highlighted the role of rotamerization of a highly conserved glutamine residue (Q513 in AsPhot1-LOV2). In a series of simulations on LOV domains from various organisms, Baeurle and coworkers have highlighted the importance of a light-induced hydrogen bond between Q513 and N492 (using AsPhot1-LOV2 numbering) in AsPhot1-LOV2 44,46 and N. crassa Vivid 45 (but not in a LOV1 domain from C. reindardtii 47. In the specific case of AsPhot1-LOV2, they concluded that disruption of the Jα-LOV2 interface was triggered by stress due to the increased coupling of the Hβ and Iβ strands 44,46, and subsequent breakage of Q497-D540 and Q479-E518 hydrogen bonds 44. An alternative proposal is that Jα dissociation is caused by changes in the conformation and motility of inter-strand loops in the LOV domain core 19. These prior studies suffered, however, from the relatively short duration of the simulations used (20 ns or less in all cases but Peter et al.46, which used a kinetic Monte Carlo approach and thus is difficult to pinpoint an effective duration for) and, in one case, absence of the Jα helix due to the unavailability of a structure at the time 19. In addition, the specific suggestion that breakage of the Q497-D540 and Q479-E518 hydrogen bonds plays a role in Jα dissociation is inconsistent with recent experimental data on AsPhot1-LOV2 mutants removing one partner from each of these interactions 62.
To better understand the atomic-scale mechanisms of the allosteric switch in LOV2, we performed a series of long-timescale molecular dynamics (MD) simulations on A. sativa LOV2 domain. We find evidence for a link between conformational switching of Q513 (a conserved glutamine residue near the FMN binding site) and Jα dissociation, mediated both through conformational changes in the Iβ strand and through a dynamical coupling of A′α, Aβ, and Bβ with the Jα helix. Our results are consistent with a variety of biophysical and biochemical data 30,41,62, while at the same time they provide previously unavailable atomistic-level insight into the mechanisms of LOV2 allostery.
Methods
The initial structures for all simulations were taken from the room-temperature dark state (PDB code 2V1A) and light state (PDB code 2V1B) crystal structures of Halavaty and Moffat 23. The N- and C- terminii were blocked with N-acetyl and N-methylamide groups, respectively. All crystallographic water and cofactors (with the exception of FMN) were discarded and the structures were initially solvated using SOLVATE1.2 22 with default settings to add water to a distance of 15 Å from the protein. A solvent cube was then completed with the solvate plugin of VMD 1.8.6 27 and a minimal neutralizing set of six sodium ions placed randomly with the autoionize plugin of VMD.
All molecular dynamics simulations were performed using NAMD 2.748 with CHARMM27 parameters 38 and CMAP corrections 39; the TIP3P water model 31 was employed for water in the system. Newly derived parameters were used for the FMN and FMN-cysteinyl adduct (see Supplementary Material for details). Where simulations were carried out in a constant temperature ensemble (see below), the temperature was maintained at 298 K using Langevin dynamics, with a damping constant of 5.0 ps−1 during equilibration and 1.0 ps−1 during production runs. During equilibration, NPT ensemble simulations were employed, with a pressure of 1 atm maintained using a Nosé-Hoover Langevin piston 48. The simulations used multiple timestepping, with a base timestep of 2 fs, short-range interactions calculated every step, and long-range electrostatics every 2 steps. All hydrogen bond lengths were constrained using RATTLE or SETTLE as appropriate. Short range interactions were cut off at 10 Å, with switching starting at 9 Å. Full electrostatics were performed using PME 48 with a grid density of 1.0 Å-3. All MD trajectories were saved once per two ps.
The dark and light state structures were equilibrated in multiple steps. Initially, all protein backbone atoms were constrained to their initial coordinates, and all other protein and FMN atoms were restrained to their initial positions using a harmonic potential with a spring constant of 10.0 kcal / (mol Å2). The system was subjected to 3000 steps of conjugate gradient minimization, equilibrated for 200 ps, and then the spring constants on the harmonic restraints were reduced to zero in 10 steps over an additional 200 ps of simulation. The constraints on the protein backbone were then converted to harmonic restraints and the system equilibrated for 200 ps, followed by an additional 200 ps removal of the restraints as before. The unrestrained system was then equilibrated for an additional 100 ps. For each state, we subsequently ran five independent 200 ns simulations in the NVT ensemble, beginning from different random velocities and pseudorandom number generator seeds. With this simulation length, one can expect to observe events corresponding to short-timescale relaxation following the photoreaction, and perhaps early events leading up to Jα dissociation, but this duration is not sufficient to observe the complete dissociation event.
Network analysis
Dynamic network analysis was performed using the networkview plugin of VMD 1.9.1 and the accompanying gncommunities and subopt tools, using CARMA 1.1 21 as a backend for correlation calculations. For each of the dark and light states, a network was constructed with a node at the alpha carbon of each amino acid residue and four representative nodes at the FMN chromophore. The correlations Cij between motions of each pair of nodes i and j were calculated using the last 120 ns of each trajectory, and edges between connected nodes were assigned weights equal to - log(Cij), as in Sethi et al. 54, using trajectories aligned by the core β sheet.
For any path connecting a non-adjacent pair of nodes i and j, a distance was assigned to that path equal to the sum of weights of all edges along the path. The shortest distance between each pair of nodes was identified using the Floyd-Warshall algorithm18, and then each edge assigned a “betweenness” equal to the number of shortest paths passing through it. Each network was then partitioned into communities using the Girvan-Newman algorithm20 to maximize the difference in betweenness between inter- and intra-community edges.
Results
For both the dark and light states of the LOV domain, five separate 200 ns equilibrium MD trajectories were run to analyze differences in structure and dynamics between the two states. A summary of all simulations performed is presented in Table 1.
Table 1.
Summary of simulations performed. RMSDs compare a MD trajectory and the appropriate room-temperature crystal structure 23; core RMSDs include only residues 414-516, omitting the A′α and Jα helices.
| Name | Type | Structure | Duration | RMSD | RMSD(core) |
|---|---|---|---|---|---|
| DEQ-1 | NVT | Dark | 200.0 ns | 2.00 | 0.88 |
| DEQ-2 | NVT | Dark | 200.0 ns | 3.04 | 0.85 |
| DEQ-3 | NVT | Dark | 200.0 ns | 1.90 | 0.96 |
| DEQ-4 | NVT | Dark | 200.0 ns | 2.14 | 0.95 |
| DEQ-5 | NVT | Dark | 200.0 ns | 3.55 | 0.86 |
| LEQ-1 | NVT | Light | 200.0 ns | 2.45 | 1.16 |
| LEQ-2 | NVT | Light | 200.0 ns | 3.04 | 1.17 |
| LEQ-3 | NVT | Light | 200.0 ns | 2.30 | 1.05 |
| LEQ-4 | NVT | Light | 200.0 ns | 2.36 | 1.13 |
| LEQ-5 | NVT | Light | 200.0 ns | 2.53 | 0.99 |
The most obvious feature of all 10 trajectories is that the A′α helix shifts from its initial crystallographic conformation to a position where it interacts more closely with Jα, frequently leading to formation of a salt bridge between residues R410 and E545. In all cases, repositioning of A′α occurs less than 80 ns into the trajectory, and thus unless otherwise noted the analysis below is restricted to the last 120 ns of each trajectory. In no case did we observe the A′α helix to substantially repopulate the crystallographic conformation once it had undergone the shift.
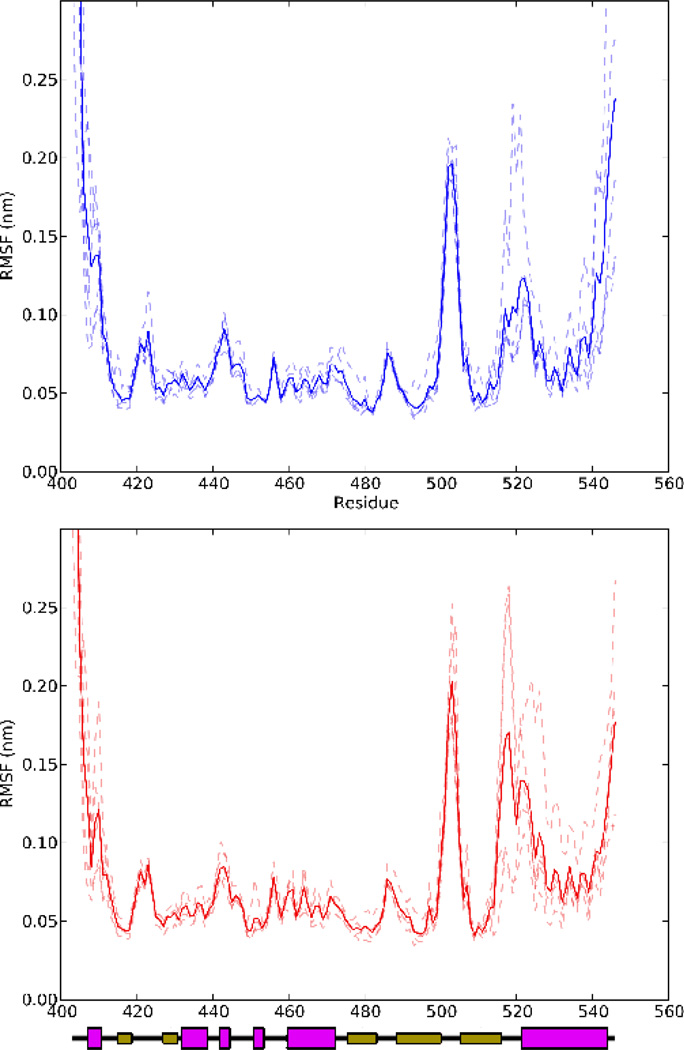
The Cα root mean square fluctuations (RMSFs) of all residues in the LOV domain for all ten equilibrium simulations are plotted in Fig. 2. The relatively low RMSFs of all residues in the core, particularly those involved in secondary structure elements, indicate that the domain core (residues 414-516, found among all LOV and PAS domains) is stable in all of these trajectories. On the other hand, significant fluctuations occur in all trajectories in the terminal regions, Jα helix, and Hβ-Iβ loop. Systematic differences are apparent between light and dark state simulations in the Iβ-Jα loop and Jα helix, where the linker region and top two turns of the helix are clearly more flexible in the light state than in the dark state. Substantial inter-trajectory variability is present in both cases; in the dark state, only one trajectory (DEQ-2) shows a particularly high RMSF in the Iβ-Jα loop and Jα helix, whereas in the light state trajectories LEQ-2, LEQ-3, and LEQ-5 all show higher mobility than in the dark state simulations.
Figure 2.
RMSF of all Cα atoms for each of the equilibrium MD runs in this study. The top panel shows the dark state simulations, and the bottom panel the light state simulations; in both cases traces are shown for each trajectory as dashed lines, and an average shown as a solid line.
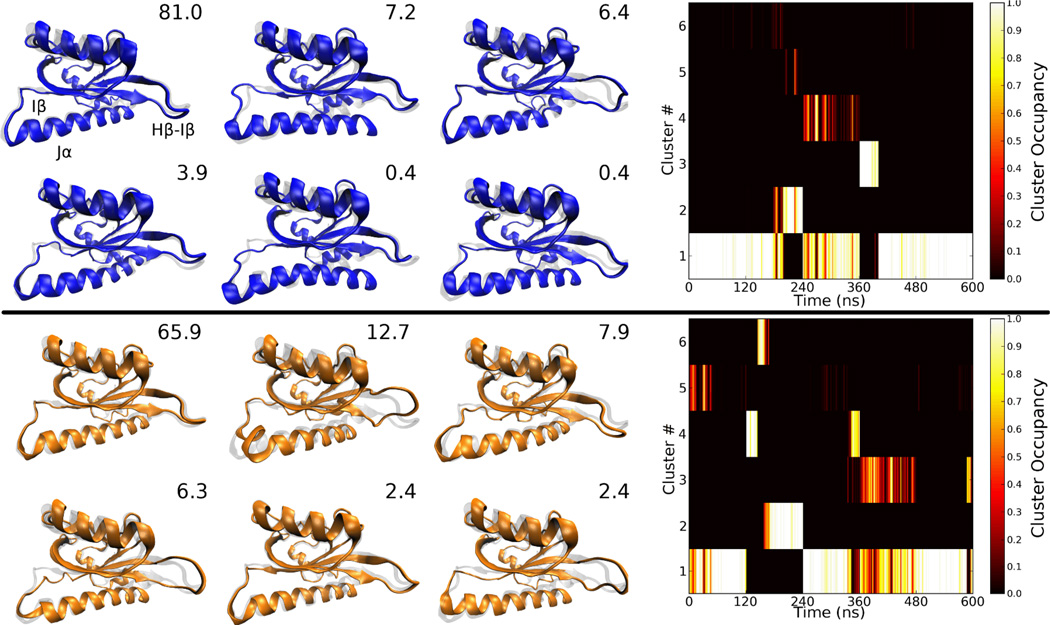
To further characterize the motion of the LOV domain during equilibrium MD, clustering analysis was performed on the dark and light state trajectories (excluding the first 80 ns of each run), taking frames once per 20 ps, with the LOV domain core aligned in both cases and then RMSDs for clustering calculated using all non-symmetry related heavy atoms except those in the N-terminal tail (prior to residue 414). Clusters were assigned using the Gromos clustering method 15 of GROMACS 3.3 58 with a cutoff of 0.16 nm; these RMSDs correspond to the first local minimum in the pairwise RMSD distribution for the light state, and the mode for the dark state (which showed only a single peak). Applying the clustering method to the complete dark and light state trajectories yields 17 and 23 clusters, respectively. The clusters are sorted in descending order of occupancy; we focus on the top six clusters for each case, which comprise 99.3% and 97.7% of timesteps for the dark and light states, respectively. The average conformations for each of the top six clusters from the dark and light states are compared to the dark state crystal structure in Fig. 3. The major clusters observed in the dark state simulations are all similar to each other and to the equilibrated structure, differing primarily in the positioning of the last two turns of Jα; however, the majority of contacts between Jα and the core remain intact; other structural elements do not undergo systematic changes (the largest change that does occur is in cluster two, where the Jα helix rotates slightly in the plane of the main β sheet). The light state, in contrast, shows several highly occupied clusters in which Iβ is bent away from the core, partly displacing Jα (clusters 2, 3, 4, and 6), and in which the Hβ-Iβ loop is rotated to bring it closer to Jα (clusters 2, 3, and 6). Equivalent bending of the Iβ strand occurs in the dark state only in trajectory DEQ-4, where the bent conformation (cluster 4) interchanges repeatedly with the dominant crystal-like conformation.
Figure 3.
Conformations of the first six clusters from clustering analysis on the dark (top) and light state equilibrium MD trajectories, with population in order of descending population. The dark state crystal structure of the appropriate state is shown in a grey transparent representation for reference. Inset numbers indicate the percentage of timesteps belonging to that cluster. To the right of the structures, the occupancy density of each cluster throughout each trajectory is shown (averaged over 2 ns windows). Results for the five trajectories for each state are concatenated for simplicity of display. Due to culling of the first 80 ns of each trajectory, the 600 ns time period shown corresponds to 5 × 120 ns since each trajectory contributes only 120 ns to the plot.
Correlated motion of the LOV domain
In order to better characterize the structural changes noted based on clustering analysis, correlated motions in the equilibrium trajectories of the LOV domain were analyzed using both conventional Pearson coefficients 28 and Lange and Grubmüller’s mutual information based general correlation method 35. The combination of these methods is particularly useful: Pearson coefficients provide a signed measure of the collinear component of correlation between two atoms, and thus their sign can be directly interpreted in terms of whether atoms tend to move in lockstep (positive) or show opposed motion (negative) while the magnitude indicates the strength of the correlation; this method will not detect correlations that are not collinear or occur out of phase with each other. The mutual information-based correlation matrix provides a more sensitive method for detecting any type of dependence in the motion of two atoms, regardless of (for example) the direction of motion, but cannot itself provide information on the direction or nature of the correlation; the unsigned value provided by this method is simply a normalized measure of how much information on one atom’s position is provided by that of another atom.
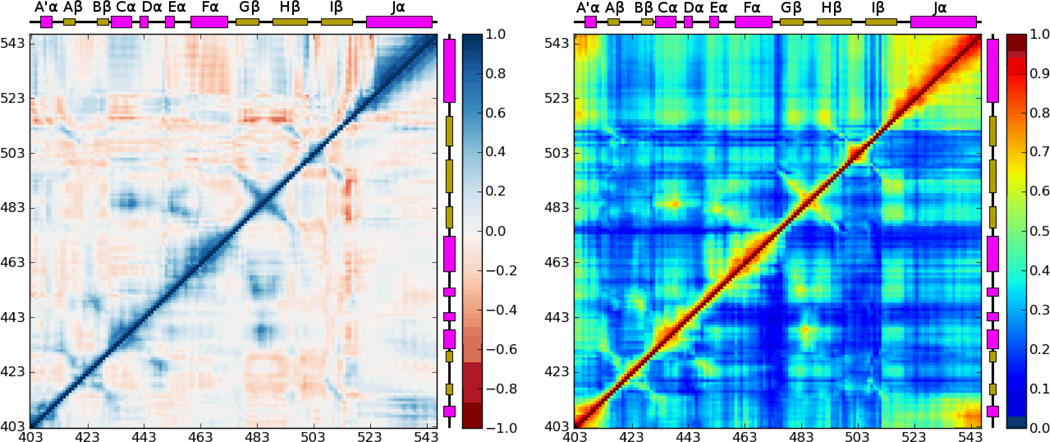
The two types of correlation matrices were calculated for the combined trajectories of simulations DEQ-1-5 and LEQ-1-5, and are shown in Fig. 4. Both the dark and light states show a fairly typical pattern of correlations corresponding to secondary structure elements, with lines perpendicular to the main diagonal indicating interacting β-strands and a thickened region of relatively high correlation along the diagonal indicating α-helices. Other off-diagonal correlations such as that between the Gβ-Hβ loop and the Eα helix generally correspond to strong interactions between neighboring regions of the structure, such as (in this case) the E444-K485 salt bridge.
Figure 4.
Covariance (left) and correlation (right) matrices for the equilibrium MD simulations in this study. In each case, the combined results for the five dark state trajectories are shown in the lower triangle and results for the light state trajectories are shown in the upper triangle. Regions showing particularly strong differences in correlation structure, the A′α/Jα and Hβ-Iβ/Jα interactions, are highlighted with purple boxes.
Two particularly significant differences between the correlated motions of the dark and light state are readily apparent. While in both the dark and light states the motion of the A′α helix is correlated with the C-terminal portion of Jα, in the light state the A′α helix and adjacent loops are also strongly coupled to the C-terminal portion of Iβ and the Iβ-Jα linker. At the N-terminal end of the Iβ strand, in the dark state the Hβ-Iβ loop shows little correlation with other regions of the protein, and the linker region and Jα helix show strong internal correlations, but only weaker correlation with each other; in the light state the motion of the linker and entire Jα helix are strongly correlated with both each other and the Hβ-Iβ loop. The sign of the Pearson correlation for these regions indicates that at least the collinear component of the motion of the linker is anticorrelated with that of the Jα helix; the Hβ-Iβ loop shows opposed patterns for the N- and C-terminal halves of the loop, consistent with the structures observed in clustering analysis in which the loop twists relative to the crystal structure such that its C-terminal end and the beginning of Iβ approach Jα.
Formation of the C450-FMN photoadduct triggers coupled motions in the A′α helix and Jα helix
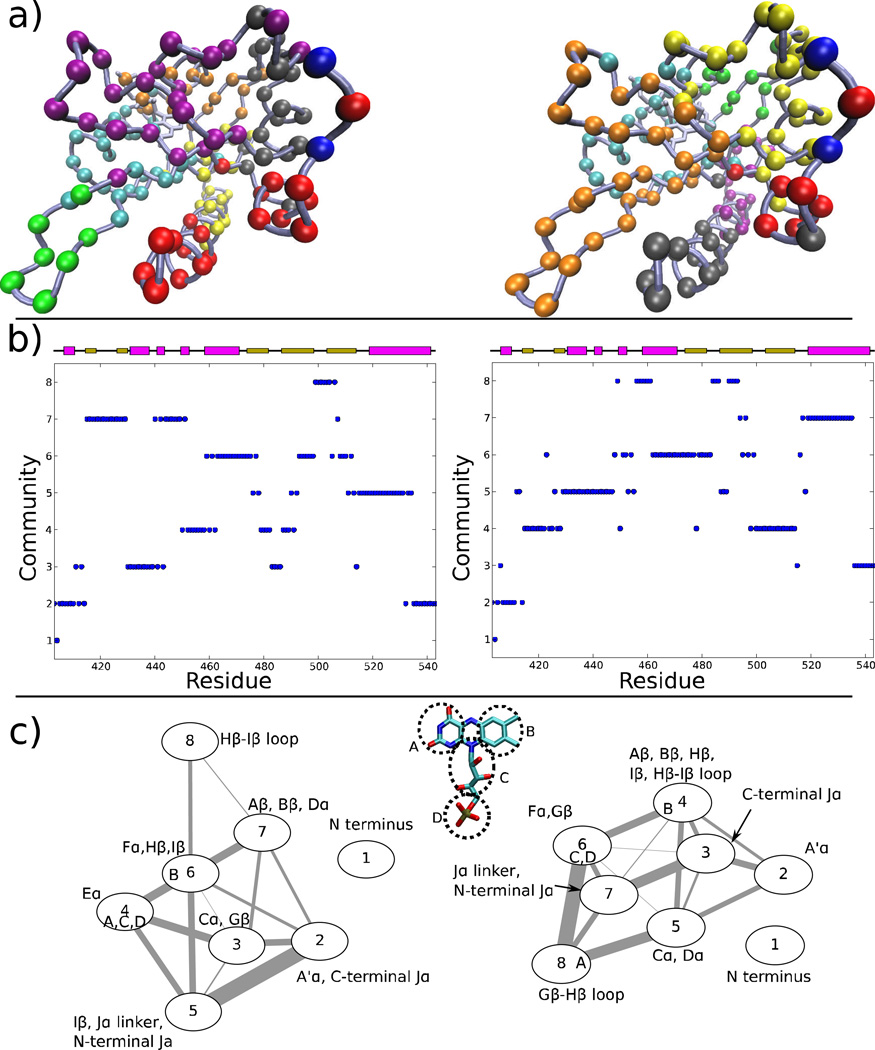
The analysis of the information flow in a coarse-grained network with connectivities determined based on the correlations between motions of different residues has recently proven to be a powerful tool in identifying the pathways mediating allosteric interactions in tRNA synthetases 2,54. In order to identify the paths of information flow altering the behavior of the Jα helix in AsPhot1-LOV2, we generated such a network separately for the dark and light state equilibrium trajectories described above. In this network, each amino acid residue is represented as a single node, FMN is divided into 4 nodes (see Fig. 5a), and edges are determined based on the proximity and correlated motion of the nodes (see Methods for details). We then partitioned the nodes into strongly connected communities using the Girvan-Newman algorithm, and examined community membership (Fig. 5a–b) and the strength of coupling between communities (Fig. 5c).
Figure 5.
Network analysis of correlated motions in the LOV domain. (a) Partition of residues from the dark (left) and light (right) states into communities; the communities are colored in numerical order: blue, red, gray, orange, yellow, tan, purple, green, white. (b) Community identity of each residue in the dark (left) and light states. (c) Interactions of communities in the dark (left) and light states; the width of each edge is proportional to the number of shortest paths passing through the interface of those communities (see Sethi et al. 54). Membership of nodes from the chromophore is indicated by letters within the circle for each node, based on the division shown in an inset.
Two differences between the community interactions in the dark and light states are particularly striking. First, whereas in the dark state motion of the A′α helix is sufficiently coupled to the C-terminus of Jα such that these secondary structure elements form a community (community 2), in the light state, only one A′α residue (T407) interacts strongly enough with Jα to be included in the same community. Second, whereas in the dark state the Hβ-Iβ loop acts more or less independently from the β sheet core, in the light state changes in the dynamics of the Hβ and Iβ strands occur that couple the motions of the Hβ-Iβ loop, Hβ and Iβ strands with the Jα helix (compare community 8 of the dark state with community 4 of the light state). It is also notable that the two light state trajectories that show partly dissociated states (LEQ-2 and LEQ-4) also show very little interaction between A′α and Jα when network analysis is run independently on the individual trajectories, whereas the other three light state simulations show a community assignment pattern more similar to the dark state (data not shown); LEQ-2 is also the only trajectory showing substantial interaction between the community containing the Hβ-Iβ loop and Jα.
The results of the network analysis, combined with the correlation analysis shown in Fig. 4, point toward two changes in allosteric coupling between the FMN binding site and the Jα helix. Formation of the FMN photoadduct causes the formation of a rigid region of the LOV domain core consisting of the Hβ-Iβ loop and proximal portions of the Hβ and Iβ strands (communities 6 and 8 in the dark state; community 4 the light state), which disrupts the native contacts with Jα due to tilting of the Iβ strand toward Jα. At the same time, the A′α helix loses interactions with Jα and shifts away from it (compare community 2 of the dark state with 2 and 3 in the light state; see also Fig. 4). The combination of these structural changes causes the partial Jα dissociation observed in some of our light state trajectories, and presumably leads to complete dissociation over longer timescales (which could not be consistently captured by our 200 ns simulations).
A recent comparison of methods for network analysis showed that at least in the case of imidazole glycerol phosphate synthase, better correspondence with experimental results was obtained by using the center of mass of each residue in the needed calculations, rather than each residue’s alpha carbon position 59. We thus repeated the above network analysis using this residue-level method; the results are shown in Supplementary Figure SI2. The results from the residue-level analysis are almost completely equivalent to the Cα-based analysis presented above; most notably, the strong coupling in the light state between the Hβ-Iβ loop, Jα, and the Aβ and Bβ strands in the light state is still apparent. The only notable difference is that the residue-level network suggests somewhat stronger interaction between A′α and the C-terminal portion of Jα in the light state than the Cα-based analysis. This likely reflects the side chain-side chain interactions between these portions of the protein present in some light state trajectories; their significance for Jα dissociation is unclear given that these very contacts are generally least stable in the light state trajectories showing the most dissociation.
The FMN binding site
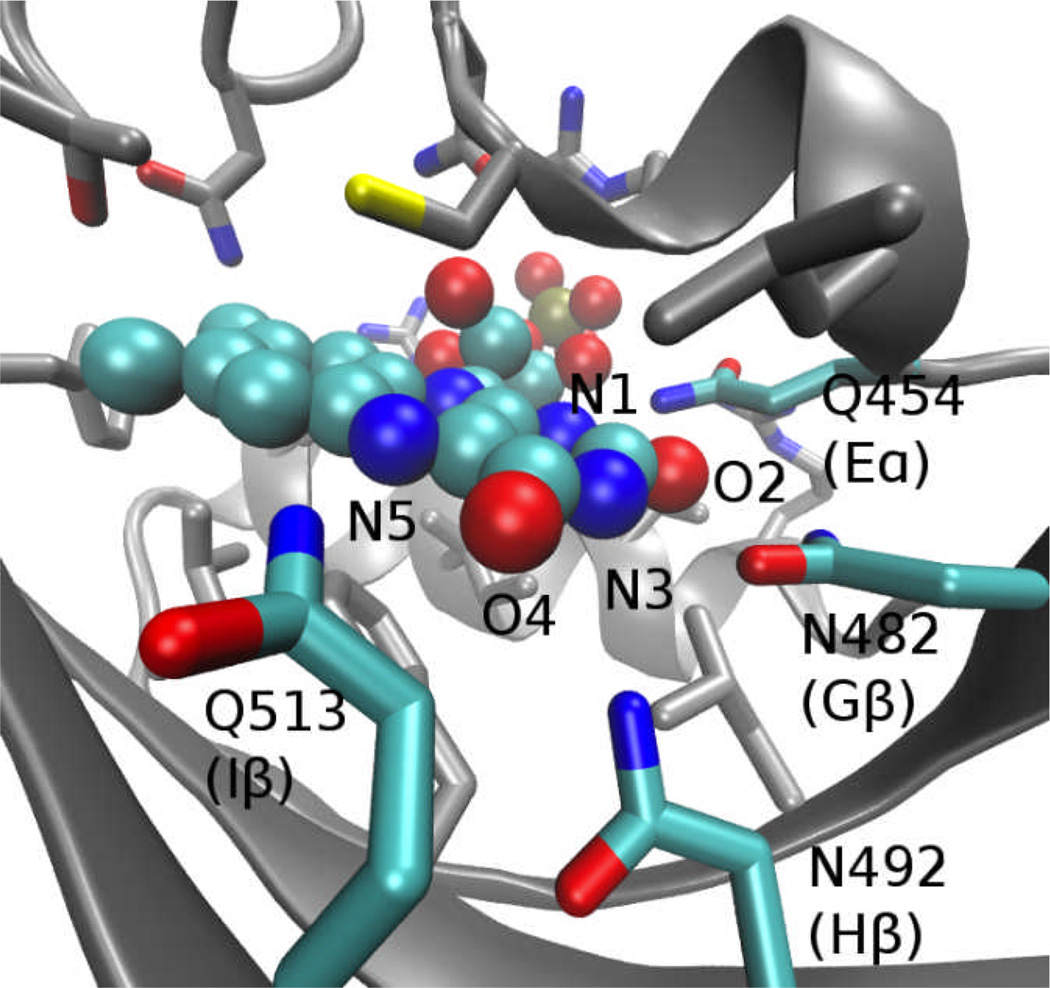
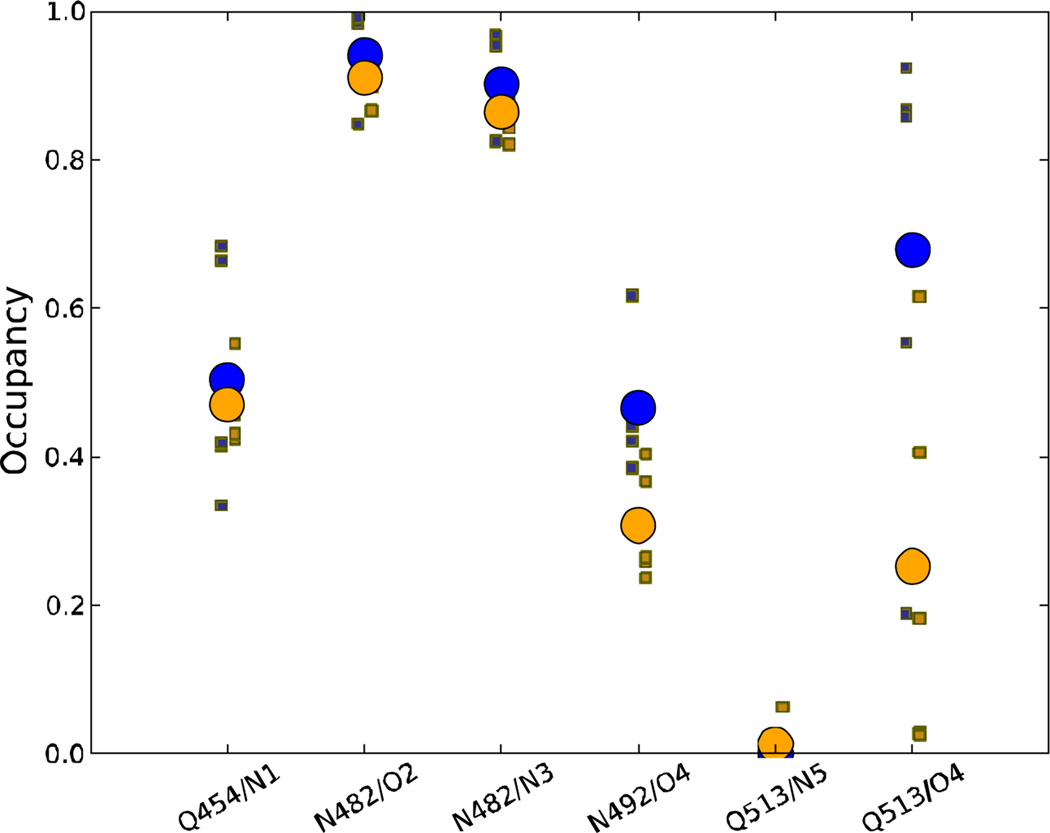
Given the differences in behavior of the Hβ-Iβ loop, Iβ strand, and Jα helix in the dark vs. light state trajectories noted above, the question arises how the small chemical change associated with the LOV domain photoreaction gives rise to the large scale structural rearrangements needed for switching the domain between resting and signaling states. As in previous simulations 19, no major qualitative changes were apparent in the interactions between the LOV domain and the ribose moiety of FMN or the hydrophobic half of the isoalloxazine ring; however, we observed substantial changes in the network of hydrogen bonds between the protein and the polar half of the ring (see Fig. 6). The occupancy frequencies of these hydrogen bonds throughout the trajectories in the present study are shown in Fig. 7. The most substantial differences between trajectories occur in the behavior of Q513, which shows higher inter-trajectory variation than other contacts, and which has a substantially lower occupancy of the O4-Q513 hydrogen bond in the light state trajectories relative to the dark state. The N492-O4 hydrogen bond also shows a smaller, but still significant, drop in occupancy in the light state trajectories (p=0.017, Welch’s t-test, treating the occupancy of each trajectory as a single data point).
Figure 6.
Key residues and FMN ring atoms involved in polar contacts at the FMN binding site.
Figure 7.
Occupancies of major FMN-LOV hydrogen bonds over the last 120 ns of the equilibrium trajectories in this study. Hydrogen bonds are defined by a donor-acceptor distance of less than 3.5 Å. Dark state data are shown in blue and light state in orange; the occupancies of the individual trajectories are shown as small squares, and the average over the 5 trajectories as a large circle.
As discussed below, a number of previous experimental 24,30,37,43, structural 13,14,17,23,63, and computational studies 16,19 implicated the residue at the position of Q513 in the activation of LOV domains from phototropins of several organisms, as well as N. crassa Vivid 63. In contrast with the usual inference from structural studies that formation of the photoadduct leads to reorientation of Q513 to form a hydrogen bond with the (newly protonated) N5 atom of FMN (e.g., ref. 23), hydrogen bonding with N5 was nearly nonexistent in the trajectories shown here. The lowered occupancy of the Q513-O4 hydrogen bond in our trajectories thus cannot result from competition with hydrogen bonding to N5.
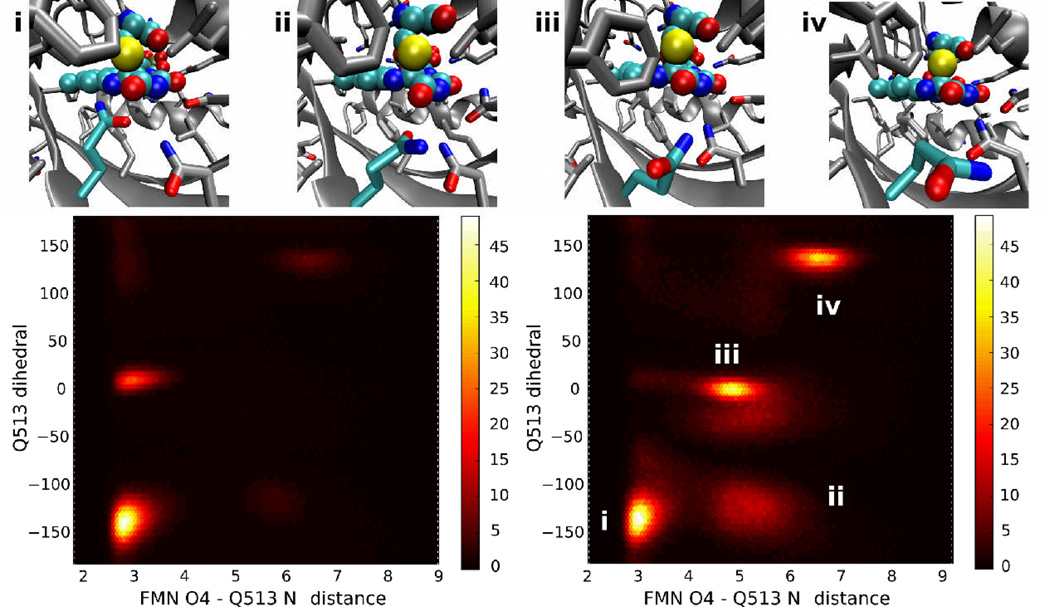
Instead, investigation of the behavior of Q513 throughout our simulations reveals the presence of several heavily occupied conformations of the Q513 side chain, with differing occupancies between the dark and light states. The dominant conformers are shown in Fig. 8; conformation i is the most occupied one in both the dark and light states, and forms a hydrogen bond between Q513 and the O4 atom of FMN similar to that observed in the dark state crystal structure. However, whereas in the dark state only one other (unlabeled) conformation with a similar hydrogen bond but different orientation of Q513 shows any substantial occupancy, in the light state three other major conformations occur. In conformation ii, the interactions between Q513 and FMN are disrupted by a bound water interacting with the protonated FMN N5 atom, and Q513 instead interacts with the water molecule. Conformation iii is similar to ii in the presence of a site-bound water molecule, but differs in the degree to which Q513 is rotated away from the FMN ring system. In conformation iv, even indirect contact with FMN is lost, and Q513 is instead rotated toward N414 and D515. As noted previously 55, in cases where Q513 is no longer hydrogen bonded to the N5 atom of FMN, we frequently observe the presence of a site-bound water hydrogen bonded to N5, O4, and the Q513 amide nitrogen.
Figure 8.
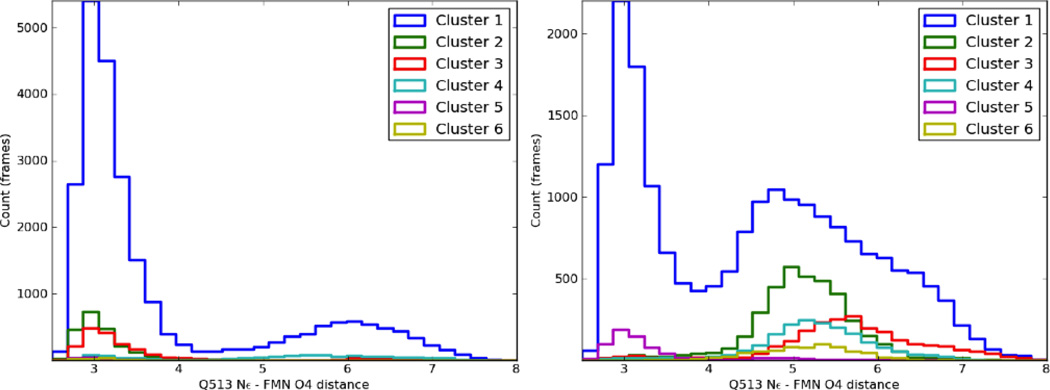
Common conformations of Q513 observed during equilibrium trajectories. Along the top of the figure, representative structures from the four most common Q513 orientations are shown; the chromophore and Q513 are colored cyan. Below, density plots of the occupancy throughout all dark (left) or light (right) state trajectories for different values of the Q513 Nε-FMN O4 distance and the dihedral formed by the N, Cα, Cγ, and Cδ atoms of Q513 (Q513 dihedral); the color scale shows the number of snapshots in each bin, with snapshots taken at 2 ps intervals over the complete trajectories. The regions corresponding to the four snapshots are shown in the light state density plot.
Even in conformation i, minor variations exist in both the dark and light state, in that we observe an equilibrium between two conformations differing at the Q513 χ2 dihedral (compare conformation i in Fig. 8 with the crystallographic conformation shown in Fig. 6). The alternate Q513 rotamer, which occurs in approximately 80% of dark state timesteps and 84% of light state timesteps, allows Q513 to maintain interaction with the FMN O4 while at the same time hydrogen bonding with N414 or the antechamber site water (see Song et al.55). We did not, however, observe any correlation between the χ2 rotamerization state and larger-scale changes in LOV domain structure except in cases where the Q513-O4 interaction was lost as described above.
The overall importance of the conformation of Q513, located in the Iβ strand, becomes readily apparent when its behavior is compared to overall structural deformations in the LOV domain. As noted above, clustering analysis of the dark state trajectories showed few differences between major conformations, whereas in the light state several clusters (2, 3, 4, and 6) showed tilting of Iβ to disrupt the interaction surface with Jα, and rearrangement of the Hβ-Iβ loop and nearby regions of Hβ/Iβ to interact more strongly with Jα. The status of the Q513-FMN O4 hydrogen bond for timesteps corresponding to different clusters in each set of trajectories is shown in Fig. 9. Crucially, whereas little correlation is observed between cluster occupancies and the status of the Q513-FMN interaction in the dark state, our corresponding light state simulations show disruption of the core-Jα interaction (clusters 2, 3, 4, and 6) only when the Q513 interaction is broken, with or without replacement by a site-bound water.
Figure 9.
Histograms showing the occupancy of different Q513 Nε-FMN O4 heavy atom distances for the dark (left) and light (right) simulations, divided by the cluster assigned at each timestep (see above). As with the clustering itself, only one frame is considered per 20 ps.
The fact that the Iβ strand interacts directly with the Aβ strand, and that these strands are strongly coupled in both the dark and light states, provides a ready explanation for the change in behavior of A′α/Aβ/Bβ in the light state: tilting of Iβ induced by conformational changes in Q513 also triggers a change in the conformation of this N-terminal portion of the protein through their direct interaction in the β sheet, giving rise to the very different behavior of A′α observed in the light state. When rotated away from FMN, Q513 also frequently hydrogen bonds with N414. This presents an intriguing alternative possibility for propagation of information from the FMN photoreaction to A′α, Aβ, and Bβ: hydrogen bonding between N414 and Q513 in the rotated conformation of Q513 could alter the dynamics of the A′α-Aβ-Bβ region and allowing it to interact more strongly with Jα rather than the LOV domain core. This possibility is supported by the fact that Sosnick and colleagues found an N414V mutation to yield a protein with somewhat deficient Jα dissociation that was also aggregation prone 62.
Discussion
Despite the fact that the equilibrium MD simulations performed here were significantly shorter than the expected times for Jα dissociation (on the order of hundreds of µs 7,40,61), putative pre-dissociation states were observed in several of the light-state trajectories; these states likely represent an intermediate conformation on the complete dissociation pathway. The pre-dissociation states show characteristic structural changes to the Iβ strand and Hβ-Iβ loop, leading to tilting of the Iβ strand (such that it forces a corresponding movement of Jα and disrupts the native interface between the secondary structure elements) and rearrangement of the Hβ-Iβ loop to interact more closely with Jα. In addition, network analysis of correlated motions in the equilibrium trajectories shows that both this shift in the Hβ-Iβ loop and a structural change in the A′α helix, Aβ, and Bβ strands are strongly correlated with the motion of Jα in the light state, suggesting that both changes are involved in Jα dissociation.
In seeking a trigger for the observed conformational change, closer inspection of the FMN binding site in the light state simulations indicates that only one residue, Q513, shows qualitative changes in its conformation and interactions with FMN between the light and dark states. This residue exhibited four major conformations, differing both in their interactions with FMN and the surrounding portions of the protein. While in the dark state mainly the initial structure was populated, in the light state all four Q513 conformations arose. Q513 has been noted as a possible mediator of activation in numerous previous studies 13,14,16,17,19,23,24,30,37,43,44,63; in the present simulations it is also clearly linked to a structural transition in the light state.
In one particularly pertinent experimental study, Nash et al. found that a Q513L mutation locked the LOV domain into a pseudo-dark state with only a minor structural response to illumination, whereas a Q513N mutation put the protein into a pseudo-lit state even without illumination 41. The effect of the Q513N mutation is readily rationalized in light of our findings on conformational transitions of Q513 in the wild type protein: the Q513N mutation likely forces this glutamine to assume a light-like conformation in the dark due to its shortened side chain. The fact that any light-induced conformational change occurs in the Q513L mutant, however, deserves further exploration. We obtained two 160 ns trajectories of the Q513L mutation in each of the light and dark state (using methods identical to those for the wild type trajectories, with the Q513L substitution imposed using the VMD psfgen plugin). Neither dark state trajectory showed any notable change in the relative positions of secondary structure elements compared with the wild type protein, but in both light state trajectories, the C-terminal end of the Iβ sheet bent slightly toward Jα, similar to the behavior observed in the wild type light state simulations. Unlike the wild type trajectories, however, no larger scale effects (Jα dissociation or Hβ-Iβ bending) occurred in either trajectory. These results suggest that in the Q513L mutant, the LOV domain photoreaction causes a local conformational change that is superficially similar to that occurring in the wild type protein, but due to the inability of the leucine to participate in the alternate hydrogen bonding patterns of Q513 (with N414, N492, O4, and site bound waters), this local structural change does not propagate to the rest of the protein. The shift in L513 that does occur in this mutant appears to be the result of conformational frustration in the FMN binding site: as shown in Supplementary Figure SI3, in the dark state of the Q513L mutant, both F434 and L513 are much more constrained in their motion than in the wild type protein. Formation of the FMN photoadduct causes both of these residues to shift further toward the main beta sheet, causing a slight bend in Iβ. It is also notable that, unlike in the wild type light state simulations, we did not observe site bound water interacting with N5 in our Q513L light state trajectories, presumably due to the absence of Q513 as a hydrogen bonding partner. This absence may contribute to the lack of larger-scale structural changes in the Q513L mutant, and also likely accounts for the extremely slow recovery kinetics of the Q513L mutant (more than tenfold slower than the wild type 41), given our previous findings that solvent at this site contributes to the recovery reaction 55.
The role of Q513 in overall structural changes of the LOV domain should not be surprising, particularly given the nature of the changes identified in our simulations; Q513 resides on the Iβ strand, and thus changes to its conformation can easily translate both to tilting of Iβ and rearrangement of the Hβ-Iβ loop. Based on our simulations it is apparent that the formation of a LOV domain-FMN photoadduct simultaneously changes the conformation of Q513 to break the Q513-FMN O4 hydrogen bond present in the dark state, and generates a partially occupied water binding site adjacent to the newly protonated N5 atom of FMN (this water occupancy also appears to play a key role in the reversion of the photoadduct to the dark state 55). The reconfiguration of Q513, in turn, propagates to cause changes in Iβ and the Hβ-Iβ loop which alter the overall interaction with Jα. The shifting conformation of Iβ also triggers a rearrangement in the dynamics of A′α and the Aβ and Bβ strands, coupling them to Jα. These two changes in information flow are apparent both from correlation analysis, which reveals substantial changes in correlations between A′α, Hβ-Iβ, and Jα; from the network analysis shown in Fig. 5, which illustrates that the dynamics of Aβ,Bβ, Hβ, Iβ, and the Hβ-Iβ loop become coupled in the light state; and from the finding that the partially dissociated states observed in our light state simulations almost uniformly show substantial tilting of Iβ (which, as noted above, is strongly correlated with several other secondary structure elements).
Our analysis has thus revealed two paths through which the LOV domain photoreaction is coupled to Jα dissociation, which apparently act in parallel: formation of the photoadduct causes a shift in Iβ through rotamerization of Q513, altering the dynamics of Jα both through coupling with the A′α helix, and a shift of the Hβ-Iβ loop and nearby portions of the connected β strands toward Jα. The A′α helix was recently found to play a key role in Jα dissociation of AsPhot1-LOV2 based on extensive mutagenesis, circular dichroism, and NMR data 62. The Hβ-Iβ loop was also predicted to play a role in Jα dissociation based on previous computational results on a truncated phototropin LOV domain 19, but to our knowledge its role has not yet been directly tested experimentally. The present set of trajectories do not show substantial interactions between the Hβ-Iβ loop itself and Jα; instead, it appears likely that the shift of this loop is symptomatic of the tilting of Iβ that destabilizes the LOV-Jα interface. We must also caution that although both the data presented here and recent experimental results suggest roles for changing interactions between A′α-Jα and Iβ-Jα in causing Jα dissociation, as the identity and order of intermediate states have not yet been established, it would be premature to denote any one interaction as a primary or initial “driver” of dissociation, and it appears likely that both interactions contribute substantially to the thermodynamics of the dissociated state. As noted above, the duration of the present simulations was still insufficient to observe complete Jα dissociation, and thus it remains unclear how the altered dynamics of the A′α-Aβ-Bβ region stabilize conformations in which Jα is dissociated from the LOV domain core (this may be as simple as the loss of interactions between A′α and Jα stabilizing dissociated conformations in the light state). Future computational results in which dissociation has been induced using applied forces may provide additional insight into this facet of AsPhot1-LOV2 activation and reveal the most common structural paths followed during dissociation.
In revealing these paths of information flow from the FMN binding site to Jα dissociation, our results provide an atomic-level interpretation for a variety of experimental data, most recently the results of an extensive series on LOV domain mutants from the Sosnick group 62. We have identified the specific residues coupling the LOV domain photoreaction to Jα, which involves both interactions through the A′α helix, and through the core β sheet of the LOV domain. Analysis of the most substantially dissociated states observed in our trajectories (particularly those from LEQ-2) permits us to propose several follow-up experiments to test specific aspects of our mechanistic hypothesis. Our finding that activation requires rotation of Q513 away from FMN O4 and toward N414 suggests that mutation of Q513 to a glutamic acid residue would push the LOV domain toward the light state (by destabilizing the interaction with FMN); the effect would be expected to be stronger if complemented with mutation of N414 to a basic residue. The peculiar conformation of the Hβ-Iβ loop observed in our partially dissociated states allows formation of a salt bridge between D501 and K544 that is not otherwise heavily occupied in the dark or light states. Mutation of D501 to a basic residue should thus delay or weaken the light-induced Jα dissociation, as this conformation of the Hβ-Iβ loop accompanies the Iβ tilting that triggers dissociation. Our proposal that Iβ tilting ultimately plays a major role in Jα dissociation has already been tested to some extent by Zayner and colleagues 62, who designed mutations at this interface based on a brief description of the mechanism in our previous work 55 and observed substantial effects on light-induced structural changes. We may add to this set of mutations two additional proposals: first, in the partially dissociated states, a salt bridge forms between E518 and R521, which is not otherwise strongly occupied; removal of this salt bridge should reduce the strength of light-induced structural changes. Second, we observe that dissociation begins with breakage of Jα at G528 (in agreement with simulations from the Baeurle group suggesting that dissociation begins in the N-terminal portion of Jα 46); replacement of G528 with a helix-stabilizing residue such as alanine should hinder light-induced Jα dissociation.
Supplementary Material
Acknowledgments
This work was supported by grant No. PHS-5-P41-RR05969 from the National Institutes of Health (to K.S.), grant No. MCB02-34938 from the National Science Foundation (to K.S.), and grant No. R01-GM081875 (to K.H.G.), and grant I-1424 from the Robert A. Welch Foundation (to K.H.G.). Computer time was provided by the Turing cluster at UIUC, an Illinois Advanced Computing Applications and Technologies grant on the Abe cluster at UIUC, and LRAC MCA93S028. P.L.F. gratefully acknowledges support from the National Science Foundation Graduate Research Fellowship Program. We thank Dr. Jérôme Hénin, Dr. Chris Harrison, Dr. Elizabeth Villa, and Dr. Abigail Nash for many helpful discussions.
References
- 1.Bittl R, Kay CWM, Weber S, Hegemann P. Characterization of a flavin radical product in a C57M mutant of a LOV1 domain by electron paramagnetic resonance. Biochemistry. 2003;42:8506–8512. doi: 10.1021/bi034123i. [DOI] [PubMed] [Google Scholar]
- 2.Black Pyrkosz A, Eargle J, Sethi A, Luthey-Schulten Z. Exit strategies for charged tRNA from GluRS. J Mol Biol. 2010;397(5):1350–1371. doi: 10.1016/j.jmb.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briggs WR, Beck CF, Cashmore AR, Christie JM, Hughes J, Jarillo JA, Kagawa T, Kanegae H, Liscum E, et al. The phototropin family of photoreceptors. Plant Cell. 2001;13:993–997. doi: 10.1105/tpc.13.5.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Briggs WR, Christie JM. Phototropins 1 and 2: versatile plant blue-light receptors. Trends Plant Sci. 2002;7:204–210. doi: 10.1016/s1360-1385(02)02245-8. [DOI] [PubMed] [Google Scholar]
- 5.Briggs WR, Christie JM, Salomon M. Phototropins: a new family of flavin-binding blue light receptors in plants. Antioxid Redox Signal. 2001;3:775–788. doi: 10.1089/15230860152664975. [DOI] [PubMed] [Google Scholar]
- 6.Briggs WR, Huala E. Blue-light photoreceptors in higher plants. Annu Rev Cell Dev Biol. 1999;15:33–62. doi: 10.1146/annurev.cellbio.15.1.33. [DOI] [PubMed] [Google Scholar]
- 7.Chen E, Swartz TE, Bogomolni RA, Kliger DS. A LOV story: the signaling state of the phot1 LOV2 photocycle involves chromophore-triggered protein structure relaxation, as probed by far-UV time-resolved optical rotatory dispersion spectroscopy. Biochemistry. 2007;46(15):4619–4624. doi: 10.1021/bi602544n. [DOI] [PubMed] [Google Scholar]
- 8.Christensen R. The electronic states of carotenoids. In: Frank H, Young A, Britton G, Cogdell R, editors. The photochemistry of carotenoids. The Netherlands: Kluwer; 1999. pp. 137–159. [Google Scholar]
- 9.Christie JM, Briggs WR. Blue light sensing in higher plants. J Biol Chem. 2001;276:11 457–11460. doi: 10.1074/jbc.R100004200. [DOI] [PubMed] [Google Scholar]
- 10.Christie JM, Reymond P, Powell GK, Bernasconi P, Raibekas AA, Liscum E, Briggs WR. Arabidopsis NPH1: a flavoprotein with the properties of a photoreceptor for phototropism. Science. 1998;282(5394):1698–1701. doi: 10.1126/science.282.5394.1698. [DOI] [PubMed] [Google Scholar]
- 11.Christie JM, Swartz TE, Bogomolni RA, Briggs WR. Phototropin LOV domains exhibit distinct roles in regulating photoreceptor function. Plant J. 2002;32(2):205–219. doi: 10.1046/j.1365-313x.2002.01415.x. [DOI] [PubMed] [Google Scholar]
- 12.Corchnoy SB, Swartz TE, Lewis JW, Szundi I, Briggs WR, Bogomolni RA. Intramolecular proton transfers and structural changes during the photocycle of the LOV2 domain of phototropin 1. J. Biol. Chem. 2003;278(2):724–731. doi: 10.1074/jbc.M209119200. [DOI] [PubMed] [Google Scholar]
- 13.Crosson S, Moffat K. Structure of a flavin-binding plant photoreceptor domain: insights into light-mediated signal transduction. Proc Natl Acad Sci U S A. 2001;98:2995–3000. doi: 10.1073/pnas.051520298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crosson S, Moffat K. Photoexcited structure of a plant photoreceptor domain reveals a light-driven molecular switch. Plant Cell. 2002;14:1067–1075. doi: 10.1105/tpc.010475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daura X, Gademann K, Jaun B, Seebach D, van Gunsteren WF, Mark AE. Peptide folding: When simulation meets experiment. Angewandte Chemie International Edition. 1999;38:236–240. [Google Scholar]
- 16.Dittrich M, Freddolino PL, Schulten K. When light falls in LOV: a quantum mechanical/molecular mechanical study of photoexcitation in Phot-LOV1 of Chlamydomonas reinhardtii. J Phys Chem B. 2005;109(26):13 006–13 013. doi: 10.1021/jp050943o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fedorov R, Schlichting I, Hartmann E, Domratcheva T, Fuhrmann M, Hegemann P. Crystal structures and molecular mechanism of a light-induced signaling switch: The Phot-LOV1 domain from Chlamydomonas reinhardtii. Biophys J. 2003;84:2474–82. doi: 10.1016/S0006-3495(03)75052-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Floyd R. Algorithm 97. Comm. ACM. 1962;5–6:345. [Google Scholar]
- 19.Freddolino PL, Dittrich M, Schulten K. Dynamic switching mechanisms in LOV1 and LOV2 domains of plant phototropins. Biophys J. 2006;91(10):3630–3639. doi: 10.1529/biophysj.106.088609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Girvan M, Newman MEJ. Community structure in social and biological networks. Proc Natl Acad Sci U S A. 2002;99(12):7821–7826. doi: 10.1073/pnas.122653799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glykos NM. Software news and updates. Carma: a molecular dynamics analysis program. J Comput Chem. 2006;27(14):1765–1768. doi: 10.1002/jcc.20482. [DOI] [PubMed] [Google Scholar]
- 22.Grübmuller H. SOLVATE 1.0 Manual. 1996. [Google Scholar]
- 23.Halavaty A, Moffat K. N- and C-Terminal Flanking Regions Modulate Light-Induced Signal Transduction in the LOV2 Domain of the Blue Light Sensor Phototropin 1 from Avena sativa. Biochemistry. 2007;46:14 001–14 009. doi: 10.1021/bi701543e. [DOI] [PubMed] [Google Scholar]
- 24.Harper SM, Christie JM, Gardner KH. Disruption of the LOV-Jalpha helix interaction activates phototropin kinase activity. Biochemistry. 2004;43(51):16 184–16 192. doi: 10.1021/bi048092i. [DOI] [PubMed] [Google Scholar]
- 25.Harper SM, Neil LC, Day IJ, Hore PJ, Gardner KH. Conformational changes in a photosensory LOV domain monitored by time-resolved NMR spectroscopy. J. Am. Chem. Soc. 2004;126:3390–3391. doi: 10.1021/ja038224f. [DOI] [PubMed] [Google Scholar]
- 26.Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 27.Humphrey W, Dalke A, Schulten K. VMD – Visual Molecular Dynamics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 28.Ichiye T, Karplus M. Collective motions in proteins: A covariance analysis of atomic fluctuations in molecular dynamics and normal mode simulations. Proteins: Struct.,Func.,Gen. 1991;11:205–217. doi: 10.1002/prot.340110305. [DOI] [PubMed] [Google Scholar]
- 29.Jarillo JA, Gabrys H, Capel J, Alonso JM, Ecker JR, Cashmore AR. Phototropin-related NPL1 controls chloroplast relocation induced by blue light. Nature. 2001;410:952–954. doi: 10.1038/35073622. [DOI] [PubMed] [Google Scholar]
- 30.Jones MA, Feeney KA, Kelly SM, Christie JM. Mutational Analysis of Phototropin 1 Provides Insights into the Mechanism Underlying LOV2 Signal Transmission. J. Biol. Chem. 2007;282(9):6405–6414. doi: 10.1074/jbc.M605969200. [DOI] [PubMed] [Google Scholar]
- 31.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 32.Kagawa T, Sakai T, Suetsugu N, Oikawa K, Ishiguro S, Kato T, Tabata S, Okada K, Wada M. Arabidopsis NPL1: a phototropin homolog controlling the chloroplast high-light avoidance response. Science. 2001;291:2138–2141. doi: 10.1126/science.291.5511.2138. [DOI] [PubMed] [Google Scholar]
- 33.Kay CWM, Schleicher E, Kuppig A, Hofner H, Rdiger W, Schleicher M, Fischer M, Bacher A, Weber S, et al. Blue light perception in plants. Detection and characterization of a light-induced neutral flavin radical in a C450A mutant of phototropin. J Biol Chem. 2003;278(13):10 973–10 982. doi: 10.1074/jbc.M205509200. [DOI] [PubMed] [Google Scholar]
- 34.Kinoshita T, Doi M, Suetsugu N, Kagawa T, Wada M, Shimazaki K. Phot1 and Phot2 mediate blue light regulation of stomatal opening. Nature. 2001;414:656–660. doi: 10.1038/414656a. [DOI] [PubMed] [Google Scholar]
- 35.Lange OF, Grubmüller H. Generalized correlation for biomolecular dynamics. Proteins: Struct. Func. Bioinf. 2006;62:1053–1061. doi: 10.1002/prot.20784. [DOI] [PubMed] [Google Scholar]
- 36.Losi A. Flavin-based Blue-Light photosensors: a photobiophysics update. Photochem Photobiol. 2007;83(6):1283–1300. doi: 10.1111/j.1751-1097.2007.00196.x. [DOI] [PubMed] [Google Scholar]
- 37.Losi A, Ghiraldelli E, Jansen S, Gärtner W. Mutational effects on protein structural changes and interdomain interactions in the blue-light sensing LOV protein YtvA. Photochem Photobiol. 2005;81(5):1145–1152. doi: 10.1562/2005-05-25-RA-541. [DOI] [PubMed] [Google Scholar]
- 38.MacKerell AD, Jr, Brooks B, Brooks CL, III, Nilsson L, Roux B, Won Y, Karplus M. CHARMM: The energy function and its parameterization with an overview of the program. In: Schleyer P, editor. The encyclopedia of computational chemistry. Chichester: John Wiley & Sons; 1998. pp. 271–277. [Google Scholar]
- 39.MacKerrell AD, Jr, Feig M, Brooks CL., III Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comp. Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 40.Nakasone Y, Eitoku T, Matsuoka D, Tokutomi S, Terazima M. Dynamics of conformational changes of arabidopsis phototropin 1 LOV2 with the linker domain. J Mol Biol. 2007;367(2):432–442. doi: 10.1016/j.jmb.2006.12.074. [DOI] [PubMed] [Google Scholar]
- 41.Nash AI, Ko W-H, Harper SM, Gardner KH. A conserved glutamine plays a central role in LOV domain signal transmission and its duration. Biochemistry. 2008;47(52):13 842–13 849. doi: 10.1021/bi801430e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neiss C, Saalfrank P. Ab initio quantum chemical investigation of the first steps of the photocycle of phototropin: a model study. Photochem Photobiol. 2003;77:101–109. doi: 10.1562/0031-8655(2003)077<0101:aiqcio>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 43.Nozaki D, Iwata T, Ishikawa T, Todo T, Tokutomi S, Kandori H. Role of Gln1029 in the photoactivation processes of the LOV2 domain in adiantum phytochrome3. Biochemistry. 2004;43(26):8373–8379. doi: 10.1021/bi0494727. [DOI] [PubMed] [Google Scholar]
- 44.Peter E, Dick B, Baeurle SA. Mechanism of signal transduction of the LOV2-Jα photosensor from Avena sativa. Nat Commun. 2010;1:122. doi: 10.1038/ncomms1121. [DOI] [PubMed] [Google Scholar]
- 45.Peter E, Dick B, Baeurle SA. Illuminating the early signaling pathway of a fungal light-oxygen-voltage photoreceptor. Proteins. 2011 doi: 10.1002/prot.23213. [DOI] [PubMed] [Google Scholar]
- 46.Peter E, Dick B, Baeurle SA. A novel computer simulation method for simulating the multiscale transduction dynamics of signal proteins. J. Chem. Phys. 2012;136(12):124 112–124 114. doi: 10.1063/1.3697370. [DOI] [PubMed] [Google Scholar]
- 47.Peter E, Dick B, Baeurle SA. Signals of lov1: a computer simulation study on the wildtype lov1-domain of chlamydomonas reinhardtii and its mutants. J Mol Model. 2012;18(4):1375–1388. doi: 10.1007/s00894-011-1165-6. [DOI] [PubMed] [Google Scholar]
- 48.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, et al. Scalable molecular dynamics with NAMD. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sakai T, Kagawa T, Kasahara M, Swartz TE, Christie JM, Briggs WR, Wada M, Okada K. Arabidopsis NPH1 and NPL1: blue light receptors that mediate both phototropism and chloroplast relocation. Proc. Natl. Acad. Sci. USA. 2001;98(12):6969–6974. doi: 10.1073/pnas.101137598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salomon M, Christie JM, Knieb E, Lempert U, Briggs WR. Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, phototropin. Biochemistry. 2000;39(31):9401–9410. doi: 10.1021/bi000585+. [DOI] [PubMed] [Google Scholar]
- 51.Salomon M, Eisenreich W, Dürr H, Schleicher E, Knieb E, Massey V, Rüdiger W, Müller F, Bacher A, et al. An optomechanical transducer in the blue light receptor phototropin from. Avena sativa. Proc. Natl. Acad. Sci. USA. 2001;98(22):12 357–12 361. doi: 10.1073/pnas.221455298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salomon M, Lempert U, Rüdiger W. Dimerization of the plant photoreceptor phototropin is probably mediated by the LOV1 domain. FEBS Lett. 2004;572:8–10. doi: 10.1016/j.febslet.2004.06.081. [DOI] [PubMed] [Google Scholar]
- 53.Schleicher E, Kowalczyk RM, Kay CWM, Hegemann P, Bacher A, Fischer M, Bittl R, Richter G, Weber S. On the reaction mechanism of adduct formation in LOV domains of the plant blue-light receptor phototropin. J Am Chem Soc. 2004;126:11 067–11 076. doi: 10.1021/ja049553q. [DOI] [PubMed] [Google Scholar]
- 54.Sethi A, Eargle J, Black AA, Luthey-Schulten Z. Dynamical networks in tRNA:protein complexes. Proc Natl Acad Sci U S A. 2009;106(16):6620–6625. doi: 10.1073/pnas.0810961106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Song S-H, Freddolino PL, Nash AI, Carroll EC, Schulten K, Gardner KH, Larsen DS. Modulating LOV domain photodynamics with a residue alteration outside the chromophore binding site. Biochemistry. 2011;50(13):2411–2423. doi: 10.1021/bi200198x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Swartz TE, Wenzel PJ, Corchnoy SB, Briggs WR, Bogomolni RA. Vibration spectroscopy reveals light-induced chromophore and protein structural changes in the LOV2 domain of the plant blue-light receptor phototropin 1. Biochemistry. 2002;41(23):7183–7189. doi: 10.1021/bi025861u. [DOI] [PubMed] [Google Scholar]
- 57.Taylor BL, Zhulin IB. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 1999;63(2):479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. Gromacs: Fast, flexible, and free. J. Comp. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 59.VanWart AT, Eargle J, Luthey-Schulten Z, Amaro RE. Exploring residue component contributions to dynamical network models of allostery. J. Chem. Theory Comput. 2012;8(8):2949–2961. doi: 10.1021/ct300377a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vreede J, van der Horst MA, Hellingwerf KJ, Crielaard W, van Aalten DMF. PAS domains. Common structure and common flexibility. J.Biol. Chem. 2003;278:18 434–18 439. doi: 10.1074/jbc.M301701200. [DOI] [PubMed] [Google Scholar]
- 61.Yao X, Rosen MK, Gardner KH. Estimation of the available free energy in a LOV2-Jα photoswitch. Nat Chem Biol. 2008;4(8):491–497. doi: 10.1038/nchembio.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zayner JP, Antoniou C, Sosnick TR. The amino-terminal helix modulates light-activated conformational changes in AsLOV2. J Mol Biol. 2012;419(1–2):61–74. doi: 10.1016/j.jmb.2012.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zoltowski BD, Schwerdtfeger C, Widom J, Loros JJ, Bilwes AM, Dunlap JC, Crane BR. Conformational switching in the fungal light sensor Vivid. Science. 2007;316(5827):1054–1057. doi: 10.1126/science.1137128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.