Abstract
HOTAIR is a long noncoding RNA (lncRNA) that is transcribed from the antisense strand of HOXC gene locus in chromosome 12. HOTAIR coordinates with chromatin modifying enzymes and regulates gene silencing. It is overexpressed in various carcinomas including breast cancer. Herein, we demonstrated that HOTAIR is crucial for cell growth and viability and its knockdown induced apoptosis in breast cancer cells. We also demonstrated that HOTAIR is transcriptionally induced by estradiol (E2). Its promoter contains multiple functional estrogen-response-elements (EREs). Estrogen receptors (ERs) along with various ER-coregulators such as histone methylases MLL1 and MLL3 and CBP/p300 bind to the promoter of HOTAIR in an E2-dependent manner. Level of histone H3K4-trimethylation, histone acetylation and RNA polymerase II recruitment is enriched at the HOTAIR promoter in presence of E2. Knockdown of ERs and MLLs down regulated the E2-induced HOTAIR expression. Thus, similar to protein coding gene transcription, E2-induced transcription of antisense transcript HOTAIR is coordinated via ERs and ER-coregulators and this mechanism of HOTAIR over expression potentially contributes towards breast cancer progression.
Introduction
Long non-coding RNA (lncRNA) are emerging class of key regulatory RNA that do not code for protein and are not translated. LncRNAs are crucial players in various key biological processes that include dosage compensation, genomic imprinting, chromatin organization, gene regulation, and alternative splicing. Mammalian genome possesses a large number of lncRNAs that exceed the fraction of protein coding genes 8. In mammals, lncRNAs comprise at least half the total number of RNAs transcribed by RNA polymerase II (RNAP II) and, like other protein coding mRNAs, many lncRNAs are capped, spliced, and polyadenylated. In addition, a number of them are transcribed from both strands (sense and antisense) of the genome 10. Studies indicate that lncRNAs are dysregulated in various devastating human diseases including cancer 10. Misregulation of lncRNAs has been correlated with poor patient outcome, prognosis and cancer metastasis 5. Although lncRNAs appear to be critical for various biological processes, their mechanism of action and transcriptional regulation still remains elusive.
HOTAIR (HOX antisense intergenic RNA) is an example of lncRNA that is localized on chromosome 12 within the homeobox C (HOXC) gene cluster 11. HOTAIR, a 2.2 kb long transcript possessing six exons, is transcribed by RNAP II from the antisense strand and thus HOTAIR is an antisense transcript 11. HOTAIR is co-expressed with the HOXC gene cluster and participates in down regulation of various genes in a genome-wide fashion, in trans 11. For example, HOTAIR suppresses the expression of HOXD gene cluster present on chromosome 2 that are tightly regulated during development 12. In vitro studies demonstrated that HOTAIR interacts with various chromatin modifying enzymes regulating gene expression. For example, HOTAIR, through its 5′-end, interacts with histone H3 lysine 27 (K3K27) specific methyl-transferase complex, PRC2 (polycomb repressive complex 2) that is involved in gene silencing 12. HOTAIR, through its 3′ end, interacts with LSD1 (histone demethylase)/CoREST/REST complex, leading to HOTAIR-mediated assembly of PRC2 and LSD1 complexes 12. HOTAIR acts as a bridge coordinating the targeting of PRC2 and LSD1 complexes to chromatin for coupled histone H3K27 methylation and H3K4 demethylation processes and that in turn aides in silencing of gene in HOXD cluster and various other target genes. HOTAIR is highly expressed in primary breast tumors 5, hepatocellular carcinoma 13, colorectal cancer 14, and gastrointestinal stromal tumors 15. Recent studies showed that lncRNAs in the HOX loci become systematically dysregulated during breast cancer progression 5. HOTAIR expression is augmented in primary breast tumors and metastases, and HOTAIR expression level in primary tumors is a powerful predictor of metastases and death. Enforced expression of HOTAIR in epithelial cancer cells induced genome wide retargeting of PRC2 complex to an occupancy pattern more resembling embryonic fibroblasts. This retargeting of PRC2 complex led to altered histone H3K27 trimethylation, gene expression, and increased cancer invasiveness and metastasis in a manner dependent on PRC2 5. Loss of HOTAIR expression inhibits cancer invasiveness, particularly in cells that possess excessive PRC2 activity 17.
Thus, lncRNA plays active roles in modulating the cancer epigenome and may be important targets for cancer diagnosis and therapy. As HOTAIR is over expressed in primary breast tumors, we hypothesize that HOTAIR expression is potentially regulated by estrogen in breast cancer cells. Herein we have investigated the importance of HOTAIR in breast cancer cells and examined its transcriptional regulatory mechanisms especially in the presence of breast cancer associated steroid hormone estradiol. Our studies revealed that HOTAIR is essential for the survival of breast cancer cells and it is transcriptionally regulated by estrogen.
Results
HOTAIR siSENSE oligonucleotide design and knockdown of HOTAIR in breast cancer cells
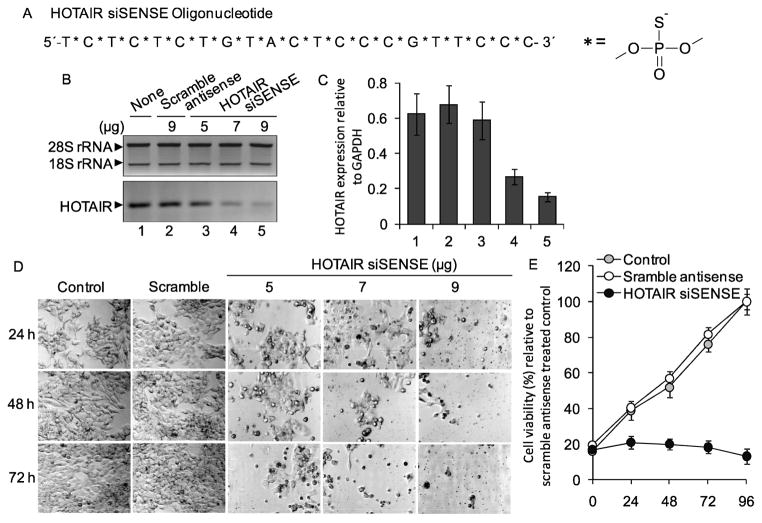
The lncRNA HOTAIR is an antisense transcript that is over expressed in primary breast tumors. To investigate the significance of HOTAIR overexpression in breast cancer, we aimed to knockdown HOTAIR expression in breast cancer cells (MCF7) and analyze its impact on cell growth and viability. Initially, for knocking down HOTAIR, we designed a small interfereing oligonucleotide DNA (21 nt long) complementary to the 773–794 nt region of HOTAIR transcript (Figure 1A, Table 1). As HOTAIR is an antisense transcript that is transcribed from antisense strand of HOXC gene locus present in chromosome 12, the complementary sequence of HOTAIR lncRNA is identical to the sense strand. Therefore, we termed the synthetic oligonucleotide DNA that is complementary to HOTAIR transcript as small interfering sense (siSENSE) oligonucleotide and this term have been used throughout the manuscript. The normal phosphodiester bonds of HOTAIR-siSENSE DNA molecule were replaced with phosphorothioate linkage to minimize the nuclease digestion and enhance its in vivo stability (Figure 1A).
Figure 1.
Knockdown of HOTAIR and its impact on cell growth and viability. (A) Sequence of HOTAIR-siSENSE containing phosphorothioate bonds. (B–C) Knockdown of HOTAIR. MCF7 cells were transfected with varying concentrations of HOTAIR-siSENSE (5–9 μg) or scramble antisense for 72 h, RNA was reverse-transcribed, and analyzed by RT-PCR using primers specific to HOTAIR. rRNA was used as a loading control. Real-time PCR quantification of HOTAIR transcript was performed and the transcript levels relative to GAPDH were plotted (panel C). Each experiment was performed in three replicates for at least two times. Bars indicate standard error (n = 3; p ≤ 0.05). (D–E) Impact of HOTAIR knockdown on cell growth. HOTAIR-siSENSE transfected (varying concentrations and time) MCF7 cells were visualized under a microscope (panel D). Scramble antisense transfection was performed as control. For the growth rate measurement, MCF7 cells were transfected with HOTAIR-siSENSE (9 μg) or scramble antisense for varying time periods (24 – 96 h), stained with trypan blue, viable cells were counted using a hematocytometer and plotted (panel E). Each experiment with three replicates was repeated for at least two times. Bars indicate standard errors (n = 8; p ≤ 0.05).
Table 1.
Nucleotide sequence of antisense and primers
| Gene | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| PCR primers | ||
| MLL1 | GAGGACCCCGGATTAAACAT | GGAGCAAGAGGTTCAGCATC |
| MLL2 | AGGAGCTGCAGAAGAAGCAG | CAGCCAAACTGGGAGAAGAG |
| MLL3 | CATATGCACGACCCTTGTTG | ACTGCTGGATGTGGGGTAAG |
| MLL4 | CCCTCCTACCTCAGTCGTCA | CAGCGGCTACAATCTCTTCC |
| GAPDH | CAATGACCCCTTCATTGACC | GACAAGCTTCCCGTTCTCAG |
| HOTAIR | GGGGCTTCCTTGCTCTTCTTATC | GGTAGAAAAAGCAACCACGAAGC |
| HOXD10 | GCTCCTTCACCACCAACATT | AAATATCCAGGGACGGGAAC |
| HOXC10 | ACCACAGGAAATTGGCTGAC | GATCCGATTCTCTCGGTTCA |
| PCDHB5 | ATAGAAGAGCTGTTGGATGAGGAC | CACTAATACTGTCACACACGCTCA |
| Bcl1 | CGGAACAGCTACTCCTCCTG | GATCCGATTCTCTCGGTTCA |
| Bcl2 | GGATGCCTTTGTGGAACTGT | TCACTTGTGGCCCATTTCTGGCTA |
| BID | AAGAAGGTGGCCAGTCACAC | GTCCATCCCATTTCTGGCTA |
| ERα | AGCACCCTGAAGTCTCTGGA | GATGTGGGAGAGGATGAGGA |
| ERβ | AAGAAGATTCCCGGCTTTGT | TCTACGCATTTCCCCTCATC |
| HOTAIR ERE1 | CCTGCAATAATCCCTTTAGTATGC | CTGCTGCAGAGAATTTCAGGT |
| HOTAIR ERE2 | CTCCAGGTGGCTTATTTGTATCTT | CTGCTGCAGAGAATTTCAGGT |
| HOTAIR ERE3 | TCAAGGAAAGAAGGCCCTGGC | AGGTATTGATGCTGTGGCCAG |
| HOTAIR ERE4 | TATGGCTTAGTTTTTCAACAA | TCAGTGGCCAGGGCCTTCTTT |
| Cloning primers | ||
| HOTAIR ERE1 | GGTACCCTTGCCTATATTTCTCTCCCTTACAGa | CTCGAGGCTCGTAAAATAGGGCTTTTATGGa |
| HOTAIR ERE2 | GGTACCCTCCAGGTGGCTTATTTGTATCTTAa | CTCGAGCTGCCTTAACTTTGGTCCAGCTACa |
| HOTAIR ERE3 | GGTACCTCAAGGAAAGAAGGCCCTGGCa | CTCGAGAGGTATTGATGCTGTGGCCAGa |
| HOTAIR ERE4 | GGTACCTATGGCTTAGTTTTTCAACAAa | CTCGAGTCAGTGGCCAGGGCCTTCTTTa |
| Antisense oligonucleotides (DNA) | ||
| HOTAIR-siSENSE | TCTCTCTGTACTCCCGTTCCCb | |
| MLL1 | TGCCAGTCGTTCCTCTCCACb | |
| MLL2 | ACTCTGCCACTTCCCGCTCAb | |
| MLL3 | CCATCTGTTCCTTCCACTCCCb | |
| MLL4 | CCTTCTCTTCTCCCTCCTTGTb | |
| ERα | CATGGTCATGGTCAGb | |
| ERβ | GAATGTCATAGCTGAb | |
| Scramble | CGTTTGTCCCTCCAGCATCTb | |
cloning primers flanked by appropriate restriction sites
Phosphodiester linkages replaced by phosphorothioate linkages.
To examine the knockdown efficacy of HOTAIR siSENSE, we transfected MCF7 cells with varying concentration of the HOTAIR siSENSE oligonucleotide for 72 h. Cells were also transfected in parallel with a scramble-antisense that has no complementarity to HOTAIR RNA. RNA from the control, siSENSE/antisense-treated cells were reverse-transcribed and subjected to PCR-amplification using HOTAIR specific primers (Figure 1B, Table 1). Ribosomal RNA (rRNA) was used as loading control (Figure 1B). Our results showed that application of HOTAIR siSENSE knocked down specifically and effectively (~ 80% knockdown) HOTAIR transcript levels in a dose dependent manner, 9 μg siSENSE being the most effective in HOTAIR knockdown (lane 5, Figure 1B). The same cDNA was also analyzed by real-time PCR (qPCR) for quantification of HOTAIR expression (Figure 1C, GAPDH was used as control). This analysis showed that our HOTAIR siSENSE is effective in knocking down HOTAIR in MCF7 cells.
HOTAIR is essential of growth and viability of MCF7 cells
To investigate the importance of HOTAIR in breast cancer cells, we knocked it down using HOTAIR-siSENSE oligonucleotide and examined its impact on cell growth and viability. Briefly, we transfected MCF7 cells with varying concentrations of HOTAIR siSENSE or scramble antisense oligonucleotides separately for varying time periods. Microscopic analysis showed that HOTAIR knockdown has suppressed the growth of MCF7 cells in comparison to the control or scramble-antisense treated cells even at 24 h of post HOTAIR siSENSE transfection (Figure 1D). Longer time (48 –72 h) and higher doses of incubation with HOTAIR-siSENSE affected further on cell growth and induced cellular fragmentation indicating death (Figure 1D). We also examined the rate of cell growth after transfection with HOTAIR-siSENSE. Growth analysis showed that HOTAIR knockdown has suppressed the growth of MCF7 cells significantly in comparison to the control (untreated) or scramble-antisense treated cells (Figure 1E). Populations of the control as well as the scramble antisense treated cells were doubled at 48 h while HOTAIR-siSENSE treated cells growth was significantly inhibited. At longer time periods (72 h) of HOTAIR knockdown, cells did not grow further and populations were even decreased due to cell death (Figure 1E). This analysis demonstrated that expression of HOTAIR is important for the survival and maintenance of breast cancer cells (MCF7).
HOTAIR knockdown induces apoptosis in MCF7 cells
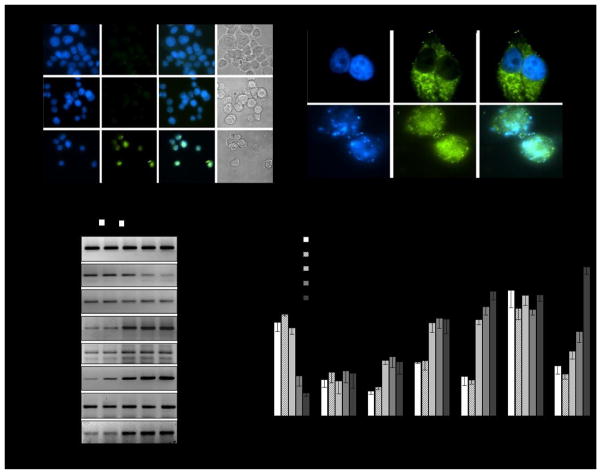
To examine the type of cell death induced by HOTAIR knockdown, we analyzed the HOTAIR knocked down cells using terminal dUTP-nicked-end labeling (TUNEL) assay. In brief, MCF7 cells were transfected with HOTAIR-siSENSE or scramble antisense and then subjected to terminal dUTP-nicked-end labeling and DAPI staining (nuclear stain) and then visualized under fluorescence microscope. Our analysis showed that in comparison to the control or scramble-antisense treated cell, large number of cell nuclei DNA were fragmented in the HOTAIR-siSENSE transfected environment, as shown by the terminal dUTP-nicked-end labeling (green colored nuclei, Figure 2A, bottom panel) indicating nuclear DNA fragmentation and apoptotic cell death. Green colored cell nuclei were also condensed (intense DAPI staining and smaller nuclei) under HOTAIR knockdown condition (Figure 2A, bottom panel). Light microscopic images also indicated that cellular morphologies were deformed and fragmented under HOTAIR knockdown environment indicating cell death (Figure 2A). Scramble antisense had no significant impact on nuclear integrity and cellular morphologies and most cells remained healthy (Figure 2A).
Figure 2.
HOTAIR knockdown induced apoptosis. (A) TUNEL assay: MCF7 cells were transfected with 9 μg of HOTAIR-siSENSE and scramble antisense seperately for 48 h, fixed with formaldehyde and subjected to terminal dUTP nicked end labeling and stained with DAPI (blue staining). The green speckles are showing apoptotic cells containing fragmented DNA. (B) Immunostaining with cytochrome-c: Control and HOTAIR-siSENSE transfected (9 μg for 48 h) MCF7 cells were immunostained with cytochrome-c antibody followed by FITC-labeled secondary antibody, stained with DAPI and visualized under fluorescence microscope. (C–D) HOTAIR misregulated apoptotic genes and affected target gene expression. MCF7 cells were transfected with HOTAIR-siSENSE (9 μg) for 72 h. RNA from HOTAIR-knocked down or control cells were analyzed by RT–PCR by using primers specific to Bcl1, Bcl2 and BID, and HOTAIR target genes such as HOXD10 and PCDHB5. HOXC10 was used as non-HOTAIR target gene as control. GAPDH expression was used as a loading control. Real-time PCR analysis data is shown in panel D.
Perturbation in mitochondrial membrane potential followed by cytochrome-c translocation is an important indication of apoptosis 20. To examine the impacts of HOTAIR knockdown on mitochondrial membrane potential and integrity, we immunostained HOTAIR-siSENSE treated MCF7 cells with cytochrome-c antibody and then analyzed by immuno-fluorescence microscopy. DAPI staining was performed to stain the nuclei. As seen in Figure 2B, in the untreated control cells, cytochrome-c is localized inside the mitochondria as evidenced by the distinct speckles outside the nucleus. However, in the HOTAIR-siSENSE treated cells, cytochrome-c was released from mitochondria to cytosol and spread all over the cells (Figure 2B). Cell nuclei are clearly fragmented (DAPI staining). The release of cytochrome-c from the mitochondria to cytosol indicated the perturbation in the mitochondrial membrane potential and apoptotic cell death.
To understand further the mechanism of apoptotic cell death induced by HOTAIR knockdown, we analyzed the expression level of various apoptosis related genes such as Bcl1, Bcl2 and BID. Notably, BID (BH3 interacting domain death agonist) protein heterodimerizes with either agonist BAX or antagonist Bcl2 21. BID protein is a mediator of mitochondrial damage induced by caspase-8 (CASP8) that ultimately triggers cytochrome c release 21. Although, Bcl2 is an antiapoptotic gene, various studies have demonstrated that BAX/Bcl2 ratio, than the Bcl2 alone, is important for the survival of drug-induced apoptosis in cancer cells 22. Our analysis demonstrated that transfection with HOTAIR-siSENSE effectively knocked down HOTAIR and that resulted in upregulation of Bcl2 and BID expression (Figures 2C, real-time PCR data in figure 2D). Bcl1 level was mostly unaffected. The misregulation of Bcl2 and BID may have contributed towards the apoptotic cell death under HOTAIR knockdown environment.
Notably, previous studies showed that HOX genes present in the HOXD locus are potential targets of HOTAIR 11. These include HOXD8, HOXD9, HOXD10, and HOXD11 11. HOTAIR also have other known target genes such as PCDHB5 (protocadherin B5, PCDH10 (protocadherin 10) and JAM2 (junction adhesion molecule 2) that are involved in intercellular adhesion and signaling 5. PCDHB5, PCDH10 and JAM2 are known to be associated with breast cancer progression. Importantly, HOTAIR containing PRC2 complex, which is involved in histone H3K27-methylation, binds to the promoter of these target genes and induce gene silencing. Herein, to further examine and confirm the specificity of HOTAIR siSENSE, we performed real-time PCR analysis of several HOTAIR target gene such as PCDHB5, HOXC10, and HOXD10. Our results demonstrated that upon transfection with HOTAIR siSENSE, along with HOTAIR knockdown, the HOTAIR target genes such as HOXD10 and PCDHB5 were upregulated (Figures 2C–D). HOXC locus gene such as HOXC10 were not affected (Figure 2C–D). These results further demonstrated that HOTAIR siSENSE specifically knocked down HOTAIR transcript in MCF7 cells and in agreement with previous studies, our analysis demonstrated that HOTAIR knockdown induced the expression of HOXD10 and PCDHB5 indicating roles of HOTAIR in down regulating these target genes (Figures 2C–D).
HOTAIR gene is transcriptionally induced by estradiol in MCF7 cells
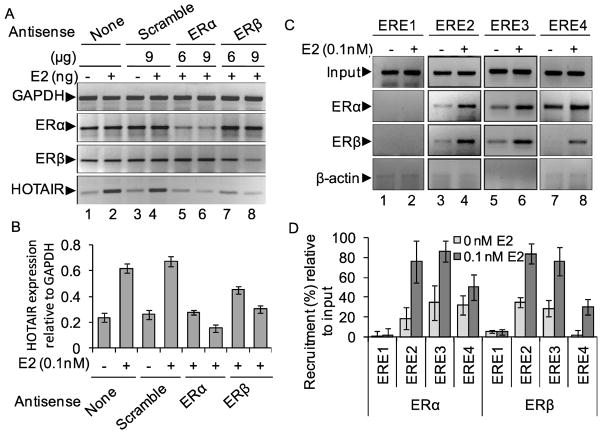
As HOTAIR is overexpressed in primary breast cancer tissue and found to be critical for viability of MCF7 cells, we examined its potential regulation by estrogen. We exposed MCF7 cells with 17β-estradiol (E2) and analyzed the levels of HOTAIR transcript in the absense and presence of E2. In brief, MCF7 cells were initially grown in phenol-red free media containing charcoal stripped FBS for at least 3 generations and then treated with varying concentrations of E2 for 6 h 26. RNA was isolated from the control and E2-treated cells, reverse-transcribed into cDNA and analyzed using regular RT-PCR and real-time PCR (Figures 3A–B). GAPDH was used as loading control. Interestingly, these analyses demonstrated that HOTAIR expression is induced by E2 in a dose-dependent manner (lanes 1–4, Figures 3A–B). qPCR analysis showed that HOTAIR was induced by ~ 5 fold at 0.1 nM E2 (compare lanes 1 and 4, Figure 3A–B). The E2-induced expression of HOTAIR was suppressed at higher concentration of E2, likley due to squelching (compare lane 5, Figures 3A–B). To examine the specificity of E2, we examined the E2-induced expression of HOTAIR in presence of an anti-estrogen such as tamoxifen. Our results showed that treatment with tamoxifen alone has no significant impact on expression of HOTAIR (lanes 1 and 8, Figure 3A–B). However, tamoxifen treatment suppressed the E2-induced HOTAIR expression (lanes 6–7, Figures 3A–B). HOTAIR expression was insensitive to E2-treatment in an estrogen-receptors (ER)-neagtive breast cancer cell line such as MDA-MB-231 (data not shown), indicating involvement of ERs in E2-mediated expression of HOTAIR. Time course analysis demonstrated that E2-induced HOTAIR expression was increased with the increase in incubation time with a maximum at around 4 h and this induction was decreased at longer time incubation indicating squelching (Figures 3C–D). These results demonstrated that HOTAIR is an estrogen-responsive gene and therefore transcriptionally induced by E2 in ER-positive breast cancer cells such as MCF7.
Figure 3.
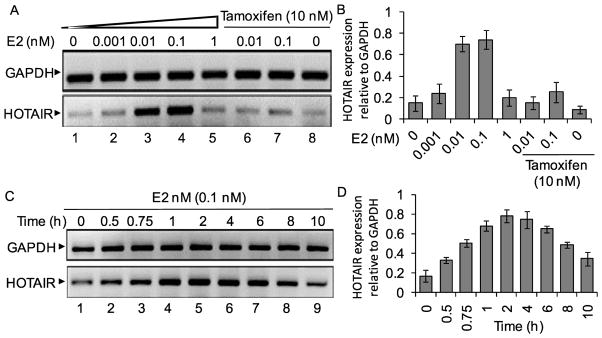
Effect of 17β-estradiol (E2) on HOTAIR gene expression. (A–B) MCF7 cells were grown in phenol red free DMEM-F-12 media and treated with varying concentrations of E2 and also in the absence and presence of tamoxifen. RNA from the control and E2-treated cells were analyzed by RT-PCR using primers specific to HOTAIR. GAPDH was used as a loading control. The real-time quantification of HOTAIR expression (relative to GAPDH) is in panel B. Each experiment was repeated for two times with three parallel replicates. Bars indicate standard errors (n = 3; p ≤ 0.05). (C–D) MCF7 cells were treated with 0.1 nM E2 for varying time periods (0–10 h). RNA was analyzed by regular RT-PCR (panel C) and real-time PCR (panel D) as above.
HOTAIR promoter contains estrogen-response-elements (EREs)
As HOTAIR is found to be an E2-responsive gene, we investigated the potential mechanism of E2-induced expression of HOTAIR. Initially, we examined the HOTAIR promoter for the presence of putative estrogen-response-elements (EREs) close to the transcription start site. We found that HOTAIR promoter contains multiple ERE1/2 sites (GGTCA elements) within −2000 nt upstream of the transcription start site (Figure 4A). The ERE2 (GGTGCnnnTGACC) and ERE3 (GGTCA nnnAGACA) that are located at −1486 nt and −1721 nt respectively, appear to be imperfect full EREs with two bases mismatch in the palindromic regions (consensus full ERE sequence is TGACCnnnGGTCA) regions (Figure 4A). Irrespective of the sequences, we cloned each ERE region in a luciferase based reporter construct pGL3 and examined E2-response of each ERE region by luciferase assay 26. Each ERE-pGL3 construct was co-transfected with renilla luciferase construct (supplied by the manufacturer) into MCF7 cells and then treated with E2 (0.1 nM for 4 h) and subjected to luciferase analysis using a dual-luciferase assay kit. Luciferase activities of EREs were normalized using renilla expression and plotted (Figure 4B). These analyses revealed that ERE2 and ERE3 regions were highly induced upon treatment with E2 (about 16 and 18 fold, respectively) in comparison to control cells (transfection with pGL3 alone) (Figure 4B). ERE1 was mostly non-responsive to E2-treatment while ERE4 was stimulated by about 8 fold in the presence of E2 (Figure 4B). These analyses demonstrated that HOTAIR promoter contains functional EREs. ERE2 and ERE3 regions, being more senstive to E2-treatment, may potentially contribute towards E2-mediated transcriptional activation of HOTAIR.
Figure 4.
HOTAIR promoter contains functional EREs (A) HOTAIR gene promoter EREs (termed as ERE1, ERE2, ERE3 and ERE4, locations and the neighboring sequences are shown in Panel A). were cloned individually (clones 1–4) into a luciferase based reporter construct, pGL3, used for transfection and reporter assay. (B) Luciferase based reporter assay. ERE-pGL3 or empty pGL3 (vector control) constructs were transfected into MCF7 cells separately for 24 h. A renilla luciferase construct was also co-transfected along with ERE-pGL3 constructs as an internal transfection control. Cells were then treated with 0.1 nM E2 and subjected to luciferase assay by using dual-Glo Luciferase Assay kit. The luciferase activities (normalized to renilla activity) were plotted. The experiment with four replicate treatments was repeated at least twice. Bars indicate standard errors (n = 3; p ≤ 0.05).
E2-induced HOTAIR expression is coordinated via estrogen-receptors (ERs)
ERs are key players in estrogen signaling. ERα and ERβ are two major ERs that play crucial roles in estrogen-mediated gene regulation and signaling. In general, in genomic ER-signaling pathway, upon binding to estrogen, either ERα or ERβ or both ERs, get activated and bind to target gene promoter ERE regions 29. Then various ER-coregulators are recruited to the promoter and induce chromatin remodeling followed by activation of target gene expression. Herein, to examine potential involvement of ERs during E2-induced HOTAIR, we knocked down ERα and ERβ independently using ER-specific antisenses in MCF7 cells and subsequently exposed to E2 followed by examination of HOTAIR expression using regular RT-PCR and real-time PCR (Figures 5A-B, GAPDH was used as control). Our analysis demonstrated that transfection with ERα-antisense sepcifically knocked down ERα and the knockdown of ERα downregulated the E2-induced expression of HOTAIR transcript (compare lanes 2 with 5–6, Figure 5A, qPCR data is shown in figure 5B). Similarly, knockdown of ERβ also downregulated the expression of HOTAIR in presence of E2 (Figures 5A–B). Knockdown of ERα and ERβ were also confirmed in the protein level (data not shown). These results demonstrated that both ERα and ERβ are necessary for the E2-dependent activation of HOTAIR in MCF7 cells.
Figure 5.
Roles of ERs on E2-induced expression of HOTAIR. (A–B) Effects of ER-knockdown. MCF7 cells were transfected with ERα, ERβ (6 and 9 μg each) or scramble antisense (9 μg) separately for 48 h and treated with E2 (0.1 nM) for additional 4 h. RNA was isolated and subjected to regular RT-PCR (analyzed by agarose gel, panel A) analysis by using primers specific to HOTAIR, ERα, and GAPDH (loading control). Real-time quantification of HOTAIR expression under ER-knockdown environment was shown in panel B. Each experiment was repeated at least thrice. Bars indicate standard errors (n = 3; p ≤ 0.05). (C–D) E2-induced binding of ERs in the HOTAIR promoter EREs. MCF7 cells were treated with 0.1 nM E2 for 4 h and subjected to ChIP assay using antibodies specific to ERα, ERβ and β-actin (control IgG). The immuno-precipitated DNA fragments from E2-treated and control cells were PCR-amplified using primers specific to ERE1, ERE2, ERE3 and ERE4 of HOTAIR promoter (panel C). ChIP DNA fragments were analyzed by real-time PCR and shown in the panel D. Each experiment was repeated at least thrice. Bars indicate standard errors (n = 3; p ≤ 0.05).
To understand further the involvement of ERs, we examined the binding of ERs in the HOTAIR promoter regions using chromatin immuno-precipitation (ChIP) assay 35. Briefly, cells were treated with E2, fixed with formaldehyde, sonicated to shear the chromatin and then subjected to immuno-precipiation using ERα, ERβ and β-actin (control) antibodies. The immunoprecipitated chromatins were reverse-crosslinked and DNA were subjected to PCR analysis using primers specific to different ERE regions of the HOTAIR promoter. Our analysis showed that the binding of both ERα and ERβ were enriched in ERE2 and ERE3 regions in presence of E2 (Figure 5C, real-time PCR data in 5D). Some amount of enrichment of ERβ was observed in the ERE4 region in the presence of E2 (Figures 5C–D). There was no significant binding of ERs in the ERE1 region irresepctive of E2 (Figures 5C–D). No binding of β-actin was observed both in absence and presence of E2 (Figure 5C). These results further demonstrated that both ERα and ERβ are involved in E2-dependent HOTAIR gene activation, via binding to the promoter regions (especially ERE2 and ERE3) in an E2-dependent manner. Notably, luciferase based reporter assay (Figure 4B) also showed the higher E2-responses of ERE2 and ERE3 regions supporting the present observation.
ER-coactivators such as MLL histone methylases, CBP/p300 (histone acetylase) are recruited to the HOTAIR promoter in presence of E2
ER-coactivators are crucial integral components of estrogen-dependent gene activation. During activation of E2-responsive genes, ERs interact with various coregulators which bridge ERs to chromatin proteins and basal transcription machinery. Many ER-coactivators are discovered that include SRC1 family protein, CREB-binding protein (CBP/p300), p/CAF, ASCOM (activating signal cointegrator-2 (ASC2) complexes). Recent studies demonstrated that mixed lineage leukemia (MLL) histone methylases act as ER-coactivators and regulate various E2-resposnisve genes. MLLs are well recognized as histone H3 lysine-4 (H3K4) specific histone methyl transferases that play key roles during gene activation. MLLs interact with ERs via their LXXLL domains and participate in E2-mediated gene activation 33.
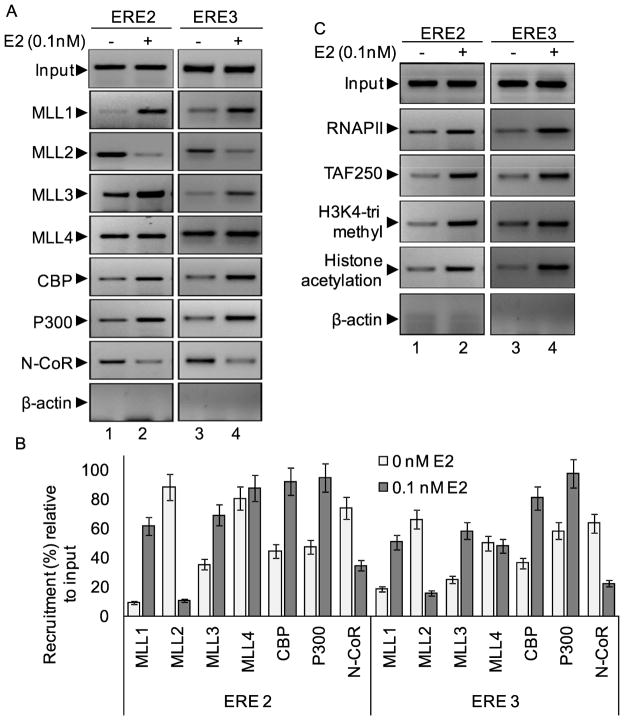
Herein, we examined if any ER-coactivators are associated with E2-induced HOTAIR expression. Using ChIP assay, we analyzed the recruitment of CBP/p300, MLL family of HMTs (MLL1–MLL4) in the HOTAIR promoter in the absence and presence of E2. Interestingly, we observed that E2-treatment resulted in enrichment of ER-coregulators such as CBP, p300, histone methylases MLL1 and MLL3 to the ERE2 and ERE3 regions of HOTAIR promoter. These observations demonstrated that E2-dependent HOTAIR transcription is assisted by ER-coregulators such as MLL histone methylases and CBP/p300 acetyl-transferase. On the other hand, we observed the constitutive binding of histone methylase MLL2 and nuclear receptor corepressor N-CoR in both ERE2 and ERE3 regions in the absence of E2, and their binding were decreased upon treatment with E2, indicating their dissociation from the repressed chromatin. This observation indicate that MLL2 and N-CoR are associated with maintenance of repressed basal transcription state of HOTAIR and they are released during E2-mediated HOTAIR promoter activation followed by gene expression. Notably similar exchange of transcription factors were previsouly observed during transition from basal to activated transcription state in protein coding genes such as HOXC6 46.
As histone acetylases (HATs) and methyl-transferases (HMTs) are involved in E2-mediated HOTAIR gene activation, we also analyzed the levels of histone acetylation and histone H3K4-trimethylation and RNA polymerase II (RNAP II) into the HOTAIR promoter in the presence of E2. Notably histone H3K4-trimethylation and histone acetylation are essential post-translation modifications in the histone tails that are closely associated with gene activation. MLLs are well known to be histone H3K4-specific trimethylases and critical players in gene activation. Our ChIP analysis demonstrated that the level of histone H3K4-trimethylation, histone acetylation and levels of RNAPII were enriched at the HOTAIR promoter ERE2 and ERE3 regions in presence of E2 (Figure 6C). General transcription factor TFIID component such as TAF250 was also recruited upon addition of E2 (Figure 6C).
Figure 6.
Roles of ER-coregulators: (A–C) Recruitment of MLLs (MLL1-MLL4) and CBP, p300 and N-CoR: MCF7 cells were treated with 0.1 nM E2 for 4 h and subjected to ChIP assay using antibodies specific to MLL1, MLL2, MLL3, MLL4, CBP, p300 and N-CoR. ChIP DNA fragments were PCR-amplified using primers specific to ERE2 and ERE3 of HOTAIR promoter (panel A). ChIP DNA fragments were analyzed by real-time PCR and shown in panel B. Recruitment of RNAPII, TAF250 (TFIID component) and levels of histone methylation and acetylation in the HOTAIR promoter are shown in panel C. β-actin antibody was used as control IgG.
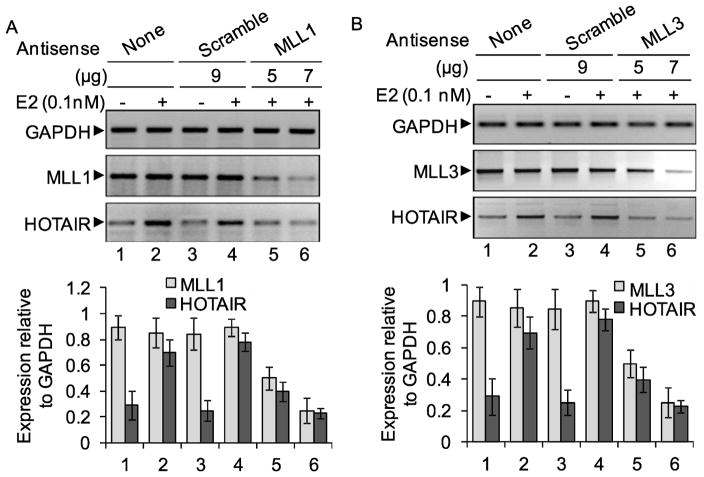
To further confirm the involvement of ER-coactivators, we knocked down some of the ER-coregulators such as MLL1, MLL2, MLL3, MLL4 and then examined the E2-dependent expression of HOTAIR. This analysis demonstrated that knockdown of MLL1 and MLL3, suppressed the E2-induced HOTAIR expression significantly indicating crucial roles of MLL1 and MLL3 as ER-coactivators during E2-mediated HOTAIR induction (Figures 7A–B, qPCR data in the respective bottom panels). MLL2 and MLL4 knockdown has no significant impact on E2-induced HOTAIR expression (data not shown) and these observations are in agreement with ChIP data shown in figure 6A. These observations demonstrated that, similar to transcription of protein coding genes, ER-coregulators, histone acetylases, histone methylases, and general transcription factors along with RNAPII, play critical roles in transcription activation of antisense-transcript lncRNA HOTAIR under E2-environment.
Figure 7.
Role of MLL histone methylases in E2-dependent expression of HOTAIR. MCF7 cells were transfected with MLL or scramble antisense for 48h and then treated with 0.1 nM E2 for additional 4 h and subjected to RT-PCR analysis using primers specific to HOTAIR, MLL1 (panel A) and MLL3 (panel B). cDNA were analyzed by real-time PCR and data is shown in respective bottom panel (n = 3; p ≤ 0.05).
Discussion
It is increasingly being recognized that a large proportion of the mammalian transcriptome does not code for proteins. LncRNA are major class of non-coding RNAs that appeared to be closely associated with chromatin function, genome organization and various human diseases. It is estimated that human and mouse genomes produce as many lncRNAs as mRNAs, though their functional significance still remains elusive 50. Among others, Xist/Tsix, H19, HOTAIR, and AIR are few examples of lncRNAs whose functions are extensively being investigated. H19 gene expresses a lncRNA that acts as a tumor suppressor 51. It also possesses a microRNA (miR-675) in its exon-1 that is involved in alternative splicing of insulin-like growth factor 2 (IGF2). Mutations in H19 gene are associated with Beckwith-Wiedemann syndrome and Wilms tumorigenesis 54,55. Tsix lncRNA plays critical roles in X-inactivation 56. Tsix, like HOTAIR, is an antisense transcript lncRNA that is transcribed from the 3′-end of the XIST locus and is co-expressed with Xist, from the inactive X chromosome 56. Tsix represses Xist through modification of the chromatin structure in the Xist promoter region and preventing recruitment of polycomb complexes that is required for the maintenance of active X chromosomes in females57. AIR lncRNA is imprinted, mono-allelically and is expressed from the paternal allele and interacts with histone methyl-transferase G9a 58. Alterations in expression of lncRNAs is associated with various human disorders such as cancer, myotonic dystrophies 59, spinocerebellar ataxia 60, alzheimer’s disease 61 and thalassemia 62.
LncRNA, HOTAIR, is transcribed from the antisense strand of the chromosome 12 at the HOXC gene cluster 11. Though it is an antisense transcript, like other protein coding genes, HOTAIR is transcribed by RNA polymerase II (RNAPII) 11. It undergoes 5′-end capping and 3′-end polyadenylation. HOTAIR transcript undergo alternative splicing producing three splice variants. HOTAIR plays critical roles in transcriptional regulation of many genes. Importantly, though HOTAIR gene is encoded in the antisense-strand of HOXC gene cluster, microarray and genome-wide ChIP sequencing based studies demonstrate that HOTAIR does not affect the expression of genes present in HOXC locus 11. Rather, HOTAIR regulate genes in the HOXD cluster such as HOXD8, HOXD9, HOXD10, HOXD11 etc in “trans” manner 11. For example, 40 kb of HOXD gene locus is repressed via HOTAIR mediated recruitment of PRC2 complex and LSD1-CoREST-REST complex 12. HOTAIR aids in promoting genome wide chromatin reprogramming and coordinates in long-range gene expression and regulation. Genome wide analysis showed that knockdown of HOTAIR affects expression of diverse type of genes associated with apoptosis, tumor suppression, tumorigenesis, cell differentiation and development. Our results demonstrated that siSENSE-mediated HOTAIR knockdown in MCF7 cells, upregulated its targets genes such as HOXD10, PCDHB5 etc. However, HOXC locus genes such as HOXC9 (data not shown), HOXC10 (Figure 2C–D), and HOXC11 (data not shown) were not affected. These results are in agreement with previous studies 11.
Beyond it roles in gene regulation, HOTAIR is found to be over expressed in hepatocellular carcinoma, pancreatic cancer, colorectal cancer, and in primary and metastatic breast tumors. Over expression of HOTAIR in epithelial cancer cells induces the genome-wide re-targeting of PRC2, altered H3K27 methylation pattern and increased cancer invasiveness and metastasis. It is shown to be an independent prognostic marker for death and metastasis in breast cancer patients. Expression levels of HOTAIR along with another lncRNAs such as MALAT1 are repressed in bleomycin treated HeLa and MCF7 cells, while levels of other lncRNAs are also affected, indicating the presence of differential regulation of lncRNAs under genotoxic stress 63.
In an effort to understand the biochemical function of HOTAIR and its transcriptional regulation, initially we designed a small interfering oligonucleotide complementary to the HOTAIR transcript. As HOTAIR is an antisense transcript, we termed this synthetic olionucleotide as siSENSE (small interfering SENSE) oligonucleotide. We knocked down HOTAIR in human breast cancer cells MCF7 using HOTAIR siSENSE and then examined its impact on cell growth and viability. Our results demonstrated that HOTAIR-siSENSE efficiently knocked down HOTAIR expression in MCF7 cells. Knockdown of HOTAIR suppressed the growth of MCF7 cells and ultimately induced apoptotic cell death. HOTAIR is known to suppress the expression of various target genes including HOXD10 and procadherins and JAM2. Our analysis demonstrated that knockdown of HOTAIR induced expression of HOXD10 and PCDHB5 along with other apoptotic genes further confirming the target specificity of HOTAIR siSENSE in MCF7 cells.
As HOTAIR is over expressed in breast carcinomas, we hypothesized that it may potentially be regulated by estrogen. To understand potential mechanism of transcriptional regulation of HOTAIR, we exposed estrogen-receptor positive human breast cancer cell MCF7 with 17β-estradiol (E2) and examined its impact on HOTAIR gene expression. Our studies demonstrated that HOTAIR is transcriptionally induced upon treatment with E2 and that was suppressed upon exposure to an anti-estrogen tamoxifen. HOTAIR expression was found to be not E2-responsive in ER-negative human breast cancer cells MDAMB231, indicating involvement of estrogen receptors in E2-induced HOTAIR expression. These observations demonstrated that HOTAIR is indeed an estrogen responsive gene and potentially regulated by estrogen-receptors in breast cancer cells.
Sequence analysis demonstrated that HOTAIR promoter contains multiple functional estrogen-response-elements (ERE) located near transcription start sites. ERE2 and ERE3 that are located at −1486 and −1721 nt upstream of transcription start site, have two base pair mismatches from the consensus full ERE sites (GGTCAnnnTGACC). Indeed these two EREs showed most response to E2-treatment in luciferase based reporter assay. Furthermore, estrogen receptors ERα and ERβ were bound to these ERE regions in the HOTAIR promoter in presence of E2. Knockdown of both ERα and ERβ resulted in downregulation of E2-induced activation of HOTAIR in MCF7 cells, indicating crucial roles of ERs during E2-induced HOTAIR expression.
Along with ERs, many ER-coregulators such as SRC-family of proteins, CBP/P300, MLL-histone methylases are known to functionally interact with ERs during E2-induced activation of protein coding genes. Our analysis demonstrated that, similar to protein coding gene activation, CBP/p300, histone methylases MLLl and MLL3 were recruited to the promoter of HOTAIR in the presence of E2. H3K4-trimethylation and histone acetylation levels that are marks of RNAPII mediated protein coding gene activation were also enriched at the HOTAIR promoter upon treatment with E2. Knockdown of either MLL1 or MLL3 down regulated E2-induced HOTAIR expression. In contrast, significant amount of constitutive binding of MLL2 as well as nuclear receptor corepressor N-CoR was observed in the absence of E2 and they were dissociated upon treatment with E2. These observations suggested that HOTAIR gene was repressed in the basal state via coordination of MLL2 and N-CoR, and upon treatment with E2, HOTAIR promoter went into activated state via exchange of transcription activators and coregulators leading to chromatin remodeling and gene activation.
Taken together our results demonstrated that HOTAIR is an estrogen-responsive gene. Like other protein coding E2-responsive genes, HOTAIR being transcribed from the antisense strand is also activated by E2 and this is coordinated by ERs, ER-coregulators, and general transcription factors (GTFs) associated with RNAP II transcription. Chromatin modification marks such as histone H3K4-trimethylation and histone acetylation are also associated with activation of antisense-transcript HOTAIR promoter. Our studies provide novel mechanistic insight of antisense-strand promoter activation and gene transcription. We also demonstrated that lncRNA, like HOTAIR are potentially regulated by RNAPII machineries under E2-environments. Finally, as HOTAIR is an estrogen-responsive gene, it’s over expression in breast cancer is likely associated with E2-indcued gene activation. HOTAIR, being crucial for the growth and viability of breast cancer cell, is a potential target for novel therapy for the treatment of breast cancer.
Materials and Methods
Cell culture and treatment with estrogen
Human breast adenocarcinoma (MCF7) cell was purchased from American Type Cell Culture Collection (ATCC). Cells were grown and maintained in DMEM (Dulbecco’s modified Eagle’s media; Sigma) supplemented with 10 % heat inactivated fetal bovine serum (Sigma), 2 mM L-glutamine, 100 units/ml penicillin and 0.1 mg streptomycin/ml. All cells were maintained in a humidified incubator with 5 % CO2 at 37 °C.
For 17β-estradiol (E2) treatment, MCF7 cells were grown and maintained for at least 3 generation in phenol-red free DMEM-F12 media (Sigma) supplemented with 10 % charcoal stripped FBS, 2 mM L-glutamine and 100 units/ml penicillin and 0.1 mg/ml streptomycin. Cells were grown up to 60 % confluency in 60 mm culture plates and treated with varying concentrations of E2, incubated for 4 h (or varying time periods). RNA was isolated and subjected to RT-PCR or qPCR analysis as previously described 46.
HOTAIR knockdown, siSENSE and antisense transfection
For HOTAIR knockdown experiments, initially, a siSENSE oligonucleotide was designed using IDT-DNA antisense design software. Based on the GC content (about 50%) and melting temperature consideration (~ 40 °C), HOTAIR-siSENSE sequence was selected, custom synthesized with all the phosphorothioate linkages instead of regular phosphodiester bonds. As it is complementary to the antisense-transcript HOTAIR, we termed it as small interfering sense (siSENSE) oligonucleotide.
For HOTAIR knockdown, MCF7 cells were grown in DMEM media with appropriate supplements, then transfected with HOTAIR siSENSE in DMEM devoid of FBS and antibiotics for 24 h and then supplemented with 20 % FBS containing DMEM and other supplements and then allowed the cells to grow for additional 48 h prior to harvesting cells for RNA/protein extraction 26. Transfection with scramble-antisense (Table 1) was also performed in parallel as control.
For MLL and ER knockdown experiments, MCF7 cells were grown up to 60 % confluency in 60 mm culture plates and then transfected with ERα, ERβ, MLL1, MLL2, MLL3, MLL4 and scramble antisense oligonucleotides (Table 1) using ifector transfection reagent (KD Medical Inc) as previously described. Specifically, cocktails of different concentrations of various antisenses (5 – 9 μg) and transfection reagents were made in 300 μL DMEM (without supplements) and incubated in dark for 30 min. Cells were washed thrice with blank DMEM-F12 media and then 1.7 mL of supplement free DMEM-F-12 was added to each cell culture plate. Antisense-transfection reagents cocktail was applied to the cells and incubated for 24 h, then 2 mL of DMEM-F12 media with all supplements and 20% FBS was added to all cell plates. The cells were incubated for additional 48 h then treated with 0.1 nM 17β-estradiol for 4 h and then harvested for RNA extraction and/or fixed for microscopic analysis 35.
RNA extraction, cDNA synthesis, RT-PCR and real-time PCR (qPCR)
Cells were harvested and centrifuged at 500 g for 5 min at 4 °C, then resuspended in diethyl pyrocarbonate (DEPC) treated buffer A (20 mM Tris-HCl, pH 7.9; 1.5 mM MgCl2; 10 mM KCl and 0.5 mM dithiothreitol (DTT); 0.2 mM phenylmethanesulfonyl fluoride (PMSF)), for 10 min on ice and then centrifuged at 3500 g for 5 min at 4 °C. Supernatant was subjected to phenol-chloroform extraction followed by ethanol precipitation. The RNA pellets were air dried and dissolved in DEPC treated water containing 1 mM EDTA and quantified using nanodrop spectrophotometer.
For cDNA synthesis, a cocktail of 2.4 μM of oligo dT (Promega), 100 units of MMLV reverse transcriptase, 1 x MMLV RT buffer (Promega), 100 μM each of dNTPs (dATP, dGTP, dCTP and dTTP, Promega), 1 mM DTT and 20 units of RNaseOut (Invitrogen) were added to 500 ng of the RNA extract and subjected to reverse transcription reaction. Each cDNA product was diluted to 100 μL, and 5 μL of the diluted cDNA was subjected to PCR using specific primer pairs (Table 1). Each experiment was repeated thrice (n = 3).
For the real-time PCR experiments, cDNA (500 ng) was diluted to 50 μL final volume. The cDNA was amplified using SsoFast EvaGreen supermix (Bio-Rad) using specific primers and CFX96 real-time PCR detection system. GAPDH was used as control. The results were analyzed using the CFX Manager. Expressions of gene relative to GAPDH were plotted. Each real-time PCR experiments was done in three parallel replicates and repeated at least twice (n = 3). The real-time PCR analysis of the ChIP DNA fragments were done with primers specific to ERE1, ERE2, ERE3 and ERE4 regions of HOTAIR promoter. Expressions relative to input were plotted. Each PCR reaction was done in triplicates and repeated at least twice.
Impacts of HOTAIR knockdown on cell viability and growth
MCF7 cells (upto 60 % confluency) were transfected with HOTAIR-siSENSE and scramble antisense separately and incubated for varying time periods (0 h, 24 h, 48 h and 72 h). Cells were harvested, stained with trypan blue and viable cells were counted using hematocytometer and plotted as a function of time. Each experiment was performed in 3 replicates and repeated at least twice. Cells at different stages were visualized under microscope (NIKON TE200) to examine their morphologies.
Tunnel assay and cytochrome c immunostaining
For TUNEL assay, HOTAIR-siSENSE transfected MCF7 cells were fixed in 1% formaldehyde, permeabilized in 0.2% Triton X-100, and subjected to dUTP-end labeling (ApoAlert DNA Fragmentation Assay Kit; Clontech, Mountain View, CA, USA). The MCF7 cells were also stained with DAPI and visualized under fluorescence microscope (NIKON TE200-U, Tokyo, Japan).
For immune-fluorescence staining, MCF7 cells were transfected with HOTAIR siSENSE or scramble-antisenses for 72 h and subjected to immunostaining with anti-cytochrome c antibody as described by us previously 35. Briefly, cells were fixed by 4% formaldehyde, washed twice with cold PBS, permeabilized by 0.2 % Triton X-100 in PBS for 15–20 min, blocked with goat serum and then incubated with cytochrome c antibody (Upstate) for at least 3–4 h at room temperature. Samples were washed twice with 0.2% tween 20 in PBS and incubated with FITC (fluorescein isothiocyanate) conjugated anti-mouse secondary antibody for 1 h at room temperature. DAPI staining of the cells were done to visualize nuclear integrity. Samples were mounted on the microscopic slides and subjected to fluorescence microscopy.
Chromatin immuno-precipitation assay (ChIP)
For ChIP assays MCF7 cells were treated with 0.1 nM 17β-estradiol for 4 h and fixed in 4% formaldehyde, lysed in SDS lysis buffer followed by sonication to shear the chromatins. The fragmented chromatin was pre-cleaned with protein-G agarose beads and subjected to immuno-precipitation with antibodies specific to ERα, ERβ, MLL1, MLL2, MLL3, MLL4, H3K4-trimethyl, histone acetylation, NCoR, CBP, P300, TAF250, β-actin and RNAPII overnight. Immuno-precipitated chromatins were washed and de-proteinized to obtain purified DNA fragments that were used as templates in PCR amplifications using various primers corresponding to different EREs of HOTAIR promoter (Table 1). Antibodies were purchased from commercial sources that are as follows: MLL1 (Abgent, AP6182a), MLL2 (Abgent, AP6183a), MLL3 (Abgent, AP6184a), MLL4 (Sigma, AV33704), ERα (D-12, Santa Cruz, sc-8005), ERβ (H-150, Santa Cruz, sc-8974), H3K4-trimethyl (Upstate, 07–473), RNA pol II (RNAPII, Abcam, 8WG16), TAF250 (Upstate, 05–500), NCoR(C-20, sc-1609), β-actin (Sigma, A2066), CBP (A22, Santa Cruz Biotechnology, Sc369) and P300 (N15, Santa Cruz Biotechnology, Sc584).
Dual luciferase reporter assay
HOTAIR promoter spanning ERE1 (−536 to −924 nt), ERE2 (−1315 to −1531 nt), ERE3 (−1623 to −1755 nt) and ERE4 (−1731 to −1833 nt) regions were cloned and inserted upstream of the promoter of firefly luciferase gene in pGL3-promoter vector (Promega) (cloning primers in Table 1). MCF7 cells (60% confluency) were grown in 6 well plate in DMEM-F-12 media, and co-transfected with 1500 ng of these ERE-pGL3 constructs along with 150 ng of a reporter plasmid containing renilla luciferase (pRLTk, Promega) as an internal transfection control using FuGENE6 transfection reagent. Control transfections were done using pGL3 promoter vector without any ERE insertion. At 24 h post transfection, cells were treated with 0.1 nM E2 and incubated for additional 4 h and then subjected to luciferase assay using dual luciferase reporter assay kit (Promega) as instructed. Firefly luciferase activities were assayed and normalized to those of renilla luciferase. Each treatment was done in four replicates and the experiment was repeated at least twice26.
Statistical analysis
Each experiment was done in 2–3 replicates and then cells were pooled (and treated as one sample), subjected to RNA extraction, RT-PCR and ChIP analysis and each experiment was repeated at least thrice (n=3). For luciferase assay each treatment was done in replicates of 4 and the experiment was repeated at least twice. The real time PCR analysis of such samples were done in three replicate reactions and repeated so in all three independent experiments (n = 3). Normally distributed data were analyzed by ANOVA and non-normally distributed data were analyzed using student-t tests (SPSS) to determine the level of significance between individual treatments. The treatments were considered significantly different at p ≤ 0.05.
Highlights.
Long noncoding RNA HOTAIR is critical for cell growth and viability
Knockdown of HOTAIR induce apoptosis in breast cancer cells.
HOTAIR is transcriptionally induced by estradiol (E2).
Estrogen receptors coordinates with MLLs during E2-induced HOTAIR expression
Results provide novel mechanistic insight of antisense-strand gene expression.
Acknowledgments
We thank all the Mandal lab members for helpful discussions. Research in Mandal laboratory is supported in parts by grants from NIH (1R15 ES019129-01, 2R15 CA113747-02), NSF (0821969), and American Heart Association (0765160Y).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes & development. 2011;25:1915–27. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carninci P, Hayashizaki Y. Noncoding RNA transcription beyond annotated genes. Current opinion in genetics & development. 2007;17:139–44. doi: 10.1016/j.gde.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–26. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 4.Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, Willard HF. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–42. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 5.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niland CN, Merry CR, Khalil AM. Emerging Roles for Long Non-Coding RNAs in Cancer and Neurological Disorders. Frontiers in genetics. 2012;3:25. doi: 10.3389/fgene.2012.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, Yamanaka I, Kiyosawa H, Yagi K, Tomaru Y, Hasegawa Y, Nogami A, Schonbach C, Gojobori T, Baldarelli R, Hill DP, Bult C, Hume DA, Quackenbush J, Schriml LM, Kanapin A, Matsuda H, Batalov S, Beisel KW, Blake JA, Bradt D, Brusic V, Chothia C, Corbani LE, Cousins S, Dalla E, Dragani TA, Fletcher CF, Forrest A, Frazer KS, Gaasterland T, Gariboldi M, Gissi C, Godzik A, Gough J, Grimmond S, Gustincich S, Hirokawa N, Jackson IJ, Jarvis ED, Kanai A, Kawaji H, Kawasawa Y, Kedzierski RM, King BL, Konagaya A, Kurochkin IV, Lee Y, Lenhard B, Lyons PA, Maglott DR, Maltais L, Marchionni L, McKenzie L, Miki H, Nagashima T, Numata K, Okido T, Pavan WJ, Pertea G, Pesole G, Petrovsky N, Pillai R, Pontius JU, Qi D, Ramachandran S, Ravasi T, Reed JC, Reed DJ, Reid J, Ring BZ, Ringwald M, Sandelin A, Schneider C, Semple CA, Setou M, Shimada K, Sultana R, Takenaka Y, Taylor MS, Teasdale RD, Tomita M, Verardo R, Wagner L, Wahlestedt C, Wang Y, Watanabe Y, Wells C, Wilming LG, Wynshaw-Boris A, Yanagisawa M, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–73. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 10.Tahira AC, Kubrusly MS, Faria MF, Dazzani B, Fonseca RS, Maracaja-Coutinho V, Verjovski-Almeida S, Machado MC, Reis EM. Long noncoding intronic RNAs are differentially expressed in primary and metastatic pancreatic cancer. Molecular cancer. 2011;10:141. doi: 10.1186/1476-4598-10-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geng YJ, Xie SL, Li Q, Ma J, Wang GY. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. The Journal of international medical research. 2011;39:2119–28. doi: 10.1177/147323001103900608. [DOI] [PubMed] [Google Scholar]
- 14.Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, Miyano S, Mori M. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer research. 2011;71:6320–6. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 15.Niinuma T, Suzuki H, Nojima M, Nosho K, Yamamoto H, Takamaru H, Yamamoto E, Maruyama R, Nobuoka T, Miyazaki Y, Nishida T, Bamba T, Kanda T, Ajioka Y, Taguchi T, Okahara S, Takahashi H, Nishida Y, Hosokawa M, Hasegawa T, Tokino T, Hirata K, Imai K, Toyota M, Shinomura Y. Upregulation of miR-196a and HOTAIR drive malignant character in gastrointestinal stromal tumors. Cancer research. 2012;72:1126–36. doi: 10.1158/0008-5472.CAN-11-1803. [DOI] [PubMed] [Google Scholar]
- 16.Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F, Zheng SS. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Annals of surgical oncology. 2011;18:1243–50. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 17.Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, Kim S, Safe S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2012 doi: 10.1038/onc.2012.193. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ansari KI, KS, Mishra BP, Mandal SS. Mixed lineage leukaemia-4 regulates cell-cycle progression and cell viability and its depletion suppresses growth of xenografted tumour in vivo. British journal of cancer. 2012;107(2):315–324. doi: 10.1038/bjc.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ansari KI, Kasiri S, Mandal SS. Histone methylase MLL1 has critical roles in tumor growth and angiogenesis and its knockdown suppresses tumor growth in vivo. Oncogene. 2012 doi: 10.1038/onc.2012.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woldemariam GA, Mandal SS. Iron(III)-salen damages DNA and induces apoptosis in human cell via mitochondrial pathway. J inorganic biochemistry. 2008;102:740–7. doi: 10.1016/j.jinorgbio.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 21.Li H, Zhu H, Xu C-j, Yuan J. Cleavage of BID by Caspase 8 Mediates the Mitochondrial Damage in the Fas Pathway of Apoptosis. Cell. 1998;94:491–501. doi: 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- 22.Wall NR, Mohammad RM, Al-Katib AM. Bax:Bcl-2 ratio modulation by bryostatin 1 and novel antitubulin agents is important for susceptibility to drug induced apoptosis in the human early pre-B acute lymphoblastic leukemia cell line, Reh. Leukemia Research. 1999;23:881–888. doi: 10.1016/s0145-2126(99)00108-3. [DOI] [PubMed] [Google Scholar]
- 23.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–8. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 24.Novak P, Jensen T, Oshiro MM, Watts GS, Kim CJ, Futscher BW. Agglomerative epigenetic aberrations are a common event in human breast cancer. Cancer research. 2008;68:8616–25. doi: 10.1158/0008-5472.CAN-08-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naik MU, Naik TU, Suckow AT, Duncan MK, Naik UP. Attenuation of Junctional Adhesion Molecule-A Is a Contributing Factor for Breast Cancer Cell Invasion. Cancer research. 2008;68:2194–2203. doi: 10.1158/0008-5472.CAN-07-3057. [DOI] [PubMed] [Google Scholar]
- 26.Ansari KI, Hussain I, Kasiri S, Mandal SS. HOXC10 is overexpressed in breast cancer and transcriptionally regulated by estrogen via involvement of histone methylases MLL3 and MLL4. Journal of Molecular Endocrinology. 2012;48:61–75. doi: 10.1530/JME-11-0078. [DOI] [PubMed] [Google Scholar]
- 27.Kok M, Linn SC. Gene expression profiles of the oestrogen receptor in breast cancer. The Netherlands journal of medicine. 2010;68:291–302. [PubMed] [Google Scholar]
- 28.Kok M, Linn SC. Gene expression profiles of the oestrogen receptor in breast cancer. Neth J Med. 2010;68:291–302. [PubMed] [Google Scholar]
- 29.Nilsson S, Gustafsson JA. Estrogen receptor action. Crit Rev Eukaryot Gene Expr. 2002;12:237–57. doi: 10.1615/critreveukaryotgeneexpr.v12.i4.10. [DOI] [PubMed] [Google Scholar]
- 30.Denger S, Reid G, Brand H, Kos M, Gannon F. Tissue-specific expression of human ERalpha and ERbeta in the male. Molecular and cellular endocrinology. 2001;178:155–60. doi: 10.1016/s0303-7207(01)00417-8. [DOI] [PubMed] [Google Scholar]
- 31.Lebeau A, Unholzer A, Amann G, Kronawitter M, Bauerfeind I, Sendelhofert A, Iff A, Lohrs U. EGFR, HER-2/neu, cyclin D1, p21 and p53 in correlation to cell proliferation and steroid hormone receptor status in ductal carcinoma in situ of the breast. Breast cancer research and treatment. 2003;79:187–98. doi: 10.1023/a:1023958324448. [DOI] [PubMed] [Google Scholar]
- 32.Nettles KW, Gil G, Nowak J, Metivier R, Sharma VB, Greene GL. CBP Is a dosage-dependent regulator of nuclear factor-kappaB suppression by the estrogen receptor. Molecular endocrinology. 2008;22:263–72. doi: 10.1210/me.2007-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ansari KI, Kasiri S, Hussain I, Mandal SS. Mixed lineage leukemia histone methylases play critical roles in estrogen-mediated regulation of HOXC13. The FEBS journal. 2009;276:7400–11. doi: 10.1111/j.1742-4658.2009.07453.x. [DOI] [PubMed] [Google Scholar]
- 34.Ansari KI, Mishra BP, Mandal SS. MLL histone methylases in gene expression, hormone signaling and cell cycle. Frontiers in bioscience: a journal and virtual library. 2009;14:3483–95. doi: 10.2741/3466. [DOI] [PubMed] [Google Scholar]
- 35.Shrestha B, Ansari KI, Bhan A, Kasiri S, Hussain I, Mandal SS. Homeodomain-containing protein HOXB9 regulates expression of growth and angiogenic factors, facilitates tumor growth in vitro and is overexpressed in breast cancer tissue. The FEBS journal. 2012;279:3715–26. doi: 10.1111/j.1742-4658.2012.08733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dreijerink KM, Varier RA, van Beekum O, Jeninga EH, Hoppener JW, Lips CJ, Kummer JA, Kalkhoven E, Timmers HT. The multiple endocrine neoplasia type 1 (MEN1) tumor suppressor regulates peroxisome proliferator-activated receptor gamma-dependent adipocyte differentiation. Mol Cell Biol. 2009;29:5060–9. doi: 10.1128/MCB.01001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hermanson O, Glass CK, Rosenfeld MG. Nuclear receptor coregulators: multiple modes of modification. Trends Endocrinol Metab. 2002;13:55–60. doi: 10.1016/s1043-2760(01)00527-6. [DOI] [PubMed] [Google Scholar]
- 38.Lee S, Lee J, Lee SK, Lee JW. Activating signal cointegrator-2 is an essential adaptor to recruit histone H3 lysine 4 methyltransferases MLL3 and MLL4 to the liver X receptors. Mol Endocrinol. 2008;22:1312–9. doi: 10.1210/me.2008-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–9. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horwitz KB, Jackson TA, Bain DL, Richer JK, Takimoto GS, Tung L. Nuclear receptor coactivators and corepressors. Mol Endocrinol. 1996;10:1167–77. doi: 10.1210/mend.10.10.9121485. [DOI] [PubMed] [Google Scholar]
- 41.Lonard DM, O’Malley BW. Nuclear receptor coregulators: judges, juries, and executioners of cellular regulation. Mol Cell. 2007;27:691–700. doi: 10.1016/j.molcel.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 42.McKenna NJ, Cooney AJ, DeMayo FJ, Downes M, Glass CK, Lanz RB, Lazar MA, Mangelsdorf DJ, Moore DD, Qin J, Steffen DL, Tsai MJ, Tsai SY, Yu R, Margolis RN, Evans RM, O’Malley BW. Minireview: Evolution of NURSA, the Nuclear Receptor Signaling Atlas. Mol Endocrinol. 2009;23:740–6. doi: 10.1210/me.2009-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mo R, Rao SM, Zhu YJ. Identification of the MLL2 complex as a coactivator for estrogen receptor alpha. J Biol Chem. 2006;281:15714–20. doi: 10.1074/jbc.M513245200. [DOI] [PubMed] [Google Scholar]
- 44.Moggs JG, Orphanides G. Estrogen receptors: orchestrators of pleiotropic cellular responses. EMBO Rep. 2001;2:775–81. doi: 10.1093/embo-reports/kve185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ansari KI, Hussain I, Das HK, Mandal SS. Overexpression of human histone methylase MLL1 upon exposure to a food contaminant mycotoxin, deoxynivalenol. The FEBS journal. 2009;276:3299–307. doi: 10.1111/j.1742-4658.2009.07055.x. [DOI] [PubMed] [Google Scholar]
- 46.Ansari KI, Hussain I, Shrestha B, Kasiri S, Mandal SS. HOXC6 Is transcriptionally regulated via coordination of MLL histone methylase and estrogen receptor in an estrogen environment. Journal of molecular biology. 2011;411:334–49. doi: 10.1016/j.jmb.2011.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dreijerink KM, Mulder KW, Winkler GS, Hoppener JW, Lips CJ, Timmers HT. Menin links estrogen receptor activation to histone H3K4 trimethylation. Cancer Res. 2006;66:4929–35. doi: 10.1158/0008-5472.CAN-05-4461. [DOI] [PubMed] [Google Scholar]
- 48.Guenther MG, Jenner RG, Chevalier B, Nakamura T, Croce CM, Canaani E, Young RA. Global and Hox-specific roles for the MLL1 methyltransferase. Proc Natl Acad Sci U S A. 2005;102:8603–8. doi: 10.1073/pnas.0503072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hess JL. MLL: a histone methyltransferase disrupted in leukemia. Trends Mol Med. 2004;10:500–7. doi: 10.1016/j.molmed.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 50.Huttenhofer A, Schattner P, Polacek N. Non-coding RNAs: hope or hype? Trends in genetics: TIG. 2005;21:289–97. doi: 10.1016/j.tig.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 51.Runge S, Nielsen FC, Nielsen J, Lykke-Andersen J, Wewer UM, Christiansen J. H19 RNA binds four molecules of insulin-like growth factor II mRNA-binding protein. The Journal of biological chemistry. 2000;275:29562–9. doi: 10.1074/jbc.M001156200. [DOI] [PubMed] [Google Scholar]
- 52.Bartolomei MS, Zemel S, Tilghman SM. Parental imprinting of the mouse H19 gene. Nature. 1991;351:153–5. doi: 10.1038/351153a0. [DOI] [PubMed] [Google Scholar]
- 53.Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA. 2007;13:313–6. doi: 10.1261/rna.351707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Poole RL, Leith DJ, Docherty LE, Shmela ME, Gicquel C, Splitt M, Temple IK, Mackay DJ. Beckwith-Wiedemann syndrome caused by maternally inherited mutation of an OCT-binding motif in the IGF2/H19-imprinting control region, ICR1. European journal of human genetics: EJHG. 2012;20:240–3. doi: 10.1038/ejhg.2011.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frevel MA, Sowerby SJ, Petersen GB, Reeve AE. Methylation sequencing analysis refines the region of H19 epimutation in Wilms tumor. The Journal of biological chemistry. 1999;274:29331–40. doi: 10.1074/jbc.274.41.29331. [DOI] [PubMed] [Google Scholar]
- 56.Lee JT, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nature genetics. 1999;21:400–4. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- 57.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–6. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nie L, Wu HJ, Hsu JM, Chang SS, Labaff AM, Li CW, Wang Y, Hsu JL, Hung MC. Long non-coding RNAs: versatile master regulators of gene expression and crucial players in cancer. American journal of translational research. 2012;4:127–50. [PMC free article] [PubMed] [Google Scholar]
- 59.Ranum LP, Cooper TA. RNA-mediated neuromuscular disorders. Annual review of neuroscience. 2006;29:259–77. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- 60.Moseley ML, Zu T, Ikeda Y, Gao W, Mosemiller AK, Daughters RS, Chen G, Weatherspoon MR, Clark HB, Ebner TJ, Day JW, Ranum LP. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nature genetics. 2006;38:758–69. doi: 10.1038/ng1827. [DOI] [PubMed] [Google Scholar]
- 61.Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G, 3rd, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nature medicine. 2008;14:723–30. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tufarelli C, Stanley JA, Garrick D, Sharpe JA, Ayyub H, Wood WG, Higgs DR. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nature genetics. 2003;34:157–65. doi: 10.1038/ng1157. [DOI] [PubMed] [Google Scholar]
- 63.Ozgur E, Mert U, Isin M, Okutan M, Dalay N, Gezer U. Differential expression of long non-coding RNAs during genotoxic stress-induced apoptosis in HeLa and MCF-7 cells. Clinical and experimental medicine. 2012 doi: 10.1007/s10238-012-0181-x. [DOI] [PubMed] [Google Scholar]
- 64.Ansari KI, Grant JD, Woldemariam GA, Kasiri S, Mandal SS. Iron(III)-salen complexes with less DNA cleavage activity exhibit more efficient apoptosis in MCF7 cells. Organic & biomolecular chemistry. 2009;7:926–32. doi: 10.1039/b816858j. [DOI] [PubMed] [Google Scholar]
- 65.Ansari KI, Kasiri S, Grant JD, Mandal SS. Apoptosis and anti-tumour activities of manganese(III)-salen and -salphen complexes. Dalton transactions. 2009;40:8525–31. doi: 10.1039/b905276c. [DOI] [PubMed] [Google Scholar]