Abstract
Type I galactosemia is a genetic disorder that is caused by the impairment of galactose-1-phosphate uridylyltransferase (GALT; EC 2.7.7.12). Although a large number of mutations have been detected through genetic screening of the human GALT (hGALT) locus, for many it is not known how they cause their effects. The majority of these mutations are missense, with predicted substitutions scattered throughout the enzyme structure and thus causing impairment by other means rather than direct alterations to the active site. To clarify the fundamental, molecular basis of hGALT impairment we studied five disease-associated variants p.D28Y, p.L74P, p.F171S, p.F194L and p.R333G using both a yeast model and purified, recombinant proteins. In a yeast expression system there was a correlation between lysate activity and the ability to rescue growth in the presence of galactose, except for p.R333G. Kinetic analysis of the purified proteins quantified each variant’s level of enzymatic impairment and demonstrated that this was largely due to altered substrate binding. Increased surface hydrophobicity, altered thermal stability and changes in proteolytic sensitivity were also detected. Our results demonstrate that hGALT requires a level of flexibility to function optimally and that altered folding is the underlying reason of impairment in all the variants tested here. This indicates that misfolding is a common, molecular basis of hGALT deficiency and suggests the potential of pharmacological chaperones and proteostasis regulators as novel therapeutic approaches for type I galactosemia.
Keywords: GALT, yeast model, disease associated mutation, stability, substrate binding, protein misfolding
1. Introduction
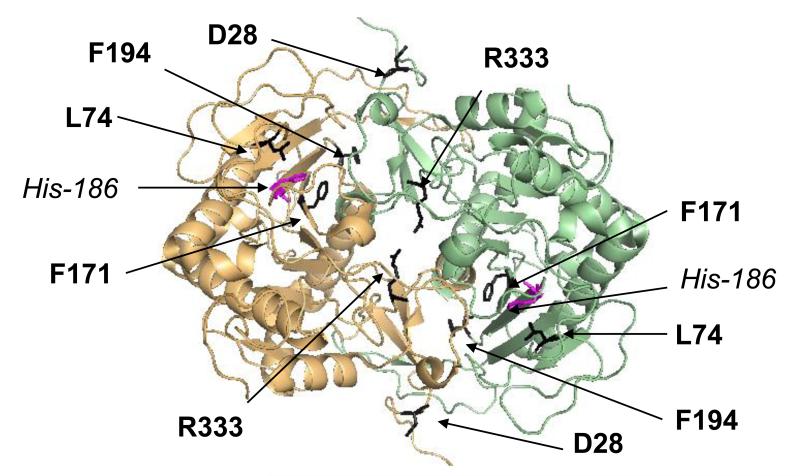
Type I galactosemia (OMIM #230400) is a genetic disorder that is caused by impairment of galactose-1-phosphate uridylyltransferase (GALT; EC 2.7.7.12) [1]. Two other forms of galactosemia are also recognized: galactokinase deficiency (type II; OMIM #230200) and UDP-galactose 4′-epimerase deficiency (type III; OMIM #230350) [2;3]. GALT is involved in the metabolism of galactose and it catalyses the reversible conversion of UDP-glucose and galactose-1-phosphate to UDP-galactose and glucose-1-phosphate via an uridylated enzyme intermediate [4;5]. Deficiency of human GALT (hGALT) is detected through newborn screening in many developed countries minimizing the acute pathology that can otherwise include jaundice, cataracts, vomiting, diarrhea, hepatomegaly, sepsis and neonatal death [6]. Galactose restriction in the diet can immediately mitigate or prevent these acute manifestations, but does not appear to prevent longer-term complications that include ovarian failure and disabilities in learning and speech, among other problems [7]. The underlying mechanism of these long-term pathologies is not fully known, and the role of accumulated galactose-1-phosphate in the process remains controversial [8]. In addition, understanding phenotype-genotype correlations is difficult as compound heterozygosity plays a role in disease [9]. This is because the hGALT protein functions as a dimer (Figure 1) [10-12]. However, recently the level of predicted residual GALT activity associated with genotype of a cohort of school-age children with type I galactosemia was demonstrated to influence the level of scholastic achievement of those students in mathematics [13].
Figure 1. Structure of hGALT and location of mutants analysed in relation to active site.
The homology-modelled structure of the hGALT dimer is presented as a cartoon. Each monomer is coloured light orange or light green. The residues altered by the disease-associated mutations are highlighted in dark grey and presented in stick representations. The active site residue, His-186 is also represented in a stick model and is highlighted in purple. This figure was visualised in PyMol (www.pymol.org) using PDB entry 1R3A [41]. Readers are directed to the online version of this article for the colour version of this figure.
To date, 264 variants have been reported from genetic screening of the hGALT gene. Of these, 159 are missense mutations and for the majority it is not known how they cause their effects [14;15]. The most commonly detected severe mutant, Q188R, and selected others have been studied using a yeast model [16-21] which has provided useful information about the severity of each mutation in vivo. However, detailed functional and structural analyses have been lacking, as only a small number of variants have been studied in any detail in vitro [12;22-27].
It is interesting that although a number of mutations are located in the active site of hGALT and therefore are predicted to affect catalysis directly, the majority are located elsewhere throughout the enzyme’s structure [28;29]. Computational analysis using a homology model of hGALT has suggested that these mutations alter hydrogen bond networks and hydrophobic interactions. Decreased monomer stability was predicted for over half of the studied variants, which suggests that they may cause protein misfolding [28;29]. More recently it has been shown that disease-associated mutants affect the expression and solubility of hGALT in an E. coli expression system. Molecular dynamics simulations predicted that these mutations affect the overall flexibility of the enzyme thus altering substrate affinity [30]. Similarly, previous studies have shown that some mutants can cause temperature sensitivity and decreased levels of expression in yeast [20;21]. Effects on dimer formation have also been detected which further supports the hypothesis that alterations in overall structure are involved [12;25].
Since misfolding has not been experimentally verified for the majority of hGALT mutants [15] five representative variants, p.D28Y, p.L74P, p.F171S, p.F194L and p.R333G were studied here with the aim of establishing whether, or not, this is a common feature of variants associated with type I galactosemia. These variants have been previously found to be associated with type I galactosemia (Table S1) and all five variants are classified as pathogenic in the hGALT mutant database [14]. Only p.F171S and p.L74P are located at the active site (Figure 1) and both have been shown to severely impair enzyme activity (Table S1) [19;20;31]. The remaining three variants are located away from the active site and all five have been included in a recent molecular modelling study of variant GALT enzymes [29]. Thus the studied set represents a diverse group of mutants, which have previously been clinically characterised (Table S1) and subject to, at least, some theoretical analysis. Each of the five mutants was studied in terms of their effects in vivo using an established yeast model and in vitro with the recombinant, purified variant proteins from a bacterial expression system to determine their stability, substrate binding, ability to dimerise and enzyme kinetics in the forward and reverse directions.
2. Materials & Methods
2.1 Expression of hGALT alleles in yeast
Each hGALT allele was recreated by site-directed mutagenesis of the centromeric yeast vector pMM22.hGALT as described previously [20;21] and confirmed by dideoxy sequencing of the entire GALT open reading frame. Creation and analysis of the F171S substitution has been described previously in the context of other studies [19;20]. The primers used to generate these alleles are listed in Table S2.
Each plasmid was transformed into each of two haploid strains of Saccharomyces cerevisiae: JFy3747 [21], which is deficient in GAL7, the gene encoding endogenous yeast GALT [32], and JFy5555, which is deficient in GAL7 and also deficient in GAL1 and GAL10, which encode the endogenous yeast GALK and GALE enzymes, respectively [32]. JFy3747 was used as the host for all growth curve experiments, and JFy5555 was used as the host for all biochemical studies performed using yeast lysates. All yeast strains were grown on medium lacking tryptophan to maintain selection for the MM22-based plasmids.
2.2 Enzyme activities from soluble yeast lysates
GALT activity assays were performed using soluble protein lysates from JFy5555 expressing each of the desired GALT alleles, essentially as described previously [21] except that progress of the reaction was quantified by monitoring the appearance of UDP-galactose (in nmol UDP-gal/μg protein/min). Because the host yeast were deficient in GALK and GALE as well as endogenous GALT there was essentially no background conversion of UDP-glc to UDP-gal by GALE in the absence of GALT activity. The average ± SD (n=3) of GALT enzyme activity for yeast expressing each allele was normalized to the activity level observed in yeast expressing wild-type hGALT from the same plasmid backbone.
2.3 Yeast growth studies
Colonies of JFy3747 yeast expressing the desired alleles of hGALT were cultured and assessed for growth in the wells of 96 well plates using SGE-trp medium with and without 0.01% galactose, as described previously [21]. OD600 readings from any wells that showed evidence of air bubbles or clumping were excluded from analysis. The average ± SD of OD600 readings from 3 separate wells representing yeast expressing each GALT allele and galactose condition were plotted.
2.4 Expression and purification of recombinant proteins
The gene encoding hGALT was amplified by PCR from the IMAGE clone [33] number 3922902 and was cloned into the NdeI and EcoRI sites of pET43a using primers, which incorporated sequence encoding a hexahistidine tag at the 5′ end. The insertion of the gene into the recombinant expression vector was verified by sequencing and this plasmid was then transformed into E. coli Rosetta(DE3) (Merck, Nottingham, UK). Single colonies resulting from this transformation were picked and grown in 5 ml of LB (supplemented with 100 μg.ml−1 ampicillin, 34 μg.ml−1 chloramphenicol, 50 μM ZnCl2), shaking at 30°C overnight. This culture was then diluted into 1 L of LB (supplemented with 100 μg.ml−1 ampicillin and 34 μg.ml−1 chloramphenicol, 50 μM ZnCl2) and grown, shaking at 30 °C until A600nm was between 0.6 and 1.0 (typically 6 h). At this point the culture was induced with 1 mM IPTG at 15 °C and grown for a further 20 h. Cells were harvested by centrifugation at 4,200μg for 20 min and cell pellets were resuspended in buffer R (50 mM HEPES, 5 mM imidazole, pH 7.5, 150 mM NaCl, 10 % (v/v) glycerol, 5 mM DTT). These suspensions were frozen at −80 °C until required.
The cell suspensions were thawed and the cells broken by sonication on ice (three 30 s pulses of 100 W with 30 s gaps in between for cooling). The extract was centrifuged at 20,000μg for 20 min to remove insoluble material and the supernatant applied to a 1 ml nickel agarose (Sigma, Poole, UK) column. Once this solution had passed through, the column was washed with 20 ml buffer W (as buffer R, expect with 500 mM NaCl and 20 mM imidazole) and the protein eluted with a 2 ml wash of buffer E (buffer W supplemented with 250 mM imidazole). The eluate was further purified by size exclusion chromatography on a Sephacryl S-300 (Pharmacia) column (55 ml) at 4 °C with a mobile phase that consisted of 50 mM HEPES, pH 7.5, 150 mM NaCl, 10 % (v/v) glycerol, 5 mM DTT. A flow rate of 1 ml.min−1 was used and 1 ml fractions were collected. Control proteins of known molecular mass were used to construct a standard curve and, thus, determine the oligomeric state of hGALT. Protein containing fractions, (judged by absorbance at 280 nm) corresponding to the molecular mass of hGALT dimers (87 kDa), were pooled together. These pooled fractions were then concentrated using Amicon Ultra-4 (Millipore) centrifugal filtration devices (cut-off of 3 kDa) at 4 °C to a final volume of ≈ 600 μl. The protein solution was then divided into 30 μl aliquots and stored frozen at −80 °C.
The Quick Change protocol [34] was used to change the appropriate codons in the expression vector. Successful mutagenesis was verified by sequencing (MWG-Biotech, Ebersburg, Germany). These mutated plasmids were used to express p.D28Y, p.L74P, p.F171S, p.F194L and p.R333G-hGALT using the same protocol as used with the wild-type protein.
Recombinant human UDP-glucose dehydrogenase was expressed and purified as described [35]. The expression and purification of all proteins was monitored by 10 % SDS-PAGE. All protein concentrations were estimated using the Bradford assay [36] with bovine serum albumin as standard.
2.5 Spectroscopic measurements
Intrinsic fluorescence of each hGALT variant was measured using 5.5 μM protein in 10 mM HEPES, pH 8.8 in a total volume of 180 μl. The binding of 1-anilinonaphthalene-8-sulphonic acid (ANS-1) was used to determine the degree of surface hydrophobicity with each variant at 5 μM in 10 mM HEPES, pH 8.8 with 100 μM ANS-1 in a total volume was 200 μl. Samples with ANS-1 were incubated at room temperature in the dark for 30 min before measurement. Fluorescence spectra were measured (in triplicate) at room temperature using a Spectra Max Gemini X plate-reader fluorimeter (Molecular Devices. CA, USA) with excitation at 280 nm, emission 300-500 nm, and a slit width of 10 nm for intrinsic fluorescence. Excitation at 370 nm, emission 420-580 nm, and a slit width of 5 nm was carried out for ANS-1 binding. Emission spectra were averaged for each variant and corrected for the emission of buffer only or ANS-1 in buffer only, as appropriate.
2.6 Measurement of the steady state kinetic parameters for galactose-1-phosphate uridylyltransferase
Enzymatic activities of the hGALT variants in the forward reaction were determined using a spectrophotometric coupled assay based on that described previously [19] which couples the production of glucose 1-phosphate to its isomerization to glucose 6-phosphate and subsequent NADP+-dependent oxidation of this compound. The standard reaction was performed at 37 °C and contained 10 mM HEPES, pH 8.8, 5 mM DTT, 5 mM glucose 1,6-bisphosphate, 5 mM MgCl2, 0.8 mM NADP+, 0.03 mg glucose-6-phosphate dehydrogenase, and 0.4 mg of phosphoglucomutase. Assays were performed in triplicate in a 96 well plate format each with a total volume of 150 μl. Kinetic constants were determined for UDP-Glc by varying its concentration from 0.01 to 1.0 mM while the concentration of Gal-1P was held at a constant 1.0 mM. Conversely the kinetic constants for Gal-1P were determined by varying its concentration from 0.01 to 2.0 mM while UDP-Glc was held constant at 0.5 mM. The amount of NADPH produced (detected by absorption at 340 nm) is equivalent to the amount of Glc-1P formed.
Enzymatic activities of the hGALT variants in the reverse reaction were also determined using a spectrophotometric coupled assay based on that described previously [37] which couples the production of UDP-glucose to the NAD+-dependent oxidation of this compound. The standard reaction was performed at 37 °C and contained 10 mM HEPES, pH 8.8, 5 mM DTT, 5 mM MgCl2, 10 mM NAD+, 1.2 μM human UDP-glucose dehydrogenase. Assays were performed in triplicate in a 96 well plate format each with a total volume of 150 μl. Kinetic constants were determined for UDP-gal by varying its concentration from 0.01 to 1.0 mM while the concentration of Glc-1P was held at a constant 1.0 mM. Conversely the kinetic constants of Glc-1P were determined by varying its concentration from 0.01 to 2.0 mM while UDP-gal was held constant at 0.5 mM. The amount NADH produced, measured at 340 nm, is equivalent to twice the amount of UDP-Glc formed [38].
All reactions were monitored at 340 nm for 40 min at 37 °C using a Multiskan Spectrum spectrophotometer (Thermo Scientific). Controls lacking either one or both substrates were routinely included, for both forward and reverse kinetic assays, and always gave the expected negative results.
The initial rate of product formation was plotted against substrate concentration and analyzed using non-linear curve fitting of GraphPad Prism (GraphPad Software, CA, USA). The data was fitted to either Michaelis-Menten (1), Michaelis-Menten with substrate inhibition (2) or sigmoidal kinetics (3).
| (1) |
where is the apparent maximum, limiting rate and is the apparent Michaelis constant.
| (2) |
where is the apparent dissociation constant.
| (3) |
where h is the Hill coefficient and is the concentration of substrate to give a rate equal to half of . [S] is the concentration of the varied substrate for all equations. The goodness of fit to these equations was compared using the F test and results are reported for the best fit to the data
2.7 Chemical cross-linking
Before the addition of a chemical cross-linker, hGALT variants (5 μM in 10 mM HEPES, pH 8.8) were incubated at 37 °C for 5 min with and without ligands (1 mM). BS3 (Sigma) or glutaraldehyde (Sigma) was then added to final concentrations of 100 μM and 0.25 % (v/v) respectively. Cross-linking was allowed to proceed for 30 min and was then halted by the addition of an equal volume of SDS loading buffer (125 mM tris-HCl, pH 6.8, 4% (w/v) SDS, 20% (v/v) glycerol, 1% (w/v) dithiothreitol, 0.002% (w/v) bromophenol blue). Samples were denatured at 95 °C for 5 min before analysis by 10 % SDS-PAGE.
2.8 Limited proteolysis
hGALT variants (5 μM in 10 mM HEPES, pH 8.8) were incubated at 37 °C for 5 min with and without ligands (1 mM). Thermolysin, trypsin or chymotrypsin (Sigma), as indicated, was then added as final concentrations of 240 nM, 120 nM, and 24 nM respectively. Digestion was carried out for 30 min and was stopped by the addition of an equal volume of SDS loading buffer. Samples were denatured at 95 °C for 5 min. before analysis by 15 % SDS-PAGE.
2.9 Thermal Inactivation of hGALT
Thermal inactivation of hGALT variants was judged kinetically using the forward reaction setup described in section 2.8. Aliquots (100 μl) of each active variant hGALT at 0.5 μM were incubated in 10 mM HEPES, pH 8.8 for 15 min at temperatures ranging from 30 to 70 °C (5 °C increments). These aliquots were chilled in ice immediately after incubation and the residual activity was determined at 0.5 mM UDP-glucose and 1.0 mM galactose-1-phosphate with 50 nM hGALT. Measurements were carried on three independent assays for each temperature and the average activity was calculated with standard deviations. These were normalized to the activity of each variant at 30 °C.
2.10 Differential scanning fluorimetry assay
Differential scanning fluorimetry was carried out essentially as previously described [39;40]. Protein samples were diluted in 10 mM HEPES, pH 8.8 to a final concentration of 5 μM and any ligands used were added at a final concentration of 1 mM. Sypro orange (Sigma, Poole, UK) was diluted from a 5000μ solution (manufacturer’s concentration definition) into a 50μ solution with 10 mM HEPES, pH 8.8 and was mixed well prior to each use before 1 μl was added to each mixture. Reactions were set up in a total volume of 20 μl in 0.2 ml PCR tubes and controls of no protein added were always included.
Reaction mixtures were loaded into a Rotor-Gene Q cycler (Qiagen) and the following protocol was used: High resolution melt run (460 nm source, 510 nm detector), 25 °C to 95 °C ramp with a 1 °C rise for each step and no gain optimisation. The melting temperatures, (Tm), were calculated using the inbuilt analysis software. The shift in stability corresponding to the change of melting temperature, ΔTm, for each variant and ligand binding were calculated using equations (4) and (5) respectively.
| (4) |
| (5) |
To determine the significance of the differences in Tm the one way ANOVA with Dunnett comparison test was used.
2.11 In silico analysis of variants
The homology model of hGALT, PDB 1R3A [41] was used to determine the location of altered residues. Additional homology models of p.D28Y, p.L74P, p.F171S, p.F194L and p.R333G hGALT, based on 1R3A, were obtained from the hGALT mutant structure database [28] (http://bioinformatica.isa.cnr.it/GALT/). Structures were viewed using PyMol (http://www.pymol.org/).
Sequence alignment was carried out using ClustalW2 [42] and all sequences were obtained from the UniProt database (http://www.uniprot.org/). Conserved residues, and those involved in cofactor and metal binding were identified with ClustalW2.
In determining the effects on stability of hGALT mutants the following programs were used: Dmutant [43], PoPMusic 2.1 [44], Cupsat [45], SDM [46], Eris [47], Concoord/PBSA [48], I-Mutant 2.0 [49], MuPro [50], and Mustab [51]. When appropriate, the structure 1R3A was used. Both thermal and denaturation options were used of the Cupsat server. Additionally both flexible and inflexible backbone options were used of the Eris server. When using the SDM server the mutant structures obtained from the hGALT mutant structure database were used. The overall consensus of stability change was determined with the percentage of agreeing predictions. All predictions were determined from the A chain contained in the coordinate files.
Prediction of intrinsically disordered regions in hGALT was carried out using metaPrDOS [52] and Spine-D [53]; metaPrDOS uses a consensus based approach using multiple predictors. Both predict the probability of disorder and those residues with a probability of 0.5 and higher are deemed disordered. The regions predicted from these two servers were mapped onto the homology model (1R3A) using PyMol.
The FTMap server [54] was used to predict any allosteric sites in hGALT. This server predicts potential binding sites of proteins, which can act as the starting pointing of identifying ’druggable hotspots’. In addition, this server can predict potential substrate binding and allosteric sites [55]. FTMap uses a fragment-based approach that uses sixteen small organic molecules to map these potential binding sites. This is based on a crystallographic approach (Multiple Solvent Crystal Structures or MSCS) where structures are solved in a number of different solvents containing organic solvents [54]. In addition to FTMap, both Q-SiteFinder and Pocket-Finder [56] were used to predict potential binding pockets. The homology structure 1R3A was submitted to these servers and the resulting sites were visualised using PyMol.
3. Results
3.1 In silico analysis suggests alteration of overall protein charge, surface hydrophobicity and monomer stability due to each amino acid substitution
Previously, a computational approach was used to understand how 107 missense mutations cause their effects on hGALT structure and this suggested changes in residue interactions, surface area and stability [28;29]. Here we extended this work by using a number of different protein analysis servers to predict how p.D28Y, p.L74P, p.F171S, p.F194L and p.R333G affect hGALT structure and function. This was done to improve the confidence of our predictions since each of the algorithms has different strengths and weaknesses [57].
Initial analysis using PolyPhen-2 [58] and SIFT [59] suggested that all the mutations were “probably damaging” and “damaging” respectively. In contrast only p.D28Y and p.R333G were predicted to have altered overall charges and pI values of the linear protein chain as predicted using the Protein Calculator version 3.3 (www.scripps.edu/~cdputnam/protcalc.html). However, using the POPS server [60] and the hGALT mutant dimer structures [28] all were predicted to have increases in surface hydrophobicity (Table S3A). Further analysis using a number of different protein stability prediction servers suggested that each mutation results in stability changes of hGALT monomer. Interestingly, the overall consensus predicted both p.D28Y and p.F194L to be stabilised whereas p.L74P, p.F171S and p.R333G were predicted to be destabilised (Table S3B). Previous studies using only two servers suggested that all are destabilised except p.D28Y where no definite prediction could be made [28].
Taken together these analyses suggest that these residues are important in maintaining the overall structure of hGALT. Furthermore protein sequence alignment revealed L74, F171 and R333 were strictly conserved across species, whereas D28 and F194 were not. However, it was revealed that a hydrophilic residue (e.g. aspartate, glutamate or serine) is always at position 28 and that F194 is conserved in all except yeast, where it is a serine (Figure S1).
3.2 Impact of disease-associated substitutions on hGALT activity measured in soluble yeast lysates
As a first assessment of the functional consequence of each of the patient GALT alleles described here, we expressed each in the context of a haploid strain of Saccharomyces cerevisiae (baker’s yeast), JFy5555, which is deficient in the entire endogenous Leloir pathway, and monitored GALT activity measured in vitro in soluble cell lysates. Budding yeast represents a good, eukaryotic model for inherited metabolic diseases due to the organism’s short generation time and the ease with which it can be genetically modified [61-64]. However, it cannot recapitulate multicellular or tissue level consequences. Here, we used it to understand how the disease-associated mutations affect GALT activity in a cellular context. Two of the patient alleles tested, L74P and F171S, each demonstrated no detectable activity above background (Table 1). For L74P this was a new finding; for F171S this result had been observed previously as part of another study [19]. One allele, R333G, demonstrated detectable, albeit residual (<1%), GALT activity above background, and finally two alleles, D28Y and F194L, each demonstrated >10% wild-type activity (Table 1).
Table 1.
GALT activity levels observed in null-background yeast expressing each hGALT allele from a centromeric plasmid.
| hGALT variant | nucleotide change | % wild-type activity (mean ± SD) n=3 |
|---|---|---|
| WT | N.A. | 100 ± 0.00 |
| no GALT | N.A. | 0.01 ± 0.03 |
| p.D28Y | c.82G>T | 13.55 ± 5.72 |
| p.L74P | c.221T>C | 0.01 ± 0.09 |
| p.F171S | c.512T>C | −0.03 ± 0.01 |
| p.F194L | c.580T>C | 11.93 ± 2.00 |
| p.R333G | c.997C>G | 0.60 ± 0.07 |
3.3 Effect of each substitution on the ability of hGALT to rescue galactose stressed yeast
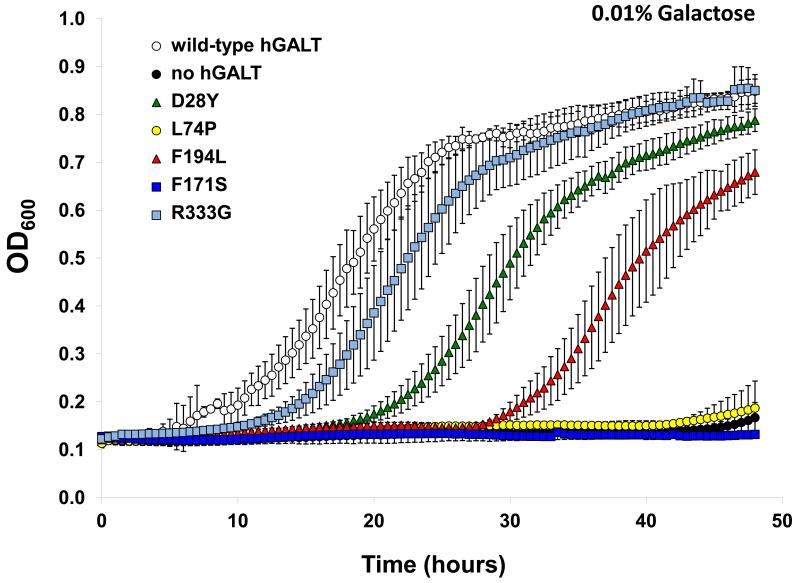
As a test of function in vivo each patient allele was expressed in the haploid yeast strain JFy3747 [21] which is missing endogenous GALT but expresses endogenous galactokinase (Gal1p) and UDP-galactose 4′-epimerase/galactose mutarotase (Gal10p). JFy3747 expressing each hGALT allele were inoculated into medium containing 2% glycerol and 2% ethanol in the presence vs. absence of 0.01% galactose. In the absence of galactose all of the cultures grew well (data not shown), but in the presence of 0.01% galactose clear distinctions were evident. As expected from prior studies [16;20;21] yeast expressing wild-type hGALT grew well, and yeast expressing empty plasmid with no hGALT, completely failed to grow (Figure 2). Also as expected, yeast expressing each of the two hGALT alleles (D28Y and F194L) that demonstrated >10% residual GALT activity in vitro demonstrated intermediate growth in the presence of 0.01% galactose (Figure 2). What was surprising, however, was that the allele that demonstrated only marginal residual activity in vitro, R333G, supported growth in the presence of galactose that was more robust than that seen with either D28Y or F194L. The explanation for this apparent disparity in yeast between in vitro and in vivo function for R333G-hGALT remains unclear but underscores the complexity of the relationship between mutation, expression, and function in different contexts.
Figure 2. Yeast growth curves expressing each hGALT variant in the presence of 0.01% galactose.
Cultures of gal7-null yeast expressing either WT, no hGALT, p.D28Y-hGALT, p.L74P-hGALT, F171S-hGALT, F194L-hGALT or R333G-hGALT were inoculated into 96-wellplates in SGE-trp medium with the indicated amount of galactose added at t = 0. Growth of each culture was monitored at OD600. Plotted values represent means ± SD; n = 3.
3.5 Expression and purification of wild-type and mutant hGALT variants
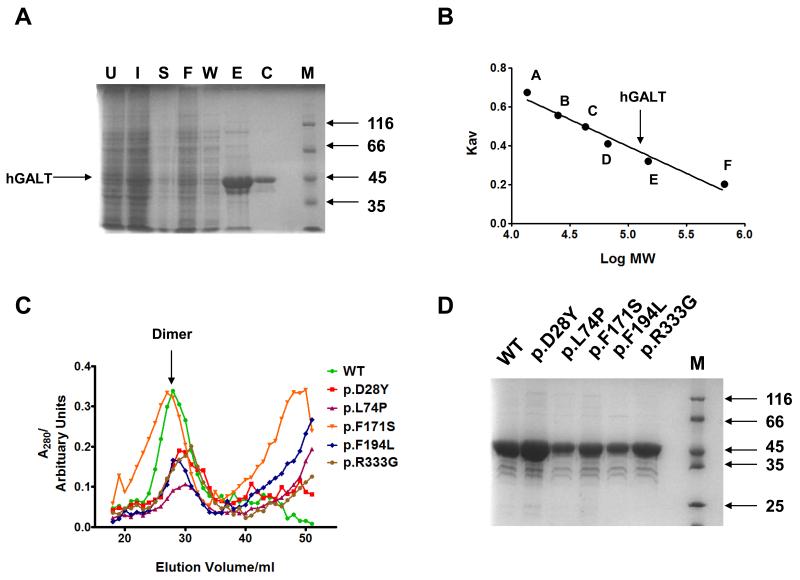
The His-tagged wild-type and five mutant proteins were expressed and purified using a modified E. coli expression system along with affinity and size exclusion chromatography. Initial attempts at expression using a previous protocol [26;27] resulted in small amounts of poor quality purified protein, which were not amenable to study. Using the E. coli Rosetta strain coupled with decreasing the induction temperature to 15 °C and supplementing the growth media with ZnCl2 increased the amount of expressed protein as judged by SDS-PAGE (data not shown). During initial purification attempts precipitation also occurred frequently during dialysis, especially when the media or buffers were supplemented with iron (II) ions. However, decreasing the pre-induction temperature from 37 °C to 30 °C prevented precipitation with ZnCl2-supplemented media, but not those with FeCl2. This is likely to be due to oxidation of Fe2+ to Fe3+. In the E. coli enzyme it has been shown that Zn2+ is essential for maintenance of the structure and that Zn2+ can substitute for Fe2+ at a second divalent cation binding site [65]. In addition, only Zn2+ has been confirmed to be present in hGALT [24]. For these reasons iron supplementation was discontinued. Furthermore size-exclusion chromatography was used instead of dialysis to decrease the purification time; this allowed the oligomerisation state of each hGALT variant to be judged (Figure 3A, B, C). This protocol resulted in roughly 1.0 mg of highly purified hGALT per litre of initial bacterial culture (Figure 3D).
Figure 3. Expression and purification of recombinant hGALT from E. coli using size exclusion chromatography.
(A) WT hGALT purification was resolved by SDS-PAGE (10%) and visualised by staining with Coomassie blue. M = molecular mass markers (kDa). (B) Size exclusion chromatography standard curve. The proteins used for standards are as follows A: Ribonuclease (13.7 kDa); B: Chymotrypsinogen A (25 kDa); C: Ovalbumin (43 kDa); D: Serum albumin (67 kDa) E: Alcohol dehydrogenase (149.5 kDa); F: Thyroglobulin (669 kDa). (C) Chromatograms of wild-type and mutant hGALT proteins. All the proteins eluted mainly at the expected dimer molecular mass. However p.L74P, p.F171S and p.F194L showed increases in the absorbance at approximately one column volume suggesting some degradation to smaller peptides had occurred. (D) Purified hGALT variants resolved by SDS-PAGE (10 %) and visualised by staining with Coomassie blue. M = molecular mass markers (kDa). Readers are directed to the online version of this article for the colour version of this figure.
All hGALT variants were purified as dimers as judged by size-exclusion chromatography (Figure 3C) and each was expressed and purified successfully using the modified protocol (Figure 3D). Notably both F171S and F194L demonstrated some lower molecular weight contaminants, which were likely to be degradation products.
3.6 Kinetic analysis of the forward and reverse reactions on the recombinant hGALT variants reveals perturbed kinetic constants
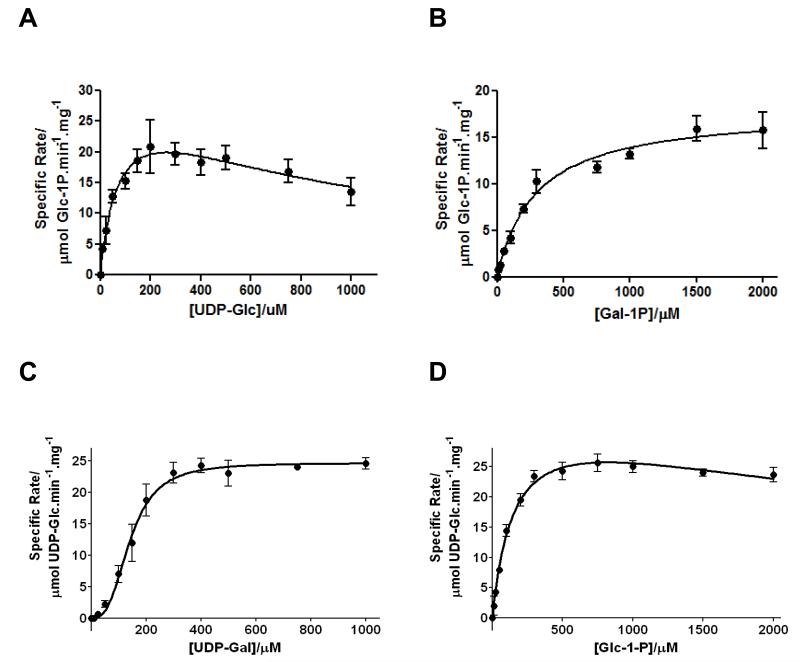
Kinetic analysis of both the forward (UDP-Glc + Gal-1P → UDP-Gal + Glc-1P) and reverse reactions (UDP-Gal + Glc-1P → UDP-Glc + Gal-1P) was carried out on all hGALT variants. The wild-type protein demonstrated kinetics that fitted to three different kinetic models and this depended on the variable substrate (Figure 4, Tables 3, 4). In the forward reaction, varying UDP-Glc resulted in Michaelis-Menten kinetics with substrate inhibition, whereas varying Gal-1P caused the enzyme to demonstrate classical Michaelis-Menten kinetics. Kinetic analyses of the reverse reaction revealed that varying UDP-Gal resulted in sigmoidal kinetics and that varying Glc-1P fitted best to Michaelis-Menten kinetics with substrate inhibition. However, the kinetic parameters determined for the wild-type protein are well within the range of values reported in the literature [23;26;27;37] and substrate inhibition from both UDP-Glc and Glc-1P has been reported previously [19;66]. Interestingly hGALT demonstrated positive cooperativity in the reverse reaction with varied UDP-Gal (Hill coefficient of 2.9 ± 0.3) and although there have been no definitive reports of cooperativity for wild-type hGALT, there have been suggestions of this in purified heterodimers of hGALT [12;67]. Submission of the wild-type homology model structure to three binding site predictors [54;56], resulted in the prediction of an allosteric site at the dimer interface on the opposite side of the enzyme to the active sites (Figure S2).
Figure 4. The steady state enzyme kinetics for WT hGALT in the forward and reverse reaction.
(A) The steady state enzyme kinetics for WT hGALT in the forward reaction with varied UDP-Glc and 1 mM Gal-1P. The kinetic data fitted best to the Michaelis-Menten equation with substrate inhibition with the following constants: , , μmol Glc-1P.min−1.mg−1. (B) The steady state enzyme kinetics in the forward reaction with varied Gal-1P and 500 μM UDP-Glc. The kinetic data fitted best to the Michaelis-Menten equation with the following constants: , μmol Glc-1P.min−1.mg−1. (C) The steady state enzyme kinetics for WT hGALT in the reverse reaction with varied UDP-Gal and 1 mM Glc-1P. The kinetic data fitted best to the Hill equation (sigmoidal kinetics) with the following constants: , μmol UDP-Glc.min−1.mg−1, h = 2.9. (D) The steady state enzyme kinetics in the reverse reaction with varied Glc-1P and 500 μM UDP-Gal. The kinetic data fitted best to the Michaelis-Menten equation with substrate inhibition with the following constants: , , μmol UDP-Glc.min−1.mg−1. WT hGALT (10 nM) was used for both forward and reverse reactions and controls with no GALT always gave no activity. Each point represents the mean of three independent determinations of the rate and the error bars represent the standard deviations of these means.
Table 3.
Steady state kinetic constants for the reverse reaction of recombinant wild-type and active variant hGALT enzymes.
| hGALT Variant | Apparent Km or K0.5 μM |
Glc-1P Ki mM |
UDP-Gal h | Apparent kcat s−1 |
Apparent kcat/Km or kcat/K0.5 × 105 l.mol−1.s−1 |
||
|---|---|---|---|---|---|---|---|
| UDP-Gala | Glc-1Pb | UDP-Gala | Glc-1Pb | ||||
| WT | 141 ± 5.2 | 159 ± 17 | 4.3 ± 0.9 | 2.9 ± 0.3 | 17.7 ± 0.4 | 1.26 ± 0.05 | 1.11 ± 0.12 |
| p.D28Y | 179 ± 12 | 121 ± 24.9 | 2.3 ± 0.6 | 1.4 ± 0.1 | 9.3 ± 0.3 | 0.52 ± 0.03 | 0.77 ± 0.16 |
| p.F194L | 89.7 ± 5.7 | 118 ± 14.4 | 3.2 ± 0.7 | 1.8 ± 0.2 | 5.5 ± 0.1 | 0.61 ± 0.04 | 0.47 ± 0.06 |
| p.R333G | 324.4 ± 33.6 | 202.1 ± 17.9 | 5.9 ± 1.2 | 1.6 ± 0.2 | 5.5 ± 0.3 | 0.16 ± 0.02 | 0.27 ± 0.03 |
[Glc-1P] = 1000 μM
[UDP-Gal] = 500 μM
Table 4.
Change of melting temperatures ΔTm of recombinant wild-type and variant hGALT enzymes in the presence of various substrates.
| ΔTm (K) | ||||||
|---|---|---|---|---|---|---|
| hGALT Variant |
+Gal-1P | +UDP-Glc | +Glc-1P | +UDP-Gal | +Gal-1P & UDP-Glc |
+Glc-1P & UDP-Gal |
| WT | 6.5 ± 0.6*** | 2.5 ± 0.8*** | 6.5 ± 0.4*** | 2.5 ± 0.6*** | 6.1 ± 0.4*** | 6.6 ± 0.8*** |
| p.D28Y | 8.8 ± 1.0*** | 3.2 ± 1.4*** | 8.0 ± 1.9*** | 2.2 ± 1.5* | 8.0 ± 1.5*** | 8.2 ± 1.1*** |
| p.L74P | −0.7 ± 0.9 | −0.6 ± 1.0 | −0.3 ± 1.1 | −1.4 ± 0.9 | −1.5 ± 0.7* | −1.5 ± 0.7* |
| p.F171S | −0.3 ± 1.4 | −0.3 ± 0.9 | 0.1 ± 0.9 | −0.4 ± 1.0 | 0.0 ± 0.9 | 0.5 ± 1.0 |
| p.F194L | 1.9 ± 0.5*** | 1.6 ± 0.1*** | 1.3 ± 0.5*** | 0.5 ± 0.0 | 2.1 ± 0.2*** | 2.5 ± 0.0*** |
| p.R333G | −1.1 ± 2.0 | 1.6 ± 0.8* | −2.0 ± 0.9** | −0.2 ± 0.8 | 0.3 ± 0.7 | −2.0 ± 0.8* |
All ligands were added to a final concentration of 1 mM in 10 mM HEPES, pH 8.8. ΔTm values are reported as the means ± SD of three separate experiments.
P < 0.05,
P < 0.001,
P < 0.0001 (One way ANOVA with Dunnett comparison test).
The variant hGALT proteins demonstrated altered kinetics with two, p.L74P and p.F171S, showing no detectable activity in both the forward and reverse assays in the protein concentration range studied (0 to 0.5 μM). This is in agreement with the yeast lysate activities (Table 1) along with previous reports that these variants are inactive [19;20;31]. All other variants showed decreased activity with altered kinetic parameters and decreased Hill coefficients (Tables 3, 4). p.D28Y (Figure S3) had the least altered kcat with only slightly lower activity than the wild-type protein. Previously the specificity constants have been used as an estimate of the rates of the formation and decay of the uridyl intermediate [23]. Thus, p.D28Y was impaired in terms of uridylylation, for both reactions, as judged from the specificity constants (Tables 3,4). This variant was more prone to substrate inhibition from UDP-Glc with a Ki value nine times lower than that of the wild-type. p.F194L (Figure S4) and p.R333G (Figure S5), however, did not show substrate inhibition by UDP-Glc and had much lower activity than the wild-type and p.D28Y. For both p.F194L and p.R333G, when UDP-Gal was varied, non-Michaelis-Menten kinetics were observed, with large increases in activity at 700 μM UDP-Gal. This might be due to these proteins’ stabilities during storage; however, in the lower substrate range both fitted well to the Michaelis-Menten equation. These variants’ specificity constants revealed different levels of impairment in uridylylation and deuridylylation for both reactions and each had altered apparent Km and K0.5 values for all substrates (Tables 3,4).
3.7 Chemical cross-linking reveals that substrate binding alters the dimer interface
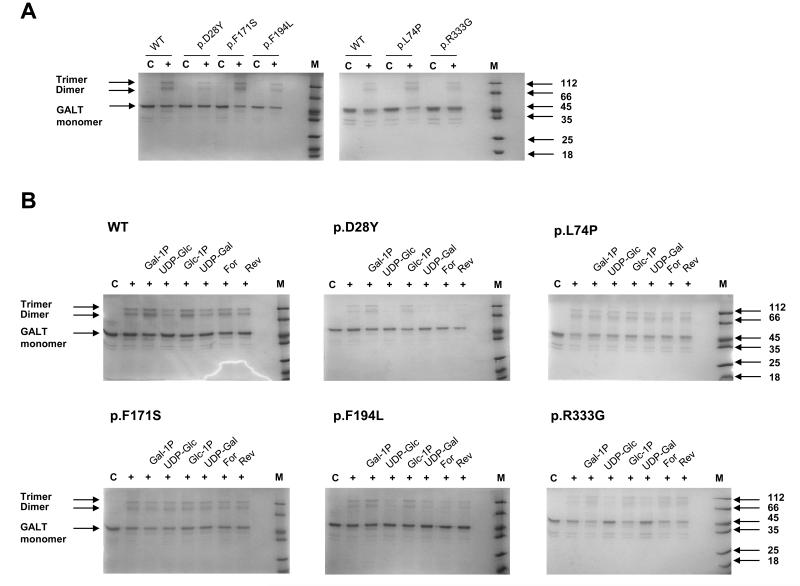
GALT only functions as a dimer and both active sites in the holoenzyme include residues from both polypeptide chains [10;68;69]; it has been shown that specific mutations can affect dimer formation [12;22;67]. Chemical cross-linking has been a useful tool in determining whether other mutant proteins, involved in disease, have the potential to oligomerise correctly [70]. Here we employed the cross-linkers BS3 and glutaraldehyde to investigate the effects of ligands and point mutations. Both cross-linkers confirmed that all hGALT variants form dimers (Figure 5A; S6A) in agreement with the size-exclusion chromatography experiments (Figure 3C). Trimers were also detected with BS3 and other higher molecular weight aggregates were detected when glutaraldehyde was used. These may be artefactual due to the high ratio of cross-linker:protein.
Figure 5. Chemical cross-linking of hGALT variants with BS3 in comparison to the wild-type hGALT and in the presence of various substrates.
(A) The hGALT variants showed little difference in their ability to form dimers in comparison to the wild-type. Higher order oligomers were also formed in the presence of cross-linker. C: control, protein (5 μM) with no cross-linker; +: protein (5 μM) with the cross-linker BS3 (100 μM). (B) Active and inactive hGALT variant show different substrate dependent effects on dimerisation as determined by cross-linking. C: control, protein (5 μM) with no cross-linker; +: protein (5 μM) with BS3 (100 μM) in the presence of various substrates (1 mM). Gal-1P: galactose 1-phosphate; UDP-glc: uridine diphosphate glucose; Glc-1P: glucose 1-phosphate; UDP-gal; uridine diphosphate galactose; For: galactose 1-phosphate and uridine diphosphate glucose; Rev: glucose 1-phosphate and uridine diphosphate galactose.
Differences in cross-linking were detected in the presence of each UDP-sugar, sugar-phosphate and appropriate UDP-sugar/sugar-phosphate pair from the forward and reverse reactions. A pattern of slightly decreased and slightly increased cross-linking was detected in the presence of a UDP-sugar and sugar-phosphate respectively (Figure 6B; S6B). Addition of each UDP-sugar/sugar phosphate pair resulted in similar levels of cross-linking to protein without substrate. Each hGALT variant showed similar responses to substrates with cross-linkers to the wild-type protein, except for the enzymatically inactive p.L74P and p.F171S. Although these variants could be cross-linked, little change in the crosslinking pattern was seen in the presence of ligands (Figure 5B; S6B). This pattern was similar for both cross-linkers suggesting these effects on dimerisation are due to protein-substrate interactions and not an effect due to the cross-linker itself.
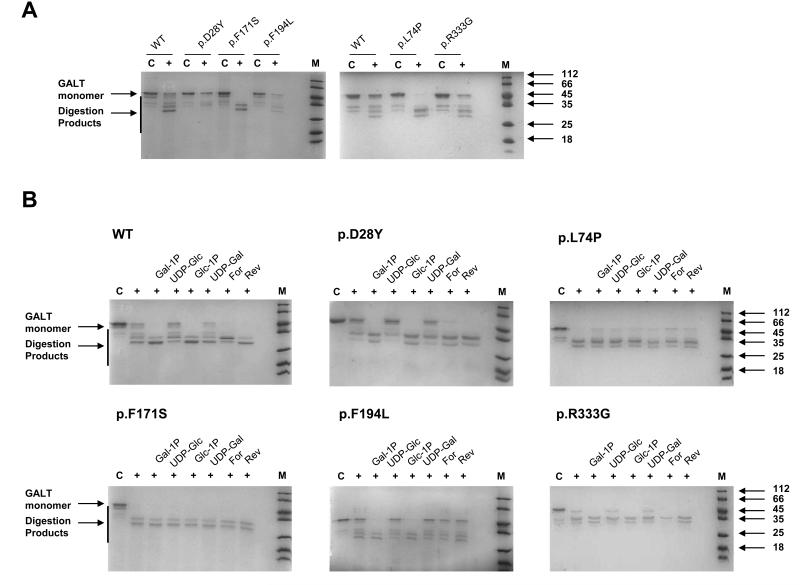
Figure 6. Limited proteolysis of hGALT variants in comparison to the wild-type hGALT and in the presence of various substrates.
(A) The hGALT variants showed differences in susceptibly to degradation by the protease thermolysin in comparison to the wild-type. C: control, protein (5 μM) with no protease; +: protein (5 μM) with thermolysin (240 nM). (B) WT hGALT showed substrate dependent effects on the susceptibility to proteolysis where sugar 1-phosphates increased degradation and uridine diphosphate sugars had little effect. Additionally this increased degradation due to sugar 1-phosphates is the most dominant affect when the protein was in the present of both a sugar 1-phosphate and uridine diphosphate sugar. Active and inactive hGALT variants show different substrate dependent effects on protease sensitivity. C: control, protein (5 μM) with no protease; +: protein (5 μM) with thermolysin (240 mM) in the presence of various substrates (1 mM). Gal-1P: galactose 1-phosphate; UDP-glc: uridine diphosphate glucose; Glc-1P: glucose 1-phosphate; UDP-gal; uridine diphosphate galactose; For: galactose 1-phosphate and uridine diphosphate glucose; Rev: glucose 1-phosphate and uridine diphosphate galactose.
3.8 hGALT mutant proteins show differences in susceptibility to proteases which is altered in presence of each substrate
Increased susceptibility to proteases can occur as a consequence of structural changes due to mutation [71]. Carrying out limited proteolysis using thermolysin and trypsin revealed that p.L74P, p.F171S and p.F194L have increased susceptibility to proteolytic degradation. p.R333G showed little change in degradation. In contrast, p.D28Y was more resistant to degradation (Figure 6A; S7A). Digestion with chymotrypsin gave less clear results, but showed that p.D28Y hGALT is slightly more resistant to degradation whereas p.L74P hGALT is slightly more susceptible (Figure S8A).
Since cross-linking was affected by the presence of each substrate, limited proteolysis was carried out under similar conditions for all variants. Changes in protease susceptibility in the presence of each substrate were detected and this followed a similar pattern to that observed in the cross-linking experiments. Increased degradation was detected in the presence of both sugar-phosphates, whereas a slight decrease in degradation occurred with both UDP-sugars. Each UDP-sugar/sugar-phosphate pair showed similar levels of degradation to protease treated protein with sugar-phosphate. This pattern occurred for all hGALT variants except for p.L74P and p.F171S, where the presence of substrate(s) conferred little, or no, change in degradation (Figure 6B; S7B). Additionally, p.F194L demonstrated no increase in degradation in the presence of both UDP-sugar/sugar-phosphate pair. Again the same results were obtained for both thermolysin and trypsin, but chymotrypsin resulted in less clear results for p.F194L and p.R333G (Figure S8B). The common patterns resulting from digestion with each protease suggest that these effects on degradation are due to protein-substrate interactions and not an effect on the proteases.
These findings of altered protease stability are in agreement with the suggestion from in silico analysis that altered protein folding has occurred, but they do not agree with the overall consensus for each variant. In addition, the decreased effect on stability from substrates for p.L74P and p.F171S demonstrates that these variants have altered substrate interactions, agreeing with the cross-linking and kinetic experiments.
3.9 hGALT mutants demonstrate altered intrinsic fluorescence and show an increased surface hydrophobicity
hGALT contains 24 tryptophan residues per dimer and their excitation resulted in a broad emission spectrum for the wild-type protein (Figure S9A). In comparison, all variants studied, except p.D28Y, had increased relative emission intensity suggesting changes in the microenvironment of the tryptophans most likely slight reorientation of one or more of these residues towards a more hydrophobic environment or away from polar quenching groups (Figure S9A).
In addition, as in silico analysis suggested changes in surface hydrophobicity, the binding of the hydrophobic fluorescent probe ANS-1 to each variant was used to investigate any misfolding in the ground state. All variants except p.D28Y had an increased ANS-1 fluorescence compared to the wild-type protein suggesting p.L74P, p.F171S, p.F194L and p.R333G hGALT all have larger accessible hydrophobic surface areas (Figure 7A; S9B). These fluorescence results suggest all variants tested, except p.D28Y hGALT, have an altered conformation compared to the wild-type.
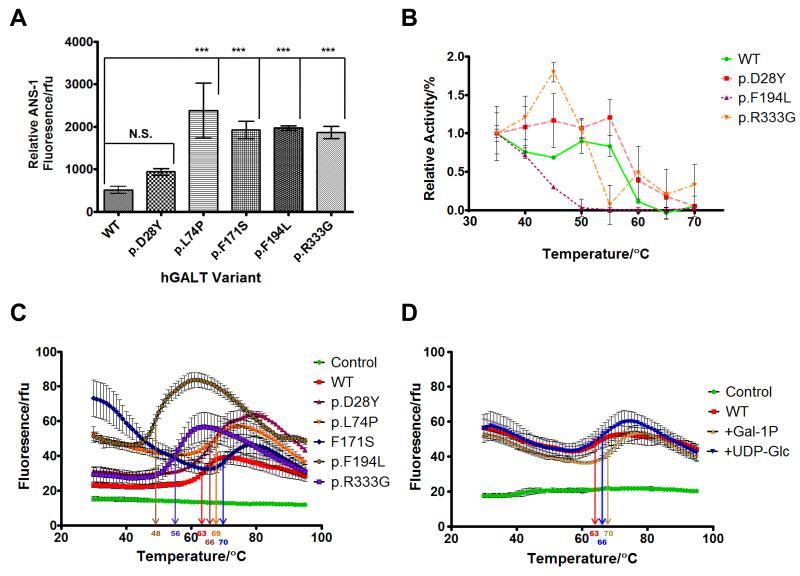
Figure 7. Surface hydrophobicity and thermal stability of hGALT variants.
(A) Relative surface hydrophobicity of each hGALT variant as judged by ANS-1 binding. The emission (at 480 nm) of ANS-1 binding was measured following excitation at 370 nm. The average fluorescence and standard deviation was calculated from three independent assays with each corrected for ANS-1 alone. The significance of change in fluorescence in comparison to the wild-type was determined using one-way ANOVA with a Dunnett’s post-test. (B) Thermal inactivation of wild-type hGALT and active variants (p.D28Y, p.F194L and p.R333G) at 50 nM. Aliquots (100 μl) of each variant hGALT (0.5 μM) were incubated in HEPES (10 mM), pH 8.8 for 15 min at temperatures ranging from 30 to 70 °C. Residual activity of the forward reaction was determined with 0.5 mM UDP-Glc and 1.0 mM Gal-1P. Activities were normalized to those at 30 °C. Three independent assays for each temperature were carried out and the points show the average activity and the error bars the standard deviations of these means. (C) Each hGALT variant showed a unique melting curve and susceptibility to thermal denaturation as judged by DSF. Melting temperatures, (Tm) are presented in table S2. (D) Melting profiles and melting temperatures of WT hGALT with and without Gal-1P or UDP-Glc. WT hGALT showed a shift in stability when Gal-1P or UDP-Glc was present consistent with binding to the substrate and consequent stabilisation of the substrate. Melting temperatures, (Tm) are presented in table S2. Reaction mixtures contained 5 μM protein, 1 mM ligand and 2.5× Sypro orange (manufacturer’s concentration definition) dissolved in 10 mM HEPES pH 8.8. Controls contained no protein. Assays were set up in triplicate and the curves were constructed from the mean fluorescence values; the error bars represent the standard deviations of these means. Readers are directed to the online version of this article for the colour version of this figure.
3.10 hGALT mutants show altered resistance to thermal denaturation
Thermal stability was first determined through thermal inactivation of enzyme activity. Wild-type hGALT lost activity around 65 °C and p.D28Y was only slightly more resistant to thermal denaturation. p.F194L and p.R333G, however, each lost activity at a much lower temperatures than the wild-type (50 °C and 55 °C respectively) with p.R333G showing an inactivation profile with increasing activity up to 45 °C (Figure 7B).
Since thermal inactivation was only applicable to those mutants that have activity, differential scanning fluorimetry was carried out on all hGALT variants. This method uses an extrinsic hydrophobic fluorophore (Sypro orange) to detect unfolding in the presence of increasing temperature [72]. The melting temperature, Tm, is then calculated as the midpoint between the maximum and initial minimum fluorescence allowing for comparison of stability between variants. There is good agreement between DSF and differential scanning calorimetry [72]. We have previously used this technique to study the stability changes in human UDP-galactose 4′-epimerase due to ligand binding and disease-associated mutations [39;40].
Figure 7C shows the unfolding curves of all six hGALT variants and Tm values are presented in table S4. p.F171S was found to have a relatively high initial fluorescence signal at 30 °C as did p.L74P and p.F194L, to lesser extents. This agrees with the respective ANS-1 fluorescence results; however, p.R333G showed little difference in initial fluorescence from the wild-type protein and this may be due to differences in the binding of ANS-1 and Sypro orange or the starting temperature of this assay.
Thermal denaturation revealed that each hGALT variant had different Tm values and unfolding profiles with the wild-type having a Tm of 63 °C. p.F194L and p.R333G were less resistant to denaturation with lower Tm values of 48 °C and 56 °C respectively. In contrast, p.D28Y was slightly more resistant with a Tm of 66 °C. p.L74P and p.F171S were also both more resistant to denaturation with Tm values of 69 °C and 70 °C respectively. These findings correlate well with the thermal inactivation results. Taken together these results demonstrate that each substitution causes alterations to the thermal stability of the hGALT protein and that some show misfolding in the ground state.
3.11 Variant hGALT proteins show altered substrate binding
In addition to determining stabilities of proteins, DSF can also provide information about substrate binding. Here the presence of a substrate can induce conformational changes which can be inferred from the change in Tm (ΔTm) [73]. Tables 5 and S4 present the ΔTm and Tm values of each hGALT variant in the presence of various substrates. Figure 7D shows the unfolding curves of WT hGALT in the presence of UDP-Glc and Gal-1P. Wild-type and p.D28Y hGALT showed similar, statistically significant increases in stability in the presence of all substrates. Here both Glc-1P and Gal-1P resulted in similar ΔTm values of approximately 6.5 K whereas UDP-Glc and UDP-Gal resulted in ΔTm values of approximately 2.5 K. Both substrate pairs resulted in similar increases of stability of 6.5 K. p.L74P and p.F171S differed, showing no significant changes in stability with each substrate individually, although p.L74P appeared to be slightly destabilised in the presence of both substrate pairs (ΔTm of −1.5 K). p.F194L also showed increases in stability for all substrates, but not to the same extent as the wild-type protein. This variant showed decreased stability with both sugar phosphates (ΔTm of 1.9 K), UDP-Gal (ΔTm of 0.5 K) and both substrate pairs (ΔTm of 2.2 K). Interestingly, p.R333G showed the most different behaviour when compared to the wild-type protein. The presence of UDP-Gal and Gal-1P resulted in no significant changes, whereas UDP-Glc resulted in an increase of 1.6 K and Glc-1P caused a decrease of −2.0 K. Additionally changes in stability in the presence of each substrate pair appeared to be the sum of the results of each substrate on its own.
Overall these results indicate that the D28Y substitution does not cause any significant changes in substrate binding, whereas both L74P and F171S appear to cause a substantially decreased ability to bind substrates. p.F194L and p.R333G hGALT still have the ability to bind substrates but not to the same extent as the wild-type protein.
4. Discussion
4.1 Structural bioinformatics analyses reveal further details of the effects of each substitution on hGALT structure and function
A previous study used a homology model of hGALT to study the structural effects of 107 disease-associated variants of hGALT (http://bioinformatica.isa.cnr.it/GALT/) [29]. The structures of the p.D28Y, p.L74P, p.F171S, p.F194L and p.R333G mutant proteins were included in this study and their predicted structures allow for a more indepth analysis of how these mutations cause their effects.
Asp-28 is located towards the N-terminus and near the dimer interface but is not part of the active site. This residue forms a salt bridge with Arg-25 (Figure 8A) and its alteration to tyrosine is predicted to have no direct effects on substrate interaction. However, Asp-28 also forms a salt bridge with His-47 and this residue is in the same loop as Arg-48 and Arg-51 that are predicted to form contacts with the sugar and phosphate moieties of the opposite subunit’s active site [29;69]. Mutating Asp-28 to tyrosine removes these salt bridges (Figure 8B). It has been shown that this loop containing equivalent residues in E. coli GALT shows different conformations in the different crystal structures, being partially disordered, and this is possibly due to interactions between it and the substrate [19;29]. It has been suggested that this loop is involved in excluding water from the active site preventing the hydrolysis of the uridyl-enzyme and participating in its energetically unfavourable formation [65].
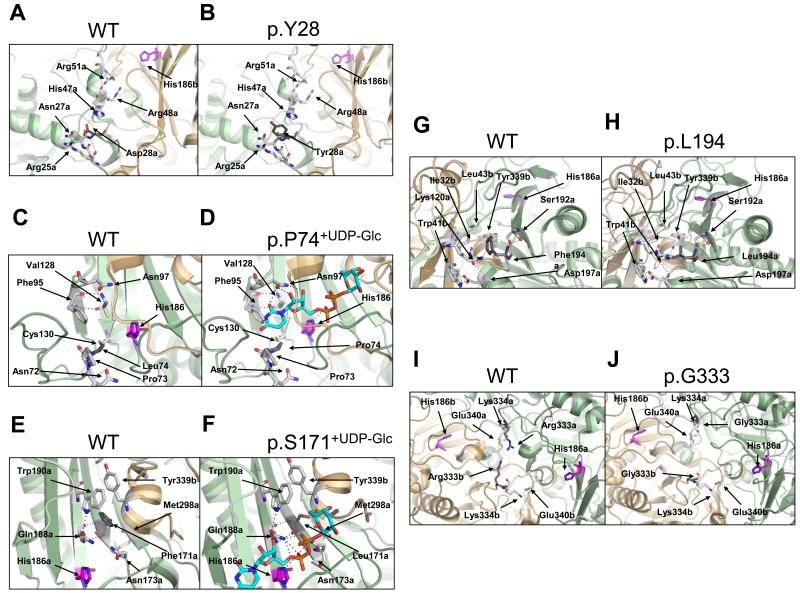
Figure 8. Amino acids affected by hGALT mutations affect side-chain and substrate interactions.
(A) Asp-28 is located in the N-terminal loop at the dimer interface and forms a number of hydrogen bonds, one of which is with His-47. This residue is on the same loop as Arg-48 and Arg-51 that form part of subunit B’s active site. (B) p.D28Y results in the removal of the interaction between this residue and His-47. (C) Leu-74 is located at the active site and is flanked by Pro-73 and Cys-75. This residue forms hydrogen bonds with Cys-130 and Tyr-89 via its backbone oxygen and with Asn-72 via the backbone nitrogen. (D) p.L74P results in the removal of the hydrogen bond with Asn-72. Leu-74 is predicted to be close to the uracil moiety of UDP-Glc and possibly interacts hydrophobically. (E) The residue Phe-171 is located at the active site and the dimer interface. It is close to Gln-188 and forms only backbone interactions with this residue. (F) p.F171S results in an additional hydrogen bond between the side chain hydroxyl and backbone oxygen of Met-298. (G) Phe-194 is located at the dimer interface and exterior of hGALT. This residue forms hydrogen bonds between its backbone oxygen with the hydroxyl of Ser-192. The side chain of Phe-194 is buried in a largely hydrophobic cleft containing residues from both polypeptide chains. (H) p.F194L results in a smaller side chain that points more towards the exterior of the protein. (I) Arg-333 is predicted to be part of a number of loops at the dimer interface between the two active sites. This residue is facing the Arg-333 of the other subunit and beside Lys-334 that is predicted to interact with the sugar moiety of either substrate [29]. (J) Alteration of arginine to glycine at residue 333 results in a void at the dimer interface due to the smaller size of the side chains. Subunit backbones are ribbon representations with subunit A in pale green and subunit B in light orange. Selected residues are shown as stick models in white with nitrogen, oxygen, phosphate and sulphur atoms in blue, red, orange and yellow respectively. Residues affected by mutation are in dark grey and hydrogen bonds are shown as black dotted lines. His-186 is shown in purple and the substrate UDP-Glc is shown in cyan. Structures are from the hGALT structure [28]. Readers are directed to the online version of this article for the colour version of this figure.
Our studies on the wild-type protein suggest that conformational changes are important in substrate binding, most likely in these active site loops (Figure 5B, 6B, S6B, S7B, S8B, Table 4). These loops in p.D28Y are likely to be less flexible due to the removal of a connection to the highly flexible N-terminus as determined from predicted regions of disorder of WT hGALT (Figure S10). This increases the enzyme’s overall stability (Figure 5A, S7A, S8A, 7C), but causes little change in the overall global conformation of the protein (Figure 7A, S9). The decreased flexibility reduces the likelihood of the release of UDP-Glc and Glc-1P thus increasing the apparent inhibition from these substrates (Table 2,3), explaining the kinetic impairment. This is not the first report of an hGALT variant showing increased inhibition and stability: a patient’s hGALT was more prone to substrate inhibition by Glc-1P and was more thermally stable than the wild-type protein [74]. The specific mutations involved were not identified but it is likely that they affected these active site loops. In addition, since p.D28Y appears to affect the other subunit’s active site it can be predicted that this mutation may be dominant negative and this further supports the hypothesis that there is communication between the active sites.
Table 2.
Steady state kinetic constants for the forward reaction of recombinant wild-type and active variant hGALT enzymes.
| hGALT Variant | Apparent Km μM |
UDP-Glc Ki μM |
Apparent kcat s−1 |
Apparent kcat/Km × 105 l.mol−1.s−1 |
||
|---|---|---|---|---|---|---|
| UDP-Glca | Gal-1Pb | UDP-Glca | Gal-1Pb | |||
| WT | 81.6 ± 16.3 | 290 ± 40 | 850 ±200 | 13.6 ± 0.5 | 1.67 ± 0.34 | 0.47 ± 0.07 |
| p.D28Y | 448.7 ± 231.4 | 288 ± 64.6 | 94 ± 48 | 9.4 ± 0.7 | 0.21 ± 0.11 | 0.33 ± 0.08 |
| p.F194L | 60.8 ± 10.9 | 45.9 ± 25.4 | N.D. | 2.5 ± 0.1 | 0.41 ± 0.08 | 0.54 ± 0.30 |
| p.R333G | 285.7 ± 87.5 | 1184 ± 216 | N.D. | 0.98 ± 0.12 | 0.03 ± 0.01 | 0.008 ± 0.001 |
[Gal-1P] = 1000 μM
[UDP-Glc] = 500 μM
Leu-74 is located at the dimer interface and active site of hGALT (Figure 8C). This residue’s carbonyl oxygen hydrogen bonds with the side chains of both Cys-130 and Tyr-89, whereas the backbone nitrogen hydrogen bonds to the side chain of Asn-72. The residue is predicted to be located close to the uracil moiety and it has been suggested that mutating this residue to proline removes van der Waal’s contacts between the residue and this part of the substrate [10]. Mutating this leucine to proline only appears to remove the hydrogen bond with Asn-72 on the same polypeptide chain (Figure 8D), as proline’s backbone nitrogen cannot hydrogen bond. Leu-74 is flanked by a conserved cysteine and proline [31] and the introduction of an additional proline would reduce the flexibility of this section of the protein (Figure S10B) resulting in a more rigid structure. Additionally this variant is also predicted to affect the flexibility of Asn-97 (Figure S10B), a residue that makes multiple contacts with the nucleotide moiety of UDP-Gal (Figure 8D). This interpretation agrees well with our in vitro findings that demonstrate that this variant is misfolded (Figure 6, 7A, S7A, S8A, S9), yet thermally stabilised (Figure 7C, Table S4). Therefore this substitution is likely to cause a conformational change at the active site resulting in improper binding of both sugar-1-phosphates and UDP-sugars (Figure 5B, 6B, S6B, S7B, S8B, Table 4).
Phe-171 is also located at the dimer interface and active site as judged from the homology model of hGALT (Figure 8E). This residue forms a hydrogen bond with Gln-188 of the same subunit and its phenyl side chain is close to both Asn-172 and Tyr-339 of the other polypeptide chain. Asn-172 forms hydrogen bonds to the phosphate moiety of the substrate and Gln-188 is predicted to favour the nucleophilic attack of sugar 1-phosphates on the uridylated enzyme and also stabilises the uridylated intermediate [27;75]. Previous studies have predicted that this mutation displaces Gln-188 and forms new bonds between it and the hydroxyl of Ser-171 [19]. In contrast, more recent modelling predicted that this hydrogen bond forms between the serine hydroxyl and the carbonyl oxygen of the backbone of Met-298 [29] (Figure 8F). This repositioning would have severe effects on activity as has been previously determined for this mutant protein [19;20]. In addition, this substitution replaces a hydrophobic residue with a polar residue, which may contribute to more general misfolding.
It has been suggested that this mutation affects the secondary structure of this region of the protein, since serine does not favour the formation of β-sheets [19]. Consistent with this, F171S is predicted to increase the disorder of this region (Figure S10B) and is thus likely to contribute to misfolding of the protein. In this study p.F171S showed similar effects on enzyme function to p.L74P, as expected considering their similar location in the enzyme (Figure 5, 6, S6, S7, S8, S9; Table 4). p.F171S is likely to cause similar conformational changes in the active site and result in a protein which is misfolded as suggested from its higher surface hydrophobicity (Figure 7A, C). This variant was suggested to be incapable of substrate binding [19] and our results are in agreement with this prediction.
Phe-194 is at the dimer interface and not at the active site (Figure 8G). The backbone of this phenylalanine forms a hydrogen bond with Ser-192 of the same monomer. Importantly, the side chain of Phe-194 is buried in a largely hydrophobic cleft containing residues from both polypeptide chains: Leu-102a, Ala-122a, Ser-192a, Pro-196a, Ile-32b, Leu-43b and Tyr-339b (where a and b represent residues from the two subunits). Substitution to leucine at this position alters these hydrophobic interactions as its side chain points away from this cleft resulting in increased solvent accessibility (Figure 8H). This is also predicted to cause a slight decrease in the flexibility of the active site residue His-186 (Figure S10B). These structural predictions agree well with our in vitro findings, which showed a higher surface hydrophobicity (Figure 7A, C) and decreased thermal stability (Figure 7B, C). Therefore this region of the protein must be important in stability and this mutation is likely to cause conformational changes as suggested from this variant’s increased intrinsic fluorescence (Figure S9) and decreased resistance to proteolysis (Figure 6, S7, S8). Unlike the active site loops this section of the protein is highly ordered forming the scaffold for the active site [29]. Misfolding of this section is likely to cause a conformational change at the active site, altering substrate binding (Figure 5B, 6B, S6B, S7B, S8B, Table 4) and decreasing activity (Table 2,3).
Arg-333 is at the dimer interface between the two active sites of the enzyme (Figure 8I). This residue faces Arg-333 of the other monomer and appears not to be involved in any interactions. Alteration of this residue to the smaller glycine results in a large void in the middle of the hGALT dimer (Figure 8J) although not directly affecting any residues in the active site. However, adjacent to this residue is Lys-334, which is predicted to form hydrogen bonds to the hydroxyl groups of the sugar moiety of each substrate. Interestingly the E. coli structure demonstrates slight conformational changes depending on the position of the 4′-hydroxyl group of the substrate’s sugar moiety. This change is predominantly at Glu-317 of the bacterial enzyme and suggests that the corresponding residue in the hGALT, Glu-340, interacts with Lys-334 (Lys-311 in bacterial GALT) only when hGALT is UDP-Gal bound. This lysine also interacts with 3′- and 4′-hydroxyl groups of both UDP-sugars [69]. Arginine side chains are large and Arg-333 possibly reduces the flexibility of this region due to the requirement to avoid steric clashes with the equivalent arginine of the other subunit. Changing either residue to glycine decreases this residue’s size and possibly alters that subunit’s flexibility at that location. Predictions, however, suggested that this alteration slightly decreases the flexibility of this region (Figure S10B). This alteration of the flexibility, and the void created, may explain this mutant’s decreased thermal stability (Figure 7; Table S4), its increased surface hydrophobicity (Figure 8A) and its altered intrinsic fluorescence (Figure S9A). Altered flexibility may also cause Lys-334 to change its interaction with Glu-340 and the substrates thus interfering with the conformational change when the enzyme binds a substrate. This may explain p.R333G’s severely impaired activity in vitro (Table 2,3) and the effects on substrate binding (Figure 5B, 6B, S6B, S7B, S8B, Table 4). The observation of non-Michaelis-Menten kinetics for both F194L and R333G (Table 2,3) may also be due to their increased flexibilities.
4.2 The hGALT variants show characteristics of protein misfolding
As a large majority of disease-associated mutants of hGALT are not located at the active site it has been suggested that they are likely to cause their affects by protein misfolding [15;29]. In this scenario alteration of the protein sequence reduces the amount of active protein by conformational changes that perturb the active site, thus impairing substrate binding, the formation and decay of the uridylated intermediate and altering the protein’s overall stability. Taken together this study of five hGALT mutants has demonstrated that each causes unique changes of these aspects of the protein. All these effects are not mutually exclusive and are caused by the removal of important interactions involved in the protein structure and in substrate binding. From this it can be concluded that protein misfolding is the underlying reason for these mutants’ enzymological impairment and adds compelling evidence to this being a common molecular mechanism of hGALT deficiency in patients.
The contribution of uridyl-enzyme intermediate also needs to be considered. hGALT has been shown to be present in vivo as a mixed population of uridylated and deuridylated enzyme [25]. As such it is likely that the purified hGALT variants in this study are a mixed population of these states. However, there is some uncertainty about the stability of the intermediate as it has been shown kinetically that UMP can dissociate to reform the free enzyme [27]. Interestingly if the uridylated enzyme is stable there are some hints of the effects of uridylation in the data presented here. Both p.L74P and p.F171S are inactive variants and probably do not form the covalent intermediate (similar to the inactive, artificial variant p.H186G [25]). p.L74P and p.F171S showed a similar level of degradation by proteases (Figure 6A, S7A) and melting temperatures (Table S4) as the wild-type in the presence of a sugar 1-phosphate. The effects seen in this study on stability from sugar-1-phosphates may be due to deuridylation and the effects of UDP-sugars could be due to dead-end binding [19]. p.L74P and p.F171S may be misfolded and trapped in a deuridylated state. Thus, those variants that are impaired in the formation of the intermediate may be more prone to protease degradation. Further investigation of the contribution of the covalent intermediate to enzyme stability is required.
4.3 Altered substrate binding: a consequence of protein misfolding
It has been shown that the ability to rescue galactose stressed null-GALT yeast correlates with each variant’s activity [20]. This was observed for all variants in this study except for p.R333G (Table S3B; Figure 2). However this is not the first time that a disparity of this relationship has been found [20] and it is likely that some variants are more sensitive to cellular environmental factors as also seen with UDP-galactose 4′-epimerase [35]. Activities in yeast lysates also correlated well with those of the purified protein and it appears that enzyme activity is roughly correlated with substrate binding and not overall stability as determined by DSF. Inactive variants p.L74P and p.F171S show little change in thermal stability whereas the least impaired variant, p.D28Y, shows similar changes to the wild-type protein. p.F194L was the second most impaired active variant and showed clear, but lower, stability changes than the wild-type protein. The least active variant, p.R333G, showed even smaller changes in stability following substrate binding. This agrees with recent molecular dynamic simulations that predicted that the wild-type protein demonstrates a conformational change, becoming more compact when bound to UDP-Gal. Mutants were predicted to be initially in a more compact structure before UDP-Gal binding and/or did not demonstrate a conformational change after binding suggesting an alteration of substrate binding is the cause of their impairment [30].
4.5 Conclusions
Since a common molecular mechanism of hGALT deficiency appears to be protein misfolding it may be possible to develop “pharmacological chaperone” treatments. Here a small ligand binds at the active site or novel binding pocket causing conformation changes that increase the stability and activity of the protein. This treatment is already used in phenylketonuria and has been suggested for other diseases [76-78]. Such small molecules usually bind to cofactor or substrate binding sites and the latter may be appropriate for hGALT. In addition, using the homology model 1R3A and three different binding site predictors [54;56] (Figure S2), we have identified an additional binding pocket at the dimer interface, on the opposite side of the active sites. The possible biological function of this predicted binding pocket is not known, but it may be involved in allosteric control of this enzyme (Figure 4C). The detection of non-Michaelis-Menten kinetics suggests the potential for allosteric activation or repression. This pocket could be targeted for pharmacological chaperone treatment. In addition the use of proteostasis regulators may also beneficial, decreasing the rate of degradation of unstable mutant proteins [79]. Such therapies for galactosemia sufferers may alleviate the severity of the disease’s acute or long-term pathology and/or allow for some relaxation of galactose restricted diets. Interestingly this mechanism of a ligand-mediated increase in stability has been used to explain the findings of increased hGALT activity in HepG2 cells in the presence of high concentrations of galactose [80].
In summary we have measured the level of impairment of five hGALT variants in a yeast model. Further research on the recombinant purified proteins demonstrated that these hGALT variants are structurally altered, which affects thermal stability and protease resistance. This, in turn, results in their decreased ability to bind substrates and lower enzymatic activity. These new insights have revealed that the molecular basis of these variants’ altered activity is due to protein misfolding and strongly suggests that this is a common, underlying cause of hGALT deficiency in type I galactosemia. In view of these findings we suggest that both pharmacological chaperone and proteostasis regulator treatments should be investigated.
Supplementary Material
hGALT mutations demonstrate a range of activity changes in vivo and in vitro
hGALT mutations alter both resistance against proteases and thermal stability
Severely impairing mutations cause the greatest changes in substrate binding
Protein misfolding is a fundamental, molecular cause of type I galactosemia
Thus, pharmacological chaperones may prove to be a viable therapy
Acknowledgements
TJM thanks the Department of Employment and Learning, Northern Ireland for a PhD studentship. We wish to thank Prof Aaron Maule (Queen’s University, Belfast) for use of a thermal cycler for the thermal scanning fluorimetry assay and Dr. Kostya Panov (Queen’s University, Belfast) for suggesting this assay. We also thank Dr. Ying Liu for her assistance with some of the HPLC analyses. Work conducted in the Fridovich-Keil lab was supported, in part, by funds from the National Institutes of Health (USA) grant DK059904 (to JLFK). The hGALT expression clone was constructed by DJT in the laboratory of Prof Richard J Reece (University of Manchester, UK).
Abbreviations
- ANS-1
1-anilinonaphthalene-8-sulphonic acid
- BS3
suberic acid bis (3-sulfo-N-hydroxysuccinimide ester)
- DSF
differential scanning fluorimetry
- GALT
galactose-1-phosphate uridylyltransferase
- Gal-1P
galactose 1-phosphate
- Glc-1P
glucose 1-phosphate hGALT: human GALT
- NAD+
oxidized nicotinamide adenine dinucleotide
- NADH
reduced nicotinamide adenine dinucleotide
- NADP+
oxidized nicotinamide adenine dinucleotide phosphate
- NADPH
reduced nicotinamide adenine dinucleotide phosphate
- DSF
Differential scanning fluorimetry
- UDP-Gal
uridine diphosphate galactose
- UDP-Glc
uridine diphosphate glucose
- UDP
uridine diphosphate
- UMP
uridine monophosphate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Isselbacher KJ, Anderson EP, Kurahashi K, Kalckar HM. Congenital galactosemia, a single enzymatic block in galactose metabolism. Science. 1956;123:635–636. doi: 10.1126/science.123.3198.635. [DOI] [PubMed] [Google Scholar]
- 2.Thoden JB, Timson DJ, Reece RJ, Holden HM. Molecular structure of human galactokinase: implications for type II galactosemia. J.Biol.Chem. 2005;280:9662–9670. doi: 10.1074/jbc.M412916200. [DOI] [PubMed] [Google Scholar]
- 3.Timson DJ. The structural and molecular biology of type III galactosemia. IUBMB Life. 2006;58:83–89. doi: 10.1080/15216540600644846. [DOI] [PubMed] [Google Scholar]
- 4.Frey PA, Wong LJ, Sheu KF, Yang SL. Galactose-1-phosphate uridylyltransferase: detection, isolation, and characterization of the uridylyl enzyme. Methods Enzymol. 1982;87:20–36. doi: 10.1016/s0076-6879(82)87004-3. [DOI] [PubMed] [Google Scholar]
- 5.McCorvie TJ, Timson DJ. The structural and molecular biology of type I galactosemia: Enzymology of galactose 1-phosphate uridylyltransferase. IUBMB Life. 2011;63:694–700. doi: 10.1002/iub.511. [DOI] [PubMed] [Google Scholar]
- 6.Jumbo-Lucioni PP, Garber K, Kiel J, Baric I, Berry GT, Bosch A, Burlina A, Chiesa A, Pico ML, Estrada SC, Henderson H, Leslie N, Longo N, Morris AA, Ramirez-Farias C, Schweitzer-Krantz S, Silao CL, Vela-Amieva M, Waisbren S, Fridovich-Keil JL. Diversity of approaches to classic galactosemia around the world: a comparison of diagnosis, intervention, and outcomes. J.Inherit.Metab.Dis. 2012;35:1037–1049. doi: 10.1007/s10545-012-9477-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waisbren SE, Potter NL, Gordon CM, Green RC, Greenstein P, Gubbels CS, Rubio-Gozalbo E, Schomer D, Welt C, Anastasoaie V, D’Anna K, Gentile J, Guo CY, Hecht L, Jackson R, Jansma BM, Li Y, Lip V, Miller DT, Murray M, Power L, Quinn N, Rohr F, Shen Y, Skinder-Meredith A, Timmers I, Tunick R, Wessel A, Wu BL, Levy H, Elsas L, Berry GT. The adult galactosemic phenotype. J.Inherit.Metab.Dis. 2012;35:279–286. doi: 10.1007/s10545-011-9372-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berry GT. Is prenatal myo-inositol deficiency a mechanism of CNS injury in galactosemia? J.Inherit.Metab.Dis. 2011;34:345–355. doi: 10.1007/s10545-010-9260-x. [DOI] [PubMed] [Google Scholar]
- 9.Boutron A, Marabotti A, Facchiano A, Cheillan D, Zater M, Oliveira C, Costa C, Labrune P, Brivet M. French Galactosemia Working Group, Mutation spectrum in the French cohort of galactosemic patients and structural simulation of 27 novel missense variations. Mol.Genet.Metab. 2012;107:438–447. doi: 10.1016/j.ymgme.2012.07.025. [DOI] [PubMed] [Google Scholar]
- 10.Wedekind JE, Frey PA, Rayment I. Three-dimensional structure of galactose-1-phosphate uridylyltransferase from Escherichia coli at 1.8 A resolution. Biochemistry. 1995;34:11049–11061. doi: 10.1021/bi00035a010. [DOI] [PubMed] [Google Scholar]
- 11.Wang BB, Xu YK, Ng WG, Wong LJ. Molecular and biochemical basis of galactosemia. Mol.Genet.Metab. 1998;63:263–269. doi: 10.1006/mgme.1998.2678. [DOI] [PubMed] [Google Scholar]
- 12.Christacos NC, Fridovich-Keil JL. Impact of patient mutations on heterodimer formation and function in human galactose-1-P uridylyltransferase. Mol.Genet.Metab. 2002;76:319–326. doi: 10.1016/s1096-7192(02)00109-9. [DOI] [PubMed] [Google Scholar]
- 13.Ryan EL, Lynch ME, Taddeo E, Gleason TJ, Epstein MP, Fridovich-Keil JL. Cryptic residual GALT activity is a potential modifier of scholastic outcome in school age children with classic galactosemia. J.Inherit.Metab.Dis. 2013 doi: 10.1007/s10545-012-9575-x. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calderon FR, Phansalkar AR, Crockett DK, Miller M, Mao R. Mutation database for the galactose-1-phosphate uridyltransferase (GALT) gene. Hum.Mutat. 2007;28:939–943. doi: 10.1002/humu.20544. [DOI] [PubMed] [Google Scholar]
- 15.McCorvie TJ, Timson DJ. Structural and molecular biology of type I galactosemia: disease-associated mutations. IUBMB Life. 2011;63:949–954. doi: 10.1002/iub.510. [DOI] [PubMed] [Google Scholar]
- 16.Fridovich-Keil JL, Jinks-Robertson S. A yeast expression system for human galactose-1-phosphate uridylyltransferase. Proc.Natl.Acad.Sci.U.S.A. 1993;90:398–402. doi: 10.1073/pnas.90.2.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fridovich-Keil JL, Quimby BB, Wells L, Mazur LA, Elsevier JP. Characterization of the N314D allele of human galactose-1-phosphate uridylyltransferase using a yeast expression system. Biochem.Mol.Med. 1995;56:121–130. doi: 10.1006/bmme.1995.1067. [DOI] [PubMed] [Google Scholar]
- 18.Fridovich-Keil JL, Langley SD, Mazur LA, Lennon JC, Dembure PP, Elsas JL., 2nd Identification and functional analysis of three distinct mutations in the human galactose-1-phosphate uridyltransferase gene associated with galactosemia in a single family. Am.J.Hum.Genet. 1995;56:640–646. [PMC free article] [PubMed] [Google Scholar]
- 19.Crews C, Wilkinson KD, Wells L, Perkins C, Fridovich-Keil JL. Functional consequence of substitutions at residue 171 in human galactose-1-phosphate uridylyltransferase. J.Biol.Chem. 2000;275:22847–22853. doi: 10.1074/jbc.M001053200. [DOI] [PubMed] [Google Scholar]
- 20.Riehman K, Crews C, Fridovich-Keil JL. Relationship between genotype, activity, and galactose sensitivity in yeast expressing patient alleles of human galactose-1-phosphate uridylyltransferase. J.Biol.Chem. 2001;276:10634–10640. doi: 10.1074/jbc.M009583200. [DOI] [PubMed] [Google Scholar]
- 21.Chhay JS, Openo KK, Eaton JS, Gentile M, Fridovich-Keil JL. A yeast model reveals biochemical severity associated with each of three variant alleles of galactose-1P uridylyltransferase segregating in a single family. J.Inherit.Metab.Dis. 2008;31:97–107. doi: 10.1007/s10545-007-0786-5. [DOI] [PubMed] [Google Scholar]
- 22.Elsevier JP, Fridovich-Keil JL. The Q188R mutation in human galactose-1-phosphate uridylyltransferase acts as a partial dominant negative. J.Biol.Chem. 1996;271:32002–32007. doi: 10.1074/jbc.271.50.32002. [DOI] [PubMed] [Google Scholar]
- 23.Quimby BB, Wells L, Wilkinson KD, Fridovich-Keil JL. Functional requirements of the active site position 185 in the human enzyme galactose-1-phosphate uridylyltransferase. J.Biol.Chem. 1996;271:26835–26842. doi: 10.1074/jbc.271.43.26835. [DOI] [PubMed] [Google Scholar]
- 24.Wells L, Fridovich-Keil JL. Biochemical characterization of the S135L allele of galactose-1-phosphate uridylyltransferase associated with galactosaemia. J.Inherit.Metab.Dis. 1997;20:633–642. doi: 10.1023/a:1005314207513. [DOI] [PubMed] [Google Scholar]
- 25.Henderson JM, Wells L, Fridovich-Keil JL. Covalent heterogeneity of the human enzyme galactose-1-phosphate uridylyltransferase. J.Biol.Chem. 2000;275:30088–30091. doi: 10.1074/jbc.M005259200. [DOI] [PubMed] [Google Scholar]
- 26.Lai K, Elsas LJ. Structure-function analyses of a common mutation in blacks with transferase-deficiency galactosemia. Mol.Genet.Metab. 2001;74:264–272. doi: 10.1006/mgme.2001.3230. [DOI] [PubMed] [Google Scholar]
- 27.Lai K, Willis AC, Elsas LJ. The biochemical role of glutamine 188 in human galactose-1-phosphate uridyltransferase. J.Biol.Chem. 1999;274:6559–6566. doi: 10.1074/jbc.274.10.6559. [DOI] [PubMed] [Google Scholar]
- 28.d’Acierno A, Facchiano A, Marabotti A. GALT protein database, a bioinformatics resource for the management and analysis of structural features of a galactosemia-related protein and its mutants. Genomics Proteomics Bioinformatics. 2009;7:71–76. doi: 10.1016/S1672-0229(08)60035-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Facchiano A, Marabotti A. Analysis of galactosemia-linked mutations of GALT enzyme using a computational biology approach. Protein Eng.Des.Sel. 2010;23:103–113. doi: 10.1093/protein/gzp076. [DOI] [PubMed] [Google Scholar]
- 30.Tang M, Facchiano A, Rachamadugu R, Calderon F, Mao R, Milanesi L, Marabotti A, Lai K. Correlation assessment among clinical phenotypes, expression analysis and molecular modeling of 14 novel variations in the human galactose-1-phosphate uridylyltransferase gene. Hum.Mutat. 2012;33:1107–1115. doi: 10.1002/humu.22093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reichardt JK, Levy HL, Woo SL. Molecular characterization of two galactosemia mutations and one polymorphism: implications for structure-function analysis of human galactose-1-phosphate uridyltransferase. Biochemistry. 1992;31:5430–5433. doi: 10.1021/bi00139a002. [DOI] [PubMed] [Google Scholar]
- 32.Johnston M. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol.Rev. 1987;51:458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lennon G, Auffray C, Polymeropoulos M, Soares MB. The I.M.A.G.E. Consortium: an integrated molecular analysis of genomes and their expression. Genomics. 1996;33:151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- 34.Wang W, Malcolm BA. Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange Site-Directed Mutagenesis. BioTechniques. 1999;26:680–682. doi: 10.2144/99264st03. [DOI] [PubMed] [Google Scholar]
- 35.McCorvie TJ, Wasilenko J, Liu Y, Fridovich-Keil JL, Timson DJ. In vivo and in vitro function of human UDP-galactose 4′-epimerase variants. Biochimie. 2011;93:1747–1754. doi: 10.1016/j.biochi.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal.Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 37.Markus HB, Wu JW, Boches FS, Tedesco TA, Mellman WJ, Kallen RG. Human erythrocyte galactose-1-phosphate uridylyltransferase. Evidence for a uridylyl-enzyme intermediate by kinetic and exchange reaction studies. J.Biol.Chem. 1977;252:5363–5369. [PubMed] [Google Scholar]
- 38.Ng WG, Donnell GN, Hodgman JE, Bergren WR. Differences in uridine diphosphate galactose-4-epimerase between haemolysates of newborns and of adults. Nature. 1967;214:283–284. doi: 10.1038/214283a0. [DOI] [PubMed] [Google Scholar]
- 39.McCorvie TJ, Liu Y, Frazer A, Gleason TJ, Fridovich-Keil JL, Timson DJ. Altered cofactor binding affects stability and activity of human UDP-galactose 4′-epimerase: implications for type III galactosemia. Biochim.Biophys.Acta. 2012;1822:1516–1526. doi: 10.1016/j.bbadis.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Friedman AJ, Durrant JD, Pierce LC, McCorvie TJ, Timson DJ, McCammon JA. The molecular dynamics of Trypanosoma brucei UDP-galactose 4′-epimerase: a drug target for African sleeping sickness. Chem.Biol.Drug Des. 2012;80:173–181. doi: 10.1111/j.1747-0285.2012.01392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marabotti A, Facchiano AM. Homology modeling studies on human galactose-1-phosphate uridylyltransferase and on its galactosemia-related mutant Q188R provide an explanation of molecular effects of the mutation on homo- and heterodimers. J.Med.Chem. 2005;48:773–779. doi: 10.1021/jm049731q. [DOI] [PubMed] [Google Scholar]
- 42.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 43.Zhou H, Zhang C, Liu S, Zhou Y. Web-based toolkits for topology prediction of transmembrane helical proteins, fold recognition, structure and binding scoring, folding-kinetics analysis and comparative analysis of domain combinations. Nucleic Acids Res. 2005;33:W193–W197. doi: 10.1093/nar/gki360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dehouck Y, Kwasigroch JM, Gilis D, Rooman M. PoPMuSiC 2.1: a web server for the estimation of protein stability changes upon mutation and sequence optimality. BMC Bioinformatics. 2011;12:151. doi: 10.1186/1471-2105-12-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parthiban V, Gromiha MM, Schomburg D. CUPSAT: prediction of protein stability upon point mutations. Nucleic Acids Res. 2006;34:W239–W242. doi: 10.1093/nar/gkl190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Worth CL, Preissner R, Blundell TL. SDM - a server for predicting effects of mutations on protein stability and malfunction. Nucleic Acids Res. 2011;39:W215–W222. doi: 10.1093/nar/gkr363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yin S, Ding F, Dokholyan NV. Eris: an automated estimator of protein stability. Nat.Methods. 2007;4:466–467. doi: 10.1038/nmeth0607-466. [DOI] [PubMed] [Google Scholar]
- 48.Benedix A, Becker CM, de Groot BL, Caflisch A, Bockmann RA. Predicting free energy changes using structural ensembles. Nat.Methods. 2009;6:3–4. doi: 10.1038/nmeth0109-3. [DOI] [PubMed] [Google Scholar]
- 49.Capriotti E, Fariselli P, Casadio R. I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005;33:W306–W310. doi: 10.1093/nar/gki375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheng J, Randall A, Baldi P. Prediction of protein stability changes for single-site mutations using support vector machines. Proteins. 2006;62:1125–1132. doi: 10.1002/prot.20810. [DOI] [PubMed] [Google Scholar]
- 51.Teng S, Srivastava AK, Wang L. Sequence feature-based prediction of protein stability changes upon amino acid substitutions. BMC Genomics. 2010;11(Suppl 2):S5. doi: 10.1186/1471-2164-11-S2-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ishida T, Kinoshita K. Prediction of disordered regions in proteins based on the meta approach. Bioinformatics. 2008;24:1344–1348. doi: 10.1093/bioinformatics/btn195. [DOI] [PubMed] [Google Scholar]
- 53.Zhang T, Faraggi E, Xue B, Dunker AK, Uversky VN, Zhou Y. SPINE-D: accurate prediction of short and long disordered regions by a single neural-network based method. J.Biomol.Struct.Dyn. 2012;29:799–813. doi: 10.1080/073911012010525022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brenke R, Kozakov D, Chuang GY, Beglov D, Hall D, Landon MR, Mattos C, Vajda S. Fragment-based identification of druggable ‘hot spots’ of proteins using Fourier domain correlation techniques. Bioinformatics. 2009;25:621–627. doi: 10.1093/bioinformatics/btp036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Landon MR, Lieberman RL, Hoang QQ, Ju S, Caaveiro JM, Orwig SD, Kozakov D, Brenke R, Chuang GY, Beglov D, Vajda S, Petsko GA, Ringe D. Detection of ligand binding hot spots on protein surfaces via fragment-based methods: application to DJ-1 and glucocerebrosidase. J.Comput.Aided Mol.Des. 2009;23:491–500. doi: 10.1007/s10822-009-9283-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Laurie AT, Jackson RM. Q-SiteFinder: an energy-based method for the prediction of protein-ligand binding sites. Bioinformatics. 2005;21:1908–1916. doi: 10.1093/bioinformatics/bti315. [DOI] [PubMed] [Google Scholar]
- 57.Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–1561. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adzhubei I, Jordan DM, Sunyaev SR. Predicting Functional Effect of Human Missense Mutations Using PolyPhen-2. Curr.Protoc.Hum.Genet. 2013 doi: 10.1002/0471142905.hg0720s76. Chapter 7. Unit7.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat.Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 60.Cavallo L, Kleinjung J, Fraternali F. POPS: A fast algorithm for solvent accessible surface areas at atomic and residue level. Nucleic Acids Res. 2003;31:3364–3366. doi: 10.1093/nar/gkg601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wells L, Fridovich-Keil JL. The yeast, Saccharomyces cerevisiae, as a model system for the study of human genetic disease. SAAS Bull.Biochem.Biotechnol. 1996;9:83–88. [PubMed] [Google Scholar]
- 62.Botstein D, Fink GR. Yeast: an experimental organism for 21st Century biology. Genetics. 2011;189:695–704. doi: 10.1534/genetics.111.130765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith MG, Snyder M. Yeast as a model for human disease. Curr.Protoc.Hum.Genet. 2006 doi: 10.1002/0471142905.hg1506s48. Chapter 15. Unit 15.6. [DOI] [PubMed] [Google Scholar]
- 64.Coughlan CM, Brodsky JL. Use of yeast as a model system to investigate protein conformational diseases. Mol.Biotechnol. 2005;30:171–180. doi: 10.1385/MB:30:2:171. [DOI] [PubMed] [Google Scholar]
- 65.Geeganage S, Frey PA. Significance of metal ions in galactose-1-phosphate uridylyltransferase: an essential structural zinc and a nonessential structural iron. Biochemistry. 1999;38:13398–13406. doi: 10.1021/bi9910631. [DOI] [PubMed] [Google Scholar]
- 66.Bertoli D, Segal S. Developmental aspects and some characteristics of mammalian galactose 1-phosphate uridyltransferase. J.Biol.Chem. 1966;241:4023–4029. [PubMed] [Google Scholar]
- 67.Elsevier JP, Wells L, Quimby BB, Fridovich-Keil JL. Heterodimer formation and activity in the human enzyme galactose-1-phosphate uridylyltransferase. Proc.Natl.Acad.Sci.U.S.A. 1996;93:7166–7171. doi: 10.1073/pnas.93.14.7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wedekind JE, Frey PA, Rayment I. The structure of nucleotidylated histidine-166 of galactose-1-phosphate uridylyltransferase provides insight into phosphoryl group transfer. Biochemistry. 1996;35:11560–11569. doi: 10.1021/bi9612677. [DOI] [PubMed] [Google Scholar]
- 69.Thoden JB, Ruzicka FJ, Frey PA, Rayment I, Holden HM. Structural analysis of the H166G site-directed mutant of galactose-1-phosphate uridylyltransferase complexed with either UDP-glucose or UDP-galactose: detailed description of the nucleotide sugar binding site. Biochemistry. 1997;36:1212–1222. doi: 10.1021/bi9626517. [DOI] [PubMed] [Google Scholar]
- 70.Coulter-Mackie MB, Lian Q. Consequences of missense mutations for dimerization and turnover of alanine:glyoxylate aminotransferase: study of a spectrum of mutations. Mol.Genet.Metab. 2006;89:349–359. doi: 10.1016/j.ymgme.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 71.Fontana A, de Laureto PP, Spolaore B, Frare E, Picotti P, Zambonin M. Probing protein structure by limited proteolysis. Acta Biochim.Pol. 2004;51:299–321. [PubMed] [Google Scholar]
- 72.Pantoliano MW, Petrella EC, Kwasnoski JD, Lobanov VS, Myslik J, Graf E, Carver T, Asel E, Springer BA, Lane P, Salemme FR. High-density miniaturized thermal shift assays as a general strategy for drug discovery. J.Biomol.Screen. 2001;6:429–440. doi: 10.1177/108705710100600609. [DOI] [PubMed] [Google Scholar]
- 73.Ericsson UB, Hallberg BM, Detitta GT, Dekker N, Nordlund P. Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal.Biochem. 2006;357:289–298. doi: 10.1016/j.ab.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 74.Lang A, Groebe H, Hellkuhl B, von Figura K. A new variant of galactosemia: galactose-1-phosphate uridylytransferase sensitive to product inhibition by glucose 1-phosphate. Pediatr.Res. 1980;14:729–734. doi: 10.1203/00006450-198005000-00005. [DOI] [PubMed] [Google Scholar]
- 75.Geeganage S, Frey PA. Transient kinetics of formation and reaction of the uridylyl-enzyme form of galactose-1-P uridylyltransferase and its Q168R-variant: insight into the molecular basis of galactosemia. Biochemistry. 1998;37:14500–14507. doi: 10.1021/bi9815546. [DOI] [PubMed] [Google Scholar]
- 76.Pey AL, Ying M, Cremades N, Velazquez-Campoy A, Scherer T, Thony B, Sancho J, Martinez A. Identification of pharmacological chaperones as potential therapeutic agents to treat phenylketonuria. J.Clin.Invest. 2008;118:2858–2867. doi: 10.1172/JCI34355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Muntau AC, Gersting SW. Phenylketonuria as a model for protein misfolding diseases and for the development of next generation orphan drugs for patients with inborn errors of metabolism. J.Inherit.Metab.Dis. 2010;33:649–658. doi: 10.1007/s10545-010-9185-4. [DOI] [PubMed] [Google Scholar]
- 78.Santos-Sierra S, Kirchmair J, Perna AM, Reiss D, Kemter K, Roschinger W, Glossmann H, Gersting SW, Muntau AC, Wolber G, Lagler FB. Novel pharmacological chaperones that correct phenylketonuria in mice. Hum.Mol.Genet. 2012;21:1877–1887. doi: 10.1093/hmg/dds001. [DOI] [PubMed] [Google Scholar]
- 79.Mu TW, Ong DS, Wang YJ, Balch WE, Yates JR, 3rd, Segatori L, Kelly JW. Chemical and biological approaches synergize to ameliorate protein-folding diseases. Cell. 2008;134:769–781. doi: 10.1016/j.cell.2008.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Elsas LJ, Webb AL, Langley SD. Characterization of a carbohydrate response element regulating the gene for human galactose-1-phosphate uridyltransferase. Mol.Genet.Metab. 2002;76:287–296. doi: 10.1016/s1096-7192(02)00106-3. [DOI] [PubMed] [Google Scholar]
- 81.Greber-Platzer S, Guldberg P, Scheibenreiter S, Item C, Schuller E, Patel N, Strobl W. Molecular heterogeneity of classical and Duarte galactosemia: mutation analysis by denaturing gradient gel electrophoresis. Hum.Mutat. 1997;10:49–57. doi: 10.1002/(SICI)1098-1004(1997)10:1<49::AID-HUMU7>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 82.Murphy M, McHugh B, Tighe O, Mayne P, O’Neill C, Naughten E, Croke DT. Genetic basis of transferase-deficient galactosaemia in Ireland and the population history of the Irish Travellers. Eur.J.Hum.Genet. 1999;7:549–554. doi: 10.1038/sj.ejhg.5200327. [DOI] [PubMed] [Google Scholar]
- 83.Ng WG, Xu YK, Wong LJ, Kaufman FR, Buist NR, Donnell GN. Two adult galactosaemia females with normal ovarian function and identical GALT mutations (Q188R/R333G) J.Inherit.Metab.Dis. 2003;26:75–79. doi: 10.1023/a:1024039916476. [DOI] [PubMed] [Google Scholar]
- 84.Gort L, Boleda MD, Tyfield L, Vilarinho L, Rivera I, Cardoso ML, Santos-Leite M, Giros M, Briones P. Mutational spectrum of classical galactosaemia in Spain and Portugal. J.Inherit.Metab.Dis. 2006;29:739–742. doi: 10.1007/s10545-006-0356-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.