Abstract
Mutations in KRAS drive the oncogenic phenotype in a variety of tumors of epithelial origin. The NF-κB transcription factor pathway is important for oncogenic RAS to transform cells and to drive tumorigenesis in animal models. Recently TAK1, an upstream regulator of IKK, which controls canonical NF-κB, was shown to be important for chemoresistance in pancreatic cancer and for regulating KRAS+ colorectal cancer cell growth and survival. Here we show that KRAS+ upregulates GSK-3α leading to its interaction with TAK1 to stabilize the TAK1/TAB complex to promote IKK activity. Additionally, GSK-3α is required for promoting critical non-canonical NF-κB signaling in pancreatic cancer cells. Pharmacologic inhibition of GSK-3 suppresses growth of human pancreatic tumor explants, consistent with the loss of expression of oncogenic genes such as c-myc and TERT. These data identify GSK-3α as a key downstream effector of oncogenic KRAS via its ability to coordinately regulate distinct NF-κB signaling pathways.
Keywords: KRAS, GSK-3α, TAK1, NF-κB, non-canonical NF-κB
Introduction
Numerous epithelial-derived cancers express mutated/activated KRAS, and many of these cancers depend upon KRAS-induced signaling for regulation of growth, survival and metabolism (1,2). For example, the great majority of pancreatic ductal adenocarcinomas (PDAC) exhibit mutations in KRAS. However, specific targeting of mutant oncogenic KRAS has been a therapeutic challenge (3,4). Thus, targeting downstream effectors has emerged as an alternative approach to inhibiting oncogenic KRAS functions. Examples of downstream signaling proteins that are important in KRAS-dependent growth and survival are the kinases TAK1 and GSK-3, in the context of colorectal (TAK1) and pancreatic cancers (TAK1, GSK-3) (5,6) (7). Interestingly, TAK1 and GSK-3 have roles in promoting the activity of components of the NF-κB transcription factor family, which is known to be important for KRAS-driven oncogenesis.
NF-κB represents a family of evolutionary conserved transcription factors consisting of RelA (p65), c-Rel, RelB, p50 (precursor p105) or p52 (precursor p100) subunits (8–11). Dimers of these subunits regulate transcription of genes associated with inflammation, proliferation, and regulation of cell death. NF-κB complexes are inhibited through interaction with IκB proteins. Two distinct pathways have been characterized that lead to activation of NF-κB. The first, and most intensively studied, is the canonical NF-κB pathway defined by activation of the p65–p50 dimer. This pathway is dependent upon the activation of the IκB kinase complex (IKK) which consists of two catalytic subunits (IKKα and IKKβ) and a regulatory subunit (IKKγ) (8). The activated IKK complex phosphorylates IκB leading to its ubiquitination and subsequent proteasome-dependent degradation, allowing NF-κB to accumulate in the nucleus and facilitate transcription of target genes. The second is the non-canonical NF-κB signaling pathway, which is dependent upon activation of NIK (NF-κB inducing kinase). Activated NIK leads to activation of IKKα homodimers, which phosphorylate p100, leading to its proteolytic processing and generation of the active p52 subunit. Consequently RelB-p52 heterodimers accumulate in the nucleus to drive transcription of target genes (12).
NF-κB activity has been linked to the progression of multiple human cancers where it suppresses cell death and promotes cell proliferation, angiogenesis and invasion (9,10). We initially showed the importance of NF-κB in RAS-induced cell transformation (13). We and others have confirmed this in animal models of RAS-driven cancer (14–17). Consistent with this, NF-κB is constitutively active in the great majority of pancreatic cancer cell lines and tumors (18). Previous work has demonstrated that constitutive NF-κB activity is dependent upon IKK in pancreatic cancer cell lines (19–22) and knockout of IKKβ suppresses tumor growth/progression in a KRAS/INK4a null animal model of pancreatic cancer (23). Moscat and colleagues showed the importance of p62 in coordinating TRAF6 to regulate IKK downstream of oncogenic KRAS-induced signaling (24). Elucidating additional signaling components in the canonical NF-κB pathway as well as understanding events associated with non-canonical NF-κB activation induced by KRAS is important in understanding KRAS-induced transformation.
Transforming growth factor-beta activated kinase 1(TAK1) is a mitogen activated protein kinase kinase kinase (MAP3K) that initiates downstream NF-κB and MAPK signaling in response to cytokines (25). TAK1 activity is dependent upon its association with TAK1-binding partners (TAB1, TAB2, TAB3 and TAB4) which facilitate auto-phosphorylation of TAK1 (Thr178, Thr184, Ser192) within the kinase activation loop. (26,27). Upon activation, TAK1 promotes the activity of p38 and JNK MAPKs (28). TAK1 plays a central role in NF-κB activation through direct phosphorylation of IKKβ (25). As described above (5,6), TAK1 has been shown to be important for survival of KRAS-dependent colorectal cancers and to play an important role in the chemoresistance of pancreatic cancers.
GSK-3 is a multifunctional serine/threonine kinase that exists as closely related isoforms (GSK-3α and GSK-3β) (29). It is a key enzyme involved in diverse biological processes such as cell cycle progression, differentiation and apoptosis (30). Moreover, GSK-3 has been implicated in as playing an oncogenic role in various human malignancies including pancreatic cancer (30–32). While most studies have focused on GSK-3β and its involvement in regulating the WNT/β-catenin pathway, a recent study implicated GSK-3α in promoting AML (33). We and others have demonstrated that GSK-3 regulates growth and survival in pancreatic cancer cell lines by driving constitutive IKK and subsequent NF-κB DNA binding and activity (21,34). However, the mechanism of how GSK-3 activates IKK and a clear distinction, if any, between the roles of the two isoforms GSK-3α and GSK-3β remains elusive.
Here we investigate the roles of GSK-3α, GSK-3β, and TAK1 downstream of mutant KRAS in driving constitutive NF-κB signaling, proliferation and survival in pancreatic cancer cells. We establish a regulatory link between GSK-3α and TAK1 and propose that constitutive canonical NF-κB activity is driven by a unique GSK-3α-TAK1-IKK signaling cascade. We also provide evidence that GSK-3α regulates non-canonical NF-κB activity in pancreatic cancer cells, independent of GSK-3β, which contributes to growth/survival of pancreatic cancer cells. Moreover, we show that acute inhibition of GSK-3 in human pancreatic tumor explants suppresses tumor growth and identify a comprehensive transcriptional profile that is changed upon GSK-3 inhibition. Collectively, these data provide new insight into constitutive NF-κB regulation in oncogenic KRAS-induced pancreatic cancer, and establish GSK-3α and TAK1 as potential therapeutic targets for this disease.
Results
GSK-3α/β promote proliferation/survival of pancreatic cancer cells
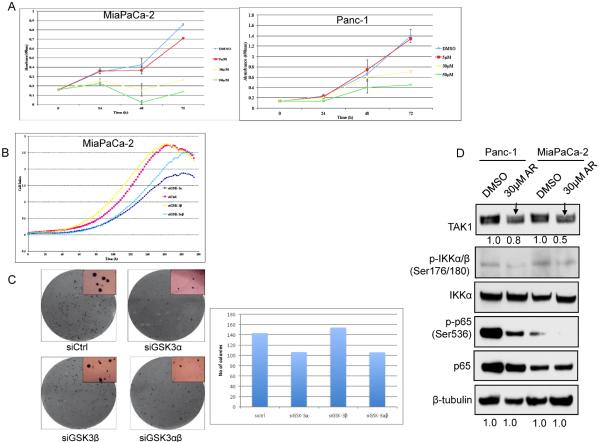
Consistent with previous reports (21,31), we observed a decrease in proliferation of two well characterized KRAS+ pancreatic cancer cell lines, Panc-1 and MiaPaCa-2, upon treatment with the selective GSK-3 inhibitor, AR-A014418 in a dose dependent manner (Fig. 1A, Supplementary S1). To exclude potential off-target effects of the drug and to determine the individual requirements for GSK-3α and GSK-3β for cell survival/proliferation, RNA interference was utilized to knock-down individual isoforms. Significant reduction in the cell index of MiaPaCa-2 cells was observed following GSK-3α RNA interference as compared with non-targeting control and GSK-3β siRNA or both GSK-3α/β siRNA (Fig. 1B).
Figure 1. GSK-3α and GSK-3β regulate growth and NF-κB activity in pancreatic cancer cells.
(A) MiaPaCa-2 and Panc-1 cells were treated with DMSO or indicated concentrations of the GSK-3 inhibitor (AR-A014418) for 24, 48 and 72 hours. Cell proliferation was measured in triplicate at each time point using a colorimetric MTS tetrazolium assay. (B) MiaPaCa-2 cells were transiently transfected with GSK-3α, GSK-3β, both GSK-3α and GSK-3β or non-targeting siRNA. 48 hours after transfection, cell growth was measured in triplicates at each time point using a cell impedance assay. (C) MiaPaCa-2 cells were transiently transfected with indicated siRNA as above. 48 hours after transfection, cells were suspended in bactoagar growth medium and 7 days later, plates were examined for colony formation. The data shown are representative of two independent experiments, each performed in triplicate. (D) Panc-1 and MiaPaCa-2 cells were treated with DMSO or 30μM of GSK-3 inhibitor, AR-A014418 for 24h. Whole cell extracts were immunobloted with specified antibodies.
To further explore the effects of GSK-3 inhibition on pancreatic cancer cell function, we analyzed the effect of GSK-3α/β knockdown on the colony formation of pancreatic cancer cells in soft agar. GSK-3α RNA interference inhibited soft agar growth of MiaPaCa-2 cells significantly as compared to non-targeting control or GSK-3β siRNA (Fig. 1C). Importantly the size of the colonies formed by GSK-3α depleted cells was significantly smaller than that of control or GSK-3β depleted cells. To determine if this abrogated soft agar growth correlated with induction of apoptosis, we measured PARP cleavage after the knockdown of GSK-3α and GKS-3β. Depletion of either GSK-3α or GSK-3β led to a modest increase in PARP cleavage (Supplementary S1 and see below).
Consistent with previous reports (21,31), GSK-3 inhibition suppressed phosphorylation of IKK and of RelA/p65 (Fig. 1D), markers of canonical NF-κB signaling. Interestingly GSK-3 inhibition also decreased the levels of TAK1, upstream IKK kinase (25). This led us to hypothesize and investigate a link between GSK-3 and TAK1 in regulating NF-κB activity in pancreatic cancer cells.
Oncogenic KRAS promotes TAK1-TAB interaction to drive canonical NF-κB activity
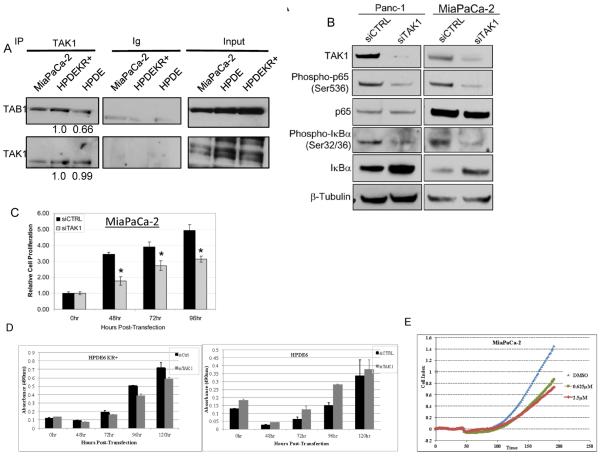
The activity of TAK1 is dependent upon its association with TAK-1 binding proteins (TAB1, TAB2, TAB3) (27). To determine whether TAK1 is active in pancreatic cancer cell lines, we examined the status of TAK1- TAB1 binding in pancreatic cancer cells and questioned if mutant KRAS affects TAK1-TAB1 interaction. TAK1-TAB1 interaction was analyzed by immunoprecipitating TAK1 from an immortalized, human pancreatic ductal epithelial cell line (HPDE6), and in the derivative KRASG12V-transformed HPDE6 cells (HPDEKR+) and from MiaPaCa-2 cells. Relative to KRAS+ MiaPaCa-2 cells and HPDE6KR+ cells, a significant (34%) decrease in TAK1-TAB1 binding was found in HPDE cells (Fig. 2A) indicating that mutant KRAS promotes TAK1-TAB1 interaction. Consistent with this hypothesis, knockdown of KRAS in HPDEKR+ cells strongly reduced TAK1 protein levels and TAK1-TAB interaction in HPDEKR+ cells. (Supplementary S2). These data demonstrate that mutant KRAS drives constitutive TAK1 activity in pancreatic cancer cell lines via stabilizing TAK1.
Figure 2. TAK1 is constitutively active and regulates NF-κB activity in pancreatic cancer cells.
(A) Endogenous TAK1 was immunoprecipitated from whole cell lysates of the indicated cell lines and blotted for TAB1. (B) Panc-1 and MiaPaCa-2 cells were transiently transfected with non-targeting or TAK1 siRNA for 48 hours and immunoblot performed using indicated antibodies. (C) MiaPaCa-2 were transfected with siRNA as indicated above and cell viability was measured at the indicated times post-transfection. Samples were measured in triplicates and normalized to untransfected cells. (D) HPDE6 and HPDEKR+ cells were transfected with siRNA targeted against TAK1 and cell viability measured as above. (E) MiaPaCa-2 cells were treated with DMSO or the TAK1 inhibitor, 5Z-7-oxozaenol (OZ) and cell impedance measured in triplicate every 2 hours.
To determine whether TAK1 activity is required for NF-κB activity in pancreatic cancer cells, the effects of transient TAK1 depletion were measured relative to canonical NF-κB activity. siRNA mediated TAK1 knockdown resulted in diminished phosphorylation of IκBα (Serine 32/36), stabilization of total IκBα levels and reduced phosphorylation of p65/RelA, indicating reduced canonical NF-κB activation (Fig. 2B). Taken together, our data indicate that TAK1 plays a key role in regulating NF-κB activity in pancreatic cancer cells, consistent with the study by Melisi et al (6), and that oncogenic KRAS promotes TAK1 activity by promoting TAK1 stabilization and hence the TAK1-TAB interaction.
To determine the functional significance of TAK1-mediated NF-κB activity, we examined the effect of TAK1 depletion on pancreatic cancer cell proliferation/survival. siRNA mediated TAK1 depletion significantly reduced the cell index of MiaPaCa-2 cells relative to the non-targeting control throughout the time-course in an MTS assay (Fig. 2C). This reduced cell index correlated with a weak PARP cleavage response (Supplementary S1). Importantly, this effect of TAK1 depletion was specific to cells harboring mutant KRAS, as knockdown of TAK1 did not decrease cell proliferation in HPDE6 cells unlike HPDEKR+ (Fig. 2D). Notably, the effect of TAK1 depletion on cell index was significantly weaker than that seen with pharmacologic inhibition of GSK-3, possibly because of incomplete knockdown of TAK1.
To achieve a greater inhibition of TAK1 kinase activity, we analyzed the effect of a selectiv e and potent TAK1 kinase inhibitor, 5 Z-7-oxozaenol, on cell survival/proliferation of pancreatic cancer cells (5). Consistent with our hypothesis, TAK1 inhibitor treatment resulted in a greater decrease in cell proliferation of MiaPaCa-2 compared to TAK1 knockdown (Fig. 2E). Again the inhibitory effect of TAK1 inhibitor on cell proliferation was specific to mutant KRAS harboring cells. (Supplementary S3). The decrease in phosphorylation of p38 (TAK1 substrate) confirms the inhibition of TAK1 activity upon 5Z-7-oxozaenol treatment (Supplementary S2).
A modest increase in PARP cleavage upon TAK1 and GSK-3 inhibition (Supplementary S2) argues against the induction of apoptosis as a major mechanism resulting in reduced cell index. We thus, examined the effect of TAK1 inhibition on cell cycle distribution. Panc-1 cells were treated with 5Z-7-oxozaenol and stained with PI. An increase in the number of cells in G2/M phase was observed with increasing concentration of the TAK1 inhibitor (Supplementary S4), suggesting that TAK1 inhibition leads to a G2/M arrest in pancreatic cancer cells leading to decreased proliferation. As shown below, the effect of GSK-3 inhibition on cell proliferation may be via TAK1 and thus cell cycle deregulation. Overall, these data suggest that TAK1 is active downstream of mutant KRAS in pancreatic cancer cells and mediates constitutive NF-κB signaling to regulate proliferation by altering cell cycle progression.
GSK-3α facilitates constitutive TAK1 activity by maintaining TAK1-TAB1 interaction
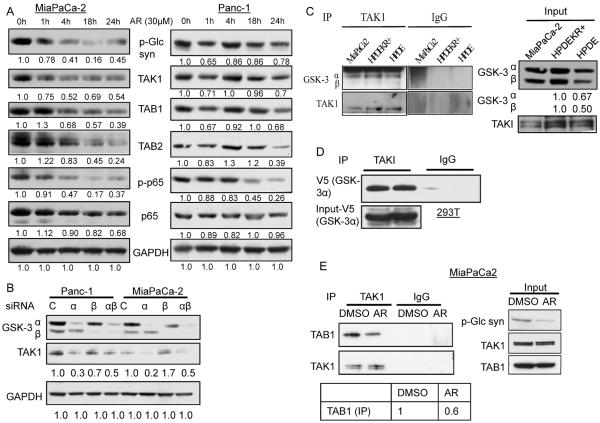
We and others previously demonstrated that GSK-3 drives constitutive IKK and NF-κB activity in pancreatic cancer cell lines (21). In Fig. 1C we observed a decrease in TAK1 levels with GSK-3 inhibition in Panc-1 and MiaPaCa-2 cells. To further analyze a role for GSK-3 in regulating TAK1-dependent NF-κB signaling, Panc-1 and MiaPaCa-2 cells were treated with GSK-3 inhibitor over a 24-hour time course. Results from this experiment revealed a time-dependent decrease in levels of TAK1, its binding partners TAB1 and TAB2, and NF-κB p65 phosphorylation (Fig. 3A). Though the kinetics of decrease in TAK1 and TABs levels were different in Panc-1 and MiaPaCa-2 cells, the levels of TAK1 were always consistent with the loss of GSK-3 activity, as seen by suppression of phospho-glycogen synthase, a substrate of GSK-3. Moreover, we also saw a decrease in TAK1 substrate phosphorylation (phospho-p38) with GSK-3 inhibition (Supplementary S5). To determine if the loss in TAK1 protein levels is due to proteasome-dependent degradation, Panc-1 and MiaPaCa-2 cells were treated with AR-A014418 in the presence of proteasome inhibitor (MG-132). The results demonstrated that MG-132 treatment restored reduced TAK1 levels in Panc-1 cells but not in MiaPaCa-2 cells (Supplementary S6). To demonstrate that the decrease in TAK1 levels was not an off-target effect of AR-A01448, siRNA was used to transiently knockdown GSK-3α and/or GSK-3β in Panc-1 and MiaPaCa-2 cells. 48 hours after siRNA transfection, a decrease in TAK1 levels was observed with knockdown of GSK-3α but not of GSK-3β (Fig. 3B).
Figure 3. GSK-3 inhibition suppresses TAK1 levels.
(A) Indicated cell lines were treated with GSK-3 inhibitor (AR-A014418) for indicated time periods and immunoblotting performed on whole cell lysates. Data is representative of 3 independent experiments. (B) Panc-1 and MiaPaCa-2 cells were transiently transfected with indicated siRNA for 48 hours, whole cell lysates harvested and immunoblotted against indicated antibodies. (C) Endogenous TAK1 was immunoprecipitated from the indicated cell lines and immunoblotted for GSK-3α and GSK-3β. (D) TAK1 and V5-tagged GSK-3α were transiently expressed in 293T cells for 24h. TAK1 was immunoprecipitated and blotted for V5. (E) MiaPaCa-2 cells were transiently transfected with TAK1 plasmid. 24h later they were treated with AR-A014418 for 24h. TAK1 was immunoprecipitated and blotted for TAB1.
To further analyze the mechanism of GSK-3-TAK1 regulation, we investigated whether GSK-3 isoforms can physically interact with TAK1. Endogenous TAK1 was immunoprecipitated from MiaPaCa-2 cells and blotted for GSK-3α and GSK-3β. We observed GSK-3α co-precipitating with TAK1 in KRAS transformed pancreatic cancer cells (MiaPaCa-2 and HPDE-KR+) as well as non-transformed HPDE (Fig. 3C), although the level of interaction was higher in the KRAS+ cells. Consistent with previous studies (35), expression of oncogenic KRAS led to the upregulation of GSK-3 isoforms (Fig. 3C). Since GSK-3α-TAK1 co-immunoprecipitated in non-transformed HPDE's as well as transformed pancreatic cancer cells, we sought to explore this interaction in other cells like 293T cells. These cells have very low levels of GSK-3 and TAK1 as compared to pancreatic cell lines, prompting us to overexpress these proteins to detect any interaction. Expression of V5-tagged GSK-3α with TAK1 in 293T cells led to an interaction as determined by co-immunoprecipitation (Fig. 3D). No interaction between over-expressed GSK-3β and TAK1 was observed indicating specificity to GSK-3α (data not shown).
To determine if GSK-3 kinase activity is required for maintaining the TAK1-TAB interaction (and not just TAK1 and TAB levels), we evaluated the effect of GSK-3 inhibition on TAK1-TAB1 complex in MiaPaCa-2 cells. Since AR-A014418 treatment affects TAK1 levels, we transiently over-expressed TAK1 in MiaPaCa-2 cells, before treating the cells with AR-A014418. As is evident in Fig. 3E, GSK-3 inhibition suppressed the co-precipitation between TAK1 and TAB1, even under conditions where TAK1 and TAB1 levels are maintained. Thus GSK-3 catalytic activity appears to be important in maintaining the TAK1-TAB1 complex. Overall, these data and those described above demonstrate that mutant KRAS induces TAK1-TAB1 interaction that is stabilized by GSK-3α leading to higher TAK1-dependent NF-κB activity in pancreatic cancer cells.
GSK-3α facilitates constitutive non-canonical signaling in pancreatic cancer cells
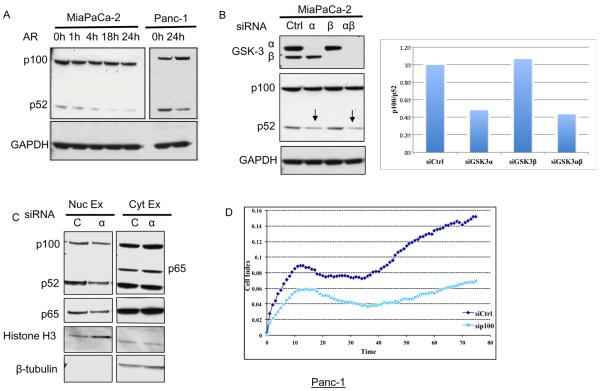
Non-canonical NF-κB pathway leading to processing of NF-κB2/p100 to p52, has been shown to be constitutively active in pancreatic cancer cells (36,37). To this point, the data links GSK-3 to constitutive canonical NF-κB signaling via TAK1. However, the effect of GSK-3 inhibition on cell survival/proliferation was much greater than that observed with depletion of TAK1, suggesting that another component regulated by GSK-3 affects pancreatic cell survival/proliferation. Thus, we asked if GSK-3 regulates the non-canonical arm of NF-κB in pancreatic cancer cells. Processing of p100 to p52 in Panc-1 and MiaPaCa-2 cells was suppressed when these cells were treated with the GSK-3 inhibitor, AR-A014418 (Fig. 4A). To account for potential off-target effects of the GSK-3 inhibitor, p100–p52 processing was analyzed upon GSK-3α/β depletion by siRNA. Depletion of GSK-3α but not GSK-3β led to a decrease in p100–p52 processing in Panc-1 and MiaPaCa-2 cells (Fig. 4B), again highlighting differences between GSK-3α and GSK-3β. Upon processing of p100, p52 accumulates in the nucleus to promote transcription of its target genes. We examined the effect of GSK-3α knockdown on the nuclear localization of p52. Surprisingly we saw that GSK-3α depletion suppressed p52 levels only in the nuclear fraction and not in the cytoplasmic extract (Fig. 4C). This suggests that either GSK-3α promotes nuclear accumulation of p52 or that GSK-3α regulates the processing of p100 in the nuclear fraction.
Figure 4. GSK-3α regulates the non-canonical NF-κB pathway in pancreatic cancer cells.
(A) Panc-1 and MiaPaCa-2 cells were treated with DMSO or AR-A014418 (30μM) for indicated time periods and whole cell lysates immunoblotted with the indicated antibodies. (B) MiaPaCa-2 cells were transiently transfected with indicated siRNA for 72 hours and whole cell lysates immunoblotted for indicated antibodies. On right is quantitation of the western blot. (C) Panc-1 cells were transiently transfected with indicated siRNA for 72 hours, nuclear and cytoplasmic fractions harvested and immunoblotted for indicated antibodies. (D) Panc-1 cells were transiently transfected with indicated siRNA for 48 hours and cell impedance measured as above.
The functional importance of the non-canonical NF-κB pathway was measured via the effect of p100 knockdown on the cell index of pancreatic cancer cells. As compared to the non-targeting control, p100 knockdown suppressed the cell index of Panc-1 cells in a cell impedance assay (Fig. 4D). This suppression of cell index was greater than that observed with TAK1 depletion. Knockdown of p100 modestly increased PARP cleavage in MiaPaCa-2 but not in Panc-1 cells (Supplementary S7). Overall, these data provide a novel link between GSK-3α and the non-canonical NF-κB pathway in pancreatic cancer cells. These results may explain why knockdown of GSK-3α (that affects both canonical and non-canonical NF-κB) leads to a greater suppression of cell growth as compared to knockdown of GSK-3β or TAK1 (Fig. 1 C, D).
GSK-3 inhibition suppresses growth of patient-derived pancreatic tumor explants
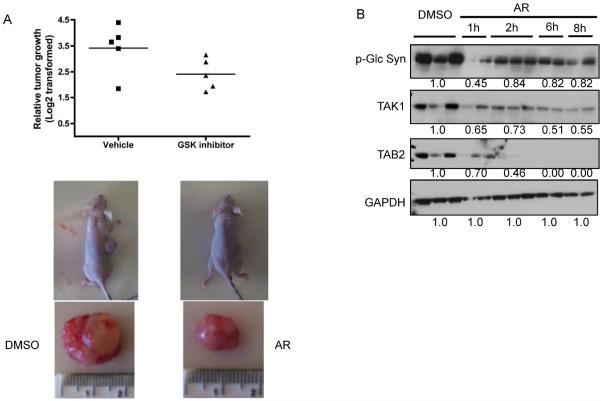
Our data and previous reports suggest that GSK-3 inhibition may be a potential target for pancreatic cancer treatment because of its effect on NF-κB activity. However, the effect of GSK-3 inhibition on human pancreatic tumors has never been evaluated. To analyze the effect of GSK-3 inhibition on tumor growth in vivo, we explanted replicates of a human pancreatic tumor in nude mice. Two weeks later, GSK-3 inhibitor, AR-A014418, was administered intraperitoneally at 120 mg/kg twice a day for two days. The growth of the tumor was then monitored over a course of 28 days. Consistent with our hypothesis and data, GSK-3 inhibition suppressed tumor growth in mice by approximately 50% (Fig. 5A). To analyze the effect of acute GSK-3 inhibition on the TAK1/NF-κB signaling in mice, we repeated the AR-A014418 treatment study on pancreatic tumor explants for shorter time courses (1h, 2h, 6h and 8h). We observed a time-dependent decrease in TAK1 and TAB levels in these tumors, consistent with our studies in pancreatic cancer cell lines (Fig. 5B). Overall these data indicate that GSK-3 inhibition suppresses pancreatic tumor growth with a concomitant decrease in TAK1-TAB activity.
Figure 5. GSK-3 inhibition suppresses tumor growth in mice.
(A) Nude mice were explanted with replicates of human pancreatic tumor and two weeks later treated with the GSK-3 inhibitor (AR-A014418) at 120mg/kg twice a day for two days (A) or the indicated time periods (B). (A). Tumor volume was measured over 28 days and is represented as the fold change in tumor volume compared to treatment start. n (vehicle)=4, n (treatment) = 5. (lower panel) Representative photograph of mice at the end of 28 days. (B) Tumors were harvested after indicated time periods and immunoblotted with the indicated antibodies.
Effect of GSK-3 inhibition on gene expression in human patient-derived pancreatic tumor explants
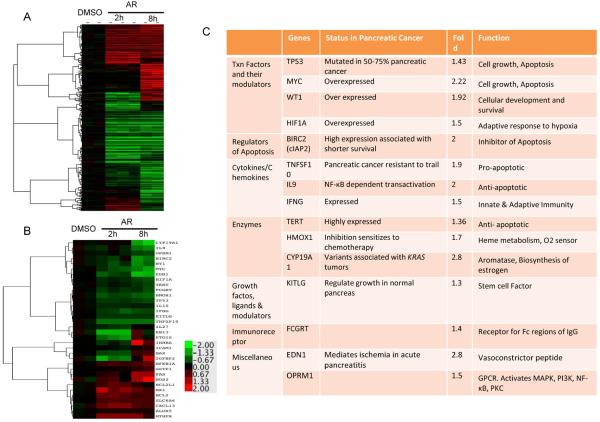
We next sought to determine the effect of GSK-3 inhibition on gene expression in the human pancreatic tumor explants in mice. Gene expression microarrays were utilized to identify global changes in gene expression that occurred in these tumors when treated with GSK-3 inhibitor. Supervised gene expression analyses were conducted to quantify gene expression changes in the tumors, after 2 or 8 hours of AR-A014418 treatment (Fig. 6A). GSK-3 inhibition led to a statistically significant change in expression (based on SAM analysis) for 470 genes, of which 155 changed more than 2 fold (Fig. 6A). We further analyzed known or suspected to be NF-κB target genes changed by GSK-3 inhibition. In this group, GSK-3 inhibition led to a statistically significant down-regulation of 17 genes after 8h and 22 genes after 2 hours of treatment (Fig. 6B). 15 out of these genes were downregulated more than 1.5 fold, with greater downregulation occurring at the 8h time point. As shown in Fig. 6C, most of these genes have been shown to have a pro-oncogenic role in pancreatic cancer. For example, we observe a decrease in the expression of pro-proliferative genes like c-Myc, TERT, and cIAP2 with GSK-3 inhibition.
Figure 6. GSK-3 inhibition leads to changes in NF-κB target gene expression.
Total RNA was isolated from human pancreatic tumor explants treated with AR-A014418 for 2h or 8h and evaluated by microarray. Gene changes that were statistically significant by SAM analysis are shown in the heat map (p≤0.01). Color key is for log2 ratio. (B) Statistically significant changes in known NF-κB target genes are shown in the heat map (p≤0.05). (C) NF-κB target genes that were downregulated >1.5 fold are listed with their known function in pancreatic cancer (p≤0.05).
Discussion
Mutant KRAS is expressed in virtually all pancreatic cancers as well as in other epithelial-derived cancers where it serves as a key oncogenic factor, promoting proliferation and survival (1,2). Despite extensive research, less than 4% of patients diagnosed with pancreatic cancer are expected to survive past 5 years (38). Since no Ras inhibitors have been effective (2,3), current research efforts are focusing on targeting deregulated signaling pathways downstream of KRAS+. The advancement of this therapeutic strategy is dependent on a detailed understanding of the complex molecular mechanisms underlying signaling events that regulate disease progression.
NF-κB is known to be constitutively active in majority of the pancreatic tumors and pancreatic cancer cell lines, where it regulates proliferation, survival, metastasis and invasion (18). NF-κB has also been shown to be activated downstream of oncogenic Ras and promote the oncogenic phenotype (14,15). Thus, the signaling cascades activating NF-κB pathway have become attractive targets for novel chemotherapeutic approaches in pancreatic and other cancers (18). There is evidence that multiple of arms of NF-κB are activated in various cancers (10), thus the ability to block NF-κB activity broadly may require multiple inhibitors unless a factor can be identified that regulates multiple NF-κB-relevant signaling pathways.
Previous studies from our lab and others have shown that GSK-3 inhibition reduces pancreatic cancer cell viability in vitro and suppresses tumor cancer cell line growth in vivo at least partly through the downregulation of NF-κB activity (21,31,34,39). However the mechanism by which GSK-3 regulates NF-κB and the distinct roles of the two isoforms, GSK-3α and GSK-3β. are not well characterized. In this report, we provide the first evidence for a role of GSK-3α. in regulating proliferation and anchorage-independent growth of pancreatic cancer cells independent of GSK-3β. We previously reported that GSK-3 signals through IKK to mediate canonical NF-κB signaling in pancreatic cancer cells (21). Here, we propose that GSK-3 and IKK are functionally linked through GSK-3α-dependent control of TAK1-TAB1 complex stability/activity, and provide the first evidence for regulation of non-canonical NF-κB by GSK-3α.
TAK1 is a central regulator of NF-κB signaling in diverse physiological processes including development, immune responses and survival (25). Its binding partner, TAB1 was recently shown to play an essential role in the maintenance of TAK1 activity in epithelial tissues (27). Here, we show that the TAK1-TAB1 complex is active in KRAS+ cancer cells and is stabilized via a GSK-3α-dependent mechanism (Fig. 2). TAK1 was recently identified as a pro-survival mediator in KRAS+-dependent colon cancer (5). While Singh et al argue against the pro-survival role of TAK1 in pancreatic cancer cells, another group showed that inhibition of TAK1 leads to a proapoptotic phenotype in pancreatic cancer cells, by suppressing NF-κB (5,6) Our results supports aspects of each of these studies, as we observe a reduction in NF-κB activity and cell proliferation with inhibition of TAK1 in pancreatic cancer cells (Fig. 2), however we do not see a strong corresponding increase in apoptosis as measured by cleavage of caspase 3 or PARP under these conditions (Supplementary S1,S2). However, inhibition of TAK1 leads to G2/M arrest in pancreatic cancer cells (Supplementary S4), which presumably causes a decrease in cell proliferation.
A mechanism to describe how TAK1 is activated downstream from oncogenic KRAS has not been described. Previously, Moscat and colleagues (24) showed the involvement of p62 in regulating TRAF6 ubiquitination downstream of oncogenic KRAS to promote IKK activity. Here, we provide the first evidence of a regulatory link between GSK-3α and TAK1. We show that pharmacological inhibition of GSK-3 in pancreatic cancer cells leads to a decrease in levels of TAK1 and its binding partners TAB1 and TAB2 (Fig 3A,B, Supplementary S8). The decrease in TAK1 and TAB levels were detected as early as 1h following GSK-3 inhibitor treatment and were maintained at low levels as long as GSK-3 activity was inhibited. The in vitro results were reproduced in vivo when human pancreatic tumor explants were treated with AR-A014418 (Fig. 5). These results are corroborated by siRNA knockdown of GSK-3α in pancreatic cancer cells. The interaction between GSK-3α and TAK1 was found in non-transformed HPDE cells, suggesting that GSK-3α can interact with TAK1 irrespective of presence or absence of mutant KRAS. However, mutant KRAS upregulates the expression of GSK-3 (35) (Supplementary S9), which then promotes the stabilization of the active TAK1-TAB1 complex. TAK1 can drive IKK-dependent NF-κB signaling leading to increased proliferation of KRAS+ transformed cells. The mechanistic link between GSK-3α and TAK1 is unclear since there is no consensus substrate motif of GSK-3 (S/T-XXX-S/T) in TAK1. However, Taelman et al have shown that TAB1 and TAB2 contain multiple GSK-3 consensus motifs (40). Thus, there is a possibility that GSK-3 phosphorylates TABs to regulate the TAK1-TAB1 complex and thus the stability of TAK1. This hypothesis will be addressed in future studies.
Although we demonstrate that GSK-3 regulates IKK through TAK1, the effect on cell proliferation with GSK-3 inhibition is much stronger as compared to TAK1 inhibition. These results suggested that additional regulatory events downstream of GSK-3 promote pancreatic cancer cell growth/survival. Non-canonical NF-κB pathway has been shown to be constitutively active and to be contributing to proliferation/survival in pancreatic cancer cells (36,37). Thus we explored the potential involvement of GSK-3 in regulating p100/NF-κB2 processing. GSK-3 inhibition, or knockdown of GSK-3α (but not GSK-3β) leads to significantly reduced p100 processing in pancreatic cancer cells (Fig. 4A,B, Supplementary S8). Importantly, we demonstrate a significant reduction in cell proliferation of pancreatic cancer cells, upon knocking down p100 subunit (Fig. 4D). We also saw a decrease in p100 and p52 levels upon TAK1 inhibition, which is consistent with p100 being a known transcriptional target of canonical NF-κB (data not shown). Interestingly we observed a decrease in nuclear accumulation of p52 subunit upon GSK-3α knockdown while, the cytoplasmic levels of p100 and p52 remained unchanged (Fig. 4C). This indicates a distinct effect of GSK-3α on nuclear p52 different from that derived via inhibition of TAK1-regulated canonical NF-κB. GSK-3 has been earlier shown to be accumulated in the nucleus in pancreatic cancers (34) which raises the potential that the GSK-3 effect on non-canonical NF-κB is via processing of nuclear p100. It is also possible that GSK-3α affects the nuclear transport of p52 subunit. We thus hypothesize that GSK-3α regulates both canonical and non-canonical NF-κB pathway in pancreatic cancer cells. Notably our results on the regulation of p100 processing by GSK-3α are likely not to relate to the recent work demonstrating the role of GSK-3β and Fbxw7α-mediated degradation of p100 in multiple myeloma (41) as we do not see p100 accumulation upon GSK-3α knockdown. However, our study does support the pro-survival role of p100 – p52 processing in a way similar to that described by Sangfelt and colleagues (42).
The human tumor explant study demonstrates a 50% inhibition of KRAS-mutant tumor growth upon a 2-day treatment with GSK-3 inhibitor (Fig. 5). This study was performed with a commercially available GSK-3 inhibitor, indicating the need for more extensive studies on a broader group of pancreatic tumors using a pharmaceutical grade inhibitor. To analyze the effect of GSK-3 inhibition on NF-κB target gene expression, we compared the gene expression profile of tumors, before and after treatment with AR-A014418. We observed a down-regulation of several established NF-κB target genes such as c-myc and TERT, which are known to be upregulated in pancreatic cancer (Fig. 6C). It is noteworthy that we saw a selective effect on NF-κB dependent gene expression. These results are consistent with previous studies that GSK-3 and Ras induce only a selective arm of the NF-κB pathway (43,44), which is different from the well characterized NF-κB regulated downstream of cytokine-induced signaling. NF-κB may also be regulated by upstream activators other than GSK-3 in these tumors, thus inhibiting GSK-3 alone will not shut down the entire NF-κB signaling.
Interestingly, significant changes were observed in long non-coding RNAs upon GSK-3 inhibition, which are now emerging as key regulators of oncogenesis (45). We understand that GSK-3 plays important roles in other signaling pathways, including Wnt, Notch, Hedgehog, which have been implicated in pancreatic cancers (46) and which may account for the rest of the observed changes in gene expression. Implications of GSK-3 inhibition on the β-catenin stabilization and Wnt pathway needs consideration (46), however our encouraging results on tumor growth inhibition and previous studies strongly support the therapeutic potential of GSK-3 inhibitors.
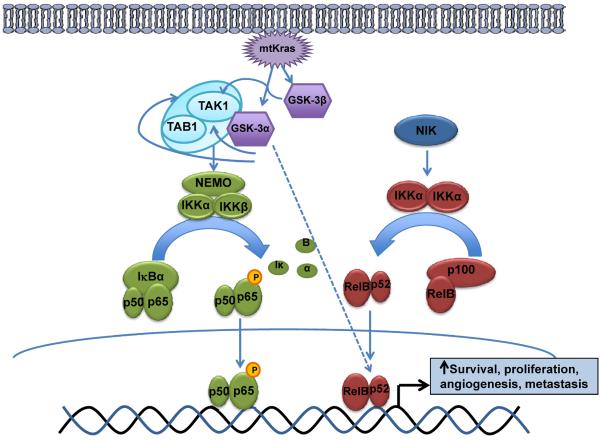
Collectively, this report provides the first evidence of GSK-3α in promoting oncogenic RAS function through regulation of TAK1-TAB activity, upstream of IKK and canonical NF-κB, and via control of non-canonical NF-κB activity in pancreatic cancer cells (Fig.7). Importantly, this data provides evidence for different roles of GSK-3α and GSK-3β in regulating NF-κB signaling in pancreatic cancer cells, and highlights the need for the development and testing of GSK-3α specific drugs.
Figure 7. Model of GSK-3-NF-κB pathway downstream of mutant Kras in pancreatic cancer.
In a normal cell, TAK1-TAB1 complex is minimally active and NF-κB pathway is dormant. However, mutant Kras leads to transcriptional upregulation of GSK-3α and GSK-3β, that can stabilize TAK1, TAB1 and TAK1-TAB1 complex leading to constitutive canonical NF-κB activation. GSK-3α also drives the non-canonical NF-κB pathway by promoting/stabilizing nuclear p52, thus leading to constitutive non-canonical NF-κB activation. NF-κB drives transcription of genes involved in survival, proliferation, metastasis that contributes to an aggressive pancreatic phenotype.
Materials and Methods
Cell culture and reagents
Panc-1, MiaPaCa-2 were purchased from American Type Culture Collection and used for no longer than 6 months before being replaced. Human pancreatic ductal epithelial cell line (HPDE6) and KRAS4BG12V–transfected HPDE (HPDEKR+) were generous gifts from Dr. Channing Der (UNC-Chapel Hill) and were maintained in keratinocyte serum-free growth medium. All cell culture reagents were obtained from Invitrogen (Carlsbad, CA). The following antibodies were purchased from Cell Signaling Techonology: phospho-Glycogen synthase (Ser641), Glycogen Synthase phospho-p65 (Ser536), p-65, TAB1, TAB2, Histone H3, Cleaved PARP (Asp214), PARP, phospho-p38 (Thr180/Tyr182), p38; Santa Cruz Biotechnology: GSK-3α/β, TAK1, GAPDH, β-tubulin; Millipore: p100/p52 and Sigma Aldrich: Anti-Flag (M2). GSK-3 inhibitor (AR-A014418) was purchased from Sigma-Aldrich. TAK1 inhibitor (5Z-7-oxozaenol) was purchased from Tocris Bioscience.
siRNA interference
The following human siRNA (siGenome SMARTpool) was purchased from Dharmacon as a pool of four annealed dsRNA oligonucleotides: MAP3K7 (M-003790-06), GSK-3α(M-003009-01), GSK-3β (M-003010-03), NF-κB2 (M-003918-02), and non-targeting control#3 (D001201-03). Dharmafect transfection reagent 1 was used to transfect 100nM siRNA according to manufacturer's instruction and cells were harvested 48 or 72 hours after transfection, as stated.
Western blot analysis
Cytoplasmic, nuclear and whole cell extracts were prepared as described previously (21). Tumor lysates were prepared by homogenizing ~1mm piece of tumor in lysis buffer (50mM Tris (pH7.6), 150mM HCL, 2mM EDTA). NP-40 was added at 1% v/v and incubated on a rocking platform for 30 minutes in cold room and cellular debris precipitated using centrifugation. Protein extracts were quantified by Bradford assay (Bio-rad Laboratories) and analyzed by SDS-PAGE (21).
MTS Assay
Cells were seeded in a 96-well plate and allowed to incubate for 24 hours. The cells were then treated with the indicated concentrations of AR-A014418 for indicated times and cellular proliferation measured using the CellTiter 96 Aqueous solution, in accordance to the manufacturer's protocol (Promega). For siRNA experiments, the cells were seeded in the plate 48 hours after siRNA transfection.
Cell impedance assay/ real time cell analysis (RTCA)
Background impedance signal was measured with 50μL cell culture medium per well of an E-Plate 16. The final volume in a single well was adjusted to 100μL cell culture medium by adding additional 50 μL medium containing 1000 cells. For siRNA experiments, cells were seeded 48 hours after transfection. For inhibitor studies, 5Z-7-oxozaenol was added one day after seeding. After plating, impedance was routinely recorded in 2-hour intervals using the xCELLigence RTCA MP instrument (Roche).
Soft Agar assay
MiaPaCa-2 cells were transiently transfected with indicated siRNA as above. 48 hours after transfection, 8000 cells were suspended in a 0.4% bacto-agar/DMEM layer (1ml) and plated over a 0.6% bacto-agar/DMEM layer (2ml) in 6-well plates. 0.5ml media was added next day and every 3–4 days thereafter for 2 weeks. After one week colonies were stained with 0.5ml of MTT (2mg/ml, Sigma Aldrich). Images were acquired and colonies counted using ImageJ 1.46 software.
Microarray analysis
Total RNA was purified using RNeasy plus RNA isolation kit (Qiagen), reverse-transcribed, labeled, and hybridized to an Agilent v2 8×60K whole human DNA microarray. Microarrays were scanned using an Agilent DNA microarray scanner and features were extracted using Agilent Feature Extraction software version 10.7.3.1. Data were uploaded to the University of North Carolina Microarray Database (UMD). Gene expression data were extracted from the UMD for each sample as log2 Cy5/Cy3 ratios, filtering for probes with Lowess normalized intensity values greater than 10 in both channels and for probes with data on greater than 70% of the microarrays. Hierarchical clustering analyses was carried out using Gene Cluster 3.0 (47), data were viewed using Java TreeView version 1.1.5r2 (48). Expression changes were determined using a linear model with terms for treatment time points (49). This model was fit to each probe, and genes corresponding to treatment effects with a q-value ≤ 0.05 (50) were considered statistically significant. The data are publicly available in Gene Expression Omnibus database (accession number GSE42559)
Tumor explant study
Tiny fragments (1.5–2mm) of a resected human pancreatic tumor were implanted subcutaneously into the flanks of nude mice. Two weeks later, GSK-3 inhibitor, AR-A014418 or vehicle control, DMSO was given intraperitonealy at 120mg/kg twice a day for two days. The growth of the tumor was measured every alternate day for 28 days. For shorter treatments, tumor was harvested immediately after the indicated time periods.
Supplementary Material
Statement of Significance.
GSK-3α functions to promote IKK/NF-κB activity downstream of oncogenic KRAS via stabilization and activation of the TAK1/TAB complex and to promote non-canonical NF-κB activity via control of nuclear levels of NF-κB2. Inhibition of GSK-3 strongly suppresses growth of human pancreatic tumor explants with downregulation of certain oncogenic NF-κB target genes such as c-myc and TERT.
Acknowledgements
We thank UNC Bioinformatics Core for help in analysis of microarray data.
Funding: This work was supported by NIH CA73756 (ASB, DB, WW), NIH CA75080 (ASB, DB, WW), Waxman Cancer Research Foundation (ASB), NIH CA140424 (JJY) and BBSP first year program of UNC-Chapel Hill (MR).
Abbreviations
- PDAC
pancreatic ductal adenocarcinoma
Footnotes
The authors disclose no potential conflicts of interest.
References
- 1.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 2.Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, et al. Oncogenic Kras Maintains Pancreatic Tumors through Regulation of Anabolic Glucose Metabolism. Cell. 2012;149:656–70. doi: 10.1016/j.cell.2012.01.058. Elsevier Inc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vaccaro V, Melisi D, Bria E, Cuppone F, Ciuffreda L, Pino MS, et al. Emerging pathways and future targets for the molecular therapy of pancreatic cancer. Expert Opin Ther Targets. 2011;15:1183–96. doi: 10.1517/14728222.2011.607438. [DOI] [PubMed] [Google Scholar]
- 4.Wolff RA. Chemotherapy for pancreatic cancer: from metastatic disease to adjuvant therapy. Cancer J. 2007 May;13:175–84. doi: 10.1097/PPO.0b013e318074e6c3. [DOI] [PubMed] [Google Scholar]
- 5.Singh A, Sweeney MF, Yu M, Burger A, Greninger P, Benes C, et al. TAK1 Inhibition Promotes Apoptosis in KRAS-Dependent Colon Cancers. Cell. 2012;148:639–50. doi: 10.1016/j.cell.2011.12.033. Elsevier Inc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Melisi D, Xia Q, Paradiso G, Ling J, Moccia T, Carbone C, et al. Modulation of pancreatic cancer chemoresistance by inhibition of TAK1. Journal of the National Cancer Institute. 2011;103:1190–204. doi: 10.1093/jnci/djr243. Oxford University Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ougolkov AV, Billadeau DD. Targeting GSK-3: a promising approach for cancer therapy? Future Oncology. 2006;2:91–100. doi: 10.2217/14796694.2.1.91. [DOI] [PubMed] [Google Scholar]
- 8.Hayden MS, Ghosh S. NF- B, the first quarter-century: remarkable progress and outstanding questions. Genes & Development. 2012;26:203–34. doi: 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karin M, Cao Y, Greten FR, Li Z-W. NF-κB IN CANCER: FROM INNOCENT BYSTANDER TO MAJOR CULPRIT. Nat Rev Cancer. 2002;2:301–10. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 10.Bassères DS, Baldwin AS. Nuclear factor-κB and inhibitor of κB kinase pathways in oncogenic initiation and progression. Oncogene. 2006;25:6817–30. doi: 10.1038/sj.onc.1209942. [DOI] [PubMed] [Google Scholar]
- 11.Perkins ND. The diverse and complex roles of NF-κB subunits in cancer. Nat Rev Cancer. 2012:19. doi: 10.1038/nrc3204. [DOI] [PubMed] [Google Scholar]
- 12.Bonizzi G, Bebien M, Otero DC, Johnson-Vroom KE, Cao Y, Vu D, et al. Activation of IKKα target genes depends on recognition of specific κB binding sites by RelB: p52 dimers. EMBO J. 2004;23:4202–10. doi: 10.1038/sj.emboj.7600391. Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayo MW, Wang CY, Cogswell PC, Rogers-Graham KS, Lowe SW, Der CJ, et al. Requirement of NF-kappaB activation to suppress p53-independent apoptosis induced by oncogenic Ras. Science. 1997;278:1812–5. doi: 10.1126/science.278.5344.1812. [DOI] [PubMed] [Google Scholar]
- 14.Meylan E, Dooley AL, Feldser DM, Shen L, Turk E, Ouyang C, et al. Requirement for NF-κB signalling in a mouse model of lung adenocarcinoma. Nature. 2009;461:104–7. doi: 10.1038/nature08462. Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang J, Splittgerber R, Yull FE, Kantrow S, Ayers GD, Karin M, et al. Conditional ablation of Ikkβ inhibits melanoma tumor development in mice. J. Clin. Invest. 2010;120:2563–74. doi: 10.1172/JCI42358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bassères DS, Ebbs A, Levantini E, Baldwin AS. Requirement of the NF-kappaB subunit p65/RelA for K-Ras-induced lung tumorigenesis. Cancer Research. 2010;70:3537–46. doi: 10.1158/0008-5472.CAN-09-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia Y, Yeddula N, Leblanc M, Ke E, Zhang Y, Oldfield E, et al. Reduced cell proliferation by IKK2 depletion in a mouse lung-cancer model. Nat Cell Biol. 2012;14:257–65. doi: 10.1038/ncb2428. Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carbone C, Melisi D. NF-κB as a target for pancreatic cancer therapy. Expert Opin Ther Targets. 2012;23 doi: 10.1517/14728222.2011.645806. [DOI] [PubMed] [Google Scholar]
- 19.Li L, Aggarwal BB, Shishodia S, Abbruzzese J, Kurzrock R. Nuclear factor-κB and IκB kinase are constitutively active in human pancreatic cells, and their down-regulation by curcumin (diferuloylmethane) is associated with the suppression of proliferation and the induction of apoptosis. Cancer. 2004;101:2351–62. doi: 10.1002/cncr.20605. [DOI] [PubMed] [Google Scholar]
- 20.Liptay S, Weber CK, Ludwig L, Wagner M, Adler G, Schmid RM. Mitogenic and antiapoptotic role of constitutive NF-κB/Rel activity in pancreatic cancer. Int. J. Cancer. 2003;105:735–46. doi: 10.1002/ijc.11081. [DOI] [PubMed] [Google Scholar]
- 21.Wilson W, Baldwin AS. Maintenance of Constitutive I B Kinase Activity by Glycogen Synthase Kinase-3 / in Pancreatic Cancer. Cancer Research. 2008;68:8156–63. doi: 10.1158/0008-5472.CAN-08-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang W, Abbruzzese JL, Evans DB, Larry L, Cleary KR, Chiao PJ. The nuclear factor-κB RelA transcription factor is constitutively activated in human pancreatic adenocarcinoma cells. Clin. Cancer Res. 1999;5:119–27. AACR. [PubMed] [Google Scholar]
- 23.Ling J, Kang Y, Zhao R, Xia Q, Lee D-F, Chang Z, et al. KrasG12D-Induced IKK2/β/NF-κB Activation by IL-1α and p62 Feedforward Loops Is Required for Development of Pancreatic Ductal Adenocarcinoma. Cancer Cell. 2012;17(21):105–20. doi: 10.1016/j.ccr.2011.12.006. Elsevier Inc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duran A, Linares JF, Galvez AS, Wikenheiser K, Flores JM, Diaz-Meco MT, et al. The Signaling Adaptor p62 Is an Important NF-κB Mediator in Tumorigenesis. Cancer Cell. 2008;13:343–54. doi: 10.1016/j.ccr.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 25.Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–51. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 26.Pickett TD, Ninomiya-Tsuji J. TAB4 Stimulates TAK1-TAB1 Phosphorylation and Binds Polyubiquitin to Direct Signaling to NF-κB. J. Biol. Chem. 2008;283:19245–54. doi: 10.1074/jbc.M800943200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Omori E, Inagaki M, Mishina Y, Matsumoto K, Ninomiya-Tsuji J. Epithelial transforming growth factor β-activated kinase 1 (TAK1) is activated through two independent mechanisms and regulates reactive oxygen species. Proc Natl Acad Sci U S A. 2012;13 doi: 10.1073/pnas.1116188109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raman M, Chen W, Cobb MH. Differential regulation and properties of MAPKs. Oncogene. 2007;26:3100–12. doi: 10.1038/sj.onc.1210392. [DOI] [PubMed] [Google Scholar]
- 29.Hoeflich KP, Luo J, Rubie EA, Tsao MS, Jin OU, Woodgett JR. Requirement for glycogen synthase kinase-3β in cell survival and NF-κB activation. Nature. 2000;406:86–90. doi: 10.1038/35017574. Nature Publishing Group. [DOI] [PubMed] [Google Scholar]
- 30.Doble BW. GSK-3: tricks of the trade for a multi-tasking kinase. Journal of Cell Science. 2003;116:1175–86. doi: 10.1242/jcs.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ougolkov AV, Fernandez-Zapico ME, Savoy DN, Urrutia RA, Billadeau DD. Glycogen Synthase Kinase-3β Participates in Nuclear Factor κB-Mediated Gene Transcription and Cell Survival in Pancreatic Cancer Cells. Cancer Research. 2005;65:2076–81. doi: 10.1158/0008-5472.CAN-04-3642. AACR. [DOI] [PubMed] [Google Scholar]
- 32.Wang Z, Smith KS, Murphy M, Piloto O, Somervaille TCP, Cleary ML. Glycogen synthase kinase 3 in MLL leukaemia maintenance and targeted therapy. Nature. 2008;455:1205–9. doi: 10.1038/nature07284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Banerji V, Frumm SM, Ross KN, Li LS, Schinzel AC, Hahn CK, et al. The intersection of genetic and chemical genomic screens identifies GSK-3α as a target in human acute myeloid leukemia. J. Clin. Invest. 2012;122:935–47. doi: 10.1172/JCI46465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ougolkov AV. Aberrant Nuclear Accumulation of Glycogen Synthase Kinase-3 in Human Pancreatic Cancer: Association with Kinase Activity and Tumor Dedifferentiation. Clinical Cancer Research. 2006;12:5074–81. doi: 10.1158/1078-0432.CCR-06-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J-S, Koenig A, Harrison A, Ugolkov AV, Fernandez-Zapico ME, Couch FJ, et al. Mutant K-Ras increases GSK-3β gene expression via an ETS-p300 transcriptional complex in pancreatic cancer. Oncogene. 2011;30:3705–15. doi: 10.1038/onc.2011.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wharry C. Constitutive non-canonical NFκB signaling in pancreatic cancer cells. Cancer Biol Therpay. 2009:1–20. doi: 10.4161/cbt.8.16.8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishina T, Yamaguchi N, Gohda J, Semba K, Inoue J-I. NIK is involved in constitutive activation of the alternative NF-κB pathway and proliferation of pancreatic cancer cells. Biochemical and Biophysical Research Communications. 2009;388:96–101. doi: 10.1016/j.bbrc.2009.07.125. [DOI] [PubMed] [Google Scholar]
- 38.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 39.Marchand B, Tremblay I, Cagnol S, Boucher MJ. Inhibition of glycogen synthase kinase-3 activity triggers an apoptotic response in pancreatic cancer cells through JNK-dependent mechanisms. Carcinogenesis. 2012;33:529–37. doi: 10.1093/carcin/bgr309. Oxford Univ Press. [DOI] [PubMed] [Google Scholar]
- 40.Taelman VF, Dobrowolski R, Plouhinec J-L, Fuentealba LC, Vorwald PP, Gumper I, et al. Wnt Signaling Requires Sequestration of Glycogen Synthase Kinase 3 inside Multivesicular Endosomes. Cell. 2010;143:1136–48. doi: 10.1016/j.cell.2010.11.034. Elsevier Inc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Busino L, Millman SE, Scotto L, Kyratsous CA, Basrur V, O'Connor O, et al. Fbxw7\alpha - and GSK3-mediated degradation of p100 is a pro-survival mechanism in multiple myeloma. Nat Cell Biol. 2012;14:1–12. doi: 10.1038/ncb2463. Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arabi A, Ullah K, Branca RMM, Johansson J, Bandarra D, Haneklaus M, et al. Proteomic screen reveals Fbw7 as a modulator of the NF-κB pathway. Nature Communications. 3:976–11. doi: 10.1038/ncomms1975. Nature Publishing Group; 1AD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanson JL. The Nuclear Factor B Subunits RelA/p65 and c-Rel Potentiate but Are Not Required for Ras-Induced Cellular Transformation. Cancer Research. 2004;64:7248–55. doi: 10.1158/0008-5472.CAN-03-3898. [DOI] [PubMed] [Google Scholar]
- 44.Steinbrecher KA, Wilson W, Cogswell PC, Baldwin AS. Glycogen Synthase Kinase 3 Functions To Specify Gene-Specific, NF- B-Dependent Transcription. Molecular and Cellular Biology. 2005;25:8444–55. doi: 10.1128/MCB.25.19.8444-8455.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Molecular Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. BioMed Central Ltd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Woodgett JR. Can a Two-Faced Kinase be Exploited for Osteosarcoma? JNCI Journal of the National Cancer Institute. 2012;104:722–3. doi: 10.1093/jnci/djs223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Hoon MJL, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–4. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 48.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–8. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 49.Smyth G. Limma: linear models for microarray data. Bioinformatics and computational biology solutions using R and Bioconductor. 2005:397–420. Springer. [Google Scholar]
- 50.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. (Series B (Methodological)).Journal of the Royal Statistical Society. 1995:289–300. JSTOR. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.