Abstract
It was appreciated early in drug discovery that the microbiota play an important role in the efficacy of therapeutic compounds. Indeed, the first antibiotic sulfa drugs were shown in the 1940s to be transformed by the bacteria that encode what we now call the intestinal microbiome. Here we briefly review the roles symbiotic bacteria play in the chemistry of human health, and we focus on the emerging appreciation that specific enzyme targets expressed by microbial symbiotes can be selectively disrupted to achieve clinical outcomes. We conclude that components of the microbiome should be considered “druggable targets,” and we suggest that our rapidly evolving understanding of the chemical biology of mammalian-microbial symbiosis will translate into improved human health.
The Microbiome and Human Health
The Human Microbiome Project has and will continue to revolutionize our appreciation of the intimate relationship between human systemic physiology and bacterial symbiosis [1]. In addition to outlining the number of microbial cells (100 trillion), microbial genes (8 million), and locations of predominant colonization, this consortium has brought into genetic granularity the gene products that enhance each side of the symbiotic equation. It is increasingly accepted that the microbiota are essential for several arenas of human health [2,3], including nutrition [4], neurobiology [5], cancer [6], immunology [4], cardiovascular disease [7], biliary function [8], irritable bowel disorders [9], and metabolic diseases like obesity [10] and diabetes [11]. Jeffrey I. Gordon at Washington University was an early [12,13]* and remains a consistently ardent contributor to our understanding of the roles specific bacterial species and bacterial genes play in mammalian health [14]. As such, his group and others continue to define the specific chemistry involved in the human-microbial axes of communication [15,16].
At the chemical level, bacterial symbiotes play necessary roles in carbohydrate metabolism, and glycosyl hydrolases and transferases are notably well represented in the microbiome [4]. In addition, the microbiota is required for the production of several essential vitamins, including B3, B5, B6, B12, K, biotin, and tetrahydrofolate, and in the absorption of iron from the intestinal lumen [4]. The processing of bile acids by intestinal bacteria has been linked to cardiovascular disease [8], and the GI microbiota produce short-chain fatty acids like acetate and butyrate that are critical to gut epithelial function and the systemic immune system [17]. Remarkably, it was recently shown that the acetates produced by intestinal bacteria find their way directly onto acetylated lysines in mammalian cells, and that bacterial-produced butyrates contribute to this process by inhibiting mammalian lysine deacetylase enzymes [18]*. The microbiome also appears to evolve in rapid and facile manner. It was found in 2010 that the enzyme beta-porphyranase encoded by marine micro-organisms had been acquired by the microbiome of Japanese individuals that consume porphyrins present in the red algae of their diet [19]. The reader is directed to the groups of Nicholson and Shanahan for their primary literature, as well as recent reviews [20,21]* that examine our growing appreciation of the chemical roles bacteria play in mammalian systems.
Two important papers that defined specific aspects of the chemical communication between the microbiota and mammalian cells were published in 2009. First, Wikoff and colleagues used mass spectrometry to elucidate how the intestinal microbiome contributes to chemical metabolites found in circulating plasma [22]**. They demonstrate in mice that there is “significant interplay between bacterial and mammalian metabolism” and point specifically to amino acid metabolites as notable, including the tryptophan-derived indole-3-propionic acid. This highlights specific chemistry performed by microbial gene products that modulates mammalian physiology. Second, Clayton and colleagues showed in 2009 that acetaminophen metabolism is directly impacted by p-cresol tyrosine metabolites produced by intestinal symbiotic bacteria [23]**. This provides a molecular link between the pharmacodynamics of a human therapeutic and the actions of specific components of the gut microbiome, and this link has been recently been deepened [24]. These are likely just a few of the firsts on what will be a long list of chemical interactions to be discovered between mammals and their microbiota.
The Microbiome and Drug Metabolism
Besides the sulfa drugs [25], at least two-dozen other therapeutic compounds have been shown to be processed by catalytic functions encoded by mammalian symbiotic bacteria. Excellent and comprehensive reviews of this topic were provided by Sousa and colleagues in 2008 [26]**, and more recently by Haiser and Turnbaugh in 2012 [7]. Because the GI contains the largest, most diverse and variable repository of bacterial species [1], this region has been the focus of past, and most likely future, studies on microbial drug metabolism. Reductions of bonds in clinical drugs performed by intestinal bacteria have been documented [26]**, as well as other transformations including hydrolysis, dehydroxylation, acetylation, deacetylation and deconjugation of glucuronides and sulfates [26]**.
Drug metabolism reactions performed by the microbiota can acutely impact human health. The bacterial transformation of the antiviral drug sorivudine, approved in Japan in 1993, was shown in 1998 to be the cause of the 18 deaths that lead to the removal of this drug from the market. Sorivudine’s key metabolite (E)-5-(2-bromovinyl)uracil (BVU) is produced by intestinal bacteria, and BVU then traffics back to the liver, where it inactivates a key liver enzyme that produces a lethal build up of 5-FU in cancer patients [27]. It has also been appreciated that the efficacy of drugs is positively impacted by their participation in entero-hepatic recirculation similar to that experienced by bilirubin. For example, the non-steroidal anti-inflammatory drug indomethacin is differentially processed by intestinal enzymes in model organisms like dogs and non-human primates relative to humans [28]. Taken together, it is clear that the intestinal microbiome plays an active role in drug metabolism, activation and reactivation. Indeed, it has been documented [26]** that this role is being incorporated as an early and essential component of the drug discovery and development pipeline.
Alleviating Microbiome-Induced Drug Toxicity
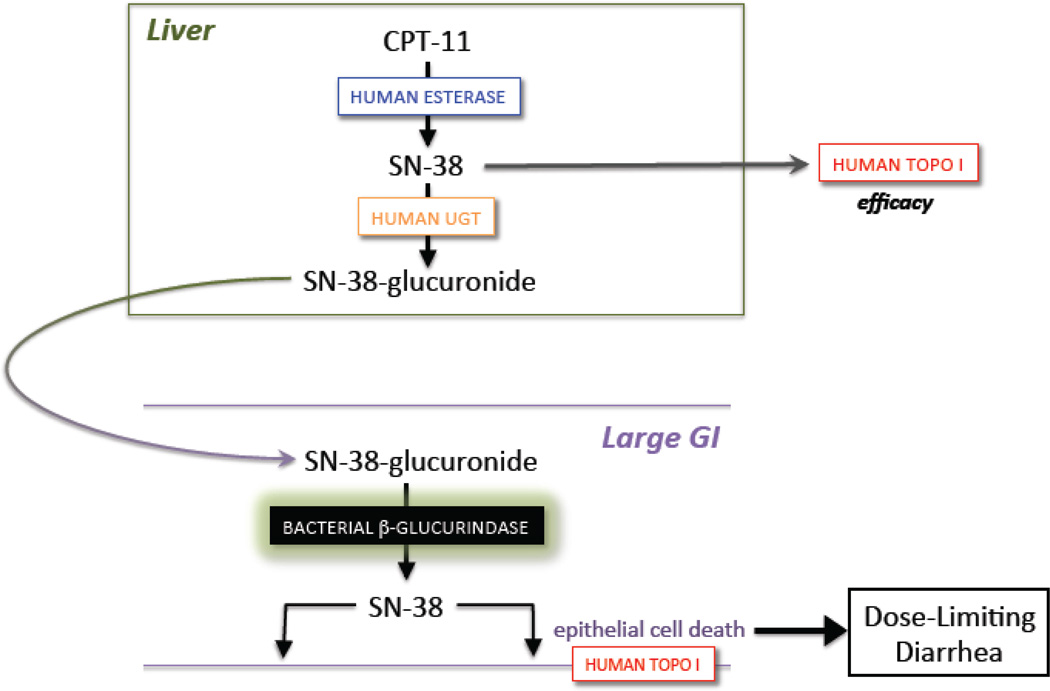
While the roles the microbiota play in therapeutic mechanisms of action and metabolism have been appreciated for decades, the selective targeting of a symbiotic bacterial factor was presented in 2010 by a team involving the authors of this commentary [29]**. The focus of this work was the anticancer drug CPT-11 (Irinotecan), which is a member of the camptothecin family of antineoplastic compounds [30]. The camptothecins target the eukaryotic type I topoisomerases [31] and trap those enzymes in a covalently linked state with DNA [32], which leads to cellular apoptosis (Figure 1A). While other drugs and sites of DNA damage can also impact the catalytic cycle of human topoisomerase I [33,34], two camptothecin derivatives, Topotecan and Irinotecan (CPT-11), are in wide-spread clinical use against a range of typically solid malignancies [30]. CPT-11 is a prodrug that is activated by non-specific esterases into SN-38, which poisons topoisomerase I. SN-38 is glucuronidated to SN-38G and cleared via the bile to the intestines as an inactive metabolite. However, it had been hypothesized that SN-38G was a substrate for bacterial β-glucuronidase enzymes encoded by the GI microbiome [35], and that this produced high lumenal concentrations of SN-38 that caused the dose-limiting toxicity of CPT-11 – severe delayed diarrhea (Figure 1A).
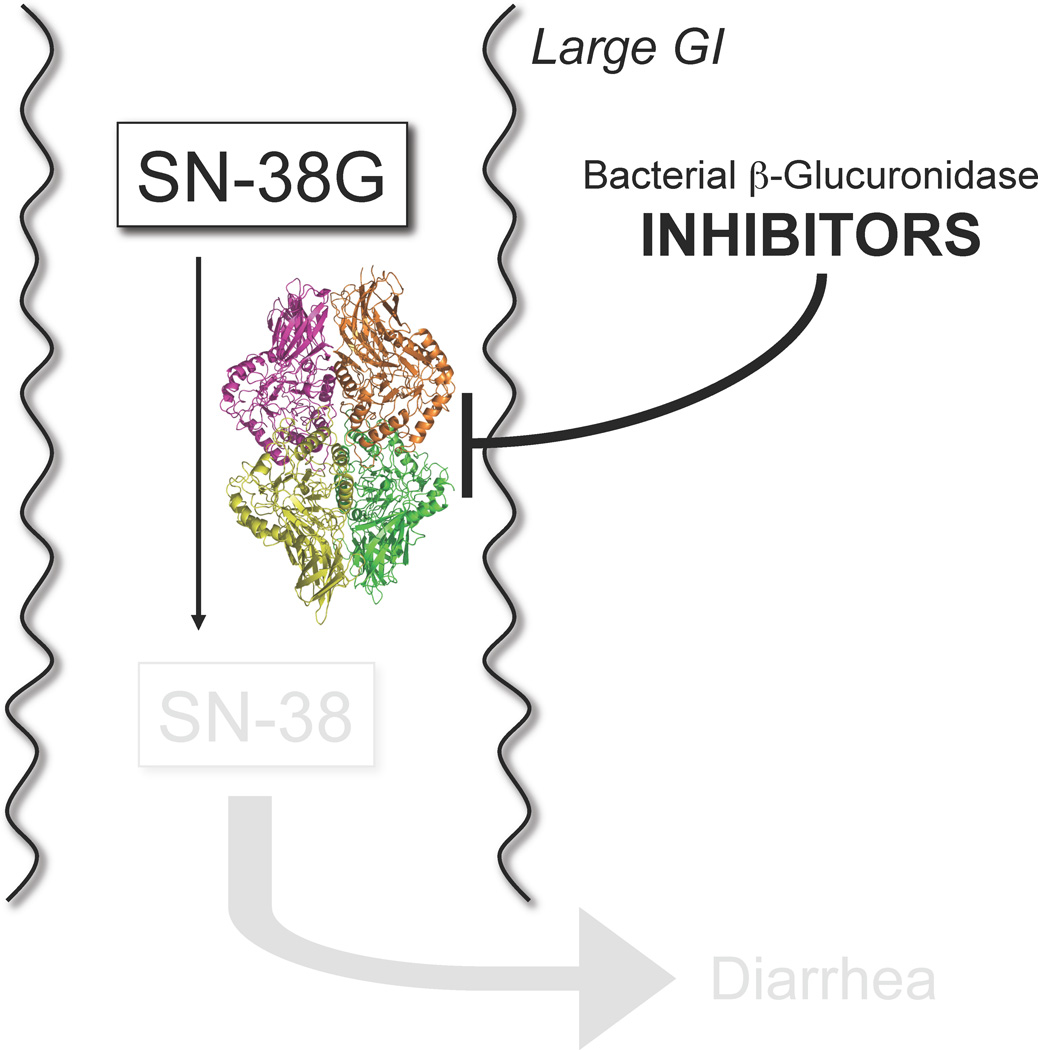
Figure 1.
A. The anticancer drug CPT-11 (Irinotecan) is a prodrug activated to the DNA topoisomerase I poison SN-38 by esterases, and SN-38 is further processed to its SN-38 glucuronide (SN-38G) metabolite by UDP-glucuronosyltransferase (UGT) enzymes. Excreted SN-38G is reactivated in the large intestines by bacterial microbiota β-glucuronidase enzymes; lumenal SN-38 contributes to the dose-limiting diarrhea associated with CPT-11 [29]**. B. Potent, selective and non-lethal bacterial β-glucuronidase inhibitors prevent the diarrhea and intestinal damage caused by the reactivation of SN-38G to SN-38 [29]**.
We identified novel, potent bacterial β-glucuronidase inhibitors that were selective for the microbial enzymes relative to the human orthologue from a high-throughput screen using purified E. coli β-glucuronidase [29]** (Figure 1B). The β-glucuronidase enzyme is non-essential to cultured E. coli, which apparently use it as a carbon-scavenging factor. The compounds reported demonstrate sub-micromolar Ki values using enzyme assays, and low nM EC50 values with a range of living bacterial species containing a β-glucuronidase gene. In addition, as demonstrated using structural and chemical biology studies, the hits examined were selective for the bacterial β-glucuronidases and not the mammalian form of the enzyme due to the presence of a 10–15 residue “bacterial loop” unique to the microbial proteins [29]**.
Importantly, these compounds were non-lethal to bacterial cells. While the use of antibiotics has been examined with variable results in efforts to reduce CPT-11-incuded toxicity [36], that approach would appear to be problematic given our emerging appreciation for the critical role symbiotic bacteria play in human health. Furthermore, the overuse of antibiotics [37] and the concomitant increase in microbial multidrug resistance [38] strongly supports the goal of employing chemical approaches that are neither bactericidal nor bacteriostatic. The β-glucuronidase inhibitors examined did not harm cultured human cell lines over several days of exposure at relatively high concentrations [29]**.
Oral dosing of one newly identified compound, Inhibitor 1, was shown to significantly reduce diarrhea and lower GI damage in mouse models of CPT-11-induced toxicity [29]**. These results help to confirm the hypothesis that the conversion of the inactive SN-38G metabolite into the active and GI epithelial damaging SN-38 compound in the intestinal lumen plays an important role in the dose-limiting toxicity of CPT-11 (Figure 1). As suggested [39], it will be important to determine what impact this approach will have on SN-38 pharmacokinetics and efficacy.
A subsequent study showed that this approach is not limited to CPT-11 [40]*. Non-steroidal anti-inflammatory drugs (NSAIDs) are some of the most widely used therapeutics in the developed world, and play important roles in treating pain and inflammation associated with lethal and non-lethal diseases. NSAIDs are typically associated with a risk of upper GI damage, particularly gastric ulcers. However, increasing data support the conclusion that NSAID use causes severe small intestinal damage and perforations that can be life-threatening [41]. The molecular bases of these effects remain unclear, but a “two hit” hypothesis has been offered that suggests that damage is first caused by the NSAID or an oxidative metabolite, and then is worsened by a secondary but acute inflammatory response [42].
The glucuronide metabolites of some NSAIDs were thought to have important roles in the small intestinal toxicity of this class of drugs (Figure 2). To test this hypothesis, a team including the authors of this review dosed mice receiving the NSAID diclofenac (DCF) with 10 µg Inhibitor 1 twice daily by oral gavage, the same scheme used for the CPT-11 animal study [29]**. This approach significantly reduced the small intestinal ulcers produced by DCF, which only appeared without Inhibitor 1 in regions of the small intestinal colonized by bacterial symbiotes [40]*. Thus, reducing the in situ reactivation of therapeutic metabolites in both the small and large intestine can play a large role in reducing GI damage. Although our understanding of NSAID processing by bacteria continues to evolve [43], these results help to confirm that hypotheses that the removal of the glucuronide sugar moieties placed on therapeutics by the mammalian UDP-glucuronosyltransferase primary phase II drug metabolism enzymes is involved in intestinal toxicity, and that this step can be selectively inhibited by targeted small molecule reagents.
Figure 2.
A. The non-steroidal anti-inflammatory drug (NSAID) diclofenac is processed to its glucuronide metabolite by UDP-glucuronosyltransferase (UGT) enzymes. Diclofenac-glucuronide is reactivated in the second half of the small intestines by β-glucuronidase enzymes expressed by the symbiotic microbiota [40]*. B. Potent, selective and non-lethal bacterial β-glucuronidase inhibitors prevent the small intestinal ulcerations caused by diclofenac’s (DCF’s) reactivation from diclofenac-glucuronide (DCF-G) [40]*.
Future Directions
It is increasingly clear that the human microbiome is an essential complement to the human genome, and that specific microbial enzyme targets can be selectively modulated for scientific and clinical purposes. As we understand the detailed roles played by the microbiota in a range of disease states related to neurobiology, immunology, heart disease, cancer, diabetes and other metabolic disorders, it is likely that several new and specific bacterial macromolecules will be identified as potential therapeutic targets. The ability to “drug” these members of the microbiome, however, will require some new capabilities. For example, targeting bacterial gene products in the GI will require drugs that preferentially remain in the intestinal lumen but are capable of reaching their molecular targets within microbial cells. These characteristics present both challenges and opportunities to drug development and delivery teams. In addition, the potential need to target gene products present only in select GI microbial species and not others may come into play in the future, which will require a deeper appreciation not only of the species present but of their detailed functional genomic and proteomic states.
Finally, we suggest the following three factors for future consideration. First, the role that the conjugative DNA transfer of genes within the microbiota, particularly those encoding virulence and antibiotic resistance properties, must be better understood. These processes play critical roles with respect to antibiotic resistance genes in the GI microbiome [44] and in clinical settings [45]. Thus, mobile plasmids and genetic elements must be clearly appreciated because they will lead to a constantly changing field of play. Indeed, evidence continues to emerge that conjugative plasmids are, like viruses, regulated by bacterial CRISPR-Cas systems [50], and furthermore that GI bacterial gene shuffling occurs based on fundamental influences like diet [19]. Second, the role that viruses, fungi and eukaryotic parasites play in the human-microbial landscape are under-appreciated to date [46]. Finally, while our understanding of the microbiome evolves, so does our ability to engineer synthetic microbial cells to serve as therapeutics, as has been demonstrated [47–49]. Thus, the future of modulating the human microbiome may include the use of designed bacterial cells that deliver the chemicals, genes or gene products necessary for disease management or treatment.
Highlights.
Microbial symbiotes significantly impact therapeutic chemistry
Specific enzyme targets in the microbiota can be potently and selectively disrupted
Inhibiting such target enzymes should be non-lethal to the microbiota
Modulating the human microbiome may improve drug efficacy and safety
ACKNOWLEDGEMENTS
The authors thank Sridhar Mani and Urs Boelsterli for our collaborations on the non-lethal targeting of microbial gene products. This work was supported by the NIH grant CA78924 (MRR).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
*papers of outstanding interest
**papers of exceptional interest
- 1.Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 3.Sonnenburg JL, Fischbach MA. Community health care: therapeutic opportunities in the human microbiome. Sci Transl Med. 2011;3:78ps12. doi: 10.1126/scitranslmed.3001626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonzalez A, Stombaugh J, Lozupone C, Turnbaugh PJ, Gordon JI, Knight R. The mind-body-microbial continuum. Dialogues Clin Neurosci. 2011;13:55–62. doi: 10.31887/DCNS.2011.13.1/agonzalez. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plottel CS, Blaser MJ. Microbiome and malignancy. Cell Host Microbe. 2011;10:324–335. doi: 10.1016/j.chom.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haiser HJ, Turnbaugh PJ. Is it time for a metagenomic basis of therapeutics? Science. 2012;336:1253–1255. doi: 10.1126/science.1224396. [DOI] [PubMed] [Google Scholar]
- 8.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marchesi JR, Holmes E, Khan F, Kochhar S, Scanlan P, Shanahan F, Wilson ID, Wang Y. Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. J Proteome Res. 2007;6:546–551. doi: 10.1021/pr060470d. [DOI] [PubMed] [Google Scholar]
- 10.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 11.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 13. Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291:881–884. doi: 10.1126/science.291.5505.881. These early data on the critical roles GI micriobiota play at the molecular level helped to establish the feasibility and importance of mammalian micriobiome research.
- 14.Gordon JI. Honor thy gut symbionts redux. Science. 2012;336:1251–1253. doi: 10.1126/science.1224686. [DOI] [PubMed] [Google Scholar]
- 15.Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, Knight R, Gordon JI. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe. 2009;6:279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koren O, Goodrich JK, Cullender TC, Spor A, Laitinen K, Backhed HK, Gonzalez A, Werner JJ, Angenent LT, Knight R, et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell. 2012;150:470–480. doi: 10.1016/j.cell.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crawford PA, Crowley JR, Sambandam N, Muegge BD, Costello EK, Hamady M, Knight R, Gordon JI. Regulation of myocardial ketone body metabolism by the gut microbiota during nutrient deprivation. Proc Natl Acad Sci U S A. 2009;106:11276–11281. doi: 10.1073/pnas.0902366106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Simon GM, Cheng J, Gordon JI. Quantitative assessment of the impact of the gut microbiota on lysine epsilon-acetylation of host proteins using gnotobiotic mice. Proc Natl Acad Sci U S A. 2012;109:11133–11138. doi: 10.1073/pnas.1208669109. Directly connects the functionally-critical post-translational acetylation of lysines to acetates and butyrates produced by the GI symbiotic bacteria.
- 19.Hehemann JH, Correc G, Barbeyron T, Helbert W, Czjzek M, Michel G. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464:908–912. doi: 10.1038/nature08937. [DOI] [PubMed] [Google Scholar]
- 20.Holmes E, Kinross J, Gibson GR, Burcelin R, Jia W, Pettersson S, Nicholson JK. Therapeutic modulation of microbiota-host metabolic interactions. Sci Transl Med. 2012;4:137rv136. doi: 10.1126/scitranslmed.3004244. [DOI] [PubMed] [Google Scholar]
- 21. Shanahan F. The gut microbiota-a clinical perspective on lessons learned. Nat Rev Gastroenterol Hepatol. 2012;9:609–614. doi: 10.1038/nrgastro.2012.145. Recent reviews on our emerging appreciation of the roles the microbiome play in mammalian physiology.
- 22. Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. Outlines specific chemistry performed by microbial gene products encoded that directly impact mammalian physiology.
- 23. Clayton TA, Baker D, Lindon JC, Everett JR, Nicholson JK. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci U S A. 2009;106:14728–14733. doi: 10.1073/pnas.0904489106. Demonstrates in quantitative detail the roles GI microbial symbiotes play in mammalian drug metabolism.
- 24.Maurice CF, Haiser HJ, Turnbaugh PJ. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell. 2013;152:39–50. doi: 10.1016/j.cell.2012.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fuller AT. Is p-aminobenzenesulphonamide the active agent of prontosil therapy? Lancet. 1937;1:194–198. [Google Scholar]
- 26. Sousa T, Paterson R, Moore V, Carlsson A, Abrahamsson B, Basit AW. The gastrointestinal microbiota as a site for the biotransformation of drugs. Int J Pharm. 2008;363:1–25. doi: 10.1016/j.ijpharm.2008.07.009. Comprehensive review covering the detailed roles symbiotic bacteria play in drug metabolism.
- 27.Okuda H, Ogura K, Kato A, Takubo H, Watabe T. A possible mechanism of eighteen patient deaths caused by interactions of sorivudine, a new antiviral drug, with oral 5-fluorouracil prodrugs. J Pharmacol Exp Ther. 1998;287:791–799. [PubMed] [Google Scholar]
- 28.Smith RV. Metabolism of drugs and other foreign compounds by intestinal microorganisms. World Rev Nutr Diet. 1978;29:60–76. doi: 10.1159/000400751. [DOI] [PubMed] [Google Scholar]
- 29. Wallace BD, Wang H, Lane KT, Scott JE, Orans J, Koo JS, Venkatesh M, Jobin C, Yeh LA, Mani S, et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science. 2010;330:831–835. doi: 10.1126/science.1191175. Demonstration that selective inhibitors of a microbial enzyme can alleviate the GI toxicity of the anticancer drug CPT-11.
- 30.Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat Rev Cancer. 2006;6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 31.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 32.Staker BL, Hjerrild K, Feese MD, Behnke CA, Burgin AB, Jr, Stewart L. The mechanism of topoisomerase I poisoning by a camptothecin analog. Proc Natl Acad Sci U S A. 2002;99:15387–15392. doi: 10.1073/pnas.242259599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chrencik JE, Burgin AB, Pommier Y, Stewart L, Redinbo MR. Structural impact of the leukemia drug 1-beta-D-arabinofuranosylcytosine (Ara-C) on the covalent human topoisomerase I-DNA complex. J Biol Chem. 2003;278:12461–12466. doi: 10.1074/jbc.M212930200. [DOI] [PubMed] [Google Scholar]
- 34.Lesher DT, Pommier Y, Stewart L, Redinbo MR. 8-Oxoguanine rearranges the active site of human topoisomerase I. Proc Natl Acad Sci U S A. 2002;99:12102–12107. doi: 10.1073/pnas.192282699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaneda N, Yokokura T. Nonlinear pharmacokinetics of CPT-11 in rats. Cancer Res. 1990;50:1721–1725. [PubMed] [Google Scholar]
- 36.Kehrer DF, Sparreboom A, Verweij J, de Bruijn P, Nierop CA, van de Schraaf J, Ruijgrok EJ, de Jonge MJ. Modulation of irinotecan-induced diarrhea by cotreatment with neomycin in cancer patients. Clin Cancer Res. 2001;7:1136–1141. [PubMed] [Google Scholar]
- 37.Blaser M. Antibiotic overuse: Stop the killing of beneficial bacteria. Nature. 2011;476:393–394. doi: 10.1038/476393a. [DOI] [PubMed] [Google Scholar]
- 38.Levy SB, Marshall B. Antibacterial resistance worldwide: causes, challenges and responses. Nat Med. 2004;10:S122–S129. doi: 10.1038/nm1145. [DOI] [PubMed] [Google Scholar]
- 39.Wu B. Use of physiologically based pharmacokinetic models to evaluate the impact of intestinal glucuronide hydrolysis on the pharmacokinetics of aglycone. J Pharm Sci. 2012;101:1281–1301. doi: 10.1002/jps.22827. [DOI] [PubMed] [Google Scholar]
- 40. LoGuidice A, Wallace BD, Bendel L, Redinbo MR, Boelsterli UA. Pharmacologic targeting of bacterial beta-glucuronidase alleviates nonsteroidal anti-inflammatory drug-induced enteropathy in mice. J Pharmacol Exp Ther. 2012;341:447–454. doi: 10.1124/jpet.111.191122. Selective targeting of a GI symbiotic microbial gene product significantly reduces small intestinal ulcers produced by an NSAID.
- 41.Scarpignato C, Hunt RH. Nonsteroidal antiinflammatory drug-related injury to the gastrointestinal tract: clinical picture, pathogenesis, and prevention. Gastroenterol Clin North Am. 2010;39:433–464. doi: 10.1016/j.gtc.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 42.Boelsterli UA, Redinbo MR, Saitta K. Multiple NSAID-induced hits injure the small intestine: Underlying mechanisms and novel strategies. Toxicol Sci. 2012 doi: 10.1093/toxsci/kfs310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu Y, Zhang QY. Role of intestinal cytochrome p450 enzymes in diclofenac-induced toxicity in the small intestine. J Pharmacol Exp Ther. 2012;343:362–370. doi: 10.1124/jpet.112.198077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sommer MO, Dantas G, Church GM. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science. 2009;325:1128–1131. doi: 10.1126/science.1176950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weigel LM, Clewell DB, Gill SR, Clark NC, McDougal LK, Flannagan SE, Kolonay JF, Shetty J, Killgore GE, Tenover FC. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science. 2003;302:1569–1571. doi: 10.1126/science.1090956. [DOI] [PubMed] [Google Scholar]
- 46.Kinross JM, Darzi AW, Nicholson JK. Gut microbiome-host interactions in health and disease. Genome Med. 2011;3:14. doi: 10.1186/gm228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nicaise C, Prozzi D, Viaene E, Moreno C, Gustot T, Quertinmont E, Demetter P, Suain V, Goffin P, Deviere J, et al. Control of acute, chronic, and constitutive hyperammonemia by wild-type and genetically engineered Lactobacillus plantarum in rodents. Hepatology. 2008;48:1184–1192. doi: 10.1002/hep.22445. [DOI] [PubMed] [Google Scholar]
- 48.Jones ML, Chen H, Ouyang W, Metz T, Prakash S. Microencapsulated Genetically Engineered Lactobacillus plantarum 80 (pCBH1) for Bile Acid Deconjugation and Its Implication in Lowering Cholesterol. J Biomed Biotechnol. 2004;2004:61–69. doi: 10.1155/S1110724304307011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takiishi T, Korf H, Van Belle TL, Robert S, Grieco FA, Caluwaerts S, Galleri L, Spagnuolo I, Steidler L, Van Huynegem K, et al. Reversal of autoimmune diabetes by restoration of antigen-specific tolerance using genetically modified Lactococcus lactis in mice. J Clin Invest. 2012;122:1717–1725. doi: 10.1172/JCI60530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Westra ER, Staals RH, Gort G, Neumann S, de la Cruz F, Fineran PC, Brouns SJJ. CRISPR-Cas systems preferentially target the leading regions of MOBF conjugative plasmids. RNA Biology. 2013;10:1–13. doi: 10.4161/rna.24202. [DOI] [PMC free article] [PubMed] [Google Scholar]