Abstract
We describe a new computer program, SnpEff, for rapidly categorizing the effects of variants in genome sequences. Once a genome is sequenced, SnpEff annotates variants based on their genomic locations and predicts coding effects. Annotated genomic locations include intronic, untranslated region, upstream, downstream, splice site, or intergenic regions. Coding effects such as synonymous or non-synonymous amino acid replacement, start codon gains or losses, stop codon gains or losses, or frame shifts can be predicted.
Here the use of SnpEff is illustrated by annotating ~356,660 candidate SNPs in ~117 Mb unique sequences, representing a substitution rate of ~1/305 nucleotides, between the Drosophila melanogaster w1118; iso-2; iso-3 strain and the reference y1; cn1 bw1 sp1 strain. We show that ~15,842 SNPs are synonymous and ~4,467 SNPs are non-synonymous (N/S ~0.28). The remaining SNPs are in other categories, such as stop codon gains (38 SNPs), stop codon losses (8 SNPs), and start codon gains (297 SNPs) in the 5′UTR. We found, as expected, that the SNP frequency is proportional to the recombination frequency (i.e., highest in the middle of chromosome arms). We also found that start-gain or stop-lost SNPs in Drosophila melanogaster often result in additions of N-terminal or C-terminal amino acids that are conserved in other Drosophila species. It appears that the 5′ and 3′ UTRs are reservoirs for genetic variations that changes the termini of proteins during evolution of the Drosophila genus.
As genome sequencing is becoming inexpensive and routine, SnpEff enables rapid analyses of whole-genome sequencing data to be performed by an individual laboratory.
Keywords: Drosophila melanogaster, Personal Genomes, next generation DNA sequencing, whole-genome SNP analysis
Introduction
When we re-sequenced the w1118; iso-2; iso-3 genome in 2009,1 bioinformatics tools were unable to rapidly categorize the ~356,660 SNPs as comparing to the y1; cn1 bw1 sp1 reference strain. The available tools at the time such as ENSEMBL’s variant web application (ensembl.org) could only analyze a few hundred to a few thousand SNPs per batch. Therefore, over the past couple of years, we have been developing a new program called SnpEff (an abbreviation of “SNP effect”) which is able to analyze and annotate thousands of variants per second and predict their possible genetic effects.
In this study, we analyze the output of SnpEff (version 1.9.6) analyses of the ~356,660 candidate SNPs that we identified in w1118; iso-2; iso-3 with respect to the y1; cn1 bw1 sp1 reference strain as reported in our previous paper.1 This is of great interest to the Drosophila community because thousands of transposon insertion stocks5 and hundreds of deficiency stocks6,7 were generated in the w1118; iso-2; iso-3 genetic background. The presence of large number and potential severity of many SNPs in the two laboratory strains was a surprising finding and the possible evolutionary implications of this are discussed.
Program description
Genomic variants comprise single nucleotide polymorphism (SNPs), insertions and deletions (INDELs), and multiple nucleotide polymorphisms (MNPs). SnpEff annotates these variants based on their genomic locations, such as intronic, untranslated region (5′ UTR or 3′UTR), upstream, downstream, splice site, or intergenic regions. It also predicts coding effects such as synonymous or non-synonymous amino acid replacement, start codon gains or losses, stop codon gains or losses, or frame shifts. Predicted effects are with respect to protein coding genes. Variants affecting non-coding genes are annotated and the corresponding bio-type is identified, whenever the information is available.
SnpEff (snpeff.sourceforge.net) is open source, platform independent and freely available for all users. The main features of SnpEff include: (1) speed - the ability to make thousands of predictions per second; (2) flexibility - the ability to add custom genomes and annotations; (3) web-based - the ability to integrate with Galaxy, an open access and web-based platform for computational bioinformatic research (gmod.org/wiki/Galaxy); (4) multi-organism - compatibility with multiple species and multiple codon usage tables (e.g., mitochondrial genomes); (5) integration with Broad's Genome Analysis Toolkit (GATK)4; and (6) ability to perform non-coding annotations.
Before using SnpEff, a variants file must be generated using sequencing information. A variants file lists all SNPs, MNPs, and INDELs found by re-sequencing a genome. A simple walk-through example on how to analyze sequencing data to calculate variants and their effects using SnpEff is shown in Listing SL1. In a nutshell, the analysis pipeline has three steps: (i) map the reads to the genome, (ii) call variants and (iii) use SnpEff to annotate variants. This example is intended for illustration purposes only since it omits many routine steps used in re-sequencing data analysis pipelines.
In addition to SnpEff, there are other recently developed programs for annotating genomic variants, most notably “Annotate Variation” (ANNOVAR)2 and “Variant Annotation, and Analysis and Search Tool” (VAAST).3 However, SnpEff differs from these programs in that it is an open source for all users, permits annotation of more genome versions, natively supports Variant Call Format (VCF)9 files and it is marginally faster (although the speeds of SnpEff, ANNOVAR, and VAAST are comparable). Table S1 shows a feature comparison of some currently available software packages.
At this time, SnpEff has been set up for annotating DNA polymorphisms of over 320 genome versions of multiple species including the human genome. Sources of information for creating these databases are ENSEMBL, UCSC as well as organism specific databases, such as FlyBase (Drosophila melanogaster), WormBase (C. elegans) and TAIR (Arabidopsis thaliana). Additional genomes can be added by the user or provided upon request.
Detailed information on how to download, install, and run, as well as usage examples of the program can be found at the website (snpeff.sourceforge.net), also a frequently asked questions (FAQ) section that addresses most issues that a user might have in operating this program.
SnpEff has already been used by over 50 institutions and universities in the bioinformatics community. Rapid analyses of whole-genome sequencing data should now be feasible to perform by an individual laboratory.
Results
Table 1 shows the beginning portion of the output generated by SnpEff when the SNPs in w1118; iso-2; iso-3 were compared with the reference genome, y1; cn1bw1sp1(Drosophila melanogaster release 5.3). A complete list of SnpEff output is shown in Table 2. We published a list of variant for w1118; iso-2; iso-3 in our previous paper1 and it was derived by comparing hundreds of millions of short sequence reads (~20-fold genome coverage)8
Table 1. Output of SnpEff.
| # SNP |
Gene_name |
Effect |
Old_AA/new_AA |
Old_codon/New_codon |
Codon_Num (CDS) |
CDS_size |
| chr2L:10006682_C/T |
CG31755 |
UPSTREAM: 541 bases |
|
|
|
|
| chr2L:10006758_G/A |
CG31755 |
UPSTREAM:465 bases |
|
|
|
|
| chr2L:10007289_G/A |
CG4747 |
SYNONYMOUS_CODING |
L/L |
TTG/TTA |
489 |
1809 |
| chr2L:10007319_G/C |
CG4747 |
SYNONYMOUS_CODING |
G/G |
GGG/GGC |
499 |
1809 |
| chr2L:10007356_A/T |
CG4747 |
INTRON |
|
|
|
1809 |
| chr2L:10007363_T/A | CG4747 | INTRON | 1809 |
SNP, a description of the single nucleotide polymorphism (SNP) indicating chromosome arm (chr2L), coordinate in genome (10006682), and nucleotide change (e.g., C/T indicates that C is replaced by T in w1118; iso-2; iso-3 at this position). Gene_name, official gene symbol of gene. Effect, description of SNP (e.g., upstream of transcription start site at position -541). Old_AA/new/AA, amino acid change, if any, in one letter code. Old_codon/New_codon, if a codon contains a SNP, the old (reference) and new (w1118; iso-2; iso-3) codons are indicated. Codon_Num(CDS), the codon number of the coding sequence (CDS). CDS_size, the size of the protein in amino acids.
Table 2. Detailed effect list from SnpEff.
| Effect |
Note |
| INTERGENIC |
The variant is in an intergenic region |
| UPSTREAM |
Upstream of a gene (default length: 5K bases) |
| UTR_5_PRIME |
Variant hits 5′UTR region |
| UTR_5_DELETED |
The variant deletes an exon which is in the 5′UTR of the transcript |
| START_GAINED |
A variant in 5′UTR region produces a three base sequence that can be a START codon. |
| SPLICE_SITE_ACCEPTOR |
The variant hits a splice acceptor site (defined as two bases before exon start, except for the first exon). |
| SPLICE_SITE_DONOR |
The variant hits a Splice donor site (defined as two bases after coding exon end, except for the last exon). |
| START_LOST |
Variant causes start codon to be mutated into a non-start codon. |
| SYNONYMOUS_START |
Variant causes start codon to be mutated into another start codon. |
| CDS |
The variant hits a CDS. |
| GENE |
The variant hits a gene. |
| TRANSCRIPT |
The variant hits a transcript. |
| EXON |
The vairant hist an exon. |
| EXON_DELETED |
A deletion removes the whole exon. |
| NON_SYNONYMOUS_CODING |
Variant causes a codon that produces a different amino acid |
| SYNONYMOUS_CODING |
Variant causes a codon that produces the same amino acid |
| FRAME_SHIFT |
Insertion or deletion causes a frame shift |
| CODON_CHANGE |
One or many codons are changed |
| CODON_INSERTION |
One or many codons are inserted |
| CODON_CHANGE_PLUS_CODON_INSERTION |
One codon is changed and one or many codons are inserted |
| CODON_DELETION |
One or many codons are deleted |
| CODON_CHANGE_PLUS_CODON_DELETION |
One codon is changed and one or more codons are deleted |
| STOP_GAINED |
Variant causes a STOP codon |
| SYNONYMOUS_STOP |
Variant causes stop codon to be mutated into another stop codon. |
| STOP_LOST |
Variant causes stop codon to be mutated into a non-stop codon |
| INTRON |
Variant hist and intron. Technically, hits no exon in the transcript. |
| UTR_3_PRIME |
Variant hits 3′UTR region |
| UTR_3_DELETED |
The variant deletes an exon which is in the 3′UTR of the transcript |
| DOWNSTREAM |
Downstream of a gene (default length: 5K bases) |
| INTRON_CONSERVED |
The variant is in a highly conserved intronic region |
| INTERGENIC_CONSERVED | The variant is in a highly conserved intergenic region |
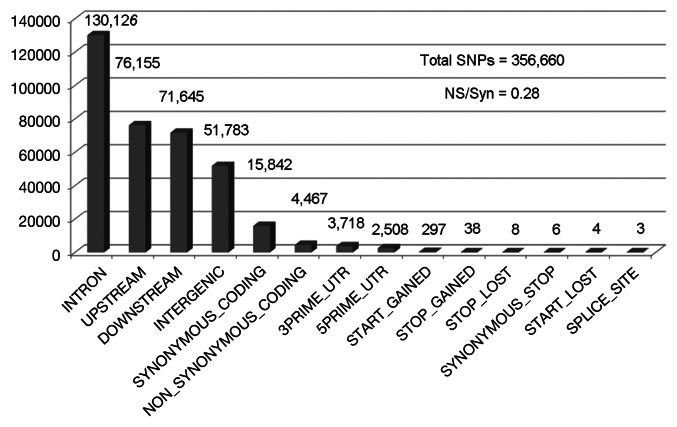
Heterozygosity is not considered in the w1118; iso-2; iso-3 sequence because the stock was isogenized and only high quality (i.e., homozygous SNPs) were used for this analysis.1 According to SnpEff (version 1.9.6), the largest number of SNPs in w1118; iso-2; iso-3 are in introns (130,126) followed by those in upstream (76,155), downstream (71,645) and intergenic (51,783) regions (Fig. 1). “Upstream” is defined as 5 kilobase (kb) upstream of the most distal transcription start site and “downstream” is defined as 5 kb downstream of the most distal polyA addition site, but these default variables can be easily adjusted. SnpEff also found thousands of SNPs within the transcribed regions of genes. For example, there are 3,718 SNPs in the 3′ untranslated regions (3′ UTR), and 2,508 SNPs in the 5′ UTR. The SNPs in the upstream, downstream, 5′ and 3′ UTR regions might affect transcription or translation, but the actual effects have to be confirmed case-by-case. In the next few sections, we present examples of several types of SNPs that might affect the protein function.
Figure 1. Classification of SNPs in w1118; iso-2; iso-3. The number of NSPs in each class is shown above the bar. The quality score was arbitrarily set at 70 and above for this graph.
SNPs that generate new start codons
There are 297 SNPs that potentially generate a new translation initiation codon in the 5′ UTR (referred to as start-gain SNPs). The most common translation initiation codon is AUG, which is coded by ATG in the genome. To be thorough, we also included CUG and UUG codons, which code for leucine, as these codons can also be used to initiate translation in rare genes in Drosophila and mammals.10,11 There are 60 genes with ATG start-gain SNPs (Table 5), 99 genes with CTG start-gain SNPs, and 120 genes with TTG start-gain SNPs in w1118; iso-2; iso-3, all by definition in 5′ UTR regions, compared with the reference genome (the reading frame is indicated on the SnpEff table). Most of the ATG start-gain SNPs are within 1 kb of the annotated translation start (Table 5), but this probably reflects the fact that most 5′ UTR sequences are less than 1 kb long. Less than expected by chance, only ~25% of the ATG start-gain SNPs are in the same open reading frame (ORF) as the annotated ORF (Table 5). Since 33% of the ATG start-gain SNPs are expected to be in-frame by chance, there might be a weak selection against this class of start-gain SNPs. Of the 60 genes with ATG start-gain SNPs, five genes have two ATG start-gain SNPs and one gene has 3 start-gain SNPs; the remaining 54 genes have a single start-gain SNP. Since SnpEff does not take into account of the Kozak consensus sequence flanking the AUG site, 5′ACC-AUG-G-3′, that is generally required for efficient translation,12 further assessment is required to determine whether a start-gain SNP is actually used.
Table 5. 60 Genes with Start-gaineded SNPs with ATGs.
| Gene_name |
bases from TSS |
Gene_name |
bases from TSS |
Gene_name |
bases from TSS |
| a |
386 (-) |
CG4766 |
367 (-) |
MESR3 |
454 (-) |
| Ace |
652 (-) |
CG4839 |
293 (-) |
Mipp2 |
67 (-) |
| Axn |
107 (-) |
CG5103 (2) |
104/17 (−/−) |
osp |
358 (-) |
| btsz |
228 (+) |
CG6024 |
269 (-) |
p120ctn |
119 (-) |
| Calx |
582 (+) |
CG7985 |
60 (+) |
Pld |
144 (+) |
| CAP |
1224 (+) |
CG8026 |
612 (+) |
Pli |
196 (-) |
| CG10186 |
402 (+) |
CG8176 |
128 (-) |
Pvr (2) |
472/915 (-/+) |
| sesn |
147 (+) |
cpo |
168 (+) |
pxb (2) |
50/76 (−/−) |
| CG12355 |
151 (-) |
dac |
103 (-) |
rib |
2 (-) |
| CG13802 (3) |
490/575/635 (−/−/-) |
dpr15 |
433 (-) |
rn |
142 (-) |
| haf |
89 (-) |
EcR |
160 (-) |
Samuel |
517 (-) |
| CG15086 |
114 |
Eip63E |
171 (+) |
sli |
307 (-) |
| CG15878 |
52 (-) |
fdl (2) |
307/437 (−/−) |
so |
5252 (-) |
| CG18522 |
40 (-) |
frtz |
196 (-) |
Sobp |
24 (+) |
| CG30419 |
253 (-) |
GC |
76 (-) |
sprt |
358 (-) |
| CG31163 |
998 (-) |
Gug |
70 (-) |
Strn-Mlck (2) |
210/228 (+/+) |
| Dscam3 |
269 (-) |
inv |
771 (+) |
tai |
203 (-) |
| CG31688 |
430 (-) |
Ipk1 |
376 (-) |
vn |
1793 (-) |
| CG32048 |
63 (+) |
klu |
576 (+) |
wg |
231 |
| CG32150 | 747 (+) | Mbs | 10 (-) | Wnt4 | 680 (-) |
Bases from TSS, bases from translation start site not including the ATG start-gained SNP. (+), in same reading frame as annotated ATG. (-), in different reading frame as annotated ATG.
Gene ontology (GO) pathway analysis of the genes affected by the 297 start-gain SNPs in w1118; iso-2; iso-3 was done using DAVID (Database for Annotation, Visualization and Integrated Discovery).13,14 We found that the GO categories “tissue morphogenesis,” “immunoglobulin like,” “developmental protein,” and “alternative splicing” are significantly enriched after multiple-comparisons correction by false-discovery rate (FDR < 0.001; Table 6). These categories are interesting because they predominantly contain proteins that show a wide degree of intra- and interspecies variability. For example, the immunoglobulin loci, which are highly divergent among humans and other vertebrates, are used for antigen recognition.15 Also, developmental proteins and proteins involved in tissue morphogenesis often have both conserved domains such as the Hox domain, and highly divergent domains such as the trans-activation domains.16,17
Table 6. Genes with Start-Gained SNP GO categories in w1118; iso-2; iso-3. Results of Gene ontology analysis for 297 start-gained SNPs in w1118; iso-2; iso-3. Bottom, the genes in the indicated gene ontology category is listed.
| Term |
Count |
% |
PValue |
List Total |
Pop Hits |
Pop Total |
Fold Enrichment |
Bonferroni |
Benjamini |
FDR |
| tissue morphogenesis |
21 |
8.898305 |
2.07E-08 |
147 |
247 |
7937 |
4.590515 |
2.37E-05 |
2.37E-05 |
3.33E-05 |
| immunoglobulin-like |
16 |
6.779661 |
3.40E-08 |
198 |
132 |
10196 |
6.241812 |
1.42E-05 |
7.08E-06 |
4.77E-05 |
| developmental protein |
29 |
12.28814 |
2.75E-07 |
229 |
540 |
12980 |
3.043992 |
3.99E-05 |
3.99E-05 |
3.27E-04 |
| alternative splicing |
31 |
13.13559 |
3.82E-07 |
229 |
616 |
12980 |
2.852464 |
5.53E-05 |
2.77E-05 |
4.53E-04 |
| | ||||||||||
| tissue morphogenesis |
FRTZ, NRX-IV, ESG, WG, PBL, SFL, MBS, RIB, TOW, WNT4, FORM3, SLI, EIP63E, PHL, YRT, FAS, SRC64B, TWI, DLG1, BTSZ, HS6ST |
|||||||||
| immunoglublin-ilke |
CG31814,DPR15, PVR, DPR16, CG14521, KLG, VN, CG12484, BEAT-IB, CG10186, DPR2, STRN-MLCK, CG34371, KEK5, FAS, CG15630 |
|||||||||
| developmental protein |
VN, ESG, DEI, INV, DAB, AWH, SCRIB, BICC, MST87F, WNT4, RIG, SLI, NUMB, PIP, INE, TWI, DLG1, FOXO, PTP10D, WG, AXN, EIP74EF, BUN, SO, FZ2, FDL, SCYL, SRC64B, POXN |
|||||||||
| alternative splicing | CPO, CPN, ECR, VN, CG11299, RN, DAB, AWH, SCRIB, INX7, SLI, PIP, NRV2, INE, DLG1, L(1)G0196, CG32048, FOXO, PTP10D, CYCT, WG, EIP74EF, BUN, CG13624, GLUT1, OSP, FDL, SSP4, PHL, SCYL, RDGC | |||||||||
An example of a start-gain SNP is in the 5′UTR of Ecdysone inducible protein 63E (Eip63E) gene, which is predicted to be a cyclin J dependent kinase required for oogenesis and embryonic development (Fig. 2).21 The potential start-gain SNP (A > G) in Eip63E changes 5′-ATA-3′ to 5′-ATG-3′ in the same reading frame with no intervening stop codons (Fig. 2a). If translation occurs at the new start-gain SNP, it would produce a protein with 57 additional N-terminal amino acids compared with the reference gene (Fig. 2b). However, the three bases prior to the new 5′-ATG-3′ sequence, 5′-AAT-3′, is a poor match to the Kozak consensus sequence, 5′-ACC-3′, discussed above.12 Therefore, it is unclear whether the start-gain SNP in Eip63E is recognized by the translational machinery.
Figure 2. Analysis of Eip63E start-gained SNP in w1118; iso-2; iso-3. (A) Location of the start-gained SNP at the Eip63E locus. Notice that the reading frame is the same as the normal translation start site (TSS). (B) Conservation of 60 amino acid N-terminal region of Eip63E in w1118; iso-2; iso-3 with Drosophila yakuba orthologous gene. The other sequenced Drosophila species do not have this N-terminal sequence (not shown).
It is interesting that a BLASTp search of the protein database reveals that the N-terminal 57 amino acids in Eip63E are 63% identical (36/57) to the 58 N-terminal amino acids of the orthologous gene in Drosophila yakuba, but not to any other Drosophila species. This suggests that the 5′ UTR of Eip63E might be a source for genetic variations encoding novel N-terminal protein sequences that potentially modulates protein function (see Discussion).
SNPs that generate new stop codons or loss of stop codons
Another surprise in our SnpEff analysis was the identification of 28 stop-gain SNPs and 5 stop-lost SNPs in w1118; iso-2; iso-3 (Table 7). A stop-gain SNP, classically called a nonsense SNP, has a coding codon changed to a stop codon, UAA, UAG, UGA.22 Three genes, oc/otd, LRP1, and trol9, have two stop-gain SNPs. Surprisingly at least 8 of the stop-gain SNPs are in genes that encode essential proteins and these are Dif, dp, ex, MESR4, mew, oc/otd, tai, and trol. It is not known whether the other stop-gain SNPs also affect essential protein-coding genes because their functions have not yet been characterized (according to www.flybase.org). We note that a stop-gain SNP in w1118; iso-2; iso-3 would be a stop-lost SNP in the reference strain and vice versa because the ancestral Drosophila melanogaster strain that gave rise to both of these strains is not known.
Table 7. Stop gained and stop lost in w1118; iso-2; iso-3.
|
stop gained |
location |
length |
phenotype |
stop gained |
location |
length |
phenotype |
||
| ade3 |
255K/* |
435 |
ND a |
ex |
693Q/* |
1428 |
Lethal d |
||
| CG10126 |
11W/* |
228 |
ND a |
lbk |
1130Y/* |
1174 |
ND d |
||
| CG15394 |
120Q/* |
186 |
ND a |
MESR4 |
1509E/* |
2072 |
lethal d |
||
| CG31145 |
27L/* |
764 |
ND a |
mew |
752Q/* |
1050 |
Lethal a |
||
| CG31784 |
1049Q/* |
1078 |
ND a |
NFAT |
12G/* |
1420 |
ND d |
||
| CG32115 |
468W/* |
476 |
ND a |
oc/otd |
389Y/*, 453Y/* |
543 |
Lethal d |
||
| LRP1 |
2917Y/*, 2918E/* |
4700 |
ND a |
Pde9 |
255C/* |
1527 |
ND a |
||
| CG34006 |
121R/* |
202 |
ND b |
rho-4 |
140W/* |
418 |
ND a |
||
| CG34326 |
49Y/* |
84 |
ND c |
Synd |
375S/* |
495 |
ND a |
||
| CG3493 |
1419E/* |
1490 |
ND a |
tai |
1420Q/* |
2048 |
Lethal d |
||
| CG3964 |
509Y/* |
983 |
ND a |
trol |
811Y/*, 808E/* |
4180 |
Lethal a |
||
| CG4068 |
379Q/* |
623 |
ND d |
stop lost |
|
|
|
||
| CG7236 |
70E/* |
502 |
ND a |
GABA-B-R1 |
*/L (+9 aa) |
837 |
ND a |
||
| Cht6 |
4175L/* |
4542 |
ND a |
CG13958 |
*/G (+8 aa) |
539 |
ND a |
||
| Cyp4s3 |
260W/* |
496 |
ND a |
CG4975 |
*/Q (+1aa) |
353 |
ND a |
||
| Dif |
263C/* |
668 |
lethal a |
bw |
*/Q (+71 aa) |
417 |
eye color c |
||
| dp | 17353L/* | 22972 | Lethal a | CG14755/pdm3 | */Q (+5 aa) | 285 | ND a | ||
Stop gained, gene with stop gained SNP. Location, amino acid number changed to a stop codon (e.g., 255K/*, indicates lysine at amino acid changed to a stop codon). Length, the length of the protein in amino acids. Phenotype, not determined (ND), withdrawn (no longer considered a gene by FlyBase), and NPC (non-protein coding, such as a rRNA). For stop lost SNPs (bottom), */L (+9 aa) indicates that the next in frame stop is after nine additional amino acids are added. a-d, refer to SNP categories 1–4 (see text).
An important consideration with stop-gain SNPs is whether the expanded C-terminal amino acids in the longest version of the protein are conserved in other Drosophila species. If the additional C-terminal amino acids are not conserved, then these amino acids might not affect the essential function of the protein but they might exert modulatory effects. If the additional C-terminal amino acids are conserved in multiple Drosophila species, then their loss might adversely affect the function of the protein. Therefore, in Table 7, we further classify the stop-gain and stop-lost SNPs into four categories: Category 1, including 23 genes, with both the N-terminal and novel C-terminal tails conserved among Drosophila species and other organisms; Category 2, including only one gene, with the entire gene sequence not conserved even among other Drosophila species. This gene is therefore probably not a functional gene; Category 3, including two genes, with novel C-termini not conserved among other Drosophila species. In this category, the N-termini are conserved among Drosophila species, but this conservation is not maintained beyond the Drosophila genus. Genes in this class are likely novel genes that arose in the Drosophila genus); and Category 4, including seven genes, with novel C-termini conserved among other Drosophila species but not beyond the Drosophila genus. In this category, the N-terminus is conserved beyond the Drosophila genus. Genes in this class probably have C-terminal domains that exert modulatory roles in the Drosophila genus but not beyond the genus).
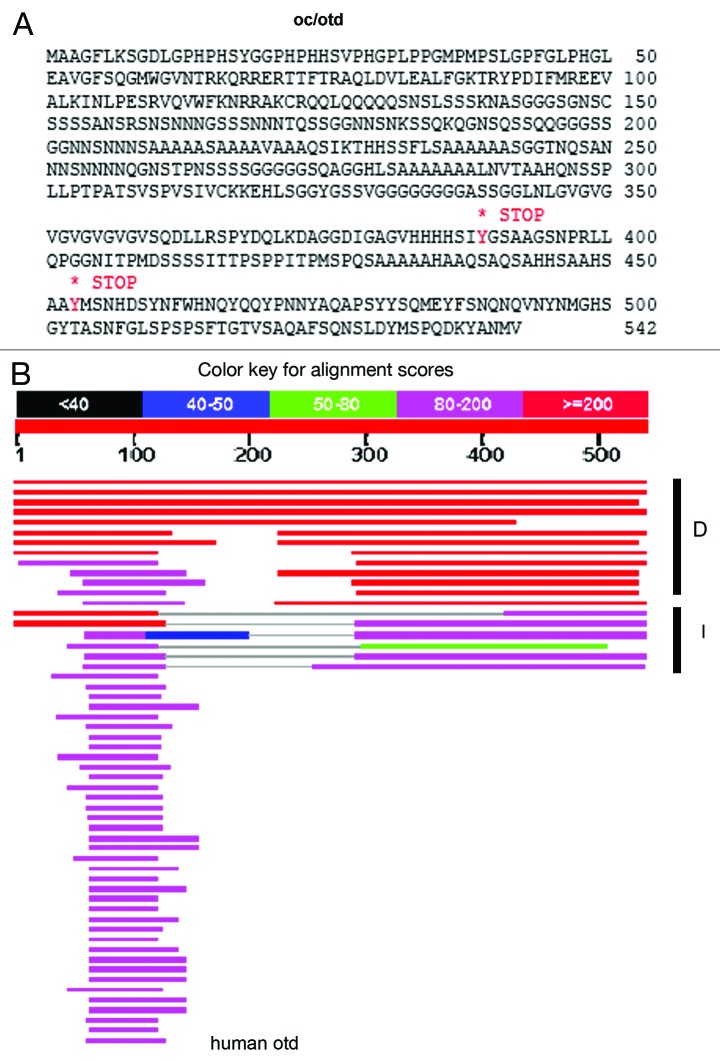
An example of an essential protein-coding gene in Category 4, where the novel C-terminus is not conserved outside the Drosophila genus, is oceliless (oc), also known as orthodenticle (otd) (Fig. 3). The oc/otd gene is a Hox-family transcription factor required for photoreceptor development in the compound eye and the light-sensing ocellus, embryonic development, and brain development segmentation.23,24 The Hox domain contains 60 amino acids, 59 of which are identical with the human Otd protein. The Hox domains, which arose before invertebrates and vertebrates split several hundred million years ago, are among the most conserved protein domains in bilaterally-symmetric organisms in evolution.25 The two stop-gain SNPs in w1118; iso-2; iso-3 are in the non-conserved C-terminal region of Oc/Otd, which is thought to have a transcriptional-regulatory function. Since both strains are viable, both oc/otd genes are apparently functional although they encode a protein with 489 amino acids in w1118; iso-2; iso-3, and a protein with 543 amino acids in the reference genome (Table 6).
Figure 3. Oc/Otd has two stop-gained SNPs in w1118; iso-2; iso-3. (A) Location of the two stop gained SNPs in oc/otd. (B) Protein BLAST of Oc/Otd against the non-redundant (nr) protein database shows that only the 60 amino Hox domain flanking amino acid 100 is conserved from Drosophila to humans. The color coding shows the alignment scores.
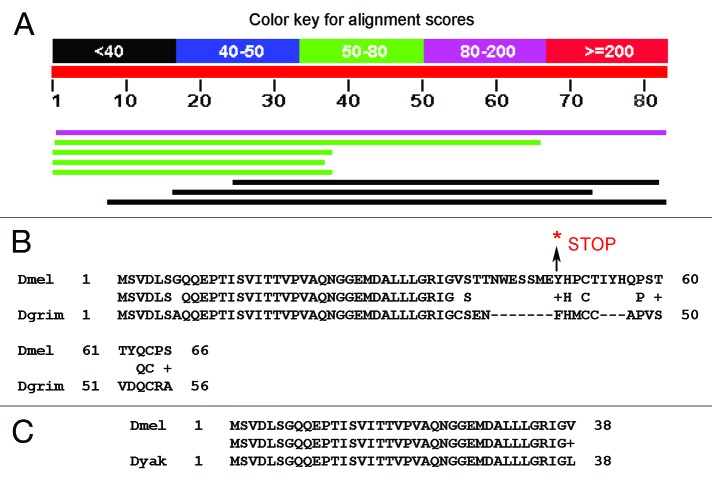
An example of a stop-lost gene in Category 3 c, where the C-terminus is not conserved among the Drosophila species, is CG34326 that encodes a protein of unknown function (Fig. 4). In w1118; iso-2; iso-3, CG34326 encodes a protein of 48 amino acids but in the reference genome it encodes a protein with 84 amino acids. When BLASTp was done with the non-redundant (nr) data set, there was not much homology beyond the 38th amino acid within the Drosophila genus. However, there was a near perfect (37/38) identity of the first 38 amino acids in four other Drosophila species: Drosophila grimshawi, Drosophila yakuba, Drosophila erecta, and Drosophila virilus (Fig. 4). This protein likely arose in the Drosophila genus since it has no known homologs outside of this genus.
Figure 4. CG34326 has one stop-gained SNP in w1118; iso-2; iso-3 in the non-conserved C-terminal region. (A) Protein BLAST of CG34326 against the non-redundant (nr) protein database shows that only the 38 N-terminal amino acids are conserved among Drosophila species and not beyond Drosophila. The colored lines represent the homologs from the following organisms: Drosophila melanogaster, Drosophila grimshawi, Drosophila yakuba, Drosophila erecta, Drosophila virilus, Ixodes scapularis, Ixodes scapularis, and Nycticebus coucang. (B) Aligment of Drosophila melanogaster CG34326 with orthologous gene from Drosophila grimshawi. (C) Aligment of Drosophila melanogaster CG34326 with orthologous gene from Drosophila yakuba.
There are also five stop-lost SNPs in w1118; iso-2; iso-3 (Table 6). All of these SNPs are in predicted protein-coding genes, metabotropic GABA-B receptor subtype 1 (GABA-B-R1), CG13958, CG4975, brown (bw), and POU domain motif 3 (pdm3). It is not known whether any of these genes are essential in Drosophila besides bw, which is not required for viability. However, the GABA-B-R1 gene is required for normal behavior in mice26 and the ortholog is therefore likely also have a function in Drosophila, although no phenotypic data are available. The bw gene is a classic gene first described in 1921 by Waaler,27 which causes the eyes to be brown rather than red and encodes an ATPase binding cassette (ABC) transporter.28 The bw1 mutation in the reference strain is a spontaneous allele with a 412-transposon repeat insertion,29 which would have been missed in our next-generation sequencing data because the input sequence we analyzed contained only short-read sequences that mapped uniquely to the reference genome.
Not much is known about the functions of several genes with in-frame stop-gain SNPs. The pdm3 gene is expressed in the larval and adult nervous system, and it encodes a highly-conserved Hox domain, but no phenotypic data are available (www.flybase.org). No phenotypic data are available for either CG13958 or CG4975. The protein encoding CG13958 has no known conserved domain, and its peak expression is observed within 6–24 h of embryogenes, during early larval stages, at stages throughout the pupal period, and in the adult male (www.flybase.org). The protein encoded by CG4975 has an Armadillo-like helical domain and an Ataxin-10 domain and has expression in the hind gut during the late larval and periods (www.flybase.org).30
Some of the stop-lost SNPs have interesting consequences. For example, a stop-lost SNP in w1118; iso-2; iso-3 is in the CG13958 gene and causes an extension of 8 amino acids before the next stop codon in 3′UTR sequence is reached (Fig. 5). Since the C-termini of CG13958 vary in w1118; iso-2; iso-3 and the reference strains of Drosophila melanogaster, it is conceivable that the C-terminus might also fluctuate in other Drosophila species. To test this idea, we investigated the C-terminal regions of CG13958 homologs in other Drosophila species.
Figure 5. CG13958 has a stop lost SNP in w1118; iso-2; iso-3. The top comparison shows the alignment of the Drosophila melanogaster reference genome with w1118; iso-2; iso-3. Notice that the stop lost causes an extension of nine amino acids. The second through sixth comparisons shows the alignment of Drosophila simulans, Drosophila erecta, Drosophila yakuba, Drosophila mojavensis, and Drosophila pseudoobscura pseudoobscura (Sbjct) with the Drosophila melanogaster reference genome (Dm-ref). The number of terminal amino acids missing or gained is shown (-1 to +3).
We found that CG13958 homologs have variable C-terminal amino acids in different species of Drosophila. When the CG13958 protein is analyzed by protein Basic Local Alignment Search Tool (BLASTp) with the non-redundant (nr) protein database (www.ncbi.nlm.nih.gov), at least two Drosophila species have extended C-terminal amino acids and at least three Drosophila species have missing amino acids at the C-termini (Fig. 5). For example, Drosophila pseudoobscura has three of the extended amino acids found in w1118; iso-2; iso-3 and Drosophila mojavenais has four of them. In contrast, Drosophila simulans is missing the last terminal amino acid, Drosophila erecta is missing the last two terminal amino acids, and Drosophila yakuba is missing the last three amino acids found in the reference strain (Fig. 5). The large number of stop-gain and stop-lost SNPs in Drosophila likely has important implications on the evolution of protein function (see Discussion).
Synonymous and Non-Synonymous SNPs in w1118; iso-2; iso-3
There are 15,842 synonymous SNPs and 4,467 nonsynonymous SNPs in annotated ORFs in w1118; iso-2; iso-3 (Fig. 1). A synonymous SNP (silent SNP) is defined as a SNP that does not change the amino acid in the protein, whereas a nonsynonymous SNP does. Evolutionary studies often compare the ratio (N/S) of nonsynonymous SNPs (N) to synonymous SNPs (S) in a gene to measure the evolutionary conservation of a particular protein or protein domain. For a highly conserved protein domain, such as the 60 amino acid Hox domain of Oc/Otd mentioned above (Fig. 3b), the number of synonymous SNPs is much higher than the number of non-synonymous SNPs. This is expected because nonsynonymous SNPs would probably disrupt the Hox protein function. The genome-wide normalized N/S ratio (dN/dS), also called ω (i.e., ω = dN/dS), is by definition normalized to 1 in most evolutionary studies.31 The non-normalized N/S ratio is ~0.28 in w1118; iso-2; iso-3 compared with the reference genome, y1; cn1 bw1 sp1 (i.e., N/S = 4,467/15,842; Table 1). For individual proteins, a dN/dS ratio < 1 (i.e., non-normalized N/S < 0.28) can indicate selection for a conserved protein that cannot withstand many amino acid substitutions. For genes with dN/dS ratio > 1, (i.e., non-normalized N/S > 0.28), the proteins or at least portions of the proteins, are able to withstand greater number of amino acid substitutions and are therefore probably lesser conserved.
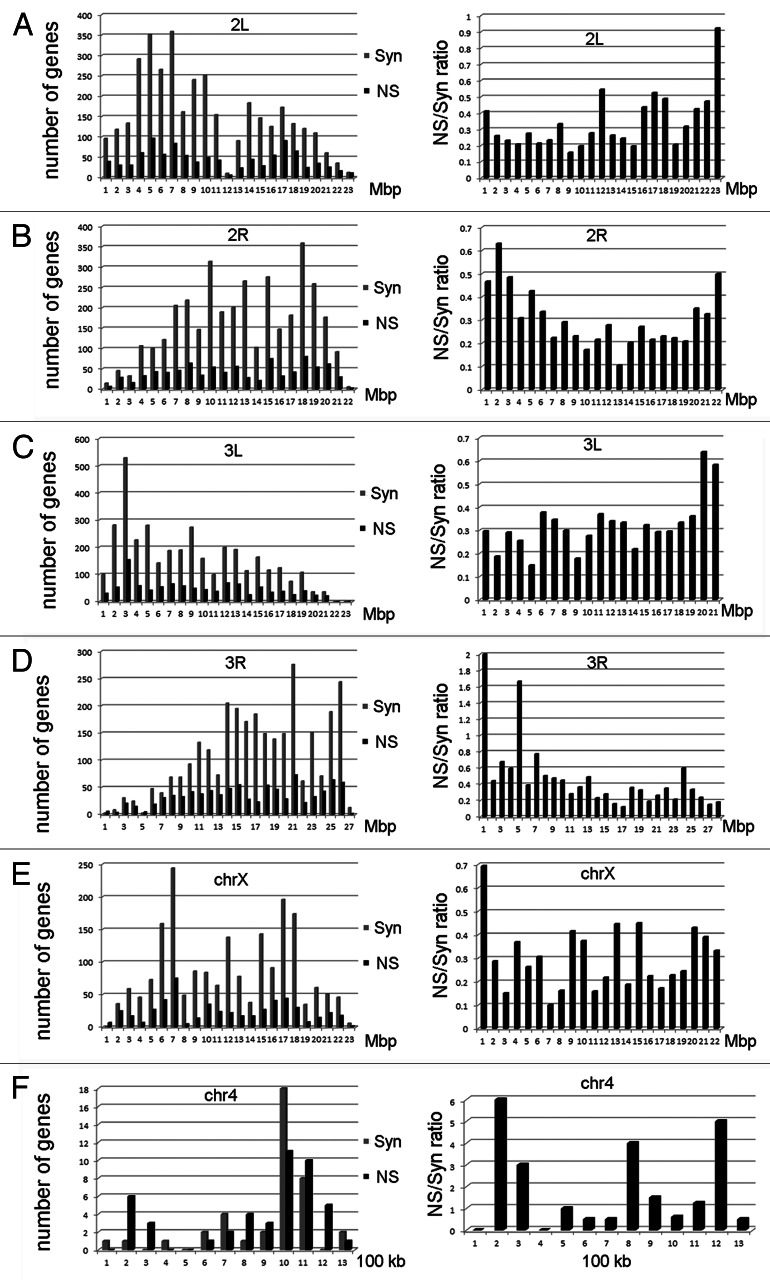
We examined the genome-wide distribution of synonymous and nonsynonymous SNPs for w1118; iso-2; iso-3 and saw higher levels of both classes of SNPs in the middle of the chromosome arms and lower levels near the centromeres and telomeres (Fig. 6, left). This was expected because the number of SNPs is proportional to the recombination frequencies at different regions of the chromosomes.32,33 Also, our previous analyses of the distribution of total SNPs revealed a similar pattern.1 We observed higher N/S ratios near the telomeres and centromeres and lower N/S ratios in the middle of the chromosome arms (Fig.6, right). We speculate that this might reflect that a majority of conserved genes are located in highly recombinogenic regions near the middle of the chromosome arms. This finding requires confirmation in natural populations of Drosophila melanogaster since the origins of the two sequenced strains discussed in this paper are not known.
Figure 6. Nonsynonymous to synonymous ratios along the chromosome arms in w1118; iso-2; iso-3. (A) Left, Nonsynonymous SNPs at 1 Mbp intervals along the 2L chromosome arm (black) and synonymous SNPs (gray). Right, N/S ratios (NS/Syn) along the chromosome arms. Notice that N/S ratios are higher near the centromere and telomere (see text). (B-F) as in (A), but for chromosome arms 2R, 3L, 3R, 4 and X.
Discussion
To illustrate the use of SnpEff, we annotated ~356,660 SNPs in w1118; iso-2; iso-3 and place them into 14 different classes based on their predicted effects on protein function. In order of prevalence, these 14 classes are intron, upstream, downstream, intergenic, synonymous, non-synonymous, 3′UTR, 5′UTR, start-gain, stop-gain, stop-lost, synonymous-stop, start-lost, and splice-site SNPs (Fig. 1). The reason for cataloging these SNPs in w1118; iso-2; iso-3 is to get a better appreciation of evolution of genome sequences and genome organization in this common laboratory strain. We appreciate the fact that both w1118; iso-2; iso-3 and y1; cn1 bw1 sp1 are derived and highly manipulated laboratory strains and do not represent natural populations. Therefore, we do not mean to imply that the analyses in this paper are representative but rather just observational. To be representative, these observations need to be followed up with natural populations. Hundreds of Drosophila natural populations have already been or are in the process of being sequenced, so this type of analyses should be feasible in the near future with a program such as SnpEff.34
Our previous analyses suggest that most of these SNPs are probably genuine and can be validated by capillary sequencing.1 A common worry about next-generation sequencing data are that SNPs are vastly over estimated. One might think that if a large fraction of the identified SNPs had the predicted “effects,” the organism would not be viable. However, since short-read next-generation sequencing, such as the short-read sequences we obtained with the Illumina platform, has high error rates, further validation of specific SNPs is needed to be absolutely certain. Further validation of SNPs is best done with long-range DNA sequencing such as with traditional capillary sequencing or sequencing with the Roche,18 Pacific Biosciences,19 and many other third generation DNA sequencing instruments that are now available20 (see ref.1 for validation examples with capillary sequencing). Many of the stop-gain and stop-lost SNPs in w1118; iso-2; iso-3 occur in essential genes that apparently still function after amino acid truncations caused by the stop-gain SNPs (Table 6). These non-critical effects of the stop codon SNPs are worth noting because nonsense SNPs generally result in nonfunctional protein products. For example, some genetic disorders, such as thalassemia and Duchenne muscular dystrophy (DMD), result from nonsense SNPs.35-37 Also, nonsense SNP-mediated RNA decay exists in yeast, Drosophila, and humans, and usually ensures that mRNAs with premature stop codons are degraded.38
The stop-gain and stop-lost SNPs in essential genes, if they are validated, could have a profound evolutionary implication in that it might suggest the involvement of prions or mutations in translational termination factors that allow read-through of stop codons in the retention and selection of these SNPs. In 1965, Brian Cox, a geneticist working with the yeast Saccharomyces cerevisiae, isolated a yeast strain auxotrophic for adenine due to a nonsense mutation and found that it was able to survive in media lacking adenine when a [PSI+] mutation is present.39 Reed Wickner showed in 1994 that the [PSI+] suppressor mutation resulted from a prion form of the translation termination factor Sup3540 that allowed a read-though of the stop codon that caused the adenine auxotrophy. Lindquist and colleagues showed in 2008 that the [PSI+] prion provides survival advantages in several stressful environments, such as high salt conditions.41 They speculate that Sup35 is an evolutionary capacitor that, when inactivated in the PSI+ form, allows translational read-through past stop codons and the expression of novel C-terminal amino acids in hundreds of proteins, some of which are beneficial in stressful environments.41
It is attractive to speculate that a similar prion-mediated evolutionary mechanism might occur in Drosophila, for both stop-loss and stop-gain SNPs, and that this might help explain the large number of SNPs that we see in these categories. We note that Drosophila has several Sup35 orthologs, some of which have N-terminal repeats known to be potentially prion-forming domains.41 While most prions are thought to not directly mutate DNA sequences, they could provide an environment that would make the retention and selection of beneficial stop codon SNPs more likely. For example, a stop-lost SNP would allow a modified protein with the new C-terminal tail to be always expressed, even when the [PSI+] prion is lost.41 Therefore, a stop-lost SNP would more likely occur in a gene with potential beneficial codons in the 3′ UTR because the cryptic C-terminal amino acids would provide a selective advantage in stressful environments when they are translated. We acknowledge that this is a highly speculative explanation for the high numbers of start-gain and stop-lost SNPs, but we believe that it is worthy of further investigation.
The many potential start-gain SNPs in Drosophila might also have evolutionary implications. Similar to the cryptic genetic variation that is revealed by stop-lost mutations in the 3′ UTR, start-gain SNPs reveal cryptic genetic variation in the 5′ UTR. Uncovering the cryptic genetic variation in times of environmental stress, such as by inducing transcription initiation at start sites upstream of the normally-used transcription start sites, could be one mechanism to facilitate the use of potential start-gain SNPs. Further genetic drift and selection of potential start-gain SNPs, such as by introducing better Kozak consensus sequences or more commonly used 5′-AUG-3′ translation initiation codons, can stabilize the cryptic genetic variation further if these SNPs lead to improved survival or reproductive fitness in stressful environment.
Methods
SnpEff overview.
The program is divided in two main parts (1) database build and (2) effect calculation.
(1) Database build
Since many databases containing genomic annotations are available with SnpEff distribution, this step is usually not run by the user. Databases are build using a reference genome and an annotation file. The reference genome must be in FASTA format. Genomic information can be parsed from four main annotation formats: GTF (version 2.2), GFF (versions 3 and 2), UCSC RefSeq tables, and tab separated text files (TXT). These annotation files are available at ENSEMBL, UCSC, or organism specific websites, such as FlyBase, WormBase and TAIR. SnpEff databases are compressed serialized objects that represent genomic annotations.
(2) Effect calculations
This can be performed once the user has downloaded or built the database. The program loads the binary database and builds a data structure called “interval forest” in order to perform an efficient interval search. Input files are parsed and each variant queries the data structures to find intersecting genomic annotations. All intersecting genomic regions are reported and whenever these regions include an exon, the coding effect of the variant is calculated. A list of the reported effects and annotations is shown in Table 2; additional information produced by the program is shown in Tables 3 and 4 for different output formats.
Table 3. Information provided by SnpEff in tab separaOutput format (TXT).
| Column |
Notes |
| Chromosome |
Chromosome name (usually without any leading 'chr' string) |
| Position |
One based position |
| Reference |
Reference |
| Change |
Sequence change |
| Change type |
Type of change (SNP, MNP, INS, DEL) |
| Homozygous |
Is this homozygous or heterozygous (Hom, Het) |
| Quality |
Quality score (from input file) |
| Coverage |
Coverage (from input file) |
| Warnings |
Any warnings or errors. |
| Gene_ID |
Gene ID (usually ENSEMBL) |
| Gene_name |
Gene name |
| Bio_type |
BioType, as reported by ENSEMBL. |
| Trancript_ID |
Transcript ID (usually ENSEMBL) |
| Exon_ID |
Exon ID (usually ENSEMBL) |
| Exon_Rank |
Exon number on a transcript |
| Effect |
Effect of this variant. See details below. |
| old_AA/new_AA |
Amino acid change |
| old_codon/new_codon |
Codon change |
| Codon_Num(CDS) |
Codon number in CDS |
| Codon_degenaracy |
Codon degenaracy |
| CDS_size |
CDS size in bases |
| Custom_interval_ID | If any custom interval was used, add the IDs here (may be more than one). |
Table 4. Information provided by SnpEff in variant call format (VCF).
| Sub-field |
Notes |
| Effect |
Effect of this variant. See details below. |
| Codon_Change |
Codon change: old_codon/new_codon |
| Amino_Acid_change |
Amino acid change: old_AA/new_AA |
| Warnings |
Any warnings or errors. |
| Gene_name |
Gene name |
| Gene_BioType |
BioType, as reported by ENSEMBL. |
| Coding |
[CODING | NON_CODING]. If information reported by ENSEMBL (e.g., has 'protein_id' information in GTF file). |
| Trancript |
Transcript ID (usually ENSEMBL) |
| Exon |
Exon ID (usually ENSEMBL) |
| Warnings | Any warnings or errors (not shown if empty). |
The information is added to the INFO fields using an tag ‘EFF’. The format for each effect is “Effect (Effect_Impact | Codon_Change | Amino_Acid_change | Gene_Name | Gene_BioType | Coding | Transcript | Exon [ | ERRORS | WARNINGS ])”
SnpEff algorithms
In order to process thousands of variants per second, we implemented an efficient data structure that allows to query for arbitrary interval overlaps. We created an interval forest, which is a hash of interval trees indexed by chromosome. Each interval tree42 is composed of nodes. Each node has five elements (i) a center point, (ii) a pointer to a node having all intervals to the left of the center, (iii) a pointer to a node having all intervals to the right of the center, (iv) all intervals overlapping the center point sorted by start position, and (v) all intervals overlapping the center point, sorted by end position. Querying an interval tree requires O(log n + m) time, where n is the number of intervals in the tree and m is the number of intervals in the result. Having a hash of trees optimizes the search by reducing the number of intervals per tree.
Input formats
Three input formats supported by SnpEff are variant call format (VCF), tab separated TXT format; and the SAMtools Pileup format.8 VCF was created by the 1000 Genomes project and it is currently the de facto standard for variants in sequencing applications. The TXT and Pileup formats are currently deprecated and being phased out.
Output formats
SnpEff also supports two output formats, TXT and VCF. The output information provided in both formats includes three main groups: (i) variant information (genomic position, the reference and variant sequences, change type, heterozygosity, quality and coverage); (ii) genetic information (gene Id, gene name, gene biotype, transcript ID, exon ID, exon rank); and (iii) effect information (effect type, amino acid changes, codon changes, codon number in CDS, codon degeneracy, etc.). Whenever multiple transcripts for a gene exist, the effect and annotations on each transcript are reported, so one variant can have multiple output lines. Table 3 shows the information provided in TXT format and Table 4 shows the information provided in VCF format. When using the VCF format, the effect information is added to the information (INFO) fields using an effect (EFF) tag. As in the case of TXT output, if multiple alternative splicing products are annotated for a particular gene, SnpEff provides this information for each annotated version (see Supplementary Data File 1 for the complete SnpEff output for w1118; iso-2; iso-3).
SnpEff accuracy
As part of our standard development cycle, we perform accuracy testing by comparing SnpEff to ENSEMBL “Variant effect predictor,” which we consider to be the “gold standard.” Current unity testing includes over a hundred test cases with thousands of variants, each to ensure that predictions are accurate.
SnpEff integration
SnpEff provides integration with third party tools, such as Galaxy,43 which creates a web based interface for bioinformatic analysis pipelines. Integration with Genome analysis tool kit4 (GATK) was provided by Broad’s GATK team.
Data access
SnpEff Data can be accessed from the supplemental data file for w1118; iso-2; iso-3 or by contacting D.M.R.
Supplementary Material
Acknowledgments
This work was supported by a Michigan Core Technology grant from the State of Michigan's 21st Century Fund Program to the Wayne State University Applied Genomics Technology Center. This work was also supported by the Environmental Health Sciences Center in Molecular and Cellular Toxicology with Human Applications Grant P30 ES06639 at Wayne State University, NIH R01 grants (ES012933) to D.M.R., and DK071073 to X.L. We thank David Roazen, Eric Banks and Mark DePristo in the GATK team at the Broad Institute who integrated SnpEff with the Genome Analysis Toolkit (GATK).
Footnotes
Previously published online: www.landesbioscience.com/journals/fly/article/19695
References
- 1.Platts AE, Land SJ, Chen L, Page GP, Rasouli P, Wang L, et al. Massively parallel resequencing of the isogenic Drosophila melanogaster strain w(1118); iso-2; iso-3 identifies hotspots for mutations in sensory perception genes. Fly (Austin) 2009;3:192–203. doi: 10.4161/fly.3.3.9652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rope AF, Wang K, Evjenth R, Xing J, Johnston JJ, Swensen JJ, et al. Using VAAST to identify an X-linked disorder resulting in lethality in male infants due to N-terminal acetyltransferase deficiency. Am J Hum Genet. 2011;89:28–43. doi: 10.1016/j.ajhg.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, et al. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet. 2004;36:283–7. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- 6.Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, et al. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet. 2004;36:288–92. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- 7.Ryder E, Ashburner M, Bautista-Llacer R, Drummond J, Webster J, Johnson G, et al. The DrosDel deletion collection: a Drosophila genomewide chromosomal deficiency resource. Genetics. 2007;177:615–29. doi: 10.1534/genetics.107.076216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H. Improving SNP discovery by base alignment quality. Bioinformatics. 2011;27:1157–8. doi: 10.1093/bioinformatics/btr076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, et al. 1000 Genomes Project Analysis Group The variant call format and VCFtools. Bioinformatics. 2011;27:2156–8. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sugihara H, Andrisani V, Salvaterra PM. Drosophila choline acetyltransferase uses a non-AUG initiation codon and full length RNA is inefficiently translated. J Biol Chem. 1990;265:21714–9. [PubMed] [Google Scholar]
- 11.Ivanov IP, Firth AE, Michel AM, Atkins JF, Baranov PV. Identification of evolutionarily conserved non-AUG-initiated N-terminal extensions in human coding sequences. Nucleic Acids Res. 2011;39:4220–34. doi: 10.1093/nar/gkr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–48. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dennis G, Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 14.Hosack DA, Dennis G, Jr., Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lazure C, Hum WT, Gibson DM. Sequence diversity within a subgroup of mouse immunoglobulin kappa chains controlled by the IgK-Ef2 locus. J Exp Med. 1981;154:146–55. doi: 10.1084/jem.154.1.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruden DM, Jamison DC, Zeeberg BR, Garfinkel MD, Weinstein JN, Rasouli P, et al. The EDGE hypothesis: epigenetically directed genetic errors in repeat-containing proteins (RCPs) involved in evolution, neuroendocrine signaling, and cancer. Front Neuroendocrinol. 2008;29:428–44. doi: 10.1016/j.yfrne.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruden DM, Ma J, Li Y, Wood K, Ptashne M. Generating yeast transcriptional activators containing no yeast protein sequences. Nature. 1991;350:250–2. doi: 10.1038/350250a0. [DOI] [PubMed] [Google Scholar]
- 18.Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–6. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 19.McCarthy A. Third generation DNA sequencing: pacific biosciences’ single molecule real time technology. Chem Biol. 2010;17:675–6. doi: 10.1016/j.chembiol.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 20.Schadt E. Genome-sequencing anniversary. First steps on a long road. Science. 2011;331:691. doi: 10.1126/science.1203235. [DOI] [PubMed] [Google Scholar]
- 21.Liu D, Finley RL., Jr. Cyclin Y is a novel conserved cyclin essential for development in Drosophila. Genetics. 2010;184:1025–35. doi: 10.1534/genetics.110.114017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brenner S, Stretton AOW, Kaplan S. Genetic code: the ‘nonsense’ triplets for chain termination and their suppression. Nature. 1965;206:994–8. doi: 10.1038/206994a0. [DOI] [PubMed] [Google Scholar]
- 23.Acampora D, Avantaggiato V, Tuorto F, Barone P, Reichert H, Finkelstein R, et al. Murine Otx1 and Drosophila otd genes share conserved genetic functions required in invertebrate and vertebrate brain development. Development. 1998;125:1691–702. doi: 10.1242/dev.125.9.1691. [DOI] [PubMed] [Google Scholar]
- 24.Younossi-Hartenstein A, Green P, Liaw GJ, Rudolph K, Lengyel J, Hartenstein V. Control of early neurogenesis of the Drosophila brain by the head gap genes tll, otd, ems, and btd. Dev Biol. 1997;182:270–83. doi: 10.1006/dbio.1996.8475. [DOI] [PubMed] [Google Scholar]
- 25.de Rosa R, Grenier JK, Andreeva T, Cook CE, Adoutte A, Akam M, et al. Hox genes in brachiopods and priapulids and protostome evolution. Nature. 1999;399:772–6. doi: 10.1038/21631. [DOI] [PubMed] [Google Scholar]
- 26.Jones KA, Borowsky B, Tamm JA, Craig DA, Durkin MM, Dai M, et al. GABA(B) receptors function as a heteromeric assembly of the subunits GABA(B)R1 and GABA(B)R2. Nature. 1998;396:674–9. doi: 10.1038/25348. [DOI] [PubMed] [Google Scholar]
- 27.Waaler GHM. The location of a new second chromosome eye colour gene in Drosophila melanogaster. Hereditas. 1921;2:391–4. doi: 10.1111/j.1601-5223.1921.tb02636.x. [DOI] [Google Scholar]
- 28.Saurin W, Hofnung M, Dassa E. Getting in or out: early segregation between importers and exporters in the evolution of ATP-binding cassette (ABC) transporters. J Mol Evol. 1999;48:22–41. doi: 10.1007/PL00006442. [DOI] [PubMed] [Google Scholar]
- 29.Dreesen TD, Johnson DH, Henikoff S. The brown protein of Drosophila melanogaster is similar to the white protein and to components of active transport complexes. Mol Cell Biol. 1988;8:5206–15. doi: 10.1128/mcb.8.12.5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39:715–20. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- 31.Stoletzki N, Eyre-Walker A. The positive correlation between dN/dS and dS in mammals is due to runs of adjacent substitutions. Mol Biol Evol. 2011;28:1371–80. doi: 10.1093/molbev/msq320. [DOI] [PubMed] [Google Scholar]
- 32.Begun DJ, Aquadro CF. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature. 1992;356:519–20. doi: 10.1038/356519a0. [DOI] [PubMed] [Google Scholar]
- 33.Charlesworth B, Coyne JA, Barton NH. The relative rates of evolution of sex chromosomes and autosomes. Am Nat. 1987;130:113–46. doi: 10.1086/284701. [DOI] [Google Scholar]
- 34.Anderson JA, Gilliland WD, Langley CH. Molecular population genetics and evolution of Drosophila meiosis genes. Genetics. 2009;181:177–85. doi: 10.1534/genetics.108.093807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flanigan KM, Dunn DM, von Niederhausern A, Howard MT, Mendell J, Connolly A, et al. DMD Trp3X nonsense mutation associated with a founder effect in North American families with mild Becker muscular dystrophy. Neuromuscul Disord. 2009;19:743–8. doi: 10.1016/j.nmd.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tran VK, Takeshima Y, Zhang Z, Habara Y, Haginoya K, Nishiyama A, et al. A nonsense mutation-created intraexonic splice site is active in the lymphocytes, but not in the skeletal muscle of a DMD patient. Hum Genet. 2007;120:737–42. doi: 10.1007/s00439-006-0241-y. [DOI] [PubMed] [Google Scholar]
- 37.Chang JC, Kan YW. beta 0 thalassemia, a nonsense mutation in man. Proc Natl Acad Sci U S A. 1979;76:2886–9. doi: 10.1073/pnas.76.6.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gatfield D, Unterholzner L, Ciccarelli FD, Bork P, Izaurralde E. Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J. 2003;22:3960–70. doi: 10.1093/emboj/cdg371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cox BS, Tuite MF, McLaughlin CS. The psi factor of yeast: a problem in inheritance. Yeast. 1988;4:159–78. doi: 10.1002/yea.320040302. [DOI] [PubMed] [Google Scholar]
- 40.Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–9. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 41.Tyedmers J, Madariaga ML, Lindquist S. Prion switching in response to environmental stress. PLoS Biol. 2008;6:e294. doi: 10.1371/journal.pbio.0060294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cormen TH. Introduction to algorithms. The MIT press, 2001. [Google Scholar]
- 43.Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–5. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.