Abstract
Myf5 is a member of the muscle-specific determination genes and plays a critical role in skeletal muscle development. Whereas the expression of Myf5 during embryonic and fetal myogenesis has been extensively studied, its expression in progenitors that will ultimately give rise to adult satellite cells, the stem cells responsible for muscle repair, is still largely unexplored. To investigate this aspect, we have generated a mouse strain carrying a CreER coding sequence in the Myf5 locus. In this strain, Tamoxifen-inducible Cre activity parallels endogenous Myf5 expression. Combining Myf5CreER and Cre reporter alleles, we were able to evaluate the contribution of cells expressing Myf5 at distinct developmental stages to the pool of satellite cells in adult hindlimb muscles. Although it was possible to trace back the origin of some rare satellite cells to a subpopulation of Myf5+ve progenitors in the limb buds at the late embryonic stage (~E12), a significant number of satellite cells arise from cells which expressed Myf5 for the first time at the fetal stage (~E15). These studies provide direct evidence that adult satellite cells derive from progenitors that first express the myogenic determination gene Myf5 during fetal stages of myogenesis.
Keywords: Myf5, satellite cells, fetal myogenesis, skeletal muscle
INTRODUCTION
Skeletal muscle is the most abundant tissue in the vertebrate body and plays a major role in physiological functions such as locomotion, breathing and energy metabolism. When damage is induced by injury or pathological conditions such as in the muscular dystrophies, skeletal muscle manifests a powerful regenerative response (Dhawan and Rando, 2005; Kuang and Rudnicki, 2008; Zammit et al., 2006). Satellite cells are considered the main stem cells responsible for adult muscle repair (Zammit et al., 2006). They owe their name to their location beneath the basal lamina in strict contact with muscle fibers (Mauro, 1961), and they are characterized by the expression of Pax7 (Seale et al., 2000). Recent studies relying on the specific ablation of Pax7-expressing cells in adult muscle demonstrate the requirement for satellite cells to achieve a productive regenerative response (Lepper et al., 2011; Murphy et al., 2011; Sambasivan et al., 2011). Upon muscle injury, satellite cells enter into the cell cycle, rapidly up-regulate the expression of myogenic regulatory factors (MRFs) such as Myf5 and MyoD, and terminally differentiate, ultimately fusing to form regenerating fibers. A fraction of activated satellite cells do not undergo myogenic differentiation but instead self-renew, thereby restoring the pool of quiescent satellite cells (Zammit et al., 2004).
Skeletal muscle formation occurs in several steps. Mesoderm-derived precursors become specified to the myogenic lineage, and these develop into myoblasts which become post-mitotic and differentiate by fusing into multinucleated fibers, expressing a set of myofibrillar proteins required for contraction. MRFs are key regulators of the myogenic program. In vertebrates, four MRFs have been identified: Myf5, MyoD, Mrf4, and Myogenin (Pownall et al., 2002). MRFs are specifically expressed in the skeletal muscle lineage and when transfected in certain other cellular types can induce them to adopt a myogenic fate (Weintraub et al., 1989). Myf5 is the first of the MRFs to be expressed during mammalian development (Ott et al., 1991). Mouse strains in which the expression of Myf5, MyoD or Mrf4 was individually genetically ablated present only relatively mild developmental phenotypes (Braun et al., 1992; Kassar-Duchossoy et al., 2004; Rudnicki et al., 1992; Zhang et al., 1995), but triple knockout strains completely lack skeletal muscle in any anatomical location (Kassar-Duchossoy et al., 2004; Rudnicki et al., 1993). Myogenin−/− mice form Myf5 and MyoD expressing muscle progenitors, but are deficient in differentiated muscle (Hasty et al., 1993; Nabeshima et al., 1993). Moreover, mice in which the expression of Mrf4, MyoD and Myogenin are simultaneously abolished present normal numbers of progenitors, but fail to form any differentiated muscle fibers (Valdez et al., 2000). Together, these observations and related cell ablation studies (Haldar et al., 2008) suggest that Myf5 and MyoD can independently initiate the myogenic program and thus act as myogenic determination genes. The role of Mrf4 is more ambiguous, as it seems to act as determination gene only during embryonic development (Kassar-Duchossoy et al., 2004). In contrast, Myogenin appears to operate downstream to the muscle determination genes by promoting muscle differentiation, a role that Myf5 cannot play alone (Valdez et al., 2000).
Pax3 and its paralogue Pax7 have been suggested to promote the progression of somitic progenitors into the myogenic lineage (Buckingham and Relaix, 2007). In contrast to MRFs, Pax3 and Pax7 are not muscle specific genes as they are expressed also in the developing central nervous system, in the neural crest cells and in the paraxial mesoderm before the establishment of definitive myogenic lineage (Buckingham and Relaix, 2007). Nevertheless, both Pax3 and Pax7 can enhance the myogenic potential of ES cells (Darabi et al., 2008;Darabi et al., 2011). Furthermore, ectopic Pax3 is able to induce MyoD and Myf5 expression in embryonic mesoderm (Maroto et al., 1997) and ablation studies indicate that Pax3 can act upstream of MyoD (Tajbakhsh et al., 1997).
Skeletal muscle is established in successive, though overlapping, phases involving different types of myoblasts (embryonic, fetal, and postnatal) which present distinct features (Biressi et al., 2007a;Murphy and Kardon, 2011;Stockdale, 1992;Tajbakhsh, 2005). Muscle formation (with the exception of the early myotome) has been attributed to a population of progenitors which are dependent on the expression of Pax3 and Pax7 (Kassar-Duchossoy et al., 2005;Relaix et al., 2005). These progenitors, initially not expressing MRFs, can induce the myogenic program and differentiate into skeletal muscle fibers during primary (E10.5–E12.5) and secondary (E14.5–E17.5) myogenesis or possibly remain as a reserve cell population within the growing muscles during peri- and post-natal stages (Kassar-Duchossoy et al., 2005;Relaix et al., 2005).
Limb satellite cells are believed to derive from Pax3+ve cells, which migrate from the dermomyotome into the developing limbs (Schienda et al., 2006). They are initially negative for Pax7, but they subsequently become dependent on Pax7 expression (Hutcheson et al., 2009;Kassar-Duchossoy et al., 2005;Lepper et al., 2009;Relaix et al., 2005;Relaix et al., 2006;Seale et al., 2000). A lineage study traced the origin of some adult limb satellite cells back to cells expressing Pax7 at E11.5 (Lepper and Fan, 2010). In contrast to the idea that adult satellite cells derive from MRF−ve precursors resident in the postnatal muscle, a recent study suggests that great majority, if not all adult satellite cells transit through a developmental stage in which the MyoD locus is active (Kanisicak et al., 2009). Nevertheless, the developmental stage at which the precursors of satellite cells first express muscle determination genes is unknown. Understanding this aspect is crucial to clarify the relationship that the precursors of satellite cells have with myogenic cells involved in embryonic, fetal and postnatal muscle growth.
In the present study, we generated a novel Tamoxifen-inducible Myf5CreER mouse line, which allowed us to monitor the fate of cells expressing Myf5 at different developmental stages. Using in vivo and in vitro analyses relying on this and other muscle-specific Cre lines, we investigated the developmental time at which the precursors of satellite cells enter the myogenic program by inducing the expression of the early muscle determination gene Myf5, expanding our knowledge of the embryonic origin of adult satellite cells.
MATERIALS AND METHODS
Mouse strains and Tamoxifen (TMX) treatments
For the Myf5CreER mouse line, a targeting vector was constructed that placed an ires-CreER™-FRT-Neo-FRT cassette in the AseI site within the 3′ untranslated region of the Myf5 gene following the stop codon in exon 3. The ires-CreER™-FRT-Neo-FRT cassette has been previously described (Nishijo et al., 2009). This strategy and integration site is identical to the previously reported non inducible Cre mouse line (Haldar et al., 2007). The linearized targeting vector was electroporated into R1 mouse embryonic stem (ES) cells, and the cells were subjected to positive selection with 200 μg/ml neomycin and negative selection with 2 μM ganciclovir. To detect positive clones upon screening, an EcoRV site was engineered within the FRT-flanked Neo cassette (Keller et al., 2004a). Positive ES cell clones were detected by EcoRV restriction endonuclease digestion of genomic DNA and Southern hybridization with a 900 base pair (bp) 3′ external probe amplified using PCR primers 5′ – TGA ACA AGA AGG AAA GTG AGA AAG G – 3′ and 5′ – CGT CAT GAC CTG AGA AGA GGA A – 3′. Cells from the identified ES cell clone were microinjected into C57BL/6 blastocysts in order to generate chimeric mice. Chimeric mice were mated to C57BL/6 dams, and their agouti offspring were confirmed to harbor the targeted allele by Southern hybridization (data not shown).
Myf5CreER/wt males were crossed to females carrying the reporter alleles R26RLacZ (Soriano, 1999) or R26RYFP (Srinivas et al., 2001) (Gt(ROSA)26Sortm1Sor/J and Gt(ROSA)26Sortm1(EYFP)Cos/J, respectively, Jackson laboratories, Bar Harbor, ME) For the experiments involving R26RLacZ mice, TMX (Sigma-Aldrich, St. Louis, MO) was dissolved in corn oil and pregnant mice were injected intraperitoneally with a single injection of 3 mg at E10.5 or two consecutive injections at E10.5 (1.5 mg) and E11.5 (2 mg). For experiments involving R26RYFP, TMX (resuspended at 50 mg/ml in 92.5% corn oil/7.5% ethanol) was administrated to pregnant mice with different protocols relying on two injection in consecutive days: E10.5 (5 mg) + E11.5 (5 mg); E12.5 (7.5 mg) + E13.5 (10 mg); E14.5 (7.5 mg) + E15.5 (10 mg). Newborn mice were injected subcutaneously with 0.5 mg TMX for five consecutive days from P7 to P11. Four-week-old mice were injected intraperitoneally with 2 mg TMX once a day for 5 days. Adult Pax7-CreERtm mice (Nishijo et al., 2009) (herein referred to as Pax7CreER/wt) received the same TMX regimen. MCre+/− (Brown et al., 2005), Myf5-NN+/− (Haldar et al., 2008) and Pax7ICNm/wt (Keller et al., 2004b) (herein referred to as Myf5Cre/wt and Pax7Cre/wt respectively) males were crossed to R26RYFP/YFP, R26RLacZ/LacZ or Z/RED+/− (Vintersten et al., 2004) female mice. To generate the Myf5CreER-NNstrain, the FRT-flanked Neo cassette was removed by breeding Myf5CreER/wt mice to transgenic mice ubiquitously expressing Flippase (Flpe) (B6.129S4-Gt(ROSA)26Sortm1(FLP1)Dym/RainJ, Jackson laboratories).
The genotype of breeders and experimental animals was identified by PCR on genomic DNA obtained by yolk sacs (prenatal) or tail clips (postnatal). Gene-targeted Myf5CreER mice were genotyped by PCR using the primer set: ck172: 5′-GGA TAG TGA AAC AGG GGC AA-3′, ck382: 5′-ACC CTC CAG CTC CAG ACT TAT C-3′, ck383: 5′-CCC TGT AAT GGA TTC CAA GCT G -3′, and ck224: 5′-ACA CTG CTC GAT GAA GTT CC-3′. Cycling conditions were: 95°C for 5 minutes, 32 cycles of 95°C for 30 seconds/59°C for 30 seconds/72°C for 120 seconds, followed by 72°C for 7 minutes. The wild type, Myf5CreER, and Myf5Cre alleles resulted in 454, 182, or 312 bp bands, respectively. Other strains were genotyped with published primers or with primers specific to the Cre transgene: cF:,5′-GCA TTT CTG GGG ATT GCT TA-3′, cR: 5′-CCC GGC AAA ACA GGT AGT TA-3′. Vaginal plug day and the first postnatal day are respectively designated as E0.5 and P1. Animal procedures were approved by the Institutional Animal Care and Use Committees.
Xgal staining and whole-mount in situ hybridization
For Xgal staining, single fibers were fixed for 5 minutes in 2% paraformaldehyde, permeabilized in PBS supplemented with 0.2% (V/V) Triton X-100, treated with DAPI (Invitrogen) to stain the nuclei and incubated for 2 days in Xgal solution (1 mg/ml 5-bromo-4-chloro-3-indolyl-β-D-galactosidase, 5 mM K4Fe(CN)6, 5 mM K3Fe(CN)6, 0.02% NP-40, 2 mM MgCl2 in PBS). Muscle cryostat sections (12 μM) were processed for immunofluroescence and subsequently incubated in Xgal solution for 2 days, washed in PBS and mounted in Fluorogel (EMS, Hatfield, PA). For whole-mount Xgal staining, embryos were fixed for 5 hours in 2% paraformaldehyde at 4°C and incubated overnight at 37°C in Xgal solution as previously described (Biressi et al., 2008).
E11.5 embryos were processed for whole-mount in situ hybridization with published probes (Kassar-Duchossoy et al., 2004), as previously described (Biressi et al., 2008).
Cell isolation, fluorescence activated cell sorting (FACS) and muscle injury
Cells were isolated by enzymatic digestion from the hindlimbs of developing and adult mice as previously described (Biressi et al., 2007b;Bjornson et al., 2012). Briefly, dissected adult hind limb muscles were digested in 0.2% type II collagenase in Ham’s-F10 medium (Cellgro, Mediatech, Manassas, VA) for 70 minutes at 37°C, followed by a second digest in 1% dispase/0.2% type II collagenase for 30 minutes at 37°C (all enzymes from Invitrogen). Satellite cells were mechanically dissociated from myofibers after the final digestion by passing the tissue suspension through a 20-gauge needle. Developing hindlimbs were dissociated with 0.15 mg/ml collagenase Type V (Sigma), 0.4 mg/ml dispase, 0.1 mg/ml DNase I (Worthington) in a buffered solution (Hank’s Balanced Salt Solution (HBSS; BioWhittaker) supplemented with 15 mM HEPES, 15 mM glucose, 1.5 mM MgSO4, 0.3% (W/V) bovine serum albumin (BSA), pH 7.4) for 45 minutes at 37°C and filtered on 40 μm cell strainers (Falcon).
FACS analysis of adult satellite cells was performed as previously described (Bjornson et al., 2012). Briefly, satellite cells were further purified using a FACSAria III (BD Biosciences, San Jose, CA). Primary antibodies used were rat anti-CD31 conjugated to allophycocyanin (APC; 1:100, BD Biosciences), rat anti-CD45 antibodies conjugated to allophycocyanin (APC;1:100, BD Biosciences), rat anti-Sca1 antibodies conjugated to Pacific Blue (1:100, BioLegend, San Diego, CA), biotinylated rat anti-CD106 antibody (VCAM; 1:100, BD Biosciences), phycoerythrin-Cy7 Streptavidin (1:100, BD Biosciences). DAPI dilactate (250 μg/μl, Invitrogen) was used to exclude dead cells.
Cell suspensions obtained from Myf5Cre/wt or Pax7Cre/wt; Z/RED−/+ mice at different developmental stage were analyzed and further purified using a Vantage Sorter SE (Becton Dickinson), as previously described (Boutet et al., 2010). Forward scatter and side scatter parameters were used to gate out cell clumps and debris. Cells dissociated from DsRED−ve littermates were used as negative controls to exclude autofluorescent cells.
Cells isolated from postnatal muscles were resuspended in Ham’s F-10 supplemented with 10% horse serum (Invitrogen) and 20 mM HEPES. Cells adhered overnight on ECM-coated slides (BioCoat BD) and fixed for 5 minutes in 4% paraformaldehyde at 4°C. To test the proliferative and differentiation ability, cells were cultured for 3 days in the same media. Prenatal myogenic cells were alternatively resuspended in Opti-MEM (Invitrogen) supplemented with 20% FBS, 20 mM HEPES, and 5 ng/ml FGF (Peprotech) to sustain proliferation or cultured for 3 days in a differentiation permissive conditions consisting in Dulbecco’s modified Eagle medium, 20% horse serum, 20 mM HEPES. When indicated, 2 μM cytochalasin D (Sigma) was added to the culture.
Tibialis anterior and gastrocnemious muscles were injured by injecting 50 μl and 100 μl, respectively, of cardiotoxin (100 μg/ml,Sigma).
Immunofluorescent staining of fibers, cultured cells and sections
Single fibers were isolated and fixed as previously described (Bjornson et al., 2012). For immunofluorescent staining, fibers were permeabilized in PBS supplemented with 0.1% (V/V) Triton X-100 for 1 hour and blocked in blocking solution (PBS, 1 mg/ml BSA, 1.5% donkey serum) for 40 minutes. Primary (overnight at 4°C) and secondary (45 minutes at room-temperature) antibody incubations were performed in blocking solution with 0.1% Tween. Cells and cryostat sections were processed as previously described (Biressi et al., 2008). For immunohistochemistry of frozen adult skeletal muscle, staining was performed using the M.O.M. Immunodetection Kit Staining Procedure (Vector Laboratories, Burlingame, CA) or the Zenon Alexa Fluor 594 Labeling Kit (Life Technologies, Grand Island, NY) following the manufacturer’s instructions. Primary antibodies used included rabbit anti-GFP (1:500, Invitrogen, Carlsbad, CA), chicken anti-GFP (1:500, Aves Labs, Tigard, OR or Chemicon, Temecula, CA), rabbit anti-Desmin (1:500, Abcam, Cambridge, MA), rabbit anti-Laminin (1:500, kindly provided by Dr. Peter D. Yurchenco, University of Medicine and Dentistry of New Jersey, NJ), rat anti-Laminin 2α (1:1000, Abcam), chicken anti-Syn4 (1:200, kindly provided by Dr. Brad Olwin, University of Colorado, Boulder), mouse anti-MyoD (1:100, Dako, Carpintaria, CA), mouse anti-Myogenin (1:200, Becton/Dickson), mouse anti-Pax7 (1:200, Developmental Studies Hybridoma Bank, Iowa City), rabbit anti-Myf5 (1:100, Santa Cruz Biotechnology, Santa Cruz, CA). Species-specific secondary antibodies (donkey) were conjugated to Alexa 488, 594, and 647 and used at a concentration of 1:1000 (Invitrogen). DAPI was added during the secondary antibody incubation to stain the nuclei.
Statistical analysis
Quantitative analyses were performed in at least 3 independent replicates. Unless otherwise stated, data are expressed as mean ± standard deviation. Unpaired t-tests were used to evaluate the statistical significance between groups.
RESULTS
Myf5CreER mice present TMX-inducible Cre activity in the muscle compartment
Limb satellite cells derive from Pax3+ve progenitors, which migrate from the dermomyotome into the developing limb buds (Schienda et al., 2006). As development proceeds, these progenitors also express Pax7; this Pax+ve/MRFs−ve cell population is detectable throughout development and is believed to give rise to adult satellite cells (Kassar-Duchossoy et al., 2005;Relaix et al., 2005). Interestingly, a recent study suggests that the great majority of adult satellite cells are derived from precursors that express the MRF MyoD at some point prenatally (Kanisicak et al., 2009). However, the developmental time at which the precursors of the satellite cells first express one of the MRFs, at that point committing to the myogenic lineage, remains unknown. To directly test this, and also to assess whether satellite cell progenitors become committed to the myogenic lineage during embryonic or fetal muscle growth, we generated a mouse strain in which Cre could be inducibly expressed from the Myf5 locus, as Myf5 is believed to be the first of the MRFs to be expressed during mouse development (Ott et al., 1991). A targeting vector was constructed that placed an ires-CreER™-FRT-Neo-FRT cassette within the 3′ untranslated region of the Myf5 gene (Fig. 1A). A correctly targeted ES clone was identified by a 4kb decrease in electrophoretic mobility by Southern hybridization using a 3′ external probe and DNA digestion by EcoRV (Fig. 1B). Germline targeted mice harboring the engineered allele were designated to have the genotype Myf5CreER/wt. Genotyping PCR was optimized to discriminate the Myf5CreER and the wild type Myf5 loci (Fig. 1C). All progeny carrying the Myf5CreER allele were found to be viable and fertile with no gross abnormalities.
Fig. 1. Generation of Myf5CreER mice.
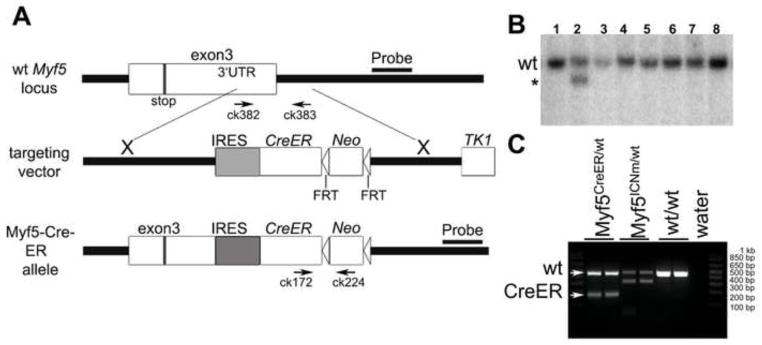
(A) A targeting vector was designed for the insertion of a multifunctional cassette into the 3′ UTR of the Myf5 gene. At the AseI site in exon 3, an IRES-CreER construct was inserted to allow bi-cistronic expression of Myf5 and CreER. An FRT-Neo-FRT cassette was inserted 3′ to IRESCreER. The positions of PCR genotyping primers ck172, ck224, ck382 and ck383 are shown. Neo; neomycin resistance gene. Stop; stop codon. AseI; AseI restriction endonuclease site. (B) Southern blot analysis of ES cell clones using a 3′ external probe. One correctly targeted clone (line 2) shows a band (asterisk), specific to the mutant Myf5CreER allele, which is 4 kb smaller than the wild type band. (C) Genotyping PCR shows Myf5-CreER (182 bp), wild type (454 bp), and Myf5-ICNm (Haldar et al., 2007) (312 bp) alleles, respectively.
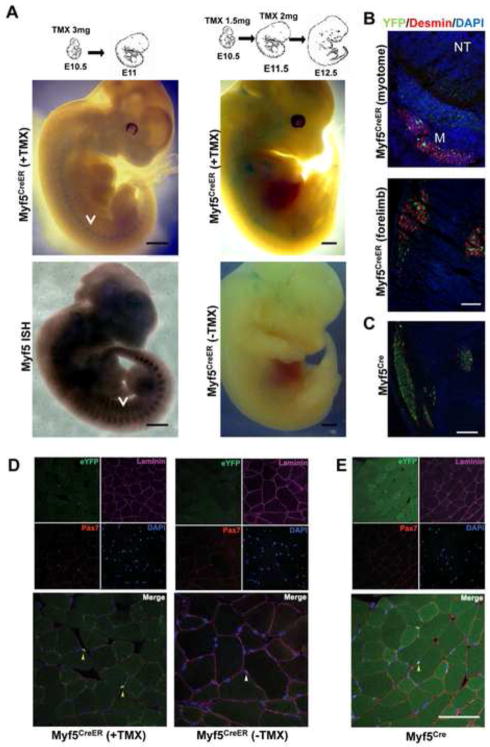
In order to test the TMX-inducible Cre activity from the engineered Myf5 locus, R26RLacZ/LacZ females were crossed with Myf5CreER/wt males and received a single injection of TMX at 10.5 days postcoitum or consecutive injections at 10.5 and 11.5 days postcoitum. Myf5CreER/wt; R26RLacZ/wt embryos were harvested 1 day after the end of the TMX treatment and were processed for whole-mount Xgal staining. β-galactosidase activity was evident in the myotomes of E11.5 embryos, paralleling the endogenous expression of Myf5, and in the developing muscles of limbs, the trunk and the head of E12.5 embryos (Fig. 2A). No expression could be observed in the absence of TMX administration (Fig. 2A). Immunostaining of E12.5 Myf5CreER/wt; R26RYFP/wt embryos treated with TMX at E10.5 and E11.5 using antibodies recognizing YFP and the myogenic marker Desmin showed that the YFP+ve cells specifically localized to the myotome and to forming muscle masses in the limbs (Fig. 2B). The pattern of YFP expression resembled that observed in non-inducible Myf5Cre/wt; R26RYFP/wt embryos, although the number of marked cells was higher in the latter (Fig. 2C).
Fig. 2. Characterization of Myf5CreER mice.
(A) Whole-mount Xgal staining of Myf5CreER/wt; R26RLacZ/wt embryos. TMX administration protocols and stages as indicated. Myf5CreER/wt; R26RLacZ/wt embryos injected with corn oil were used as negative controls. Whole-mount in situ hybridization (ISH) of E11.5 embryos with Myf5-specific probes is shown to highlight the pattern of Myf5 expression. Arrows mark the myotomes. Bar, 1 mm. (B) Transverse sections of Myf5CreER/wt; R26RYFP/wt E12.5 embryos treated with TMX (5 mg) at E10.5 and E11.5 were stained with antibodies recognizing YFP and Desmin. DAPI was used to stain the nuclei. YFP+ve cells localize to the myotome (M; Desmin+ve) lateral to the neural tube (NT) and in the forming muscle masses in the forelimbs. Bar, 50 μm. (C) Sections of the limb of E12.5 Myf5Cre/wt; R26RYFP/wt are shown as term of comparison. Bar, 50 μm. (D) Immunofluorescence analysis of skeletal muscle from adult Myf5CreER/wt; R26RYFP/wt mice 2 weeks after TMX injections, or without TMX administration. Bar, 100 μm. (E) Immunofluorescence analysis of skeletal muscle from Myf5Cre; R26RYFP/wt mice. Frozen sections of tibialis anterior muscles were immunostained for YFP, Pax7, and Laminin. Nuclei were counterstained with DAPI. Yellow arrows point to YFP+ve satellite cells (D, E). A white arrow on the merged image shows a YFP−ve satellite cell localized beneath the basal membrane (D).
To determine if the expression of Cre-recombinase from the engineered Myf5 locus paralleled the endogenous expression of Myf5 in adult muscles, we administered TMX intraperitoneally to four-week-old Myf5CreER/wt; R26RYFP/wt mice. Myf5 is expressed at variable levels among adult quiescent satellite cells (Beauchamp et al., 2000;Gayraud-Morel et al., 2012). Tibialis anterior muscles were harvested for immunohistochemistry 2 weeks after the beginning of the TMX-treatment. Muscle satellite cells, defined as Pax7+ve cells localized beneath the fiber basal laminae, were assessed for YFP expression (Fig. 2D). Out of 96 Pax7+ve satellite cells counted, YFP expression was detected in 71. To confirm the muscle specificity of the Myf5CreER allele, other organs, including brain (cerebrum and cerebellum), liver, kidney, spleen, and testis, from TMX-treated Myf5Cre/wt; R26RYFP/wt mice were also examined for YFP expression. YFP expression was not detectable in any of those tissues. In order to test if the Myf5CreER allele could induce recombination independently of TMX, muscle sections from Myf5CreER/wt; R26RYFP/wt mice without TMX administration were analyzed by immunofluorescence. In those animals, YFP expression was not detected in any of the 46 Pax7+ve satellite cells examined (Fig. 2D), suggesting a tight regulation by TMX of the recombinase activity in the Myf5CreER strain. The leakiness in absence of TMX was further quantified on cells isolated by enzymatic digestion from adult hindlimb muscles. Only one YFP+ve cell was observed when 2084 myogenic cells were analyzed (~0.05%, Suppl. Table 2). Moreover YFP was not detected by immunofluorescence in any other cells in the tissue, again indicating a high specificity of the Myf5CreER locus. We then compared the lineage tracing of the Myf5CreER allele to that of the constitutively active Myf5Cre allele. To determine the percentage of satellite cells in the Myf5 lineage using a Myf5Cre allele constructed nearly identically to the Myf5CreER allele, tibialis anterior muscles from age-matched Myf5Cre/wt; R26RYFP/wt mice were examined (Fig. 2E). YFP expression was detected in 37 out of 39 Pax7+ve satellite cells (95%). Together, these data demonstrate that the pattern of Cre expression in the Myf5CreER mice accurately reflects the activity of the Myf5 locus in muscles during embryonic development and postnatally.
Myf5CreER activity can be used to mark Myf5-expressing cells at different developmental stages
Different muscle groups exhibit different growth kinetics and distinct developmental myogenic programs (Biressi et al., 2007a). We decided to focus our attention on the muscles of hindlimbs, which represent the muscles in which satellite cells are most commonly studied. We compared different regimens of TMX administration in terms of kinetics and dosage in order to optimize Cre-mediated recombination in myogenic cells. To address this, R26RLacZ/LacZ females were crossed with Myf5CreER/wt males and given either a single injection of TMX at E10.5 or two consecutive injections at E10.5 and E11.5 to induce the expression of the reporter gene. E16 fetuses were collected and processed for whole-mount Xgal staining. Whereas both TMX administration protocols clearly induced Cre-mediated recombination in the forelimb muscles, only by administering TMX at E11.5 was robust β-galactosidase activity detectable in hindlimb muscles (Fig. 3A), probably reflecting the different activity of the Myf5 locus as myogenesis is proceeding along a rostro-caudal gradient. This observation indicates that injected TMX is effective for a narrow window of time, allowing for tight temporal control of Cre-mediated recombination in Myf5CreER mice.
Fig. 3. Temporal control and efficiency of TMX-induced labeling in developing Myf5CreER mice.
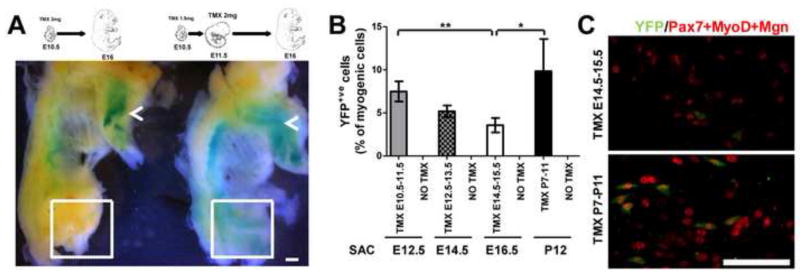
(A) Whole-mount Xgal staining of Myf5CreER/wt; R26RLacZ/wt skinned E16 fetuses. Alternative TMX administration protocols are indicated. Depending on the protocol used, Cre-mediated recombination was induced in the hindlimb muscles (dashed box) in addition to forelimb muscles (arrows). Bar, 1.2 mm. (B) Cells are isolated from hindlimb muscles of Myf5CreER/wt; R26RYFP/wt mice treated with the indicated regimen of TMX and sacrificed (Sac) at the indicated stages, and are stained with antibodies recognizing YFP and the myogenic markers Pax7, MyoD and Myogenin (Mgn). The labeling efficiency is quantified by counting the number of myogenic cells (positive for Pax7, MyoD or Mgn) that are YFP+ve. Results are expressed as mean ± standard deviation. The labeling efficacy detected at E16.5 is lower compared to that obtained postnatally and at earlier embryonic stages (*P<0.05; **P<0.01). (C) Representative images of cells obtained from E16.5 fetuses and P12 pups treated with TMX at the indicated times are shown. Bar, 50 μm.
In terms of dosage, we opted for two injections on consecutive days with different amounts of TMX depending on the developmental stage (embryonic: E10.5 (5 mg) + E11.5 (5 mg); early fetal: E12.5 (7.5 mg) + E13.5 (10 mg); mid-fetal stage: E14.5 (7.5 mg) + E15.5 (10 mg)). Newborn mice were injected subcutaneously with 0.5 mg TMX on five consecutive days from P7 to P11. Higher dosages caused a high rate of lethality. The extent of labeling with these protocols was evaluated on cells obtained by enzymatic digestion of the hindlimb muscles from Myf5CreER/wt; R26RYFP/wt mice 1 day after the end of the TMX treatment (Fig. 3B), and it ranged between approximately 4% observed at the mid-fetal stage (Fig. 3C) to approximately 10% at the neonatal stage (Fig. 3D, Suppl. Table 1). The quantification of YFP+ve cells in the absence of TMX administration indicated a negligible TMX-independent recombination at all developmental stages evaluated (<0.1%, Suppl. Table1).
Previous work showed that the removal of the Neomicin (Neo) cassette from mice engineered to express Cre-recombinase under the control of the Myf5 locus increased the labeling efficiency when compared to the parental strain containing the Neo cassette (Haldar et al., 2008). In order to maximize the number of cells we are able to mark at fetal stage (the stage presenting minor labeling), we excised the Neo cassette by breeding the Myf5CreER line with a strain ubiquitously expressing Flippase-recombinase (Suppl. Fig. 1A, 1B). We than evaluated the labeling efficiency on myogenic cells isolated from the hindlimbs of E16.5 fetuses as previously described for the mice containing the Neo cassette (Suppl. Fig. 1C). At this stage we found only a slight increase in the number of YFP+ve cells upon removal of the Neo cassette (3.8±1.6% and 3.6±0.8% in absence and presence, respectively, of the Neo cassette). This result suggests that, for the purposes of this study, the strain without the Neo cassette does not present a significant advantage over the strain with the Neo cassette. Based on this observation and our findings of the tissue-specificity of the strain containing the Neo cassette (see above), we chose to use these mice for further study.
Myf5 expression during fetal myogenesis defines the progenitors of the majority of adult satellite cells
The expression of the Cre-inducible reporter was detectable in satellite cells from adult (>2 months old) Myf5CreER/wt; R26RYFP/wt mice treated with TMX at each developmental stage evaluated, although the fraction of marked cells was low when TMX was administrated before E14.5. Experiments with single fibers from EDL muscles of Myf5CreER/wt; R26RYFP/wt mice confirmed the satellite cell identity of marked cells by co-staining with antibodies recognizing YFP and the satellite cell markers Pax7 and Syn4 (Fig. 4A). A similar result was obtained using β-galactosidase (Fig. 4B). YFP+ve satellite cells were easy to identify when TMX was administered after E14.5 (Fig. 4C). Interestingly, the adult EDL myofibers showed a weaker Xgal or YFP staining in comparison to the associated satellite. There are at least two (non-mutually exclusive) explanations for this observation. First, as the recombination of the R26R-YFP/LacZ locus in the Myf5CreER line is incomplete, it is expected that only a fraction of the myonuclei express the reporter gene, therefore diluting the signal. Second, the Rosa26 locus, which drives the expression of the reporter genes upon Cre-mediated recombination, could be less active in the terminally differentiated fiber than in the satellite cells. This second hypothesis is strongly supported by the observation that a very weak YFP signal was also observed in fibers compared to satellite cells when highly efficient Cre lines, such as the TMX-independent Myf5Cre strain (see Fig. 2E), were used. In those strains in which virtually all of the myonuclei would have recombined alleles, fibers still exhibit weak reporter gene expression from the Rosa26 locus.
Fig. 4. Adult satellite cells derive from progenitors expressing Myf5 during prenatal development.
(A) An example of a YFP+ve satellite cell (arrow) associated with an EDL fiber of an adult Myf5CreER/wt; R26RYFP/wt mouse treated with TMX at E10.5–E11.5. Staining with antibodies recognizing YFP and the satellite cell markers Pax7 and Syn4 is presented. Bar, 20 μm. (B) Xgal staining of an EDL fiber from an adult Myf5CreER/wt; R26RLacZ/wt mouse treated with TMX at E10.5–E11.5. A β-gal+ve satellite cell is shown (arrow and insets). Bar, 30 μm. (C) An example of a satellite cell expressing the reporter gene YFP (arrowhead) after administration of TMX to E14.5–E15.5 is shown. Note that due to the relatively low efficiency of recombination, most of the satellite cells are YFP−ve (asterisk). DAPI was used to stain the nuclei. Bar, 40 μm. (D) The fraction of the VCAM+ve/CD45−ve/CD31−ve/Sca1−ve satellite cells expressing the lineage marker YFP (FITC) was quantified by FACS on cells obtained from adult Myf5CreER/wt; R26RYFP/wt mice after TMX administration at different developmental stages, as indicated. Satellite cells from Pax7CreER/wt; R26RYFP/wt mice injected with TMX at adult stage were used as positive controls. Note that the analysis of the satellite cells obtained from Myf5CreER/wt; R26RYFP/wt not treated with TMX reveals a virtual absence of recombination. (E) Cells were isolated by enzymatic digestion from hindlimb muscles of Myf5CreER/wt; R26RYFP/wt mice treated with TMX at the indicated stages and allowed to adhere overnight in vitro. Cells were then stained with an antibody to YFP and with a cocktail of antibodies to Pax7, MyoD, and Mgn to reveal the total pool of myogenic cells. Bar, 25 μm. (F) Data from replicate studies shown in panel E were quantified to determine the percentage of myogenic cells that were marked by YFP. Results are expressed as mean ± standard deviation. The recombination obtained after TMX administration at E14.5–E15.5 or P7–P11 is significantly higher compared that detected at earlier stages (** P<0.01, * P<0.05). (G) Data from replicate studies shown in panel E were quantified to determine the percentage of myogenic cells that were marked by YFP and were normalized for the average efficiency of recombination observed after TMX treatment at different stages (see Fig. 3B). Each square represents the normalized value obtained in a single replicate (n=3 for each condition). Note that a minority of satellite cells traces its origin back to progenitors expressing Myf5 at the embryonic (E10.5–E11.5) or early fetal (E12.5–E13.5) stage, whereas >75% of the satellite cells derive from progenitors expressing Myf5 at the mid-fetal (E14.5–15.5) or neonatal (P7–11) stage.
Satellite cells can be prospectively isolated from cellular suspensions of adult muscles according to the surface expression of VCAM and the absence of CD45, CD31 and Sca1 (Bjornson et al., 2012). FACS analysis can be therefore used to quantify the fraction of the VCAM+ve/CD45−ve/CD31−ve/Sca1−ve satellite cells expressing the lineage marker YFP. More satellite cells were labeled when TMX injections in Myf5CreER/wt; R26RYFP/wt mice were performed at the neonatal stage than at either the embryonic or fetal stages. Moreover, TMX injection at the mid-fetal stage (E14.5–E15.5) generated more labeled satellite cells, as assessed by FACS, than TMX treatment at earlier stages (Fig. 4D). A similar trend was confirmed by quantifying the percentage of myogenic cells, which were YFP+ve using immunofluorescence microscopy (Fig. 4E, 4F, Suppl. Table 2). Notably, when the absolute numbers were normalized for the percentage of labeled myogenic cells observed after TMX treatment at each stage (see Fig. 3B), the major contribution to the adult satellite cell compartment is from cells expressing Myf5 at the mid-fetal stage (E14.5–E15.5) (Fig. 4G). These data suggest that it is at the mid-fetal stage that a large fraction (>75%) of the progenitors that will give rise to adult satellite cells commit to the myogenic lineage by inducing Myf5 expression, and that only a minority initiate the myogenic program (as defined by Myf5 expression) earlier during development (Fig. 4G). The Myf5 locus remained active also in neonatal progenitors and adult satellite cells (Fig. 2E, 4G).
We subsequently evaluated if satellite cells expressing Myf5 before adulthood could participate in muscle regeneration. To this end, injury was induced by cardiotoxin injection into hindlimb muscles of adult Myf5CreER/wt; R26RYFP/wt mice to which TMX has been administered during the newborn period. Four days after injury, the presence of labeled cells was evaluated by FACS analysis. A roughly similar proportion of cells were labeled in injured and uninjured muscles (Suppl. Fig. 2A; Fig. 4D), suggesting that satellite cells expressing Myf5 before adulthood can undergo a proliferative expansion upon injury. Moreover labeled cells expressing the myogenic activation marker MyoD and the differentiation marker Myogenin were rapidly identified in injured muscles demonstrating their ability to progress into the myogenic program in vivo (Suppl. Fig. 2B).
The early induction of Myf5 in the progenitors of satellite cells is associated with the beginning of the fetal myogenic program in the limb muscles
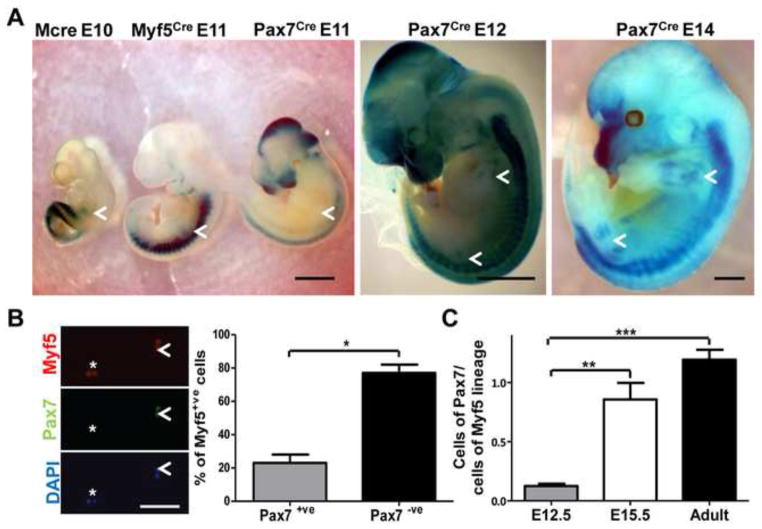
During limb muscle development Pax7+ve cells contribute negligibly to embryonic myogenesis but in their absence fetal myogenesis is dramatically compromised (Hutcheson et al., 2009). Likewise, the comparison of the expression of β-galactosidase induced in R26RLacZ/wt lines by the expression of Cre under the control of regulatory sequences of Pax3 (Mcre), Myf5 and Pax7 clearly showed that Pax7 is induced for the first time in progenitors in the limbs only at the end of primary (embryonic) myogenesis (Fig. 5A). Pax7 expression became more robust during fetal myogenesis (Fig. 5A).
Fig. 5. Enrichment in cells of the Pax7-lineage in the limb muscles at the fetal stage.
(A) Whole-mount Xgal staining of MCre+/−; R26RLacZ/wt, Myf5Cre/wt; R26RLacZ/wt and Pax7Cre/wt; R26RLacZ/wt mice at the indicated developmental stages. Limbs are indicated with arrows. Bar, 3 mm. (B) DsRED+ve cells were isolated by FACS from the hindlimb muscles of Myf5Cre/wt; Z/RED+/− E12.5 embryos, cultured for 4 hours in vitro in proliferating conditions in presence of the cytokinesis blocker cytochalasin, and stained with antibodies recognizing Pax7 and Myf5. The percentage of Myf5+ve doublets (proliferating myoblasts) positive (arrowhead) or negative (asterisk) for Pax7 is quantified. Bar, 25 μm. (C) DsRED+ve cells obtained from the hindlimb muscles of Myf5Cre/wt; Z/RED+/− and Pax7Cre/wt; Z/RED+/− mice at E12.5, E15.5 and adult stages were quantified by FACS. Data from the Pax7 line are expressed as percentage of DsRED+ve cells from the Myf5 line at the same stage.
It has been recently shown by in vivo lineage tracing that Pax7+ve cells present in the developing limbs as early as the embryonic stage E11.5 can give rise to adult satellite cells (Lepper and Fan, 2010). Our data indicate that a minority of adult satellite cells derives from cells expressing Myf5 during embryonic myogenesis (see above). Together these observations raise the possibility that Pax7+ve/Myf5+ve cells in the embryonic limbs could give rise to satellite cells. If this hypothesis holds true, a population of Pax7+ve/Myf5+ve myoblasts which is distinct from embryonic myoblasts (Pax7−ve) should be already present in the embryonic limbs. To test this, we decided to quantify the co-expression of Myf5 and Pax7 in myogenic progenitors at E12.5, extending previous observations (Kassar-Duchossoy et al., 2005). We purified myogenic cells from the hindlimb muscles of E12.5 Myf5Cre/wt; Z/RED+/− embryos by FACS (Suppl. Fig. 3A, 2B) (Boutet et al., 2010), and we quantified the percentage of Myf5+ve cells co-expressing Pax7. We restricted the analysis to proliferating cells by evaluating only the cell doublets, blocked at cytokinesis by the addition of cytochalasin. Under these conditions, ~20% of the Myf5+ve cells also expressed Pax7 (Fig. 5B). As we restricted the analysis to proliferating cells, we can exclude the possibility that Pax7−ve/Myf5+ve cells consist of differentiating cells, which have already down-regulated Pax7 expression. Rather, the presence in the pool of proliferating cells of Myf5-expressing cells, which are alternatively Pax7+ve or Pax7−ve, suggest the co-existence of two distinct populations in the hindlimbs at E12.5. As fetal, but not embryonic, myogenesis in the hindlimbs is dependent on Pax7 (see above), it is likely that the predominant population of Pax7−ve/Myf5+ve cells consists of embryonic myoblasts, whereas the subpopulation of Pax7+ve/Myf5+ve cells could include early fetal myoblasts and the ancestors of the satellite cells. In keeping with this idea, as development proceeds, the fraction of the hindlimb cells marked by the Pax7Cre lineage tracer increases when compared to the number of cells labeled in the Myf5Cre line (Fig. 5C), correlating with the increased contribution to the satellite cell compartment by cells expressing Myf5 at the mid-fetal stage (see above). Due to the absence of tools that would allow us to genetically label cells that are simultaneously expressing Myf5 and Pax7, we could not trace in time the fate of the Pax7+ve/Myf5+ve cells. Nevertheless, the data reported here suggest that some ancestors of the satellite cells are part of the pool of the Pax7+ve/Myf5+ve cells present in the late embryonic limbs and link the beginning of the fetal myogenic program with the initial induction of Myf5 in the precursors of the satellite cells.
DISCUSSION
Satellite cell ancestors and other myogenic precursors
Satellite cells in limb and body wall musculature, like embryonic and fetal myoblasts, are somitic in origin (Armand et al., 1983;Gros et al., 2005;Schienda et al., 2006). Satellite cell precursors have been reported to express Pax3 and Pax7, but not MRFs (Kassar-Duchossoy et al., 2005;Relaix et al., 2005). These Pax+ve/MRF−ve precursors were observed throughout development from early embryonic to postnatal stage. At the fetal stage, a fraction of mononucleated Pax7+ve cells could be identified beneath the fibers’ basal laminae, in the typical satellite cell position (Kassar-Duchossoy et al., 2005). However, we have no data that indicates the fate of these cells. Specifically, we do not know if they (or their progeny) were destined to remain in the sublaminar position to become the satellite cells in adult muscle, if they were destined to fuse to growing myofibers, or some combination of the two. It has been suggested that Pax+ve/MRF−ve precursors could both give rise to MRF+ve myoblasts and self-renew as MRF−ve cells (Kassar-Duchossoy et al., 2005;Relaix et al., 2005). On the other hand, a recent lineage study relying on mice expressing Cre recombinase under the control of the regulatory regions of MyoD suggests that the great majority (~98%) of satellite cells derive from cells MyoD-expressing progenitors present pre- or peri-natally (Kanisicak et al., 2009). Therefore, it remains unclear to what extent the Pax+ve/MRF−ve population is a stable reservoir of MRF−ve progenitors. Likewise, the lineage relationship between MRF−ve precursors, developmental myoblasts and satellite cells remains undetermined.
Skeletal muscle in mammals is formed through successive phases of fiber formation involving different generations of myoblasts (embryonic, fetal, and postnatal) (Biressi et al., 2007a;Murphy and Kardon, 2011;Stockdale, 1992;Tajbakhsh, 2005). Myoblasts participating in skeletal muscle development are highly heterogeneous in terms of morphology, differentiation kinetics, response to growth factors, gene expression before and after differentiation and active signaling pathways (Biressi et al., 2007a). A genome-wide expression analysis carried out on myoblasts purified at embryonic and fetal stages strongly suggest that they represent intrinsically distinct cell populations (Biressi et al., 2007b). Furthermore, a specific myogenic program in fetal muscle dependent on the Pax7-Nfix axis (Messina et al., 2010) and on Wnt signaling (Hutcheson et al., 2009) was identified. Differences in the ability to respond to chemicals in vitro have been reported between fetal and postnatal myoblasts (Cossu et al., 1983). Moreover, a gene inactivation study revealed that Pax7 is required for the regeneration of juvenile (before P21) but not adult muscles (Lepper et al., 2009). These observations suggest that fetal myoblasts, neonatal myoblasts, and adult satellite cells could also be intrinsically different lineages.
The observations presented here suggest that the majority of adult satellite cell precursors express Myf5 at the fetal stage. Only a minority (< 25%) of satellite cell precursors had expressed Myf5 at the embryonic stage. Furthermore, we show here (Fig. 5) that the early appearance of Myf5+ve satellite cell ancestors at embryonic stages overlaps with the initial appearance of the fetal and satellite cell marker Pax7 in the limbs (Hutcheson et al., 2009), indicating that the activation of Myf5 expression in the precursors of the satellite cells in the embryonic state could in fact be linked with the beginning of fetal myogenesis. Intriguingly, recent studies revealed the presence of non-cycling (Ki67−ve) Pax7+ve/Myf5+vecells at the fetal (E15) but not at the embryonic (E12) stage (Picard and Marcelle, 2013). Based on the genetic models used in that study, those investigators did not provide direct evidence that these quiescent cells expressing Myf5 at the fetal stage could give rise to the satellite cells in the adult. Nevertheless, those data indirectly support the results presented here that adult satellite cells preferentially derive from fetal Myf5+ve cells, whereas the embryonic Myf5+ve cells have limited, if any contribution. Therefore although the majority of fetal myoblasts is terminally differentiating at the fetal stage, some of their progeny remain as committed but undifferentiated cells until the end of development to give rise to satellite cells. The mechanisms regulating this fate decision are completely unknown and could possibly depend on an intrinsic heterogeneity in the pool of fetal myoblasts. The identification of molecular markers (if they exist) that can be employed to discriminate satellite cell precursors from other lineages will possibly clarify this point in future.
A recent study demonstrates that pericytes resident in post-natal growing muscle can contribute to the adult satellite cell pool during normal growth (Dellavalle et al., 2011). The authors of this study employed an alkaline phosphatase CreER mouse strain to mark pericytes with an efficiency of ~70%. They were able to trace back to pericytes the origin of a fraction of satellite cells. Intriguingly, the contribution of pericytes to the satellite cell pool is different in muscles with distinct developmental origins, ranging from almost 25% in the pectoralis to less than 10% in the hindlimb muscles. These results are not in contradiction with those reported here, showing that a large number of the satellite cells resident in adult hindlimb muscles derive from cells expressing Myf5 at a fetal stage. Intriguingly it has been recently shown that a fraction of pericytes transiently induce Myf5 during fetal development (Cappellari et al., 2013). These pericytes are likely to maintain myogenic features, as essentially all the YFP+ve cells we could isolate from the Myf5CreER/wt; R26RYFP/wt hindlimb muscles treated with TMX at different developmental stages expressed Pax7, MyoD or Myogenin (Suppl. Table 3).
Over the past decade, other MRF−ve cells which have myogenic potential but are distinct from satellite cells have been identified during development or regeneration (Tedesco et al., 2010). It has also been reported that some of them can give rise to satellite cells and participate in muscle regeneration in transplantation studies in irradiated mice or when muscle is challenged by injury or pathological conditions. However, it remains unclear if they generate satellite cells during normal development (Asakura et al., 2002;Dreyfus et al., 2004;LaBarge and Blau, 2002;Mitchell et al., 2010;Sampaolesi et al., 2003;Torrente et al., 2004). Understanding how these populations of putative myogenic cells relate not only to adult satellite cells but also to each other and to embryonic and fetal myoblasts from a lineage point of view will be crucial for the comprehension of skeletal muscle development.
Myf5-independent lineage and considerations related to normalization
The pool of satellite cells is itself heterogeneous (Biressi and Rando, 2010). Particularly relevant for the purposes of this study is a report based on the use of a non-inducible Myf5Cre strain, which shows that approximately 10% of the total population of adult satellite cells associated with EDL fibers do not appear to have a history of Myf5 expression (Kuang et al., 2007). Intriguingly, Myf5 is actively expressed, although at varying levels, in approximately 90% of quiescent satellite cells resident in the adult muscles (Beauchamp et al., 2000;Gayraud-Morel et al., 2012). A recent report showed that approximately 98% of the satellite cells associated with EDL fibers expressed MyoD at some point in their history before reaching adulthood (Kanisicak et al., 2009). These observations suggest the possibility that a small fraction of satellite cells could have a history of MyoD expression, but not of Myf5 expression, in their ancestry. This idea is also supported by the identification through conditional ablation of Myf5 expressing cells of a Myf5 independent lineage, which can participate in muscle formation (Gensch et al., 2008;Haldar et al., 2008). For obvious reasons, this small subpopulation of satellite cells would not be the object of the lineage study presented here. Performing a study similar to the one presented here, but employing an inducible MyoDCreER strain, could possibly give insight as to when this subpopulation of satellite cells is determined to the myogenic lineage.
The data presented here suggest that most satellite cells progenitors with a history of Myf5 expression have already induced its expression by the fetal stage (Fig. 4G). This conclusion depends on the normalization for the labeling efficiency we have quantified in developmental cells (Fig. 3B).
Theoretically, there are limitations in this approach that could lead to an over- or underestimation of the proportion of satellite cells derived from progenitors expressing Myf5 at a specific developmental stage. First, the normalization we have used is based on the assumption that the activity of the Myf5 locus is fairly homogeneous in myogenic cells throughout development. Studies employing the Myf5nLacZ mouse strain suggest that in the great majority of Pax7+ve progenitors in the developing limbs, the Myf5 locus is transcriptionally active (Kassar-Duchossoy et al., 2005). A recently published immunofluorescence study suggests that the fraction of Pax7+ve cells that co-express Myf5 protein, although still representing the majority of the cells, could be reduced during the fetal stage (Picard and Marcelle, 2013). These observations and any possible future observation related to the assumptions made for the normalization should be taken into account during the interpretation of the model we are presenting here. Moreover, the quantification of the extent of labeling after TMX injections is affected by a certain degree of variability, ranging from 5.8 to 8.3% at E10.5–11.5 from 4.6 to 5.9% at E12.5–13.5, from 3 to 4.5% at E14.5–15.5, and from 5.6 to 12.5% at P7–P11. Furthermore, the identification of labeled cells relies on the productive excision of the stop-sequence necessary for the induction of the reporter gene and implies that the expression of the reporter gene from the Rosa26 locus reaches detectable levels (Soriano, 1999;Srinivas et al., 2001). To have an accurate quantification of the efficiency of labeling, the temporal window between TMX administration and the analysis should allow for maximal recombination to occur. In this study, labeling was quantified using cells isolated one day after the end of the TMX treatment (see above). It is conceivable that, under these conditions, our analysis could have missed some late recombining cells. Nevertheless, several observations indicate that they should represent only a small fraction of the marked cells: 1) TMX has been reported to have a relatively short half-life (t1/2 < 12 hours) in mouse tissues (Robinson et al., 1991); 2) previous studies relying on different CreER mouse lines indicate that most of the recombination occurs in embryos between 12 and 24 hours after a single TMX injection to the pregnant mother (Hayashi and McMahon, 2002;Nakamura et al., 2006); 3) after 24 hours, the nuclear accumulation of CreER is dramatically reduced and CreER activity is negligible after 24–48 hours depending on the experimental setting, indicating that effective TMX levels are maintained for a short time window in the embryonic circulation (Hayashi and McMahon, 2002;Nakamura et al., 2006); 4) although the Myf5 locus is active from E11 onward in the hindlimbs (Ott et al., 1991), we observed only rare labeled cells when a single dose of TMX was administered the previous day, whereas an intense labeling was observed in developing muscles after an additional TMX injection at E11.5 (Fig. 3A). For these reasons, although it is possible that a fraction of the satellite cell ancestors induce Myf5 expression for the first time after the fetal stage, the data presented here suggest that the majority of them have already expressed Myf5 before birth. Moreover, the quantification of the efficiency of labeling was performed in the same way at all developmental stages considered. Therefore, the normalized data reported here (Fig. 4G) should truly reflect the dramatic increase (> 8-fold) in the induction of the myogenic program in satellite cell ancestors as development proceeds from the embryonic to the fetal stage.
Applications of the Myf5CreER mouse
In this report, we describe studies using a novel mouse in which the cDNA coding for a TMX-inducible CreER-recombinase is inserted in the 3′ UTR of the Myf5 locus. We employed this strain to genetically mark Myf5-expressing cells of the hindlimbs at different developmental stages and to evaluate the contribution of their progeny to the adult satellite cell compartment. There are, however, many developmental studies for which this strain will be of great value. For example, studies with mice expressing CreER under the control of Pax7 (Lepper and Fan, 2010) and En1 (Atit et al., 2006) suggest that the central dermomyotome contains multi-potent progenitors for muscle, dorsal dermis and brown adipose tissue (BAT). Intriguingly, it has been shown that BAT arises from cells transiently expressing Myf5 (Seale et al., 2008). The Myf5CreER strain could be used to determine at which developmental stage the progenitors giving rise to BAT are expressing Myf5 and to investigate the relationship between these Myf5-expressing cells and the progenitors in the dermomyotome.
The Myf5CreER strain represents not only a useful tool for lineage studies in which the fate of Myf5-expressing cells can be evaluated in vivo, but also in studies in which the effects of the conditional ablation of specific genes is evaluated. As opposed to strains expressing CreER under the control of the Pax7 locus (Brack et al., 2007;Lepper and Fan, 2010;Nishijo et al., 2009), the Myf5CreER mice presented here could be employed to specifically ablate gene expression only in the subpopulation of satellite cells expressing Myf5 (Beauchamp et al., 2000;Gayraud-Morel et al., 2012) or with a history of Myf5 expression (Kuang et al., 2007). This could be particularly important to evaluate the molecular basis of aspects of satellite cell heterogeneity. In this regard, as a phenotype has been reported in mice with one mutated Myf5 allele (Gayraud-Morel et al., 2012), it is important to note that the strategy adopted to generate the Myf5CreER strain does not disrupt the coding region of the gene. Moreover, we were able to obtain relatively high efficiencies of recombination in the satellite cell compartment. Approximately 74% of satellite cells were marked by 5 consecutive injections of 2 mg of TMX in adult mice. The expression of Myf5 has been reported only in approximately 90% of the satellite cells. The difference between the number of satellite cells expressing Myf5 and those expressing the label is likely resulting from incomplete CreER-mediated recombination. Therefore, the efficiency of recombination appears to be more than 80%, which is comparable to the recombination obtained in Pax7CreER mice (Nishijo et al., 2009), which have been successfully employed to abolish gene expression in the satellite cell compartment in several studies (Bjornson et al., 2012;Quach et al., 2009;Shea et al., 2010). Potentially the efficiency of recombination could be further improved by the removal of the neomycin–coding cassette, as shown for the corresponding non-inducible, Cre recombinase-expressing mouse lines (Haldar et al., 2008).
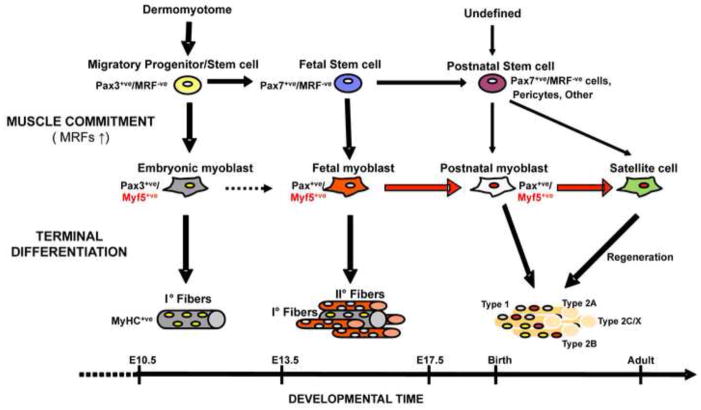
In conclusion, we present data derived using a newly generated Myf5CreER mouse strain to trace back the developmental time at which the precursors of the satellite cells enter the myogenic program through the induction of Myf5. This study suggests that most of the satellite cells resident in adult hindlimb muscles are likely to have activated Myf5 expression during fetal myogenesis, and that it is the Myf5+ve fetal myoblast that is the primary progenitor present during development that gives rise to the satellite cell pool (Fig. 6).
Fig. 6. Proposed lineage scheme for hindlimb muscles.
Pax3+ve cells delaminate from the somitic dermomyotome and migrate into the limb buds, where they generate embryonic myoblasts which terminally differentiate into primary fibers. As development proceeds into the fetal stage, muscle stem cells resident in the limb activate the expression of Pax7. Pax7+ve/MRF−ve progenitors remain throughout fetal myogenesis and after birth as source of myoblasts (fetal and postnatal) and possibly satellite cells. During secondary myogenesis they generate fetal myoblasts by expressing Myf5 and other MRFs. Fetal Myf5+ve cells alternatively terminally differentiate during secondary myogenesis or remain as committed, but undifferentiated cells until successive phases of muscle development. After birth they contribute to the pool myoblasts responsible for post-natal growth and give rise to the majority of the adult satellite cells. A minority of the satellite cells resident in the adult limb muscles is derived from pericytes present in the neonatal muscles and possibly from other postnatal stem cells. The dashed arrow indicates the still poorly defined lineage relationship between embryonic and fetal myoblasts. The red color and the weight of the arrows highlight the predominance of the lineage described in this study.
Supplementary Material
Highlights.
A new Tamoxifen-inducible Myf5CreER mouse, have been generated and characterized.
The contribution of developmental myoblasts to the satellite cell pool has been studied.
A significant number of satellite cells derive from progenitors expressing Myf5 during fetal stage.
Acknowledgments
The authors thank Drs. Giulio Cossu, Brad Olwin, and Peter D. Yurchenco for reagents, Drs. Tom Cheung, Marco Quarta, Elaine Huang, and Lusijah Rott for suggestions, help with the animal care and FACS strategy. We thank the staff of the Stanford Transgenic facility for assistance in the generation of the Myf5CreER mice.
This work was supported by the Glenn Foundation for Medical Research and by grants from the NIH (R37 AG23806, R01 AR056849, and an NIH Director’s Pioneer Award) and the Department of Veterans Affairs (Merit Review) to TAR.
Abbreviations
- HL
hindlimb
- FACS
fluorescence activated cell sorting
- TMX
Tamoxifen
- MyHC
myosin heavy chain
- MRFs
myogenic regulatory factors
- Neo
Neomicin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Armand O, Boutineau AM, Mauger A, Pautou MP, Kieny M. Origin of satellite cells in avian skeletal muscles. Arch Anat Microsc Morphol Exp. 1983;72:163–181. [PubMed] [Google Scholar]
- Asakura A, Seale P, Girgis-Gabardo A, Rudnicki MA. Myogenic specification of side population cells in skeletal muscle. J Cell Biol. 2002;159:123–134. doi: 10.1083/jcb.200202092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atit R, Sgaier SK, Mohamed OA, Taketo MM, Dufort D, Joyner AL, Niswander L, Conlon RA. Beta-catenin activation is necessary and sufficient to specify the dorsal dermal fate in the mouse. Dev Biol. 2006;296:164–176. doi: 10.1016/j.ydbio.2006.04.449. [DOI] [PubMed] [Google Scholar]
- Beauchamp JR, Heslop L, Yu DS, Tajbakhsh S, Kelly RG, Wernig A, Buckingham ME, Partridge TA, Zammit PS. Expression of CD34 and Myf5 defines the majority of quiescent adult skeletal muscle satellite cells. J Cell Biol. 2000;151:1221–1234. doi: 10.1083/jcb.151.6.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biressi S, Messina G, Collombat P, Tagliafico E, Monteverde S, Benedetti L, Cusella De Angelis MG, Mansouri A, Ferrari S, Tajbakhsh S, Broccoli V, Cossu G. The homeobox gene Arx is a novel positive regulator of embryonic myogenesis. Cell Death Differ. 2008;15:94–104. doi: 10.1038/sj.cdd.4402230. [DOI] [PubMed] [Google Scholar]
- Biressi S, Molinaro M, Cossu G. Cellular heterogeneity during vertebrate skeletal muscle development. Dev Biol. 2007a;308:281–293. doi: 10.1016/j.ydbio.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Biressi S, Rando TA. Heterogeneity in the muscle satellite cell population. Semin Cell Dev Biol. 2010;21:845–854. doi: 10.1016/j.semcdb.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biressi S, Tagliafico E, Lamorte G, Monteverde S, Tenedini E, Roncaglia E, Ferrari S, Ferrari S, Cusella-De Angelis MG, Tajbakhsh S, Cossu G. Intrinsic phenotypic diversity of embryonic and fetal myoblasts is revealed by genome-wide gene expression analysis on purified cells. Dev Biol. 2007b;304:633–651. doi: 10.1016/j.ydbio.2007.01.016. [DOI] [PubMed] [Google Scholar]
- Bjornson CR, Cheung TH, Liu L, Tripathi PV, Steeper KM, Rando TA. Notch signaling is necessary to maintain quiescence in adult muscle stem cells. Stem Cells. 2012;30:232–242. doi: 10.1002/stem.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutet SC, Biressi S, Iori K, Natu V, Rando TA. Taf1 regulates Pax3 protein by monoubiquitination in skeletal muscle progenitors. Mol Cell. 2010;40:749–761. doi: 10.1016/j.molcel.2010.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brack AS, Conboy MJ, Roy S, Lee M, Kuo CJ, Keller C, Rando TA. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science. 2007;317:807–810. doi: 10.1126/science.1144090. [DOI] [PubMed] [Google Scholar]
- Braun T, Rudnicki MA, Arnold HH, Jaenisch R. Targeted inactivation of the muscle regulatory gene Myf-5 results in abnormal rib development and perinatal death. Cell. 1992;71:369–382. doi: 10.1016/0092-8674(92)90507-9. [DOI] [PubMed] [Google Scholar]
- Brown CB, Engleka KA, Wenning J, Min LM, Epstein JA. Identification of a hypaxial somite enhancer element regulating Pax3 expression in migrating myoblasts and characterization of hypaxial muscle Cre transgenic mice. Genesis. 2005;41:202–209. doi: 10.1002/gene.20116. [DOI] [PubMed] [Google Scholar]
- Buckingham M, Relaix F. The role of Pax genes in the development of tissues and organs: Pax3 and Pax7 regulate muscle progenitor cell functions. Annu Rev Cell Dev Biol. 2007;23:645–673. doi: 10.1146/annurev.cellbio.23.090506.123438. [DOI] [PubMed] [Google Scholar]
- Cappellari O, Benedetti S, Innocenzi A, Tedesco FS, Moreno-Fortuny A, Ugarte G, Lampugnani MG, Messina G, Cossu G. Dll4 and PDGF-BB Convert Committed Skeletal Myoblasts to Pericytes without Erasing Their Myogenic Memory. Dev Cell. 2013 doi: 10.1016/j.devcel.2013.01.022. [DOI] [PubMed] [Google Scholar]
- Cossu G, Molinaro M, Pacifici M. Differential response of satellite cells and embryonic myoblasts to a tumor promoter. Dev Biol. 1983;98:520–524. doi: 10.1016/0012-1606(83)90382-2. [DOI] [PubMed] [Google Scholar]
- Darabi R, Gehlbach K, Bachoo RM, Kamath S, Osawa M, Kamm KE, Kyba M, Perlingeiro RC. Functional skeletal muscle regeneration from differentiating embryonic stem cells. Nat Med. 2008;14:134–143. doi: 10.1038/nm1705. [DOI] [PubMed] [Google Scholar]
- Darabi R, Santos FN, Filareto A, Pan W, Koene R, Rudnicki MA, Kyba M, Perlingeiro RC. Assessment of the myogenic stem cell compartment following transplantation of Pax3/Pax7-induced embryonic stem cell-derived progenitors. Stem Cells. 2011;29:777–790. doi: 10.1002/stem.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellavalle A, Maroli G, Covarello D, Azzoni E, Innocenzi A, Perani L, Antonini S, Sambasivan R, Brunelli S, Tajbakhsh S, Cossu G. Pericytes resident in postnatal skeletal muscle differentiate into muscle fibres and generate satellite cells. Nat Commun. 2011;2:499. doi: 10.1038/ncomms1508. [DOI] [PubMed] [Google Scholar]
- Dhawan J, Rando TA. Stem cells in postnatal myogenesis: molecular mechanisms of satellite cell quiescence, activation and replenishment. Trends Cell Biol. 2005;15:666–673. doi: 10.1016/j.tcb.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Dreyfus PA, Chretien F, Chazaud B, Kirova Y, Caramelle P, Garcia L, Butler-Browne G, Gherardi RK. Adult bone marrow-derived stem cells in muscle connective tissue and satellite cell niches. Am J Pathol. 2004;164:773–779. doi: 10.1016/S0002-9440(10)63165-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayraud-Morel B, Chretien F, Jory A, Sambasivan R, Negroni E, Flamant P, Soubigou G, Coppee JY, Di SJ, Cumano A, Mouly V, Tajbakhsh S. Myf5 haploinsufficiency reveals distinct cell fate potentials for adult skeletal muscle stem cells. J Cell Sci. 2012;125:1738–1749. doi: 10.1242/jcs.097006. [DOI] [PubMed] [Google Scholar]
- Gensch N, Borchardt T, Schneider A, Riethmacher D, Braun T. Different autonomous myogenic cell populations revealed by ablation of Myf5-expressing cells during mouse embryogenesis. Development. 2008;135:1597–1604. doi: 10.1242/dev.019331. [DOI] [PubMed] [Google Scholar]
- Gros J, Manceau M, Thome V, Marcelle C. A common somitic origin for embryonic muscle progenitors and satellite cells. Nature. 2005;435:954–958. doi: 10.1038/nature03572. [DOI] [PubMed] [Google Scholar]
- Haldar M, Hancock JD, Coffin CM, Lessnick SL, Capecchi MR. A conditional mouse model of synovial sarcoma: insights into a myogenic origin. Cancer Cell. 2007;11:375–388. doi: 10.1016/j.ccr.2007.01.016. [DOI] [PubMed] [Google Scholar]
- Haldar M, Karan G, Tvrdik P, Capecchi MR. Two cell lineages, myf5 and myf5-independent, participate in mouse skeletal myogenesis. Dev Cell. 2008;14:437–445. doi: 10.1016/j.devcel.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasty P, Bradley A, Morris JH, Edmondson DG, Venuti JM, Olson EN, Klein WH. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature. 1993;364:501–506. doi: 10.1038/364501a0. [DOI] [PubMed] [Google Scholar]
- Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244:305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- Hutcheson DA, Zhao J, Merrell A, Haldar M, Kardon G. Embryonic and fetal limb myogenic cells are derived from developmentally distinct progenitors and have different requirements for beta-catenin. Genes Dev. 2009;23:997–1013. doi: 10.1101/gad.1769009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanisicak O, Mendez JJ, Yamamoto S, Yamamoto M, Goldhamer DJ. Progenitors of skeletal muscle satellite cells express the muscle determination gene, MyoD. Dev Biol. 2009;332:131–141. doi: 10.1016/j.ydbio.2009.05.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassar-Duchossoy L, Gayraud-Morel B, Gomes D, Rocancourt D, Buckingham M, Shinin V, Tajbakhsh S. Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature. 2004;431:466–471. doi: 10.1038/nature02876. [DOI] [PubMed] [Google Scholar]
- Kassar-Duchossoy L, Giacone E, Gayraud-Morel B, Jory A, Gomes D, Tajbakhsh S. Pax3/Pax7 mark a novel population of primitive myogenic cells during development. Genes Dev. 2005;19:1426–1431. doi: 10.1101/gad.345505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller C, Arenkiel BR, Coffin CM, El-Bardeesy N, DePinho RA, Capecchi MR. Alveolar rhabdomyosarcomas in conditional Pax3:Fkhr mice: cooperativity of Ink4a/ARF and Trp53 loss of function. Genes Dev. 2004a;18:2614–2626. doi: 10.1101/gad.1244004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller C, Hansen MS, Coffin CM, Capecchi MR. Pax3:Fkhr interferes with embryonic Pax3 and Pax7 function: implications for alveolar rhabdomyosarcoma cell of origin. Genes Dev. 2004b;18:2608–2613. doi: 10.1101/gad.1243904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang S, Kuroda K, Le GF, Rudnicki MA. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007;129:999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang S, Rudnicki MA. The emerging biology of satellite cells and their therapeutic potential. Trends Mol Med. 2008;14:82–91. doi: 10.1016/j.molmed.2007.12.004. [DOI] [PubMed] [Google Scholar]
- LaBarge MA, Blau HM. Biological progression from adult bone marrow to mononucleate muscle stem cell to multinucleate muscle fiber in response to injury. Cell. 2002;111:589–601. doi: 10.1016/s0092-8674(02)01078-4. [DOI] [PubMed] [Google Scholar]
- Lepper C, Conway SJ, Fan CM. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature. 2009;460:627–631. doi: 10.1038/nature08209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepper C, Fan CM. Inducible lineage tracing of Pax7-descendant cells reveals embryonic origin of adult satellite cells. Genesis. 2010;48:424–436. doi: 10.1002/dvg.20630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepper C, Partridge TA, Fan CM. An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development. 2011;138:3639–3646. doi: 10.1242/dev.067595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroto M, Reshef R, Munsterberg AE, Koester S, Goulding M, Lassar AB. Ectopic Pax-3 activates MyoD and Myf-5 expression in embryonic mesoderm and neural tissue. Cell. 1997;89:139–148. doi: 10.1016/s0092-8674(00)80190-7. [DOI] [PubMed] [Google Scholar]
- Mauro A. Satellite cell of skeletal muscle fibers. J Biophys Biochem Cytol. 1961;9:493–495. doi: 10.1083/jcb.9.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messina G, Biressi S, Monteverde S, Magli A, Cassano M, Perani L, Roncaglia E, Tagliafico E, Starnes L, Campbell CE, Grossi M, Goldhamer DJ, Gronostajski RM, Cossu G. Nfix regulates fetal-specific transcription in developing skeletal muscle. Cell. 2010;140:554–566. doi: 10.1016/j.cell.2010.01.027. [DOI] [PubMed] [Google Scholar]
- Mitchell KJ, Pannerec A, Cadot B, Parlakian A, Besson V, Gomes ER, Marazzi G, Sassoon DA. Identification and characterization of a non-satellite cell muscle resident progenitor during postnatal development. Nat Cell Biol. 2010;12:257–266. doi: 10.1038/ncb2025. [DOI] [PubMed] [Google Scholar]
- Murphy M, Kardon G. Origin of vertebrate limb muscle: the role of progenitor and myoblast populations. Curr Top Dev Biol. 2011;96:1–32. doi: 10.1016/B978-0-12-385940-2.00001-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MM, Lawson JA, Mathew SJ, Hutcheson DA, Kardon G. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development. 2011;138:3625–3637. doi: 10.1242/dev.064162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima Y, Hanaoka K, Hayasaka M, Esumi E, Li S, Nonaka I, Nabeshima Y. Myogenin gene disruption results in perinatal lethality because of severe muscle defect. Nature. 1993;364:532–535. doi: 10.1038/364532a0. [DOI] [PubMed] [Google Scholar]
- Nakamura E, Nguyen MT, Mackem S. Kinetics of tamoxifen-regulated Cre activity in mice using a cartilage-specific CreER(T) to assay temporal activity windows along the proximodistal limb skeleton. Dev Dyn. 2006;235:2603–2612. doi: 10.1002/dvdy.20892. [DOI] [PubMed] [Google Scholar]
- Nishijo K, Hosoyama T, Bjornson CR, Schaffer BS, Prajapati SI, Bahadur AN, Hansen MS, Blandford MC, McCleish AT, Rubin BP, Epstein JA, Rando TA, Capecchi MR, Keller C. Biomarker system for studying muscle, stem cells, and cancer in vivo. FASEB J. 2009;23:2681–2690. doi: 10.1096/fj.08-128116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott MO, Bober E, Lyons G, Arnold H, Buckingham M. Early expression of the myogenic regulatory gene, myf-5, in precursor cells of skeletal muscle in the mouse embryo. Development. 1991;111:1097–1107. doi: 10.1242/dev.111.4.1097. [DOI] [PubMed] [Google Scholar]
- Picard CA, Marcelle C. Two distinct muscle progenitor populations coexist throughout amniote development. Dev Biol. 2013;373:141–148. doi: 10.1016/j.ydbio.2012.10.018. [DOI] [PubMed] [Google Scholar]
- Pownall ME, Gustafsson MK, Emerson CP., Jr Myogenic regulatory factors and the specification of muscle progenitors in vertebrate embryos. Annu Rev Cell Dev Biol. 2002;18:747–783. doi: 10.1146/annurev.cellbio.18.012502.105758. [DOI] [PubMed] [Google Scholar]
- Quach NL, Biressi S, Reichardt LF, Keller C, Rando TA. Focal adhesion kinase signaling regulates the expression of caveolin 3 and beta1 integrin, genes essential for normal myoblast fusion. Mol Biol Cell. 2009;20:3422–3435. doi: 10.1091/mbc.E09-02-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relaix F, Montarras D, Zaffran S, Gayraud-Morel B, Rocancourt D, Tajbakhsh S, Mansouri A, Cumano A, Buckingham M. Pax3 and Pax7 have distinct and overlapping functions in adult muscle progenitor cells. J Cell Biol. 2006;172:91–102. doi: 10.1083/jcb.200508044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relaix F, Rocancourt D, Mansouri A, Buckingham M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature. 2005;435:948–953. doi: 10.1038/nature03594. [DOI] [PubMed] [Google Scholar]
- Robinson SP, Langan-Fahey SM, Johnson DA, Jordan VC. Metabolites, pharmacodynamics, and pharmacokinetics of tamoxifen in rats and mice compared to the breast cancer patient. Drug Metab Dispos. 1991;19:36–43. [PubMed] [Google Scholar]
- Rudnicki MA, Braun T, Hinuma S, Jaenisch R. Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell. 1992;71:383–390. doi: 10.1016/0092-8674(92)90508-a. [DOI] [PubMed] [Google Scholar]
- Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- Sambasivan R, Yao R, Kissenpfennig A, Van WL, Paldi A, Gayraud-Morel B, Guenou H, Malissen B, Tajbakhsh S, Galy A. Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development. 2011;138:3647–3656. doi: 10.1242/dev.067587. [DOI] [PubMed] [Google Scholar]
- Sampaolesi M, Torrente Y, Innocenzi A, Tonlorenzi R, D’Antona G, Pellegrino MA, Barresi R, Bresolin N, De Angelis MG, Campbell KP, Bottinelli R, Cossu G. Cell therapy of alpha-sarcoglycan null dystrophic mice through intra-arterial delivery of mesoangioblasts. Science. 2003;301:487–492. doi: 10.1126/science.1082254. [DOI] [PubMed] [Google Scholar]
- Schienda J, Engleka KA, Jun S, Hansen MS, Epstein JA, Tabin CJ, Kunkel LM, Kardon G. Somitic origin of limb muscle satellite and side population cells. Proc Natl Acad Sci U S A. 2006;103:945–950. doi: 10.1073/pnas.0510164103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, Tempst P, Rudnicki MA, Beier DR, Spiegelman BM. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki MA. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102:777–786. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- Shea KL, Xiang W, LaPorta VS, Licht JD, Keller C, Basson MA, Brack AS. Sprouty1 regulates reversible quiescence of a self-renewing adult muscle stem cell pool during regeneration. Cell Stem Cell. 2010;6:117–129. doi: 10.1016/j.stem.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockdale FE. Myogenic cell lineages. Dev Biol. 1992;154:284–298. doi: 10.1016/0012-1606(92)90068-r. [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S. Skeletal muscle stem and progenitor cells: reconciling genetics and lineage. Exp Cell Res. 2005;306:364–372. doi: 10.1016/j.yexcr.2005.03.033. [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S, Rocancourt D, Cossu G, Buckingham M. Redefining the genetic hierarchies controlling skeletal myogenesis: Pax-3 and Myf-5 act upstream of MyoD. Cell. 1997;89:127–138. doi: 10.1016/s0092-8674(00)80189-0. [DOI] [PubMed] [Google Scholar]
- Tedesco FS, Dellavalle A, az-Manera J, Messina G, Cossu G. Repairing skeletal muscle: regenerative potential of skeletal muscle stem cells. J Clin Invest. 2010;120:11–19. doi: 10.1172/JCI40373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrente Y, Belicchi M, Sampaolesi M, Pisati F, Meregalli M, D’Antona G, Tonlorenzi R, Porretti L, Gavina M, Mamchaoui K, Pellegrino MA, Furling D, Mouly V, Butler-Browne GS, Bottinelli R, Cossu G, Bresolin N. Human circulating AC133(+) stem cells restore dystrophin expression and ameliorate function in dystrophic skeletal muscle. J Clin Invest. 2004;114:182–195. doi: 10.1172/JCI20325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez MR, Richardson JA, Klein WH, Olson EN. Failure of Myf5 to support myogenic differentiation without myogenin, MyoD, and MRF4. Dev Biol. 2000;219:287–298. doi: 10.1006/dbio.2000.9621. [DOI] [PubMed] [Google Scholar]
- Vintersten K, Monetti C, Gertsenstein M, Zhang P, Laszlo L, Biechele S, Nagy A. Mouse in red: red fluorescent protein expression in mouse ES cells, embryos, and adult animals. Genesis. 2004;40:241–246. doi: 10.1002/gene.20095. [DOI] [PubMed] [Google Scholar]
- Weintraub H, Tapscott SJ, Davis RL, Thayer MJ, Adam MA, Lassar AB, Miller AD. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc Natl Acad Sci U S A. 1989;86:5434–5438. doi: 10.1073/pnas.86.14.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zammit PS, Golding JP, Nagata Y, Hudon V, Partridge TA, Beauchamp JR. Muscle satellite cells adopt divergent fates: a mechanism for self-renewal? J Cell Biol. 2004;166:347–357. doi: 10.1083/jcb.200312007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zammit PS, Partridge TA, Yablonka-Reuveni Z. The skeletal muscle satellite cell: the stem cell that came in from the cold. J Histochem Cytochem. 2006;54:1177–1191. doi: 10.1369/jhc.6R6995.2006. [DOI] [PubMed] [Google Scholar]
- Zhang W, Behringer RR, Olson EN. Inactivation of the myogenic bHLH gene MRF4 results in up-regulation of myogenin and rib anomalies. Genes Dev. 1995;9:1388–1399. doi: 10.1101/gad.9.11.1388. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.