Abstract
Immunization with vaccinia virus elicits a protective antibody response that is almost completely CD4+ T cell dependent. A recent study in a rodent model observed a deterministic linkage between antibody and CD4+ T cell responses to particular vaccinia virus proteins suggesting that CD4+ T cell help is preferentially provided to B cells with the same protein specificity (Sette A et al., Immunity 2008, 847–858). However, a causal linkage between antibody and CD4+ T cell responses to vaccinia or any other large pathogen in humans has yet to be done. In this study, we measured the antibody and CD4+ T cell responses against four vaccinia viral proteins (A27L, A33R, B5R, and L1R) known to be strongly targeted by humoral and cellular responses induced by vaccinia virus-vaccination in 90 recently vaccinated and 7 long-term vaccinia-immunized human donors. Our data indicate that there is no direct linkage between antibody and CD4+ T cell responses against each individual protein in both short-term and long-term immunized donors. Together with the observation that the presence of immune responses to these four proteins is linked together within donors, our data suggest that in vaccinia-immunized humans, individual viral proteins are not the primary recognition unit of CD4+ T cell help for B cells. Therefore, we have for the first time showed evidence that CD4+ T cells provide intermolecular (also known as non-cognate or heterotypic) help to generate robust antibody responses against four vaccinia viral proteins in humans.
Introduction
Antibody responses are essential components of protective immune responses to many pathogens, such as influenza virus (1), human immunodeficiency virus-1 (HIV-1) (2), smallpox virus (3–4), and Coxiella burnetii (5). CD4+ T cell responses are also mediators of protective immunity to pathogens (6–8). The standard model of CD4+ T cell-B cell interaction can be summarized as “any-helper-epitope-is-sufficient”. Briefly, during viral infection, B cells recognizing cognate antigen on the virion can internalize and process the whole virion for antigen presentation to CD4+ T cells specific for an epitope from any of the virion proteins. In turn, the epitope-specific CD4+ T cells provide intermolecular help to the B cells to generate antibody responses against any protein from the whole virion (9). This well-accepted viral intermolecular help model, in which CD4+ T cells provide help to B cells with different protein specificities, was established in the studies of influenza virus (10–11) and hepatitis B virus (HBV) (12), and has been confirmed in many other small virus or particle systems. Intermolecular help was also known as non-cognate or heterotypic help, in which situations T and B cell determinants are present on noncovalently linked antigens (11, 13). For example, it was found that B cells producing neutralizing antibodies recognizing viral surface proteins could utilize intermolecular help from T cells specific for an rotavirus internal protein (13), and in a study of immunization with respiratory syncytial virus antigens, covalent linkage of the B- and T-cell epitopes was not necessary for the generation of T-cell dependent antibody responses, although it did improve the affinity of the antibody response (14). Studies in a murine lupus model showed that antibodies recognizing components of the small nuclear ribonucleoprotein (snRNP) particle could utilize T cell help from other components provided that they were present in the same particle, another example of intermolecular help in generation of antibodies (15).
Despite this general concordance with the “any-helper-epitope-is-sufficient” model, several studies have identified situations where some helper epitopes function much more effectively than others. An early study of the response to influenza virus proposed a model of a hierarchy of T cell help based on the observation that B cells recognizing viral surface components could receive help from T cells specific for any of the major structural viral proteins, while B cells responding to internal viral components are restricted to receive help almost exclusively from T cells with the same protein specificity (16). The mechanism proposed was based on the idea that cell-surface antibody against a viral surface protein would be likely to capture intact viruses containing many different proteins able to provide helper epitopes, whereas cell-surface antibody against a core protein would be more likely to capture that protein only. The idea of a hierarchy of CD4+ T cell help to generate antibody responses has been investigated in other systems. In one study, B cell antibody responses to lymphocytic choriomeningitis virus surface glycoprotein were generated with help from CD4+ T cells against the surface glycoprotein, but not for the internal nucleoprotein, similarly to the case with influenza (17). The concept of intermolecular help has been utilized to design more effective subunit vaccines by including both the B cell and T cell epitopes in a single antigenically diverse structure (18).
However, the studies on linkage between CD4+ T cell responses and antibody responses for large and complex pathogens, such as poxvirus and bacteria remains very limited. Recently, this linkage for vaccinia virus was evaluated in mice by Sette and colleagues (19). Using a set of previously identified CD4+ T cell epitopes (20), they found that the antibody response to each particular protein target needs to be accompanied by a matched CD4+ T cell response against the same protein, as if the virion were perceived as a collection of individual protein specificities. Vaccinia virus is a large and complex virus with about 200 viral proteins (21) and two infectious forms called intracellular mature virus (IMV3) and extracellular enveloped virus (EEV), which are different structurally, antigenically, and functionally (22). Sette et al. suggested that the large size of vaccinia virions, ~360 nm diameter, relative to B-cell endocytotic vesicles, ~150nm in diameter (23–25), would complicate the linkage between CD4+ T cell and B cell targets because of the possibility that B cell might endocytose viral fragments but not whole virions (19). This new model of intramolecular help in responses to large and complex antigens like poxviruses and bacteria, with CD4+ T cells providing help to B cells only with the same protein specificity, is essentially an extreme variant of the hierarchy of help concept developed in studies of small viruses like influenza (80 nm) or HBV (25–40 nm), with every protein behaving as if it were a viral core antigen. Intramolecular help was also termed as cognate or homotypic help, which requires the antigenic determinants recognized by T and B cells to be covalently linked on the same antigen (11, 13). The model has received great attention for its academic and practical implications in studies of the nature of T cell help for antibody generation (26–32), in the strategy of CD4+ T cell epitope identification approaches that focus only on targets eliciting strong antibody responses (33–39), and in vaccine design studies that include proteins targeted strongly by CD4+ T cells (40–45). In spite of this interest and multiple citations, few studies have experimentally attempted to establish the linkage between CD4+ T cell and antibody responses in the response to large and complex antigens. In two follow-up studies, Sette and colleagues analyzed the human allergic response to Timothy grass antigens (38), and in rodents to a bacterial pathogen, Coxiella burnetii (46). Unlike the original study, these two publications did not observe a strong correlation between the targets of antibody and T cell responses.
In humans, CD4+ T cell (47–49), and antibody responses (50–51) against vaccinia virus are extremely diverse and heterogeneous, targeting both IMV and EEV early and late proteins. Although the idea of deterministic CD4+-antibody correlation has been applied to the identification of CD4+ epitopes by focusing on targets with strong antibody responses (36), to date, the linkage of CD4+ T cell and antibody specificities for vaccinia virus in humans has yet to be evaluated experimentally.
In this study, we evaluated the linkage between CD4+ T cell and antibody responses against vaccinia viral proteins A27L, A33R, B5R, and L1R in human donors. A27L and L1R are IMV membrane proteins, while A33R and B5R are EEV membrane proteins. A DNA vaccine composed of four genes encoding A27L, A33R, B5R and L1R showed significant protective immunity in mice (52) and non-human primates (53–54). Corresponding recombinant proteins also provided protective immunity in mice (55), while a combination of DNA prime followed by a protein boost seemed more efficacious in non-human primate (54). Strong antibody responses against A27L, A33R, B5R and L1R were observed in humans after vaccination (50–51, 56). CD4+ T cell epitopes for these four proteins in humans have also been mapped (36, 57). Here, we measured the antibody responses and CD4+ T cell responses against A27L, A33R, B5R and L1R in 90 recently vaccinia virus-vaccinated healthy donors and 7 long-term vaccinated donors. We concluded that there is no direct linkage between CD4+ T cell and antibody responses against each individual protein, and thus that the conventional intermolecular help model applies to the human immune response against vaccinia virus, at least for the four proteins tested in a vaccination trial.
Materials and Methods
Human donors
Sera and peripheral blood mononuclear cells (PBMCs) from 90 healthy vaccinia-naïve humans before (day 0) and 45 days after (day 45) vaccinia virus-vaccinated were prepared at Saint Louis University Center for Vaccine Development after approval by the Saint Louis University Institutional Review Board during a study of smallpox vaccines generated by Acambis, Inc. (Cambridge, MA) (58). Thirty donors each were vaccinated with Dryvax, ACAM1000, and ACAM2000, respectively. The ACAM1000 and ACAM2000 (Acambis, Inc., Cambridge, MA) vaccines are derived from Dryvax (Wyeth Laboratories, Marietta, PA) by plaque purification cloning in Vero cells and purified from disrupted infected cells by ultrafiltration and diafiltration, and lyophilized (58–60). Sera and PBMCs from 7 long-term vaccinia-immune donors (vaccinated with Dryvax more than 4 years before this study) and 4 non-immunized donors were collected under a protocol approved by the Medical School Institutional Review Board of University of Massachusetts.
Recombinant proteins
Recombinant vaccinia proteins A27L (gene bank ID: NR-2622), A33R (NR-2623), B5R (NR-2624) and L1R (NR-2625) from the WR strain were obtained from the Biodefense Repository (BEI) (http://www.beiresources.org/).
IFNγ-ELISPOT for CD4+ T cell responses
We measured CD4+ T cell responses against A27L, A33R, B5R and L1R by IFNγ Enzyme-linked immunosorbent spot (ELISPOT) assay. Briefly, 5×105 PBMCs from each donor were stimulated with 5ug/ml of each recombinant protein in 200ul cRPMI medium (RPMI1640 supplemented with 10% human serum, 100U/ml penicillin, 100ug/ml streptomycin, 1 mM sodium pyruvate, 2 mM L-glutamine, 50 uM of 2-mercaptoethanol and 1mM non-essential amino acids from GIBCO) in Immobilon-P 96-well MultiScreen plates (Millipore Corporation, Billerica, MA) for 48 hours. 1:800 dilution of vaccinia virus (MVA strain) -infected monkey kidney CV-1 cell lysate originally containing 1.7×107 pfu/ml or 1:800 dilution of non-infected CV-1 cell lysate or medium only were used as controls to stimulate the PBMCs. Number of IFNγ-secreting cells (spots per well) was determined using ELISPOT analyzer equipped with ImmunoSpot 5.0.3 software (CTL, Shaker heights, OH).
IFNγ and IL-2 ELISA for CD4+ T cell responses
We also measured the CD4+ T cell responses against A27L, A33R, B5R and L1R in donors 09, 22, 34 and 39 by IFNγ and IL-2 enzyme-linked immunosorbent assay (ELISA). Briefly, 5×105 PBMCs from each donor were stimulated with 5ug/ml of each recombinant protein or medium only, or VV-infected CV-1 cell lysate in 200ul cRPMI medium. 100ul supernatant from each well was collected after 48 hours. The production of IFNγ and IL-2 were measured using the Human IFNγ ELISA set and Human IL-2 ELISA set, respectively (BD Biosciences, San Diego, CA).
Human Cytokine/Chemokine 96-Well Plate Multiplex Assay
The human cytokine/chemokine production followed vaccinia virus-infection was measured by MILLIPLEX® MAP Kit containing different sizes of anti-human GM-CSF, IFNγ, IL-1β, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-10, IL-12(p70), IL-13, MCP-1, and TNFα antibody-immobilized beads (EMD Millipore Corporation, Darmstadt, Germany). Briefly, 5×105 PBMCs from each donor were stimulated with heat-inactivated vaccinia virus-infected CV-1 cell lysate or control non-infected lysate in 200ul cRPMI medium for 48 hours. 100ul supernatant from each well was collected and 25ul of each sample was added to a Microtiter Filter Plate. Subsequently 25ul of anti-human cytokines/chemokines antibody-coated premixed beads were added to each well and incubated at 4°C overnight on a plate shaker. The plate was washed and 25ul detection antibodies were added to each well and incubated at room temperature for 1 hour. Then, 25ul Streptavidin-Phycoerythrin was added and the mixture was incubated at room temperature for 30 minutes. The plate was washed and 150ul of PBS was added to all wells and the beads were resuspended on a plate shaker for 5 minutes. The amount of each cytokine/chemokine was read out on a Luminex 200 analyzer (Luminex, Austin, TX).
ELISA for antibody responses
Antibody responses against A27L, A33R, B5R and L1R were measured by enzyme-linked immunosorbent assay (ELISA). A 96-well high-binding polystyrene microtiter plate (Thermo Scientific, Rochester, NY) was coated with pre-titrated optimal concentration of each recombinant protein at 0.5ug/ml in 100ul overnight at 4°C. Control wells were coated with 0.5ug/ml bovine serum albumin (BSA). The plates were washed with PBST (1xPBS with 0.1% Tween-20), and blocked with 5% BSA at 37°C for 2 hours. Subsequently 100ul of human serum diluted in PBST + 2.5% BSA from each donor was added and the plates were incubated at 37°C for 1 hour. Binding of human antibodies was revealed by using 100ul 1:4000 dilution of peroxidase-labeled goat anti-human IgG (KPL, Gaithersburg, MD) after the washing steps and incubation at 37°C for 1 hour. Finally the plates were developed with ABTS solution (Roche Applied Science, Mannheim, Germany) and read at 405nm for absorbance using Victor plate reader (PerkinElmer, Shelton, CT).
Calculation of expected number of donors positive for each combination of four proteins assuming random independent association
The experimental number of positive donors against each protein was used to calculate the expected number of donors positive for each combination of A27L, A33R, B5R and L1R based on probability theory of independent events. For example, expected number of A27L+A33R+B5R+L1R+ donors is calculated as: (number of A27L positive/total number)*(number of A33R positive/total number)*(number of B5R positive/total number)*(number of L1R positive/total number)*(total number); and expected number of A27L+A33R+B5R+L1R− donors is calculated as: (number of A27L positive/total number)*(number of A33R positive/total number)*(number of B5R positive/total number)*((total number-number of L1R positive)/total number)*(total number). The total number is number of donors analyzed, 57 for CD4+ T cell responses and 88 for antibody responses.
Correlation coefficient analysis (CC)
CC analyses relating CD4+ T cell responses (shown as spots per well, SPW) and antibody responses (shown as Absorbance at 405nm) were done using Graphpad Prism5 (GraphPad software, San Diego, CA). Pearson’s correlation coefficient (cc) and two-tailed p-value were calculated.
Results
Vaccinia virus-vaccination induces robust cellular and humoral responses against A27L, A33R, B5R and L1R
In order to study the linkage between CD4+ T cell responses and antibody responses, we obtained PBMCs and sera from 90 healthy vaccinia-naïve donors before (day 0) and 45 days after inoculation with smallpox vaccines developed by Acambis, Inc. (ACAM1000 and ACAM2000). Thirty donors each were vaccinated with ACAM1000, ACAM2000, or Dryvax (the only previous licensed smallpox vaccine). ACAM1000 and ACAM2000 are identical at the genome level, both derived from Dryvax by plaque purification cloning in Vero cells (59–60). All these three smallpox vaccines showed similar protective immunity in mice and nonhuman primates (59–60), and in humans, with a detailed characterization of safety and efficacy (58) (study number: Acambis H-400-002). ACAM2000 was approved by U.S. Food and Drug Administration (FDA) on 31 August 2007 to replace Dryvax for smallpox vaccine.
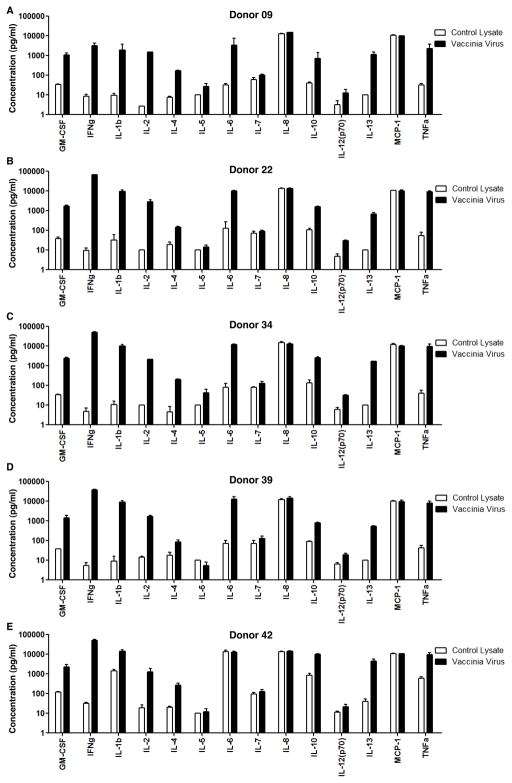
To identify which functional responses would be most useful in following the response to smallpox vaccination, we measured the production of GM-CSF, IFNγ, IL-1β, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-10, IL-12 (p70), IL-13, MCP-1 and TNFα in PBMC samples obtained 45 days after vaccination (Fig. 1). Samples were stimulated with heat-inactivated vaccinia virus-infected CV-1 cell lysate or control non-infected CV-1 cell lysate, and cytokine production was measured by multiplex bead assay. Among the 14 cytokines/chemokines measured, IL-8 and MCP-1 were nonspecifically produced in all the five donors, and IL-6 was nonspecifically produced in donor 42. IL-4, IL-5, IL-7, IL-12 and IL-13 were secreted specifically upon vaccinia stimulation but at relatively low levels. The other cytokines GM-CSF, IFNγ, IL-1β, IL-2, IL-10, and TNFα, were specifically produced at high levels in responses to vaccinia infection. The relative pattern of response did not vary greatly from donor to donor. Of the cytokines specifically produced at a high level, IFNγ had the greatest signal to background ratio (~2000-fold over background, as compared to ~700-fold for IL-2 and 10-fold to 300-fold for the others) and the lowest relative standard deviation (9%, as compared to 17% for IL-2 and 19–27% for the others). Thus we selected IFNγ for analysis of a larger set of vaccinated and non-vaccinated donors. IFNγ detection is widely used to characterize the CD4+ T cells responses against vaccinia virus and to identify CD4+ T cell epitopes in both mice (20) and humans (47–49, 61).
Figure 1. Human cytokine/chemokine production profile followed by Vaccinia virus-vaccination.
5×105 PBMCs from donors 45 days after vaccination were stimulated with heat-inactivated vaccinia virus-infected CV-1 cell lysate (solid black bar) or control non-infected CV-1 cell lysate (open bar) for 48 hours. The supernatants were collected and the production of 14 cytokines/chemokines as listed on the figure was quantified by fluorescence intensity of antibody-immobilized beads as described in the methods for (A) donor 09, (B) donor 22, (C) donor 34, (D) donor 39, and (E) donor 42. Each cytokine/chemokine was distinguished by the different sizes of the corresponding antibody-immobilized beads, and the concentration was converted from fluorescence intensity using five-parameter logistic curve model (Bio-Rad, Hercules, CA). The concentrations for IFNg were out of the range of the standard curve in the Multiplex assay, which were then determined by ELISA. Each sample had three replicates.
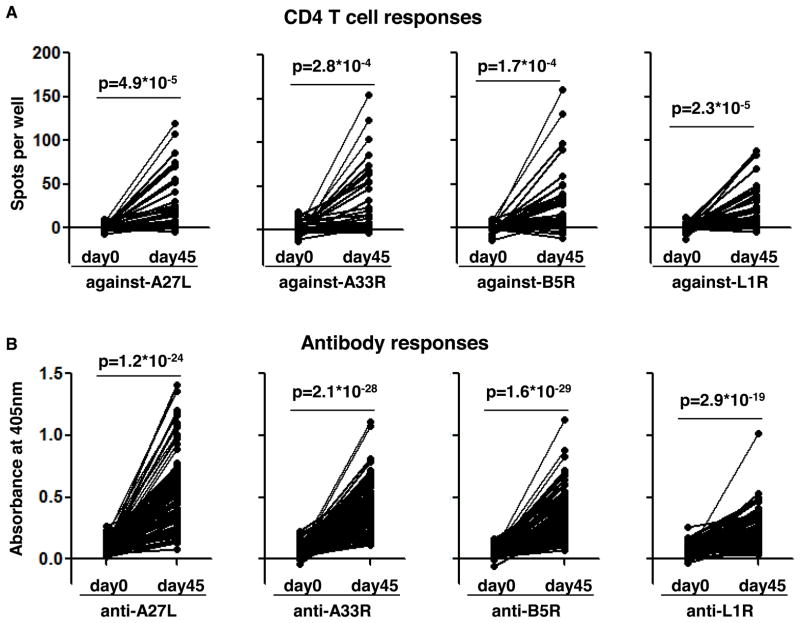
We chose the vaccinia proteins A27L, A33R, B5R and L1R for detailed analysis. These are the proteins targeted most strongly by both humoral and cellular responses during vaccinia virus infection (50–51, 56–57). Vaccination with genes encoding these four proteins and corresponding recombinant proteins provided protective immunity in mice and non-human primates (52–55), and these have been a focus of subunit vaccine development efforts. Moreover, they sample both forms of the virus, A27L and L1R from the intracellular IMV form, and A33R and B5R from the extracellular secreted EEV form. CD4+ T cell responses against A27L, A33R, B5R and L1R were measured by IFNγ-ELISPOT in PBMCs stimulated by recombinant proteins. Donors showing no responses to vaccinia virus-infected CV-1 cell lysate at day 45 (1 donor) or non-specific responses at day 0 (8 donors), or without enough PBMCs to repeat at least two times (24 donors) were excluded from the analysis and we ended up analyzing data from 57 donors (Fig. 2A and Table I). We found that vaccinia virus vaccination induces significant and diverse CD4+ T cell responses against these four proteins (Fig. 2A). More than half of the donors show positive responses and there is no preference for CD4+ T cells targeting IMV proteins (A27L and L1R, 27/57 and 29/57, respectively, Table I) or EEV proteins (A33R and B5R, 25/57 and 26/57, respectively, Table I).
Figure 2. Vaccinia virus-vaccination induces robust responses against A27L, A33R, B5R and L1R.
(A) CD4+ T cell responses (shown as spots per well) against A27L, A33R, B5R and L1R were measured by IFNγ-ELISPOT as described in the methods using PBMCs collected before (day 0) and 45 days after (day 45) vaccinia virus-vaccination. Donors showing no responses to vaccinia virus-infected CV-1 cell lysate at day 45 (n=1) or non-specific responses without stimulation (n=8), or without enough PBMCs to repeat at least two times (n=24) were excluded from the analysis and we ended up showing CD4+ T cell responses after medium background subtraction for 57 donors in (A). (B) Antibody responses (shown as Absorbance at 405nm) against the same four proteins were measured by ELISA using sera collected before (day 0) and 45 days after (day 45) vaccination. Donors showing no responses to vaccinia virus-infected CV-1 cell lysate at day 45 (n=1) or non-specific responses to BSA (n=1) and data from the left 88 donors were shown in (B) after BSA background subtraction. Significance of responses between day 0 and day 45 for each protein was indicated as p value from paired two-tailed student test. The number of positive donors and average responses were summarized in Table I.
Table I.
Immune responses summary
| Recently vaccinated donors a
| ||||||||
|---|---|---|---|---|---|---|---|---|
| CD4+ T cell response | Antibody response | |||||||
|
| ||||||||
| day 0 | day 45 | day 0 | day 45 | |||||
| # positive b | response c | # positive | response | # positive d | response e | # positive | response | |
| A27L f | 3/57 | 1.3 | 27/57 | 17.9 | 1/88 | 0.08 | 75/88 | 0.54 |
| A33R f | 6/57 | 1.4 | 25/57 | 19.8 | 2/88 | 0.06 | 75/88 | 0.41 |
| B5R f | 3/57 | 0.3 | 26/57 | 17.8 | 1/88 | 0.04 | 80/88 | 0.39 |
| L1R f | 4/57 | 0.9 | 29/57 | 16.4 | 1/88 | 0.06 | 41/88 | 0.23 |
| Any g | 12/57 | 3.9 | 43/57 | 71.9 | 5/88 | 0.24 | 87/88 | 1.57 |
|
| ||||||||
| Non-immunized and long-term immunized donors h | ||||||||
|
| ||||||||
| CD4+ T cell response | Antibody response | |||||||
|
| ||||||||
| Non-immunized | Long-term immunized | Non-immunized | Long-term immunized | |||||
| # positive | response | # positive | response | # positive | response | # positive | response | |
| A27L | 0/4 | 0.6 | 4/7 | 9.8 | 0/4 | 0.08 | 5/7 | 0.49 |
| A33R | 0/4 | 1.7 | 3/7 | 11.9 | 0/4 | 0.07 | 5/7 | 0.60 |
| B5R | 0/4 | 1.1 | 3/7 | 14.1 | 0/4 | 0.07 | 5/7 | 0.49 |
| L1R | 0/4 | 1.4 | 1/7 | 7.0 | 0/4 | 0.05 | 4/7 | 0.18 |
| Any | 0/4 | 4.8 | 5/7 | 42.8 | 0/4 | 0.27 | 6/7 | 1.76 |
Sera and PBMCs from healthy vaccinia-naïve humans before (day 0) and 45 days after (day 45) vaccinia virus-vaccination were prepared during a study of smallpox vaccines generated by Acambis, Inc. (Cambridge, MA).
The CD4+ T cell response against each protein was considered positive if SPW (spots per well) stimulated with protein > 2*SPW of medium and SPW (protein) – SPW (medium) >5.
The average spots per well in ELISPOT assay.
The antibody response against each protein was considered positive if Ab405 (absorbance at 405nm) of each protein met the criteria: Ab405 at day 45 > average+3*standard deviation of Ab405 at day 0 after BSA subtraction.
The average absorbance at 405nm in ELISA assay.
A27L and L1R are IMV proteins, and A33R and B5R are EEV proteins.
The donor was considered positive for Any if the donor showed positive responses against any of the proteins.
Sera and PBMCs from 4 non-immunized donors and 7 long-term vaccinia-immune donors were collected during a study at University of Massachusetts Medical School.
We verified the T cell responses against the four proteins in donors 09, 22, 34 and 39 using two additional assays: IFNγ-ELISA (supplemental Fig. S1A–D) and IL-2-ELISA (supplemental Fig. S1E–H), and compared them with IFNγ-ELISPOT measurement (supplemental Table SI). Significantly, in donors 09, 22 and 34, IFNγ-ELISPOT and IFNγ–ELISA resulted in the same response profile against A27L, A33R, B5R and L1R. In the other donor (donor 39), IFNγ-ELISPOT identified positive responses against B5R and L1R, while IFNγ-ELISA was negative for those (supplemental Fig. S1D). The positive responses against B5R and L1R in donor 39 were verified by the IL-2-ELISA (supplemental Fig. S1H). Consistently, IFNγ-ELISA and IL-2-ELISA resulted in similar response profile against these four proteins as IFNγ-ELISPOT (supplemental Table SI).
One caveat of this assay is that we may detect IFNγ secreted by CD8+ as well as CD4+ T cells. However, it has been shown that the potent stimulator for CD8+ T cells in ex vivo stimulating assay are peptides (62–63), instead of whole recombinant proteins, which can be processed and presented to activate CD4+ T cells (64–65). Also, multiple CD4+ T cell epitopes from A27L, A33R, B5R and L1R have been identified in humans (36, 48, 57, 61), while CD8+ T cell epitopes were only identified for B5R and A27L (61). Moreover, the same set of donors in this study has been tested for CD8+ T cell responses against all previously identified CD8+ T cell epitopes, and only one donor (donor 44) showed positive CD8+ T cell responses against B5R epitope (63). Importantly, in our measurement, that donor did not show any responses when stimulating with recombinant B5R (Fig. 4C, the arrow indicated donor). These considerations suggest that the responses after recombinant protein stimulation measured by IFNγ-ELISPOT were predominantly CD4+ T cell responses.
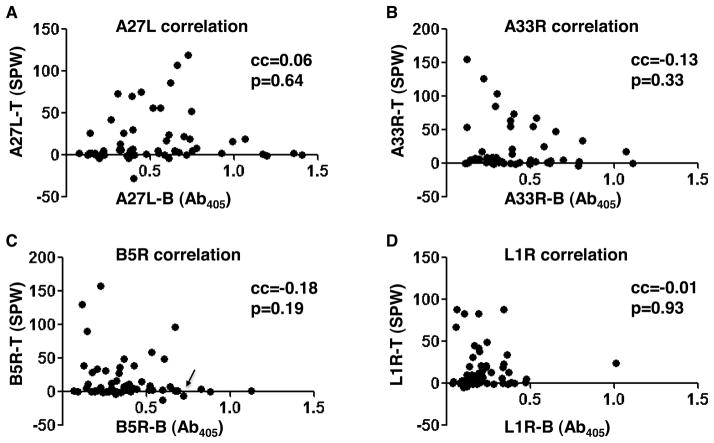
Figure 4. No direct correlation between CD4+ T cell responses and antibody responses.
Correlation between CD4+ T cell responses (shown as SPW, spots per well) and antibody responses (shown as Ab405, absorbance at 405nm) was analyzed for (A) A27L, (B) A33R, (C) B5R, and (D) L1R for the 57 donors measured for both responses. Pearson’s correlation coefficient (cc) and two-tailed p value were indicated in the upper right of each plot. In (C), the only donor (donor 44) that was shown to have positive responses against B5R-derived CD8+ T cell epitopes in reference (63) was highlighted by a black arrow.
For antibody responses, we also excluded the donor showing no response at day 45 to vaccinia virus antigen (1 donor) or showing a non-specific response to BSA (1 donor). Finally we analyzed data from 88 donors (Fig. 2B and Table I). Robust and diverse antibody responses against A27L, A33R, B5R were also found post vaccinia virus-vaccination in the majority of the donors (75, 75, and 80 respectively of 88 donors) whereas antibody responses to L1R were weaker, less variable and observed at a lower frequency (41 of 88 donors) (Fig. 2B and Table I). Previous studies on antibody responses post vaccinia virus-vaccination in humans using protein array or multiple antigens from EEV and IMV also identified A33R and B5R as the most potent targets, with A27L in the middle and least response against L1R (51, 56). The magnitude and diversity of antibody and CD4+ T cell responses that we observed are consistent with previous reports on the efficacy of the smallpox vaccines (48–49, 51, 58), and protective immunity elicited by A27L, A33R, B5R and L1R immunization (52–55).
No correlation between CD4+ T cell and antibody responses against each individual protein is observed
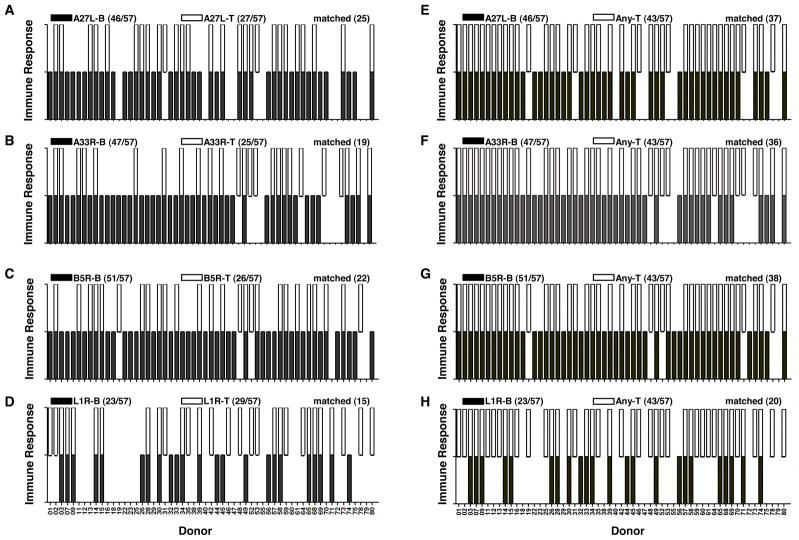
The linkage between CD4+ T cell responses and antibody responses in humans for a large and complex pathogen such as vaccinia virus is complicated by its large size and diverse responses elicited during infection (48, 51, 66). This linkage in humans has yet to be investigated in detail mainly due to the lack of availability of human donors tested for both responses. Here, we studied this linkage for the 57 donors for whom we measured both CD4+ T cell and antibody responses (Fig. 3). Our data suggest that the antibody responses against A27L, A33R, B5R and L1R are not accompanied with matched CD4+ T cell responses against the same protein (Fig. 3A–3D). For A27L, A33R and B5R, only about half of the donors showing positive antibody responses had matched positive CD4+ T cell responses against the same protein (Fig. 3A–3C and Table I). Although L1R had similar number of donors positive for antibody responses or CD4+ T cell responses, only 15 donors showed the matched pattern positive for both (Fig. 3D and Table I). Clearly, there are donors showing positive antibody responses, however, no CD4+ T cell response, and vice versa. One possibility of observing this non-matched pattern would be that some donors with overall lower responses skew the linkage. Instead, the responses were extremely diverse and randomly distributed among donors (data not shown). Another possibility is that the matched CD4+ T cell help can only be observed in donors with strong antibody responses. However, when we considered only the strong antibody responses as positive, again only about half of the donors showed matched CD4+ T cell and antibody responses (supplemental Fig. S2).
Figure 3. CD4+ T cells provide intermolecular help to generate antibody responses.
Antibody responses (B, solid black unit bar) and CD4+ T cell responses (T, open unit bar) against (A) A27L, (B) A33R, (C) B5R and (D) L1R in each donor were shown for the 57 donors measured for both responses. A unit bar was shown if the donor had positive responses against the corresponding protein. The number of donors showing positive responses for each protein, and matched antibody and CD4+ T cell responses against the same protein were summarized in parenthesis on the top of each plot. (E–H) Antibody responses against each protein (B, solid black unit bar) and CD4+ T cell responses against any of the four proteins (Any T, open unit bar) were shown for (E) A27L, (F) A33R, (G) B5R and (H) L1R. The number of donors showing positive responses, and matched antibody and any CD4+ T cell responses in the same donor were summarized in parenthesis on the top of each plot.
To test the hypothesis that CD4+ T cells provide intermolecular help to generate antibody responses, we plotted the antibody responses against each protein and CD4+ T cell responses against any of the four proteins (Any T) (Fig. 3E–3H). Additional matches were observed, but 13 donors still exhibited antibody responses in the absence of measureable CD4+ T cell responses against A27L, A33R, B5R or L1R, suggesting that CD4+ T cells specific for other vaccinia proteins might provide help for these responses. This is consistent with the finding that diverse CD4+ T cell epitopes from vaccinia virus in humans have been found (47–48, 61). Notably, all the 43 donors showing positive CD4+ T cell responses against at least one of the four proteins also were positive for an antibody response (supplemental Fig. S3).
It is possible that there might be a quantitative correlation between the strength of responses even if no qualitative linkage between presence or absence of responses were observed. We performed correlation coefficient analysis between CD4+ T cell and antibody responses (Fig. 4) Quantitatively, there was no direct correlation between CD4+ T cell and antibody responses against A27L, A33R, B5R and L1R (Fig. 4A–4D). The total CD4+ T cell responses also did not correlate with total antibody responses against these four proteins (data not shown). Previous studies also have demonstrated lack of correlation between CD4+ T cell memory and long term antibody response (67–68).
Taken together, in contrast to the observed deterministic linkage reported in mice (19), our data indicate that in humans there is no direct linkage of CD4+ T cell and antibody targets. Instead, CD4+ T cells provide intermolecular help to generate robust and diverse antibody responses.
Responses to A27L, A33R, B5R and L1R all contribute to neutralizing antibody titers
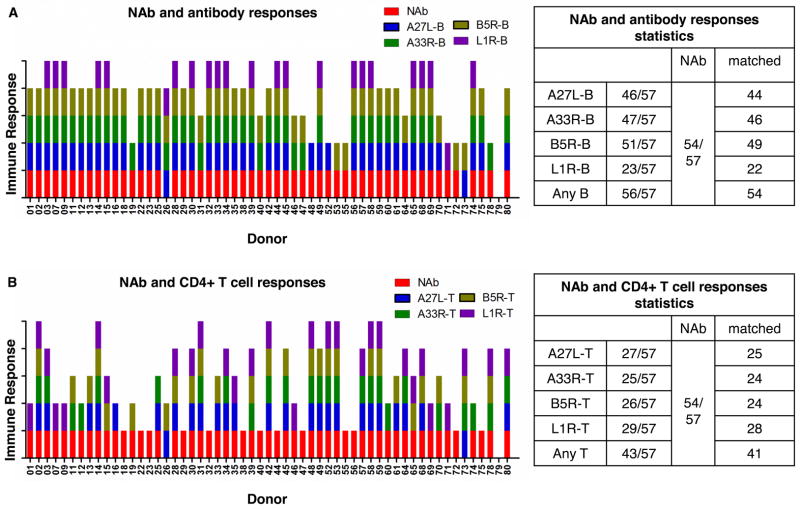
Neutralizing antibodies are of great importance in the protection from smallpox (7). EEV surface glycoproteins A33R and B5R are targets for protective antibodies in animal models (52–53, 55, 69–71), although B5R has been shown to be the major target for EEV-neutralizing antibodies in humans (56). A27L and L1R are major targets of IMV-neutralizing antibodies in both animal models (52–53, 55) and humans (56). The overall neutralizing antibodies titers for the set of human donors tested in this study were reported previously (58). By comparison, we found that all of the 54 donors with positive neutralizing antibody responses were accompanied with antibody responses against A27L, A33R, B5R or L1R (Fig. 5A), although only 41 donors had positive CD4+ T cell responses against these four proteins (Fig. 5B). It is likely that CD4+ T cell responses against other proteins also can help to generate neutralizing antibodies or that some post-translation modifications in native proteins are not represented by the recombinant proteins used in this study. Consistent with previous studies, antibodies responses against B5R contributed most to neutralizing antibody titers, as indicated by the 49 matched donors and three donors showing neutralizing antibodies with only antibody responses against B5R (donor 53, 55 and 72, Fig. 5A). Also, most of the donors showing CD4+ T cell or antibody responses against A27L, A33R and L1R were positive for neutralizing antibodies (Fig. 5). Collectively, our data indicate that responses against A27L, A33R, B5R and L1R all contribute to neutralizing antibodies in humans.
Figure 5. Responses against A27L, A33R, B5R and L1R all contribute to neutralizing antibody titers.
Total neutralizing antibody responses (NAb, red bar) and (A) antibody responses against A27L, A33R, B5R and L1R (colored unit bar for each protein in the upper right), and (B) CD4+ T cell responses against each protein in 57 donors were shown. The donor was considered having a positive neutralizing antibody response if the neutralizing antibody titer was greater than 20. Tables on the right panels showed number of donors positive for each response and matched responses. Antibody responses against any of the four proteins (Any B) and CD4+ T cell responses against any of the proteins (Any T) were also shown on the bottom row of the summary table. The neutralizing antibody data for these donors were adapted from reference (58), provided by Thomas Monath.
The presence of responses against A27L, A33R, B5R and L1R is linked together within donors
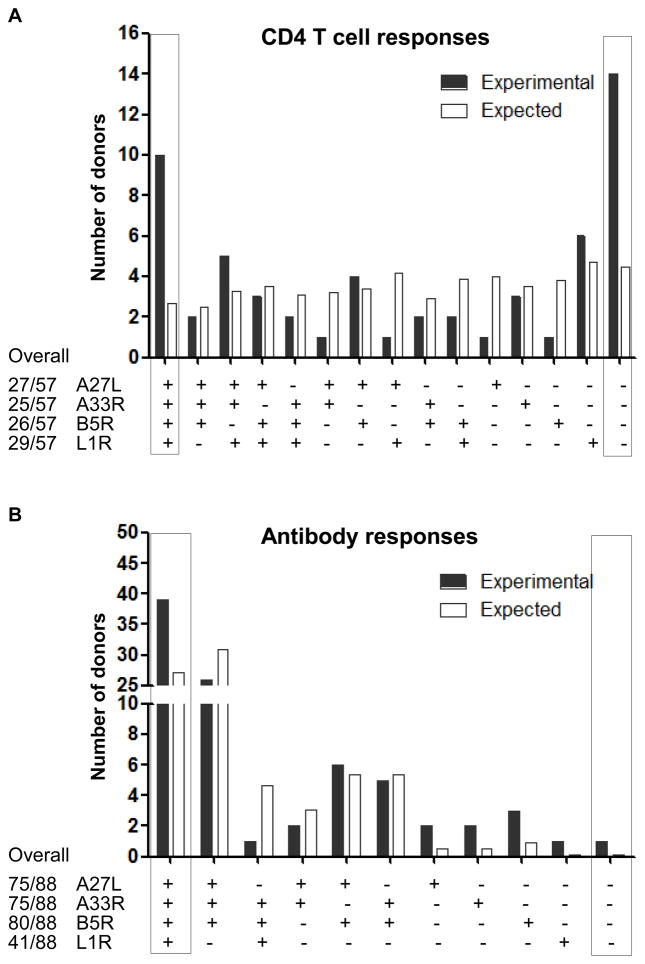
We next wanted to find whether the presence of responses against A27L, A33R, B5R and L1R was linked together within donors, which would be the case if vaccinia virions rather than individual proteins were the primary recognition unit. For CD4+ T cell responses, using the experimental number of positive donors against each protein, we calculated the expected number of donors positive for each combination of these four proteins under the assumption that the presence of responses to each protein is independent. By comparing with experimental values, we found that the observed numbers of donors positive for all four proteins (10/57) and negative for all proteins (14/57) are significantly higher than that of independently-expected values (2.7/57 and 4.5/57, respectively) (Fig. 6A). This suggests that the presence of CD4+ T cell responses against A27L, A33R, B5R and L1R is linked together within donors.
Figure 6. The presence of CD4+ T cell or antibody responses against A27L, A33R, B5R and L1R is linked within donors.
Experimental (black solid bar) and expected (open bar) number of donors showing positive responses against each combination of A27L, A33R, B5R and L1R were plotted for (A) CD4+ T cell responses and (B) antibody responses. The experimental overall number of donors showing positive responses against each protein were indicated on the bottom left of each plot. The expected number of donors showing positive responses against each combination of these four proteins was calculated using the experimental overall number of positive donors against each protein based on probability theory of independent events as illustrated in the methods. The number of donors positive for all the four proteins (tetra positive, on the very left axis) and none of the four proteins (tetra negative, on the very right axis) were highlighted with boxes.
For antibody responses, we found that 39 of 41 donors positive for L1R are also positive for A27L, A33R and B5R, which is significantly higher than the non-correlated expected values (27.1) (Fig. 6B). The experimental numbers of donors positive for two or three-protein combination matched with the non-correlated expected numbers (Fig. 6A and 6B). Consistent with CD4+ T cell responses, the presence of antibody responses against A27L, A33R, B5R and L1R is also linked together within donors.
No linkage between CD4+ T cell and antibody responses against A27L, A33R, B5R and L1R in long-term vaccinated donors
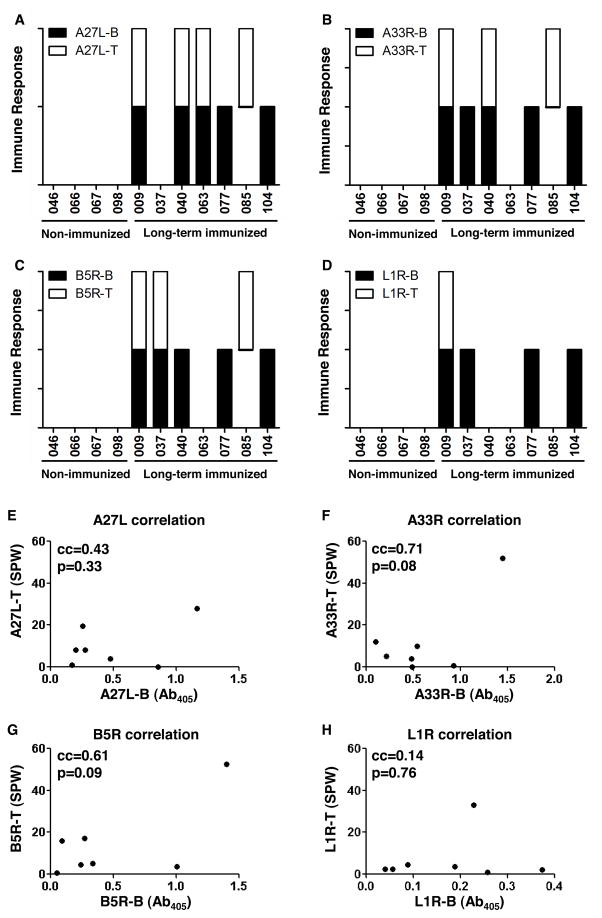
Finally, we looked at this linkage in long-term memory stage. A previous study on multiple antigens from EEV and IMV, including A27L, A33R, B5R and L1R showed that human antibody responses against these proteins decreased between 21 days and 6 months after smallpox vaccination (56). Here, we measured the antibody responses and CD4+ T cells responses against A27L, A33R, B5R and L1R in four non-vaccinated and seven long-term vaccinia virus-vaccinated healthy donors (vaccinated with Dryvax more than four years ago) (Fig. 7). The average antibody responses in the seven long-term vaccinated donors against each protein were slightly lower than those measured 45 days post vaccination, while CD4+ T cell responses were 2~3 fold lower (Table I), which confirmed that immunization with vaccinia virus can induce long-term immune responses to these four proteins (52–55) and the magnitude of responses decreases with time (56). None of the non-vaccinated donors showed any positive responses against any recombinant protein (Fig. 7A–7D). Diverse responses against A27L, A33R, B5R and L1R were elicited in the seven long-term vaccinated donors, and consistent with previous short-term vaccinated subjects, no deterministic linkage between antibody responses and CD4+ T cell responses was observed for each protein (Fig. 7A–7D). Quantitatively, there was no correlation between CD4+ T cell responses and antibody responses against the four recombinant proteins (Fig. 7E–7H).
Figure 7. No direct linkage between CD4+ T cell responses and antibody responses against A27L, A33R, B5R and L1R in long-term immunized donors.
Antibody responses (B, black solid unit bar) and CD4+ T cell responses (T, open unit bar) in against (A) A27L, (B) A33R, (C) B5R and (D) L1R for four non-vaccinated donors (046, 066, 067, 098), and seven long-term vaccinia virus-vaccinated donors (009, 037, 040, 063, 077, 085, 104). The antibody response was considered positive if the Ab405 (absorbance at 405nm in ELISA) was greater than the average+3*standard deviation of the Ab405 in the non-vaccinated donors. The CD4+ T cell responses was considered positive if the SPW (spots per well in IFNγ-ELISPOT) for protein was above 2*SPW for medium and SPW (protein) - SPW (medium) >5. A unit bar was shown if the donor had positive responses against the corresponding protein. (E–H) Correlation between CD4+ T cell responses and antibody responses in the seven vaccinated donors were analyzed for (E) A27L, (F) A33R, (G) B5R, and (H) L1R, with correlation coefficient (cc) and p value indicated in the upper left of each plot.
Discussion
Despite the eradication of smallpox by widespread vaccination with vaccinia virus, the potential use of smallpox as a bioweapon (72) and the importance of using vaccinia virus as an expression vector for immunization against other infectious diseases (73–75) and cancer (76–77), make the understanding of immune responses to vaccinia virus extremely important. In this study, we have evaluated IFNγ-CD4+ T cell responses and antibody responses against the vaccinia proteins A27L, A33R, B5R and L1R in a large set of vaccinia virus-vaccinated donors. Within this data set, no deterministic linkage between CD4+ T cell and antibody responses against each individual protein was observed, although the presence of responses against the four tested proteins seemed to be linked within donors.
The lack of direct linkage between CD4+ T cell and antibody responses would imply that in vaccinia-immunized donors B cells recognize vaccinia virion rather than individual proteins to generate MHC II epitopes for presentation to cognate CD4+ T cells. One potential argument against this model is that the vaccinia virion (~360nm in diameter) is much larger than typical endocytic vesicles (50–150nm in diameter), which would result in size exclusion at the level of cellular uptake for large and complex pathogens. However, recent studies have shown that vaccinia IMV enters by fusion with plasma membrane (78), while EEV enters cells by macropinocytosis and nonfusogenic acid-activated membrane rupture (79–81), both consistent with the model of entire virus uptake, although the mechanisms for IMV and EEV entry are different and still in debate. Our experiments support the standard model wherein the relevant particle taken up by B cells is larger than a single protein, and our observation that responses against A27L, A33R, B5R and L1R are linked together within donors also adds evidence to this model. A recent study of lymph nodes of mice injected with viruses revealed that subcapsular macrophages capture virus particles for transfer to B cells and that this transfer occurs without virus internalization or degradation. This process provides a mechanism by which B cells could encounter vaccinia viruses for uptake and processing (82).
In contrast to the results presented here, a previous study in a mouse model demonstrated a deterministic linkage, showing that each antibody response was accompanied by a matched CD4+ T cell response targeting the same protein, as if individual protein is the recognition unit for B cells (19). We consider that there are at least three plausible reasons for why the strong linkage was not observed in our human study. The first plausible reason is the difference between mouse and human immune responses to large and complex vaccinia virus. Compared with genetically homogeneous laboratory mice housed in relative free germ conditions, out-bred humans are genetically heterogeneous and have also great variability in their environmental exposure to other pathogens. The complexity of responses to large viruses, such as vaccinia virus, cytomegalovirus, herpes simplex virus, and Epstein-Barr virus, in humans compared with mice has been extensively reviewed (83). These studies suggest that many factors, including route of infection, genetic differences, and experience encountering multiple antigens, all contribute to the greater variability of antiviral responses in humans compared with mice. The second factor is that the mouse study highlighted the linkage of the CD4+ T cell and antibody responses in mice immunized with peptide and then challenged with vaccinia virus. However the linkage of the responses reported in that publication is considerably less striking in mice immunized with just virus (19), which was also the case in our human study. It is possible that the deterministic linkage observed in the mouse study might reflect an alternate mechanism for B cell –T cell interaction under conditions in which a high frequency of CD4+ T cells are present or antigen presentation is dominated by fluid-phase uptake. A final plausible reason is that the mouse and human experiments evaluated different vaccinia proteins. The four proteins characterized for linkage in the mouse study, I1L, H3L, D8L, and L4R are all IMV proteins. Instead, of the four proteins tested in our human study, A33R and B5R are EEV proteins, while A27L and L1R come from IMV, although we also did not see the linkage in either case. The hierarchy of help model specifies that antibody responses against viral surface proteins can utilize intermolecular T cell help from any antigen in the virion, whereas antibody responses against internal proteins are limited to intramolecular help involving antigens from that same protein (16). In the somewhat complicated case of vaccinia virus, IMV surface proteins (A27L and L1R) can be considered external proteins in the form of IMV and internal proteins in the form of EEV. The vaccines used in our study were likely to have both IMV and EEV forms, but the Western Reserve strain used in the mice study was prepared from the supernatant of infected HeLa cells and is likely to contain predominately EEV forms (19). Thus, at least for A27L and L1R, the lack of linkage in our human study might derive from the differences in the forms of vaccinia virus used for immunization and assay. Due to the limitations of manipulation in human donors, a study directly addressing the linkage between CD4+ T cell and antibody responses against A27L, A33R, B5R and L1R in EEV and IMV immunized mice might distinguish the different possibilities proposed above. In addition, testing the linkage for a larger set of proteins from vaccinia virus in humans would help to validate our observations.
The potential for deterministic linkage of antibody and CD4+ T cell responses in the same protein has received attention for its academic and practical implications in vaccine development and mapping of T cell epitopes (36–37). Our results show that in some human donors antibody responses can be detected without CD4+ T cell responses against the same protein, and vice versa. Thus, CD4+ T cell epitope mapping efforts directed only at antigens eliciting antibody responses might miss important immunodominant epitopes derived from antigens against which no antibody responses are made.
In summary, we have for the first time in humans studied the linkage between CD4+ T cell and antibody responses to a large and complicated virus. We observed minimal linkage between CD4+ T cell and antibody responses against A27L, A33R, B5R and L1R in vaccinated donors. However, we did observe that the presence of responses against these proteins is linked together within individual donors. These results imply that in human CD4+ T cells provide intermolecular help to generate robust antibody responses against these four abundant and immunodominant vaccinia viral proteins.
Supplementary Material
Acknowledgments
We would like to thank Loretta Lee for processing of blood samples and maintenance of cell lines, Masanori Terajima for providing CD8+ T cell responses, Thomas Monath for providing neutralizing antibody responses data for some of the donors we tested, and Jeffrey S. Kennedy for setting up the collaboration with Acambis, Inc. and MHC typing and effector assays of these samples.
Footnotes
This work was supported by National Institute of Health grant U19-57319
Abbreviations used in this paper: IMV, intracellular mature virus; EEV, extracellular enveloped virus; MHC II, major histocompatibility complex class II alleles; PBMCs, peripheral blood mononuclear cells; cc, Pearson’s correlation coefficient.
References
- 1.Gerhard W. The role of the antibody response in influenza virus infection. Curr Top Microbiol Immunol. 2001;260:171–190. doi: 10.1007/978-3-662-05783-4_9. [DOI] [PubMed] [Google Scholar]
- 2.Overbaugh J, Morris L. The Antibody Response against HIV-1. Cold Spring Harb Perspect Med. 2012;2:a007039. doi: 10.1101/cshperspect.a007039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edghill-Smith Y, Golding H, Manischewitz J, King LR, Scott D, Bray M, Nalca A, Hooper JW, Whitehouse CA, Schmitz JE, Reimann KA, Franchini G. Smallpox vaccine-induced antibodies are necessary and sufficient for protection against monkeypox virus. Nat Med. 2005;11:740–747. doi: 10.1038/nm1261. [DOI] [PubMed] [Google Scholar]
- 4.Lustig S, Fogg C, Whitbeck JC, Eisenberg RJ, Cohen GH, Moss B. Combinations of polyclonal or monoclonal antibodies to proteins of the outer membranes of the two infectious forms of vaccinia virus protect mice against a lethal respiratory challenge. J Virol. 2005;79:13454–13462. doi: 10.1128/JVI.79.21.13454-13462.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang GQ, Samuel JE. Identification and cloning potentially protective antigens of Coxiella burnetii using sera from mice experimentally infected with Nine Mile phase I. Ann N Y Acad Sci. 2003;990:510–520. doi: 10.1111/j.1749-6632.2003.tb07420.x. [DOI] [PubMed] [Google Scholar]
- 6.Xu R, Johnson AJ, Liggitt D, Bevan MJ. Cellular and humoral immunity against vaccinia virus infection of mice. J Immunol. 2004;172:6265–6271. doi: 10.4049/jimmunol.172.10.6265. [DOI] [PubMed] [Google Scholar]
- 7.Amanna IJ, Slifka MK, Crotty S. Immunity and immunological memory following smallpox vaccination. Immunol Rev. 2006;211:320–337. doi: 10.1111/j.0105-2896.2006.00392.x. [DOI] [PubMed] [Google Scholar]
- 8.Kreijtz JH, Fouchier RA, Rimmelzwaan GF. Immune responses to influenza virus infection. Virus Res. 2011;162:19–30. doi: 10.1016/j.virusres.2011.09.022. [DOI] [PubMed] [Google Scholar]
- 9.Janeway CA, Travers P, Walport M, Shlomchik MJ. Immunology: The Immune System in Health and Disease. Garland Science; New York: 2005. [Google Scholar]
- 10.Russell SM, Liew FY. T cells primed by influenza virion internal components can cooperate in the antibody response to haemagglutinin. Nature. 1979;280:147–148. doi: 10.1038/280147a0. [DOI] [PubMed] [Google Scholar]
- 11.Scherle PA, Gerhard W. Functional analysis of influenza-specific helper T cell clones in vivo. T cells specific for internal viral proteins provide cognate help for B cell responses to hemagglutinin. J Exp Med. 1986;164:1114–1128. doi: 10.1084/jem.164.4.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milich DR, McLachlan A, Thornton GB, Hughes JL. Antibody production to the nucleocapsid and envelope of the hepatitis B virus primed by a single synthetic T cell site. Nature. 1987;329:547–549. doi: 10.1038/329547a0. [DOI] [PubMed] [Google Scholar]
- 13.Esquivel FR, Lopez S, Guitierrez XL, Arias C. The internal rotavirus protein VP6 primes for an enhanced neutralizing antibody response. Arch Virol. 2000;145:813–825. doi: 10.1007/s007050050674. [DOI] [PubMed] [Google Scholar]
- 14.Shaw DM, Stanley CM, Partidos CD, Steward MW. Influence of the T-helper epitope on the titre and affinity of antibodies to B-cell epitopes after co-immunization. Mol Immunol. 1993;30:961–968. doi: 10.1016/0161-5890(93)90121-q. [DOI] [PubMed] [Google Scholar]
- 15.Fatenejad S, Mamula MJ, Craft J. Role of intermolecular/intrastructural Band T-cell determinants in the diversification of autoantibodies to ribonucleoprotein particles. Proc Natl Acad Sci U S A. 1993;90:12010–12014. doi: 10.1073/pnas.90.24.12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scherle PA, Gerhard W. Differential ability of B cells specific for external vs. internal influenza virus proteins to respond to help from influenza virus-specific T-cell clones in vivo. Proc Natl Acad Sci U S A. 1988;85:4446–4450. doi: 10.1073/pnas.85.12.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oxenius A, Bachmann MF, Zinkernagel RM, Hengartner H. Virus-specific MHC-class II-restricted TCR-transgenic mice: effects on humoral and cellular immune responses after viral infection. Eur J Immunol. 1998;28:390–400. doi: 10.1002/(SICI)1521-4141(199801)28:01<390::AID-IMMU390>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 18.Goodman-Snitkoff G, Good MF, Berzofsky JA, Mannino RJ. Role of intrastructural/intermolecular help in immunization with peptide-phospholipid complexes. J Immunol. 1991;147:410–415. [PubMed] [Google Scholar]
- 19.Sette A, Moutaftsi M, Moyron-Quiroz J, McCausland MM, Davies DH, Johnston RJ, Peters B, Rafii-El-Idrissi Benhnia M, Hoffmann J, Su HP, Singh K, Garboczi DN, Head S, Grey H, Felgner PL, Crotty S. Selective CD4+ T cell help for antibody responses to a large viral pathogen: deterministic linkage of specificities. Immunity. 2008;28:847–858. doi: 10.1016/j.immuni.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moutaftsi M, Bui HH, Peters B, Sidney J, Salek-Ardakani S, Oseroff C, Pasquetto V, Crotty S, Croft M, Lefkowitz EJ, Grey H, Sette A. Vaccinia virus-specific CD4+ T cell responses target a set of antigens largely distinct from those targeted by CD8+ T cell responses. J Immunol. 2007;178:6814–6820. doi: 10.4049/jimmunol.178.11.6814. [DOI] [PubMed] [Google Scholar]
- 21.Condit RC, Moussatche N, Traktman P. In a nutshell: structure and assembly of the vaccinia virion. Adv Virus Res. 2006;66:31–124. doi: 10.1016/S0065-3527(06)66002-8. [DOI] [PubMed] [Google Scholar]
- 22.Roberts KL, Smith GL. Vaccinia virus morphogenesis and dissemination. Trends Microbiol. 2008;16:472–479. doi: 10.1016/j.tim.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 23.Goldstein JL, Anderson RG, Brown MS. Coated pits, coated vesicles, and receptor-mediated endocytosis. Nature. 1979;279:679–685. doi: 10.1038/279679a0. [DOI] [PubMed] [Google Scholar]
- 24.West MA, Lucocq JM, Watts C. Antigen processing and class II MHC peptide-loading compartments in human B-lymphoblastoid cells. Nature. 1994;369:147–151. doi: 10.1038/369147a0. [DOI] [PubMed] [Google Scholar]
- 25.Lodish H, Berk A, Matsudaira P, Kaiser C, Krieger M, Scott M, Zipursky L, Darnell J. Molecular Cell Biology. W. H. Freeman; New York: 2003. [Google Scholar]
- 26.Cyster JG. B cell follicles and antigen encounters of the third kind. Nat Immunol. 2010;11:989–996. doi: 10.1038/ni.1946. [DOI] [PubMed] [Google Scholar]
- 27.Nayak JL, Fitzgerald TF, Richards KA, Yang H, Treanor JJ, Sant AJ. CD4+ T-Cell Expansion Predicts Neutralizing Antibody Responses to Monovalent, Inactivated 2009 Pandemic Influenza A(H1N1) Virus Subtype H1N1 Vaccine. J Infect Dis. 2013;207:297–305. doi: 10.1093/infdis/jis684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haynes L, Szaba FM, Eaton SM, Kummer LW, Lanthier PA, Petell AH, Duso DK, Luo D, Lin JS, Lefebvre JS, Randall TD, Johnson LL, Kohlmeier JE, Woodland DL, Smiley ST. Immunity to the conserved influenza nucleoprotein reduces susceptibility to secondary bacterial infections. J Immunol. 2012;189:4921–4929. doi: 10.4049/jimmunol.1201916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Snapper CM. Mechanisms underlying in vivo polysaccharide-specific immunoglobulin responses to intact extracellular bacteria. Ann N Y Acad Sci. 2012;1253:92–101. doi: 10.1111/j.1749-6632.2011.06329.x. [DOI] [PubMed] [Google Scholar]
- 30.Catron DM, Pape KA, Fife BT, van Rooijen N, Jenkins MK. A protease-dependent mechanism for initiating T-dependent B cell responses to large particulate antigens. J Immunol. 2010;184:3609–3617. doi: 10.4049/jimmunol.1000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jorgensen KW, Buus S, Nielsen M. Structural properties of MHC class II ligands, implications for the prediction of MHC class II epitopes. PLoS One. 2010;5:e15877. doi: 10.1371/journal.pone.0015877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Batista FD, Harwood NE. The who, how and where of antigen presentation to B cells. Nat Rev Immunol. 2009;9:15–27. doi: 10.1038/nri2454. [DOI] [PubMed] [Google Scholar]
- 33.Nayak JL, Richards KA, Chaves FA, Sant AJ. Analyses of the specificity of CD4 T cells during the primary immune response to influenza virus reveals dramatic MHC-linked asymmetries in reactivity to individual viral proteins. Viral Immunol. 2010;23:169–180. doi: 10.1089/vim.2009.0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richards KA, Topham D, Chaves FA, Sant AJ. Cutting edge: CD4 T cells generated from encounter with seasonal influenza viruses and vaccines have broad protein specificity and can directly recognize naturally generated epitopes derived from the live pandemic H1N1 virus. J Immunol. 2010;185:4998–5002. doi: 10.4049/jimmunol.1001395. [DOI] [PubMed] [Google Scholar]
- 35.Judkowski V, Bunying A, Ge F, Appel JR, Law K, Sharma A, Raja-Gabaglia C, Norori P, Santos RG, Giulianotti MA, Slifka MK, Douek DC, Graham BS, Pinilla C. GM-CSF production allows the identification of immunoprevalent antigens recognized by human CD4+ T cells following smallpox vaccination. PLoS One. 2011;6:e24091. doi: 10.1371/journal.pone.0024091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kennedy RB, Poland GA. The identification of HLA class II-restricted T cell epitopes to vaccinia virus membrane proteins. Virology. 2010;408:232–240. doi: 10.1016/j.virol.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valentino MD, Maben ZJ, Hensley LL, Woolard MD, Kawula TH, Frelinger JA, Frelinger JG. Identification of T-cell epitopes in Francisella tularensis using an ordered protein array of serological targets. Immunology. 2011;132:348–360. doi: 10.1111/j.1365-2567.2010.03387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oseroff C, Sidney J, Kotturi MF, Kolla R, Alam R, Broide DH, Wasserman SI, Weiskopf D, McKinney DM, Chung JL, Petersen A, Grey H, Peters B, Sette A. Molecular determinants of T cell epitope recognition to the common Timothy grass allergen. J Immunol. 2010;185:943–955. doi: 10.4049/jimmunol.1000405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Homan EJ, Bremel RD. Patterns of predicted T-cell epitopes associated with antigenic drift in influenza H3N2 hemagglutinin. PLoS One. 2011;6:e26711. doi: 10.1371/journal.pone.0026711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burton DR, Ahmed R, Barouch DH, Butera ST, Crotty S, Godzik A, Kaufmann DE, McElrath MJ, Nussenzweig MC, Pulendran B, Scanlan CN, Schief WR, Silvestri G, Streeck H, Walker BD, Walker LM, Ward AB, Wilson IA, Wyatt R. A Blueprint for HIV Vaccine Discovery. Cell Host Microbe. 2012;12:396–407. doi: 10.1016/j.chom.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Houten NE, Henry KA, Smith GP, Scott JK. Engineering filamentous phage carriers to improve focusing of antibody responses against peptides. Vaccine. 2010;28:2174–2185. doi: 10.1016/j.vaccine.2009.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swain SL, McKinstry KK, Strutt TM. Expanding roles for CD4(+) T cells in immunity to viruses. Nat Rev Immunol. 2012;12:136–148. doi: 10.1038/nri3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stern LJ, Calvo-Calle JM. HLA-DR: molecular insights and vaccine design. Curr Pharm Des. 2009;15:3249–3261. doi: 10.2174/138161209789105171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kennedy RB, I, Ovsyannikova G, Jacobson RM, Poland GA. The immunology of smallpox vaccines. Curr Opin Immunol. 2009;21:314–320. doi: 10.1016/j.coi.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richards KA, Chaves FA, Sant AJ. Infection of HLA-DR1 transgenic mice with a human isolate of influenza a virus (H1N1) primes a diverse CD4 T-cell repertoire that includes CD4 T cells with heterosubtypic cross-reactivity to avian (H5N1) influenza virus. J Virol. 2009;83:6566–6577. doi: 10.1128/JVI.00302-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen C, Dow C, Wang P, Sidney J, Read A, Harmsen A, Samuel JE, Peters B. Identification of CD4+ T cell epitopes in C. burnetii antigens targeted by antibody responses. PLoS One. 2011;6:e17712. doi: 10.1371/journal.pone.0017712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Calvo-Calle JM, Strug I, Nastke MD, Baker SP, Stern LJ. Human CD4+ T cell epitopes from vaccinia virus induced by vaccination or infection. PLoS Pathog. 2007;3:1511–1529. doi: 10.1371/journal.ppat.0030144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jing L, Davies DH, Chong TM, Chun S, McClurkan CL, Huang J, Story BT, Molina DM, Hirst S, Felgner PL, Koelle DM. An extremely diverse CD4 response to vaccinia virus in humans is revealed by proteome-wide T-cell profiling. J Virol. 2008;82:7120–7134. doi: 10.1128/JVI.00453-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jing L, Chong TM, Byrd B, McClurkan CL, Huang J, Story BT, Dunkley KM, Aldaz-Carroll L, Eisenberg RJ, Cohen GH, Kwok WW, Sette A, Koelle DM. Dominance and diversity in the primary human CD4 T cell response to replication-competent vaccinia virus. J Immunol. 2007;178:6374–6386. doi: 10.4049/jimmunol.178.10.6374. [DOI] [PubMed] [Google Scholar]
- 50.Davies DH, Liang X, Hernandez JE, Randall A, Hirst S, Mu Y, Romero KM, Nguyen TT, Kalantari-Dehaghi M, Crotty S, Baldi P, Villarreal LP, Felgner PL. Profiling the humoral immune response to infection by using proteome microarrays: high-throughput vaccine and diagnostic antigen discovery. Proc Natl Acad Sci U S A. 2005;102:547–552. doi: 10.1073/pnas.0408782102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Duke-Cohan JS, Wollenick K, Witten EA, Seaman MS, Baden LR, Dolin R, Reinherz EL. The heterogeneity of human antibody responses to vaccinia virus revealed through use of focused protein arrays. Vaccine. 2009;27:1154–1165. doi: 10.1016/j.vaccine.2008.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hooper JW, Custer DM, Thompson E. Four-gene-combination DNA vaccine protects mice against a lethal vaccinia virus challenge and elicits appropriate antibody responses in nonhuman primates. Virology. 2003;306:181–195. doi: 10.1016/S0042-6822(02)00038-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hooper JW, Thompson E, Wilhelmsen C, Zimmerman M, Ichou MA, Steffen SE, Schmaljohn CS, Schmaljohn AL, Jahrling PB. Smallpox DNA vaccine protects nonhuman primates against lethal monkeypox. J Virol. 2004;78:4433–4443. doi: 10.1128/JVI.78.9.4433-4443.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heraud JM, Edghill-Smith Y, Ayala V, Kalisz I, Parrino J, Kalyanaraman VS, Manischewitz J, King LR, Hryniewicz A, Trindade CJ, Hassett M, Tsai WP, Venzon D, Nalca A, Vaccari M, Silvera P, Bray M, Graham BS, Golding H, Hooper JW, Franchini G. Subunit recombinant vaccine protects against monkeypox. J Immunol. 2006;177:2552–2564. doi: 10.4049/jimmunol.177.4.2552. [DOI] [PubMed] [Google Scholar]
- 55.Fogg C, Lustig S, Whitbeck JC, Eisenberg RJ, Cohen GH, Moss B. Protective immunity to vaccinia virus induced by vaccination with multiple recombinant outer membrane proteins of intracellular and extracellular virions. J Virol. 2004;78:10230–10237. doi: 10.1128/JVI.78.19.10230-10237.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Putz MM, Midgley CM, Law M, Smith GL. Quantification of antibody responses against multiple antigens of the two infectious forms of Vaccinia virus provides a benchmark for smallpox vaccination. Nat Med. 2006;12:1310–1315. doi: 10.1038/nm1457. [DOI] [PubMed] [Google Scholar]
- 57.Sirven P, Castelli FA, Probst A, Szely N, Maillere B. In vitro human CD4+ T cell response to the vaccinia protective antigens B5R and A33R. Mol Immunol. 2009;46:1481–1487. doi: 10.1016/j.molimm.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 58.Frey SE, Newman FK, Kennedy JS, Ennis F, Abate G, Hoft DF, Monath TP. Comparison of the safety and immunogenicity of ACAM1000, ACAM2000 and Dryvax in healthy vaccinia-naive adults. Vaccine. 2009;27:1637–1644. doi: 10.1016/j.vaccine.2008.11.079. [DOI] [PubMed] [Google Scholar]
- 59.Weltzin R, Liu J, Pugachev KV, Myers GA, Coughlin B, Blum PS, Nichols R, Johnson C, Cruz J, Kennedy JS, Ennis FA, Monath TP. Clonal vaccinia virus grown in cell culture as a new smallpox vaccine. Nat Med. 2003;9:1125–1130. doi: 10.1038/nm916. [DOI] [PubMed] [Google Scholar]
- 60.Monath TP, Caldwell JR, Mundt W, Fusco J, Johnson CS, Buller M, Liu J, Gardner B, Downing G, Blum PS, Kemp T, Nichols R, Weltzin R. ACAM2000 clonal Vero cell culture vaccinia virus (New York City Board of Health strain)--a second-generation smallpox vaccine for biological defense. Int J Infect Dis. 2004;8(Suppl 2):S31–44. doi: 10.1016/j.ijid.2004.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tang J, Murtadha M, Schnell M, Eisenlohr LC, Hooper J, Flomenberg P. Human T-cell responses to vaccinia virus envelope proteins. J Virol. 2006;80:10010–10020. doi: 10.1128/JVI.00601-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oseroff C, Kos F, Bui HH, Peters B, Pasquetto V, Glenn J, Palmore T, Sidney J, Tscharke DC, Bennink JR, Southwood S, Grey HM, Yewdell JW, Sette A. HLA class I-restricted responses to vaccinia recognize a broad array of proteins mainly involved in virulence and viral gene regulation. Proc Natl Acad Sci U S A. 2005;102:13980–13985. doi: 10.1073/pnas.0506768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Terajima M, Orphin L, Leporati AM, Pazoles P, Cruz J, Rothman AL, Ennis FA. Vaccinia virus-specific CD8(+) T-cell responses target a group of epitopes without a strong immunodominance hierarchy in humans. Hum Immunol. 2008;69:815–825. doi: 10.1016/j.humimm.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lazarski CA, Chaves FA, Jenks SA, Wu S, Richards KA, Weaver JM, Sant AJ. The kinetic stability of MHC class II:peptide complexes is a key parameter that dictates immunodominance. Immunity. 2005;23:29–40. doi: 10.1016/j.immuni.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 65.Lazarski CA, Chaves FA, Sant AJ. The impact of DM on MHC class II-restricted antigen presentation can be altered by manipulation of MHC-peptide kinetic stability. J Exp Med. 2006;203:1319–1328. doi: 10.1084/jem.20060058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roos N, Cyrklaff M, Cudmore S, Blasco R, Krijnse-Locker J, Griffiths G. A novel immunogold cryoelectron microscopic approach to investigate the structure of the intracellular and extracellular forms of vaccinia virus. EMBO J. 1996;15:2343–2355. [PMC free article] [PubMed] [Google Scholar]
- 67.Crotty S, Felgner P, Davies H, Glidewell J, Villarreal L, Ahmed R. Cutting edge: long-term B cell memory in humans after smallpox vaccination. J Immunol. 2003;171:4969–4973. doi: 10.4049/jimmunol.171.10.4969. [DOI] [PubMed] [Google Scholar]
- 68.Hammarlund E, Lewis MW, Hansen SG, Strelow LI, Nelson JA, Sexton GJ, Hanifin JM, Slifka MK. Duration of antiviral immunity after smallpox vaccination. Nat Med. 2003;9:1131–1137. doi: 10.1038/nm917. [DOI] [PubMed] [Google Scholar]
- 69.Galmiche MC, Goenaga J, Wittek R, Rindisbacher L. Neutralizing and protective antibodies directed against vaccinia virus envelope antigens. Virology. 1999;254:71–80. doi: 10.1006/viro.1998.9516. [DOI] [PubMed] [Google Scholar]
- 70.Law M, Smith GL. Antibody neutralization of the extracellular enveloped form of vaccinia virus. Virology. 2001;280:132–142. doi: 10.1006/viro.2000.0750. [DOI] [PubMed] [Google Scholar]
- 71.Bell E, Shamim M, Whitbeck JC, Sfyroera G, Lambris JD, Isaacs SN. Antibodies against the extracellular enveloped virus B5R protein are mainly responsible for the EEV neutralizing capacity of vaccinia immune globulin. Virology. 2004;325:425–431. doi: 10.1016/j.virol.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 72.Henderson DA, Inglesby TV, Bartlett JG, Ascher MS, Eitzen E, Jahrling PB, Hauer J, Layton M, McDade J, Osterholm MT, O’Toole T, Parker G, Perl T, Russell PK, Tonat K. Smallpox as a biological weapon: medical and public health management. Working Group on Civilian Biodefense. JAMA. 1999;281:2127–2137. doi: 10.1001/jama.281.22.2127. [DOI] [PubMed] [Google Scholar]
- 73.Carroll MW, Moss B. Poxviruses as expression vectors. Curr Opin Biotechnol. 1997;8:573–577. doi: 10.1016/s0958-1669(97)80031-6. [DOI] [PubMed] [Google Scholar]
- 74.Gomez CE, Perdiguero B, Najera JL, Sorzano CO, Jimenez V, Gonzalez-Sanz R, Esteban M. Removal of vaccinia virus genes that block interferon type I and II pathways improves adaptive and memory responses of the HIV/AIDS vaccine candidate NYVAC-C in mice. J Virol. 2012;86:5026–5038. doi: 10.1128/JVI.06684-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jones BG, Sealy RE, Zhan X, Freiden PJ, Surman SL, Blanchard JL, Hurwitz JL. UV-inactivated vaccinia virus (VV) in a multi-envelope DNA-VV-protein (DVP) HIV-1 vaccine protects macaques from lethal challenge with heterologous SHIV. Vaccine. 2012;30:3188–3195. doi: 10.1016/j.vaccine.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shen Y, Nemunaitis J. Fighting cancer with vaccinia virus: teaching new tricks to an old dog. Mol Ther. 2005;11:180–195. doi: 10.1016/j.ymthe.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 77.John LB, Howland LJ, Flynn JK, West AC, Devaud C, Duong CP, Stewart TJ, Westwood JA, Guo ZS, Bartlett DL, Smyth MJ, Kershaw MH, Darcy PK. Oncolytic virus and anti-4–1BB combination therapy elicits strong antitumor immunity against established cancer. Cancer Res. 2012;72:1651–1660. doi: 10.1158/0008-5472.CAN-11-2788. [DOI] [PubMed] [Google Scholar]
- 78.Carter GC, Law M, Hollinshead M, Smith GL. Entry of the vaccinia virus intracellular mature virion and its interactions with glycosaminoglycans. J Gen Virol. 2005;86:1279–1290. doi: 10.1099/vir.0.80831-0. [DOI] [PubMed] [Google Scholar]
- 79.Law M, Carter GC, Roberts KL, Hollinshead M, Smith GL. Ligand-induced and nonfusogenic dissolution of a viral membrane. Proc Natl Acad Sci U S A. 2006;103:5989–5994. doi: 10.1073/pnas.0601025103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mercer J, Helenius A. Vaccinia virus uses macropinocytosis and apoptotic mimicry to enter host cells. Science. 2008;320:531–535. doi: 10.1126/science.1155164. [DOI] [PubMed] [Google Scholar]
- 81.Schmidt FI, Bleck CK, Helenius A, Mercer J. Vaccinia extracellular virions enter cells by macropinocytosis and acid-activated membrane rupture. EMBO J. 2011;30:3647–3661. doi: 10.1038/emboj.2011.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Junt T, Moseman EA, Iannacone M, Massberg S, Lang PA, Boes M, Fink K, Henrickson SE, Shayakhmetov DM, Di Paolo NC, van Rooijen N, Mempel TR, Whelan SP, von Andrian UH. Subcapsular sinus macrophages in lymph nodes clear lymph-borne viruses and present them to antiviral B cells. Nature. 2007;450:110–114. doi: 10.1038/nature06287. [DOI] [PubMed] [Google Scholar]
- 83.Yewdell JW. Confronting complexity: real-world immunodominance in antiviral CD8+ T cell responses. Immunity. 2006;25:533–543. doi: 10.1016/j.immuni.2006.09.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.