Abstract
TLR4 signaling must be tightly regulated to both provide effective immune protection and avoid inflammation-induced pathology. Thus, the mechanisms that negatively regulate the TLR4-triggered inflammatory response are of particular importance. Glia maturation factor-γ (GMFG), a novel actin depolymerization factor/cofilin superfamily protein that is expressed in inflammatory cells, has been implicated in mediating neutrophil and T-cell migration, but its function in macrophage immune response remains unclear. In the present study, the role of GMFG in the LPS-induced TLR4 signaling pathway was investigated in THP-1 and human primary macrophages. LPS stimulation of macrophages decreased GMFG mRNA and protein expression. We show that GMFG negatively regulates LPS-induced activation of NF-κB, MAPK, and IFN regulatory factor 3 signaling pathways and subsequent production of proinflammatory cytokines and type I IFN in human macrophages. We found that endogenous GMFG localized within multiple endosome compartments, including early and late endosomes, as well as fast and slow recycling endosomes. GMFG knockdown delayed LPS-induced TLR4 internalization and caused prolonged TLR4 retention at the early endosome, suggesting that TLR4 transport from early to late endosomes is interrupted, which may contribute to enhanced LPS-induced TLR4 signaling. Taken together, our findings suggest that GMFG functions as a negative regulator of TLR4 signaling by facilitating TLR4 endocytic trafficking in macrophages.
Introduction
TLRs, which are broadly distributed on the cells of the immune system, play a critical role in innate and adaptive immune responses through recognition of pathogenic molecules and triggering an inflammatory response (1, 2). The most extensively studied TLR in both innate immunity and signal transduction is TLR4. TLR4 senses LPS from Gram-negative bacteria and initiates two separate signaling pathways through the recruitment of MyD88 or TIR domain-containing adaptor-inducing IFN-β (TRIF), which eventually induces the production of proinflammatory cytokines and type I IFN by activation of NF-κB, MAPK, and IFN regulatory factor 3 (IRF3) (3, 4). LPS-induced TLR4 signaling is tightly controlled to prevent excessive inflammation and to allow for tissue repair and the return to homeostasis after infection. Uncontrolled or prolonged activation of TLR4 can have devastating consequences, including the development of immunopathological conditions, such as autoimmune diseases and lethal Gram-negative septic shock (5, 6). Accordingly, much attention has focused on investigating how to negatively regulate TLR4 signaling pathways (7). Although extensive studies have revealed that the negative regulation of TLR4 signaling occurs by means of various mechanisms (8), regulation of the fine-tuning of TLR4-induced signaling by endocytosis and the factors that restrict these processes remain largely unclear.
Endocytic trafficking in the TLR4 signaling pathway has recently emerged as a key feature of TLR4 function. Upon LPS stimulation, TLR4 initiates the first MyD88-dependent signaling pathway at the plasma membrane, and is subsequently internalized along with LPS into the early/sorting endosome network, where the second signaling pathway is triggered through TRIF and TRIF-related adaptor molecule (9–11). TLR4 is then either sorted to late endosomes and lysosomes for its degradation or sent to recycling compartments to be returned to the plasma membrane. Accordingly, LPS-induced endocytosis of TLR4 is essential for its signaling function. Recent studies have revealed that Rab family members regulate TLR4 trafficking. For example, Rab7b localizes to the late endosomes, where it negatively regulates TLR4 trafficking to late endosomes for degradation (12). Rab10 localizes to both the Golgi and early endosomes, where it regulates TLR4 signaling by promoting transport of TLR4 from the Golgi to the plasma membrane (13). Rab11a is enriched in pericentriolar recycling endosomes, and is essential for the trafficking of TLR4 into the Rab11a-positive endocytic recycling compartment (14). Although these GTPase Rabs facilitate vesicle intracellular trafficking, little is known about the surrounding cytoskeletal proteins that regulate endosomal trafficking and the degradation pathway of the TLR4 receptor.
Glia maturation factor-γ (GMFG), a newly recognized actin depolymerization factor/cofilin superfamily protein, is neither involved in the development of glia nor the formation of gliomas; rather, it is predominantly expressed in inflammatory cells and microendothelial cells (15). GMF is a highly conserved protein throughout the eukaryotes from yeast to mammals, with two isoforms being expressed in mammals-GMFB and GMFG. Our previous study and others have suggested that GMFG mediates neutrophil and T-lymphocyte migration via regulation of actin cytoskeletal reorganization (16, 17). Functional studies of Saccharomyces cerevisiae have shown that yeast GMF promotes debranching of actin filaments and inhibits new actin assembly through binding of the Arp2/3 complex (18). Recent studies have demonstrated that Arp2/3 complex and its regulating factors are involved in endosomal dynamics, as well as intracellular protein trafficking (19, 20).
In the present study, we have examined the role of GMFG in LPS-initiated TLR4 signaling in human macrophages. We found that GMFG could serve as a negative regulator of TLR4 signaling in macrophages via promoting TLR4 receptor trafficking from early endosomes to late endosomes.
Materials and Methods
Cells and cultures
The human monocytic leukemia cell line THP-1 was obtained from the American Type Culture Collection and cultured in RPMI 1640 with 10% heat-inactivated FBS, 50 μM β-mercaptoethanol, and 1% penicillin/streptomycin at 37°C in a humidified atmosphere consisting of 5% CO2 and passed every 2–3 d to maintain logarithmic growth. THP-1-derived macrophages were obtained by stimulating THP-1 monocytes (5 × 105/ml) with 100 ng/ml PMA (Sigma-Aldrich) for 3 d.
Human monocytes were isolated from healthy adult donors according to a protocol approved by the Institutional Review Board of the National Heart, Lung, and Blood Institute, National Institutes for Health and consistent with federal regulations. Monocytes were isolated by Ficoll-Paque (GE Healthcare) density centrifugation and magnetically sorted using CD14+ magnetic beads (MACS; Miltenyi Biotec) according to the manufacturer’s instructions. Isolated monocytes were counted and were 85%–95% CD14+ cells. Primary human macrophages were generated by differentiation of CD14+ monocytes in complete culture medium (RPMI 1640 with 10% human AB serum, 2 mM L-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin) supplemented with 10 ng/ml recombinant human GM-CSF (R&D Systems). The medium was changed 3 days after seeding, and cells developed the macrophage phenotype after 7 days in culture.
Isolation of RNA and Quantitative-PCR (Q-PCR)
Total RNA was prepared using the RNeasy Plus Mini kit (Qiagen) according to the manufacturer’s instructions. For reverse transcription, 1 μg total RNA/sample was used as a template for cDNA synthesis using Superscript III (Invitrogen) following the manufacturer’s guidelines. Q-PCR reactions (MyiQ Icycler, Bio-Rad) were performed using prevalidated TaqMan primer/probe sets purchased as Assays-on-Demand gene expression products from Applied Biosystems. Real-time PCR conditions were 5 min at 95°C and 40 cycles of 30 s at 95°C, followed by 1 min at 60°C. RNA copy numbers were calculated by comparison with standard curves generated from plasmid DNA encoding GMFG and control β-actin templates.
RNA interference and plasmid constructions
Day 1 differentiated THP-1 macrophages (2 × 106 cells) or primary human macrophages (2 × 106 cells) were transiently transfected with a GMFG small interfering RNA (siRNA) or negative-control siRNA, or GMFG-GFP plasmid or GFP empty vector, using the Nucleofector Kit V and Nucleofector I Program U-01 (Amaxa Biosystems), according to the manufacturer’s protocol. The GMFG siRNA (Silencer Select siRNA s18303) and negative-control siRNA (Neg-siRNA #2) were obtained from Applied Biosystems. The cells were continually cultured in complete RPMI 1640 medium containing 10% FBS and 100 ng/ml PMA for 48 h prior to use.
Cytokine assays
THP-1 macrophages were stimulated with 100 ng/ml LPS for the indicated time periods. The levels of TNF-α, IL-6, IFN-β, and IL-10 in culture supernatants were measured using commercial ELISA kits from BD Biosciences according to the manufacturer’s instructions.
Luciferase reporter assay
The determinations of NF-κB activity were performed as described previously (21, 22).
Immunoblotting analysis
THP-1 macrophages and primary human macrophages were lysed in RIPA protein extraction reagent (Pierce Biotechnology) as recommended by the manufacturer. Equal amounts of total protein (40 μg/lane) were separated on 4–20% SDS-polyacrylamide gels and transferred to nitrocellulose membranes using standard methods. Membranes were probed with primary Abs for GMFG (ABGENT), TLR4 (Abcam), β-actin, phosphorylated forms of Erk1/2 (Thr202/Tyr204), p38 (Thr180/Tyr182), JNK1/2 (Thr183/Tyr185), IRF3 (Ser396), NF-κB p65, and IκBα (Ser32/36), total p38, NF-κB, or IκBα (Cell Signaling Technology). HRP-conjugated anti-mouse, -rabbit, or -goat IgG secondary Abs and the Amersham ECL Western blotting system (GE Healthcare) were used for detection. Densitometric quantification of signals on blotting membranes was performed using Images J software (National Institutes of Health).
Flow-cytometry analysis
To detect cell-surface expression of TLR4, PMA-differentiated THP-1 macrophages were treated with or without 100 ng/ml LPS for 1 h, then dissociated, blocked in 5% BSA for 30 min, stained with FITC-conjugated anti-TLR4 (Imgenex) or IgG2a Ab for 30 min on ice, washed, and analyzed by EPICS ELITE ESP flow cytometry (Beckman Coulter) and the data were analyzed using CellQuest software (BD Biosciences). Cell-surface expression of TLR4 was quantified as the mean fluorescence intensity of the FITC-positive cells by FACS assay.
Immunofluorescence staining and confocal microscopy
THP-1 macrophages and primary human macrophages were transfected with control siRNA or GMFG siRNA for 48 h. The cells were then treated with 100 ng/ml LPS for the indicated time periods. Following treatment, cells were fixed in 4% paraformaldehyde/PBS for 20 min, permeabilized with 0.1% Triton X-100/PBS for 10 min, and preblocked in 10% FBS/PBS for 1 h. For TLR4 staining, after fixation with 4% paraformaldehyde, cells were further fixed with ice-cold methanol for 20 min. Cell were then stained with mouse monoclonal anti-TLR4 Ab (Abcam; 1:100) for 3 h, followed by staining with rabbit anti-Rab5, anti-Rab4, or anti-Rab7 (Cell Signaling Technology) for 1 h. After washes, the cells were stained with secondary Alexa Fluor 488 anti-mouse or Alexa Fluor 594 anti-rabbit Ab (Invitrogen), diluted 1:500, for 1 h at room temperature. Nuclear DNA was stained with DAPI (Sigma) for 5 min.
For the colocalization analysis of GMFG and organelle markers, cells were sequentially immunostained first with GMFG Ab overnight at 4oC, then stained with Ab against early endosomal-associated protein 1 (EEA1), Rab5, Rab7, Golgi marker GM130 (BD Pharmingen), or ER marker PDI (Invitrogen), then stained with the appropriate Alexa Fluor 488-conjugated or Alexa Fluor 594-conjugated secondary Ab (Invitrogen). Cells were examined under the Zeiss LSM510 META confocal microscope equipped with 405, 488, 594, and 633 nm lasers and Zen 2009 imaging software, using a 63X/1.3 NA oil-immersion objective (Carl Zeiss). Quantitative colocalization analysis was performed using Imaris 7.6 software (Bitplane). Dual-color scattergrams and the percent of co-localization were measured using the Pearsons’ coefficient to evaluate the overlap in fluorescence of TLR4 or GMFG with other endosome markers from four images taken from two independent experiments.
Subcellular fractionation
For subcellular fractionation, THP-1 macrophages were placed on ice, rinsed twice with ice-cold PBS, and subjected to Percoll-density gradient cell fractionation and centrifugation performed as described (12). A total of 12 fractions were collected starting from the top of the gradient. For immunoblotting analysis of each fraction, equal aliquots from each fraction were resolved by SDS-PAGE, and then transferred to nitrocellulose according to standard techniques.
Statistical analysis
Student’s t test was used to analyze data for significant differences. Values of p < 0.05 were regarded as statistically significant.
Results
GMFG is downregulated by LPS stimulation in macrophages
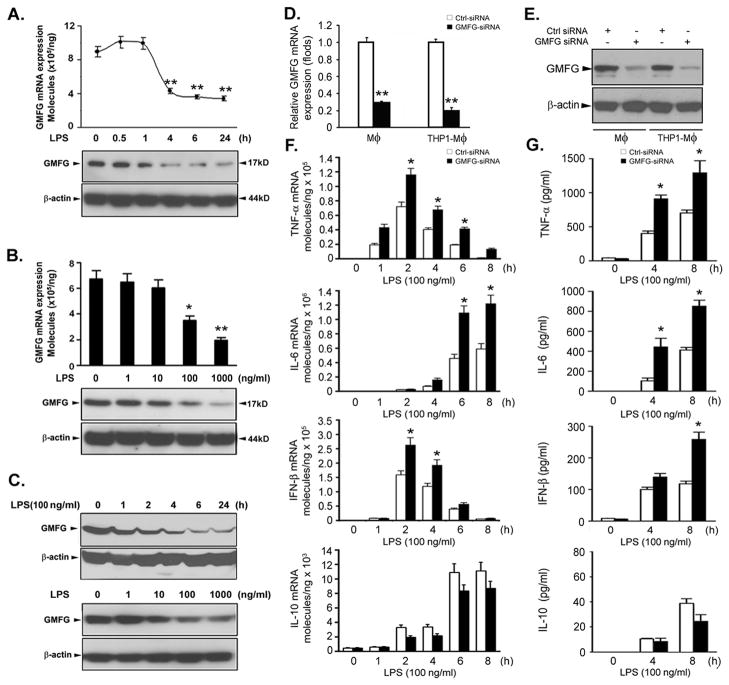
GMFG is expressed mainly in inflammatory cells and microendothelial cells, but its biological function, especially in the innate immune response in macrophages, remains unclear. To explore the possible functions of GMFG in the TLR4 signaling pathway, we first investigated whether GMFG could be regulated by LPS in human macrophages. In primary human macrophages, we found a substantial decrease in expression of GMFG mRNA and protein after stimulation with 100 ng/ml LPS for 4 h that remained low over 24 h (Fig. 1A). The inhibitory effect at 4 h was concentration-dependent and was evident at a range of LPS concentrations from 100 ng/ml to 1000 ng/ml (Fig. 1B). We further verified this effect in THP-1 macrophages produced through PMA stimulation of THP-1 monocytes. Similarly, expression of GMFG mRNA and protein decreased in these cells upon stimulation with LPS (Fig. 1C). These results suggest that GMFG may be involved in regulation of TLR4 signaling in macrophages.
Figure 1. Silencing of GMFG expression enhances LPS-induced production of proinflammatory mediators in macrophages.
(A–C) GMFG expression upon LPS stimulation. Primary human macrophages (A and B) and THP-1 macrophages (C) were treated with 100 ng/ml LPS for the indicated time periods or with the indicated doses of LPS for 24 h; GMFG mRNA expression levels were assessed by Q-PCR and protein levels were determined by immunoblotting. β-actin was used as an internal control. (D and E) Primary human macrophages and THP-1 macrophages were transfected with control negative siRNA (Ctrl siRNA) or GMFG siRNA for 48 h. The efficiency of knockdown was evaluated by Q-PCR and immunoblotting analysis. (F and G) Primary human macrophages transfected with Ctrl siRNA or GMFG siRNA were treated with 100 ng/ml LPS for the indicated time periods. TNF-α, IL-6, IFN-β, and IL-10 mRNA expression levels were assessed by Q-PCR and cytokine production was measured by ELISA. Data are presented as means ± SD of three independent experiments. * p < 0.05, ** p < 0.01.
Silencing of GMFG enhances the production of LPS-induced proinflammatory mediators in macrophages
To determine whether the downregulation of GMFG expression following LPS stimulation has any functional consequences, we silenced the expression of GMFG in human macrophages using a previously verified GMFG siRNA (16). The knockdown efficiency and specificity of the siRNA oligonucleotides were confirmed to be more than 70% in primary human macrophages and THP-1 macrophages (Fig. 1D, 1E). The effect of GMFG knockdown on LPS-induced inflammatory responses was then examined in human macrophages. Knocking down GMFG significantly enhanced the production of TNF-α, IL-6, and IFN-β in response to LPS stimulation at both the mRNA and protein level compared with control siRNA-transfected primary human macrophages (Fig. 1F, 1G). In contrast, IL-10 mRNA and protein levels were decreased in response to LPS in GMFG-knockdown macrophages compared with control siRNA-transfected cells. Similar changes were observed in THP-1 macrophages (data not shown). Because the LPS-induced production of TNF-α and IL-6 is related to LPS-initiated MyD88-mediated NF-κB or MAPK activation, whereas the LPS-induced IFN-β production is largely dependent on TRIF-mediated IRF3 activation, these data suggest that GMFG mediates regulation of the innate immune response in macrophages via MyD88-dependent and TRIF-dependent TLR4 signaling pathways.
Silencing of GMFG expression potentiates LPS-initiated signaling pathways in macrophages
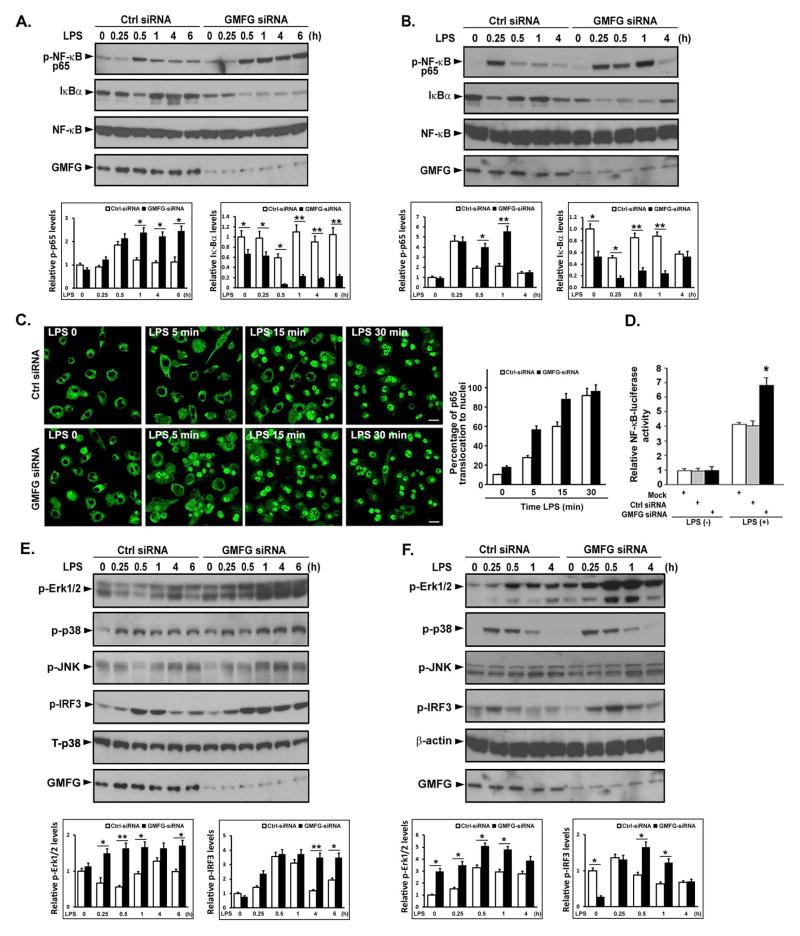
To investigate the molecular basis of the enhanced cytokine production associated with GMFG knockdown, we subsequently assessed the effect of GMFG knockdown on downstream signaling components of the TLR4 signaling pathway. We first evaluated the effect of GMFG knockdown on activation of NF-κB in THP-1 macrophages and primary human macrophages. Knockdown of GMFG led to greatly enhanced phosphorylation of NF-κB p65 and degradation of IκBα after LPS treatment for 30 min in THP-1 macrophages and 60 min in primary human macrophages, and this effect was sustained for 6 h in THP-1 macrophages (Fig. 2A, 2B). In parallel with this immunoblotting analysis, fluorescence microscopy demonstrated enhanced NF-κB activation in GMFG-knockdown THP-1 macrophages. Pronouncedly enhanced nuclear import of p65 was observed as early as after 5 min of LPS treatment of THP-1 macrophages, indicating nuclear translocation of the NF-κB p65 subunit (Fig. 2C). Further, NF-κB luciferase reporter assays of THP-1 macrophages revealed that knockdown of GMFG enhanced LPS-induced transcriptional activity of NF-κB promoters compared with activity observed in control siRNA-transfected cells (Fig. 2D). Next, we examined MAPK activation status after LPS stimulation in GMFG-knockdown THP-1 macrophages and primary human macrophages, because activation of the MAPK pathways also plays an essential role in response to LPS. Knockdown of GMFG in THP-1 macrophages and primary human macrophages revealed remarkably enhanced LPS-induced phosphorylation of Erk1/2 compared with control siRNA-transfected cells, whereas LPS-induced phosphorylation of p38 and JNK was not considerably increased in GMFG-silenced cells compared with control-transfected cells (Fig. 2E, 2F). Finally, analysis of LPS-induced activation of IRF3 also showed increased phosphorylation of IRF3 in GMFG-knockdown THP-1 macrophages and primary human macrophages compared with control-transfected cells (Fig. 2E, 2F). Collectively, these results indicate that GMFG may negatively regulate LPS-initiated activation of the NF-κB, MAPK, and IRF3 pathways.
Figure 2. Silencing of GMFG expression potentiates the LPS-induced signaling pathway in macrophages.
THP-1 macrophages and primary human macrophages were transfected with control negative siRNA (Ctrl siRNA) or GMFG siRNA. After 48 h, the cells were treated with 100 ng/ml LPS for the indicated time periods. (A and B) Cell lysates extracted from THP-1 macrophages (A) or primary human macrophages (B) were prepared and subjected to immunoblotting with the indicated Abs. NF-κB was used as an internal control. Relative quantification of phosphorylation of p65 and degradation of IκBα normalized to total NF-κB is shown in the lower panels. Data are presented as means ± SD of three independent experiments. Values for control negative siRNA (Ctrl siRNA)-transfected cells not stimulated with LPS were set to 1. * p < 0.05, ** p < 0.01. (C) Confocal-microscopy analysis of nuclear translocation of p65 after LPS stimulation in GMFG siRNA- or control negative siRNA (Ctrl siRNA)-transfected THP-1 macrophages. Scale bar, 50 μm. Quantification of p65 translocation to the nuclei is shown in the right panel. Data are presented as means ± SD of three coverslips from a representative experiment. A total of 100 cells were counted per coverslip. (D) THP-1 macrophages that had been knocked down with GMFG siRNA or control negative siRNA were transfected with pTK-Renilla-luciferase and NF-κB luciferase reporter plasmids. After 24 h, the cells were stimulated with 100 ng/ml LPS for 6 h and NF-κB luciferase activity was measured using the Dual-luciferase Reporter Assay System (Promega), normalized to the internal control renilla luciferase activity. Values for mock-transfected cells not stimulated with LPS were set to 1. Data are presented as means ± SD of three experiments. * p < 0.05. (E and F) Cell lysates extracted from GMFG siRNA- or control negative siRNA (Ctrl siRNA)-transfected THP-1 macrophages (E) or primary human macrophages (F) were subjected to immunoblotting with the indicated Abs. Total p38 and β-actin were used as internal controls. Relative quantification of phosphorylation of Erk1/2 and IκBα is shown in the lower panels. Data are presented as means ± SD of three independent experiments. Values for control negative siRNA (Ctrl siRNA)-transfected cells not stimulated with LPS were set to 1. * p < 0.05, ** p < 0.01.
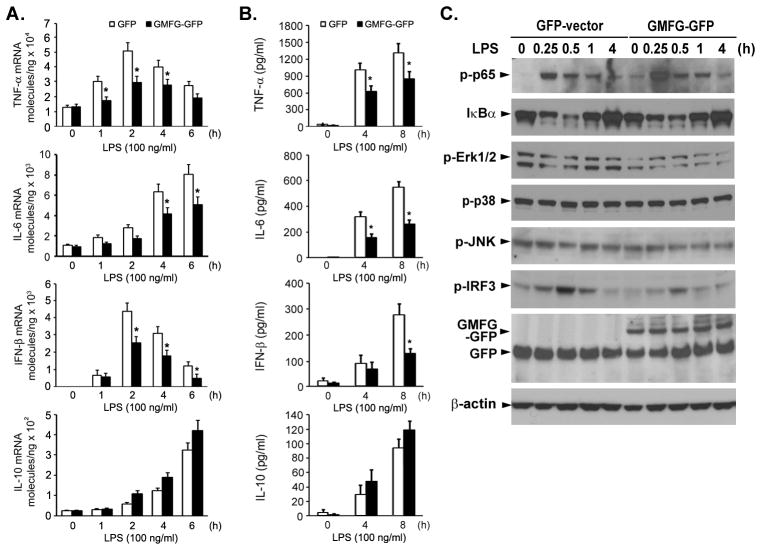
Overexpression of GMFG inhibits LPS-induced cytokine production and LPS-initiated signaling pathways in macrophages
To further support our findings that GMFG may negatively regulate the LPS-initiated signaling pathway, we overexpressed GMFG-GFP in THP-1 macrophages. GMFG-overexpressing THP-1 macrophages produced significantly less TNF-α, IL-6, and IFN-β, but not IL-10, than control-transfected cells in response to LPS (Fig. 3A, 3B), indicating that GMFG overexpression inhibited both MyD88- and TRIF-dependent TLR4 signaling in response to LPS in macrophages. We next evaluated the effect of GMFG overexpression on the LPS-induced phosphorylation state of NF-κB, Erk1/2, and IRF3 in THP-1 macrophages. We found that GMFG overexpression could inhibit LPS-induced phosphorylation of Erk1/2 and IRF3 compared with control vector transfected-cells (Fig. 3C). We also noted that GMFG overexpression had no effect on LPS-induced phosphorylation of p38 and JNK, consistent with the effect of GMFG knockdown on p38 and JNK. These data provide additional evidence that GMFG negatively regulates TLR4-initiated signaling pathways.
Figure 3. Overexpression of GMFG inhibits LPS-induced cytokine production and LPS-initiated signaling pathways in macrophages.
THP-1 macrophages were transfected with GMFG-GFP plasmid or GFP empty vector. After 24 h, the cells were stimulated with 100 ng/ml LPS for the indicated time periods. (A and B) TNF-α, IL-6, IFN-β, and IL-10 mRNA expression levels were measured by Q-PCR and cytokine production was measured by ELISA. Data are presented as means ± SD of three independent experiments. *, p < 0.05 compared with control cells. (C) Cell lysates from GMFG-GFP-transfected and GFP-vector-transfected THP-1 macrophages were prepared and subjected to immunoblotting with the indicated Abs. β-actin was used as an internal control.
Silencing of GMFG inhibits TLR4 internalization
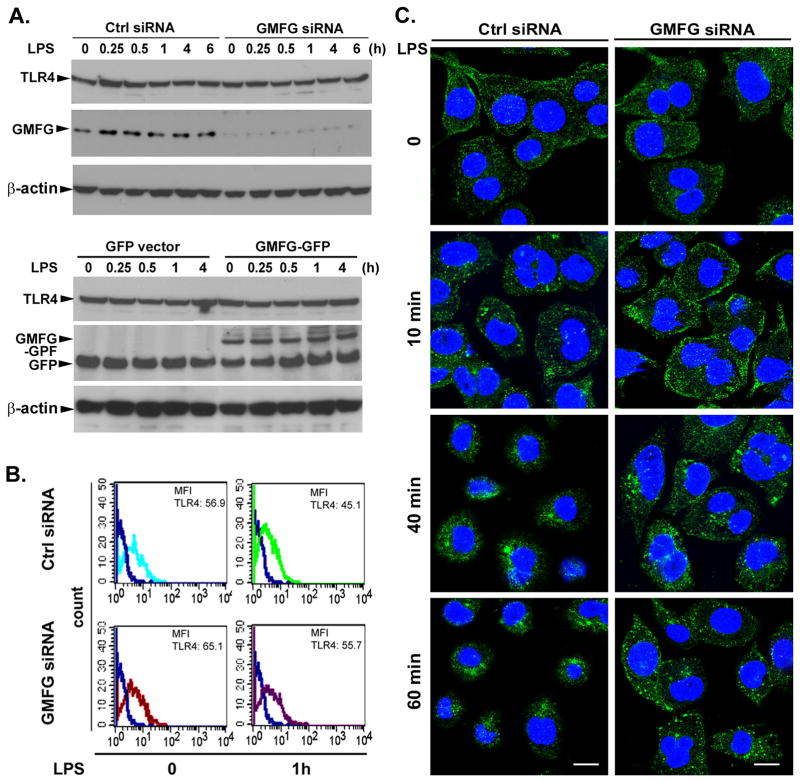
We next investigated the cellular mechanisms by which GMFG could negatively regulate TLR4 signaling. Recent studies have shown that regulation of TLR4 signaling is dependent on TLR4 expression or TLR4 degradation (12, 23, 24). We thus determined whether GMFG could modulate the expression level of TLR4. Immunoblotting analysis indicated that GMFG knockdown or overexpression did not affect total protein levels of TLR4 during the LPS stimulation in THP-1 macrophages (Fig. 4A). However, flow-cytometry analysis showed that GMFG knockdown slightly increased TLR4 surface expression in both unstimulated and stimulated cells compared with control siRNA-transfected macrophages (Fig. 4B). These results suggested that the GMFG effects on TLR4 signaling observed in our earlier results likely were not attributable to changes in cell-surface or total protein expression level of TLR4.
Figure 4. Silencing of GMFG inhibits TLR4 internalization.
(A) Immunoblotting analysis of TLR4 expression in GMFG-silenced or GMFG-overexpressed THP-1 macrophages. Cells were transfected with GMFG siRNA or control negative siRNA (Ctrl siRNA) or GFP empty vector or GMFG-GFP plasmids. After 48 h, the cells were treated with 100 ng/ml LPS for the indicated time periods, then cell lysates were prepared and subjected to immunoblotting with TLR4 or GMFG Ab. β-actin was used as an internal control. (B) Cell-surface TLR4 expression was evaluated by flow cytometry using a specific anti-TLR4 Ab in THP-1 macrophages transfected with GMFG siRNA or control negative siRNA (Ctrl siRNA). Mean fluorescence intensity is shown. (C) TLR4 localization was examined by confocal microscopy after LPS stimulation in GMFG siRNA- or Ctrl siRNA-transfected THP-1 macrophages. Cells were immunostained with anti-TLR4 Ab followed by Alexa Fluor 488-conjugated secondary Ab. Images are representative of three independent experiments. Scale bar, 100 μm.
It has been well established that activated TLR4 can be transported from the cell membrane to endosomes for ubiquitination and to lysosomes for degradation (25). We thus examined whether GMFG mediates subcellular trafficking of TLR4 after LPS stimulation by confocal-microscopy analysis. In unstimulated cells transfected with control siRNA, TLR4 was initially present at the cell surface and in tiny intracellular vesicles that were diffusely distributed throughout the cytoplasm. After 10 min of LPS treatment, TLR4 staining appeared increased intracellular vesicles staining compared with that in unstimulated cells, suggesting TLR4 was internalized into the cytoplasm. At later timepoints (40 min, 60 min) of LPS stimulation, TLR4 was predominantly accumulated in the perinuclear area (Fig. 4C, left panels). In contrast, in GMFG-knockdown cells, TLR4 staining appeared to be concentrated in the periplasma membrane area at 10 min of LPS stimulation, and was sustained in the intracellular vesicles through to the late timepoints (40 min and 60 min) (Fig. 4C, right panels). In addition, control IgG or an isotype matched (irrelevant) mAb showed minimal (or no) background fluorescence for TLR4 or GMFG (data not shown). These results indicate that GMFG is possibly involved in LPS-induced TLR4 internalization.
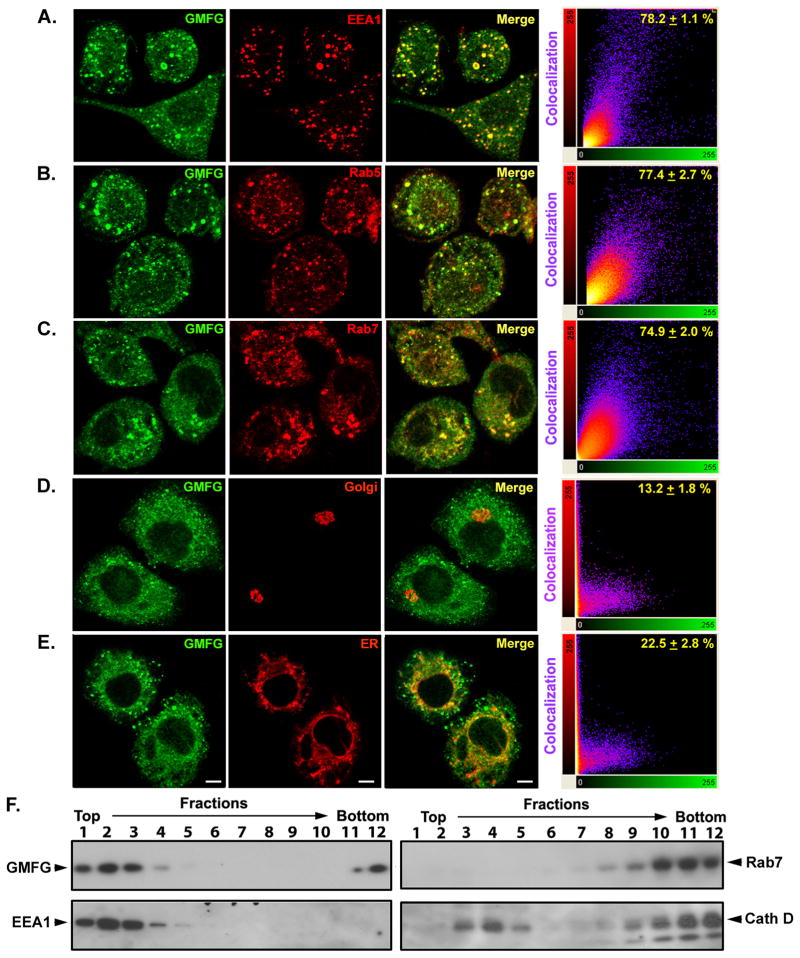
GMFG colocalizes with multiple endosome compartments
To determine the functional consequence of disrupting the LPS-induced internalization of TLR4 by GMFG knockdown in THP-1 macrophages, we examined the subcellular localization of endogenous GMFG in THP-1 macrophages by confocal-microscopy analysis. A large portion of the GMFG was displayed throughout the cytoplasm of the cells, with some high-intensity spots being readily apparent. These spots were reduced in intensity upon GMFG knockdown, confirming the staining’s specificity (data not shown). Moreover, control IgG or an isotype matched (irrelevant) mAb showed minimal (or no) background fluorescence for TLR4 or GMFG (data not shown). To characterize these spots, we attempted to colocalize them with a series of well-characterized organelle membrane markers. Costaining with different membrane-specific markers revealed that endogenous GMFG was highly colocalized with the early/sorting endosome markers EEA1 and Rab5 (Fig. 5A, 5B), as well as the late endosome marker Rab7 (Fig. 5C), but was not colocalized with the Golgi marker GM130 (Fig. 5D) or the endoplasmic reticulum marker PDI (Fig. 5E), suggesting direct linkage of GMFG to the early and late endosomes in THP-1 macrophages.
Figure 5. GMFG colocalizes with multiple endosome compartments.
(A–E) Confocal-microscopy analysis of THP-1 macrophages immunostained with anti-GMFG Ab (green) along with Abs for the indicated organelle membrane markers (red). Scale bar, 100 μm. Right, dual-color pixel analysis of the colocalization of GMFG and various organelle markers. Data are presented as means ± SD of three independent experiments. (F) Steady-state distribution of organelle marker proteins following subcellular fractionation using Percoll-density gradient centrifugation. Postnuclear supernatants of THP-1 macrophages were fractionated on 17% Percoll-density gradients and the individual fractions examined by immunoblotting with specific early endosome (EEA1), late endosome (Rab7), and lysosome (cathepsin D, Cell Signaling Technology) marker Abs, as well as GMFG Ab.
We performed subcellular fractionation using Percoll-density gradient centrifugation to confirm these steady-state findings. Consistent with our confocal-microscopy studies, we found that GMFG was mainly localized in fractions positive for the early endosome marker EEA1 (fractions 1–3), with some GMFG present in fractions positive for the late endosome marker Rab7. In addition, a partial overlap was observed between GMFG and the lysosomal marker cathepsin D in fractions 3 and 12 (Fig. 5F). Altogether, these results suggest that GMFG is an early and late endosome-associated protein in THP-1 macrophages.
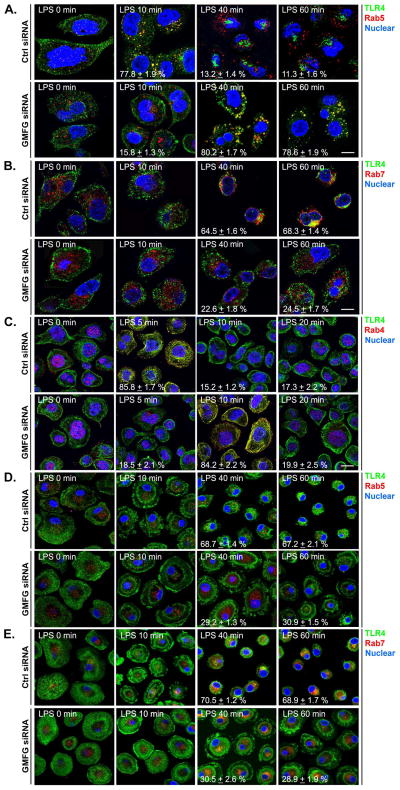
Silencing of GMFG induces abnormal TLR4 endosomal trafficking
Given that GMFG colocalized with EEA1 and Rab5 on early/sorting endosomes and with Rab7 on late endosomes, it is highly likely that GMFG may be involved in transport between these endocytic compartments. Thus, we examined the effect of GMFG knockdown on TLR4 endocytic trafficking in LPS-stimulated THP-1 macrophages and primary human macrophages. GMFG knockdown delayed TLR4 trafficking to Rab5-positive early endosomes at early timepoints (10 min) of LPS stimulation, as we observed that TLR4 did not colocalize with Rab5-positive endosomes at 10 min after LPS stimulation in GMFG siRNA-transfected cells, but could be detected in early endosomes at 10 min after LPS stimulation in control siRNA-transfected cells (Fig. 6A). At late timepoints (40 and 60 min) of LPS stimulation in GMFG-knockdown cells, TLR4 appeared to colocalize with Rab5-positive endosomes (Fig. 6A), and substantially less TLR4 transport to Rab7-positive late endosomes occurred (Fig. 6B). In control cells, following 40–60 min of LPS stimulation, TLR4 no longer colocalized with Rab5-positive early endosomes; instead, most of the TLR4 staining was accumulated in the perinuclear area, colocalized with late endosome marker Rab7 (Fig. 6B).
Figure 6. Silencing of GMFG induces abnormal TLR4 endosomal trafficking.
THP-1 macrophages and primary human macrophages were transfected with GMFG siRNA or control negative siRNA (Ctrl siRNA). After 48 h, the cells were stimulated with or without LPS for the indicated time periods. Following treatment, cells were fixed first with 4% paraformaldehyde for 20 min, then in ice-cold methanol for another 20 min. (A–C) Fixed THP-1 macrophages were immunostained with Abs against TLR4 (green) and Rab5 (early endosome marker; red) (A), Abs against TLR4 (green) and Rab7 (late endosome marker; red) (B), or Abs against TLR4 (green) and Rab4 (fast recycling endosome marker; red) (C). (D and E) Fixed primary human macrophages were immunostained with Abs against TLR4 (green) and Rab5 (red) (D) or Abs against TLR4 (green) and Rab7 (red) (E). Nuclear DNA was labeled with DAPI (blue). Scale bar, 100μm.
To further determine whether GMFG is also involved in TLR4 internalization, we examined TLR4 internalization upon LPS stimulation from very early timepoints (5, 10, and 20 min) using the fast recycling endosome marker Rab4 in GMFG-knockdown cells. Surprisingly, we found that TLR4 rapidly appeared to be colocalized with the Rab4-positive compartment within 5 min of LPS stimulation in control cells, but not with Rab5-positive early endosomes at this early timepoint (Fig. 6C). Conversely, TLR4 transport to Rab4-positive endosomes at the very early timepoint (5 min) of LPS stimulation appeared to be delayed in GMFG-knockdown cells, but TLR4 appeared to be colocalized with the Rab4-positive compartment after 10 min of LPS stimulation (Fig. 6C).
Finally, we examined the endocytic trafficking of TLR4 in GMFG-knockdown primary human macrophages. Consistent with our data from THP-1 macrophages, GMFG-knockdown primary human macrophages exhibited impaired TLR4 transport to the Rab7-positive late endosomes after 40 and 60 min of LPS stimulation (Fig. 6E) compared with control primary human macrophages. Furthermore, in GMFG-knockdown primary human macrophages, TLR4 staining appeared to be retained in the cytoplasmic punctate-and focal contact-like staining, but not accumulated perinuclear area during the late time period of LPS stimulation (Fig. 6D). However, we were unable to detect the colocalization of TLR4 with the early endosome marker Rab5 in both control siRNA-and GMFG siRNA-transfected primary human macrophages. This might represent a technical issue rather than a negative result. Taken together, these results indicate that GMFG not only mediates TLR4 transport from early to late endosomes, but also mediates TLR4 internalization.
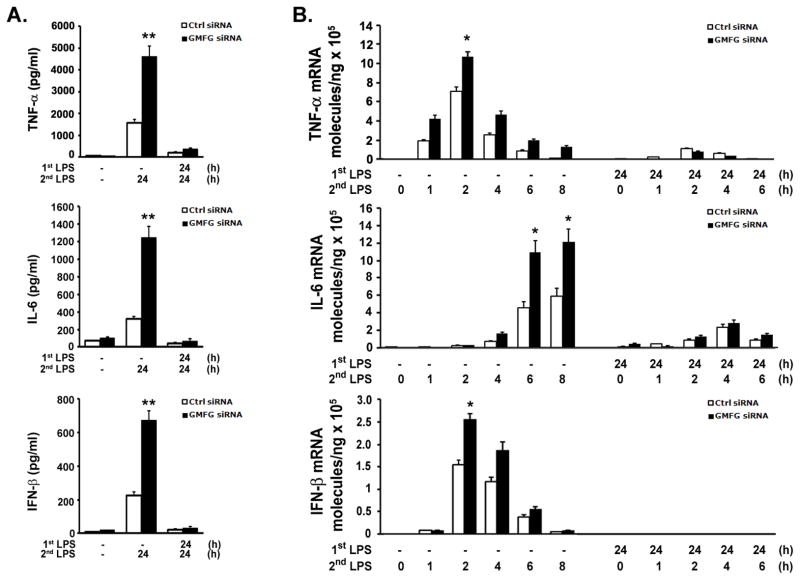
GMFG is not required for endotoxin tolerance
Because GMFG knockdown enhanced LPS-induced inflammatory response, we sought to investigate whether GMFG had an effect on endotoxin tolerance in vitro. We first pretreated THP-1 macrophages transfected with Ctrl siRNA or GMFG siRNA with 10 ng/ml LPS for 24 h and subsequently challenged them with 100 ng/ml LPS for 24 h. Consistent with published reports describing LPS-induced endotoxin tolerance, pretreatment with low-dose LPS nearly completely inhibited TNF-α, IL-6, and IFN-β production after secondary LPS challenge in THP-1 macrophages transfected with Ctrl siRNA (Fig. 7A). GMFG-knockdown cells had substantially enhanced production of TNF-α, IL-6, and IFN-β during the first 24 h after exposure to LPS in unprimed cells, whereas production of these cytokines was blocked over time in low-dose LPS-primed cells in response to secondary LPS challenge (Fig. 7A). To determine whether pretreatment with LPS altered LPS responses at the level of gene expression, we measured mRNA by real-time quantitative PCR. Pretreatment with LPS substantially suppressed subsequent LPS-induced expression of mRNA for TNF-α, IL-6, and IFN-β in both control- and GMFG-knockdown cells (Fig. 7B). These results indicate that GMFG is not required for LPS-induced downregulation of cytokine production.
Figure 7. GMFG is not required for endotoxin tolerance.
THP-1 macrophages were transfected with GMFG siRNA or control negative siRNA (Ctrl siRNA). After 48 h, the cells were treated with LPS (1st LPS; 10 ng/ml) for 24 h, then the supernatants were removed and the cells were washed and challenged with LPS (2nd LPS; 100 ng/ml) for another 24 h. (A) TNF-α, IL-6, and IFN-β levels were measured by ELISA. (B) TNF-α, IL-6, and IFN-β mRNA expression levels were assessed by Q-PCR. Data are presented as means ± SD of three independent experiments. * p < 0.05, ** p < 0.01.
Discussion
Although extensive studies have described the negative regulation of TLR4 signaling, and many negative regulators have been identified (26–30), few reports exist about the regulators that control TLR4 intracellular trafficking upon microbial recognition. In this study, we identified for the first time an important role of GMFG in TLR4 signaling–that GMFG is a novel negative regulator of the MyD88-dependent and TRIF-dependent TLR4 signaling pathways–by demonstrating that GMFG mediated regulation of TLR4 endocytosis in macrophages. Our data reveal that depletion of GMFG results in abnormal trafficking of TLR4 in endosomes in response to LPS stimulation. This abnormal accumulation of TLR4 in early endosomes may enhance the TLR4 signaling pathway.
Endocytosis has emerged as a critical mechanism for controlling the signaling pathway via sorting, internalization, and endocytic trafficking of receptors. Recent studies have defined a requirement for actin dynamics in endocytosis in mammalian cell lines (31, 32). Actin polymerization and depolymerization are tightly regulated to guarantee the correct endocytic membrane transport. Several lines of evidence have shown that actin-depolymerizing protein is involved in receptor trafficking from the endosome to the recycling compartment and degradation in normal cells. For example, cofilin (actin depolymerization factor/cofilin superfamily member)-mediated actin filaments are required for the multiple steps of endocytic trafficking (33, 34). PICK1, an inhibitor of Arp2/3-mediated actin assembly, is required for AMPA receptor endocytosis (35, 36). Therefore, regulation of actin patch disassembly plays an important role in endocytosis. GMFG, another actin depolymerization factor/cofilin superfamily protein, has been reported to function as an actin disassembly protein by inhibiting the Arp2/3 complex in yeast (37). Although GMFG’s action as a disassembly/debranching protein in mammalian cells has yet to be determined, it has a particularly interesting role in TLR4 endocytosis. Our results with GMFG are the first to characterize a role for GMFG in TLR4 trafficking in endosomes in response to LPS, as evidenced by studies in GMFG-knockdown macrophages.
Our confocal-microscopy study revealed that GMFG is found throughout the entire cytosol and colocalizes with the early and late endosome compartments, indicating that GMFG may be involved in TLR4 endocytic trafficking. TLR4 endocytosis at the early phase of LPS stimulation is predominantly clathrin dependent (25, 38), whereas recent studies show that caveolae/lipid raft-mediated endocytosis and macropinocytosis also play important roles in TLR4 endocytosis and signaling activation (39–42). These endocytosis pathways are maintained in a dynamic balance, and inhibition of one endocytosis pathway can shift receptor trafficking to another pathway. For example, siRNA knockdown of clathrin results in a significant inhibition of TLR4 uptake in the early phase of LPS stimulation, whereas TLR4 accumulates in the endosome at the late phase of LPS stimulation, indicating that TLR4 transport occurs by a clathrin-independent pathway (25). We similarly observed that knockdown of GMFG inhibited TLR4 internalization to Rab4-positive endosomes, but appears to prolong the accumulation of TLR4 in Rab5-positive early endosomes, which impairs TLR4 translocation to Rab7-positive late endosomes for further degradation. Thus, it is likely that knockdown of GMFG leads to prolonged TLR4 retention in the plasma membrane, which might enhance LPS-induced NF-κB activation. However, the mechanisms underlying GMFG-mediated TLR4 internalization and endocytic trafficking will need to be further investigated.
TLR4 translocation to the endosomes is required for TRIF-dependent IRF3 activation (11, 38). After transport to the early endosome, TLR4 is sorted and targeted to a trans-Golgi network pathway (43), and diverted to a lysosomal degradation pathway (12, 25), which is required for resolution of the inflammatory response. Minimal perturbation of any of these steps causes an abnormal inflammatory response (12, 44). Indeed, recent studies have shown that abnormal accumulation of TLR4 in early endosomes by inhibition of endocytosis or endosomal sorting leads to increased LPS-induced activation of the NF-κB, MAPK, and IRF3 pathways, which affect the downstream proinflammatory state (12, 45, 46).
Consistent with these reports, our results demonstrate that GMFG knockdown induced delayed TLR4 internalization and abnormal accumulation of TLR4 in early endosomes, which led to significantly enhanced LPS-induced TLR4 signaling. Multiple lines of evidence support this statement. First, we found that knockdown of GMFG enhanced LPS-induced phosphorylation of NF-κB, Erk1/2, and IRF3 and increased production of downstream inflammatory cytokines such as TNF-α, IL-6, and IFN-β. Second, although overexpression of GMFG did not inhibit LPS-induced activation of NF-κB, it clearly inhibited LPS-induced Erk1/2 and IRF3 phosphorylation and downstream cytokines such as TNF-α, IL-6, and IFN-β. This hyperresponsiveness to LPS under GMFG-knockdown conditions could be explained by prolonged accumulation of TLR4 in the early endosomes. Third, LPS induced downregulation of GMFG, which is consistent with a previous study finding in which transfection of Epstein-Barr virus nuclear antigen 2 in B cells also induces downregulation of GMFG (47). This finding suggested that LPS-induced GMFG downregulation is required for inducing optimal activation of the TLR4 signaling, and may be the mechanism used by the host to avoid suppression of the innate immune response aimed at thoroughly removing invading pathogenic microbes. Finally, GMFG is not required for LPS-induced endotoxin tolerance. GMFG is downregulated by stimulation with LPS at dose 100 – 1000 ng/ml. However, at lower dose of LPS stimulation (1 – 10 ng/ml), GMFG was not downregulated. Therefore, our data suggests that regulation of GMFG expression may not be a major mechanism by which the cell regulates its responses to microbial stimuli. Given all the evidence obtained so far, our results strongly suggest that GMFG may negatively regulate TLR4 signaling by the modulation of TLR4 endocytic trafficking in human macrophages.
In this study, our data showed that GMFG knockdown or overexpression did not affect total protein levels of TLR4, which differs from previous studies in which total TLR4 expression is increased along with accumulation of TLR4 in early endosomes (12, 45). Although we cannot provide the conclusive results for the role of GMFG on TLR4 expression using the present data alone, we have provided evidence that silencing of GMFG delayed TLR4 internalization and induced abnormal accumulation of TLR4 in early endosomes. These results suggest that activation of TLR4 signaling after GMFG knockdown is a direct consequence of impaired intracellular membrane trafficking, but not due to changing TLR4 expression levels.
In summary, we have identified and characterized a role for GMFG as a negative regulator of TLR4 signaling, which is facilitated by TLR4 endocytic trafficking that is evoked by its ligand LPS. Hence, further studies of the role of GMFG in the regulation of TLR family members are likely to identify useful therapeutic targets for the treatment of patients with autoimmune, chronic inflammation, or infectious diseases.
Acknowledgments
This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health.
We thank Dr. Dr. Daniela A. Malide from the NHLBI Light Microscopy Core Facility for his skillful help with confocal-microscope imaging.
Abbreviations used in this article
- EEA1
early endosomal-associated protein 1
- GMFG
glia maturation factor-γ
- IRF3
IFN regulatory factor 3
- Q-PCR
quantitative-PCR
- siRNA
small interfering RNA
- TRIF
TIR domain-containing adaptor-inducing IFN-β
Footnotes
Disclosures
The authors have no conflicting financial interests.
References
- 1.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.West AP, Koblansky AA, Ghosh S. Recognition and signaling by toll-like receptors. Annu Rev Cell Dev Biol. 2006;22:409–437. doi: 10.1146/annurev.cellbio.21.122303.115827. [DOI] [PubMed] [Google Scholar]
- 3.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 4.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 5.Barrat FJ, Coffman RL. Development of TLR inhibitors for the treatment of autoimmune diseases. Immunol Rev. 2008;223:271–283. doi: 10.1111/j.1600-065X.2008.00630.x. [DOI] [PubMed] [Google Scholar]
- 6.Beutler B, Rietschel ET. Innate immune sensing and its roots: the story of endotoxin. Nat Rev Immunol. 2003;3:169–176. doi: 10.1038/nri1004. [DOI] [PubMed] [Google Scholar]
- 7.Liew FY, Xu D, Brint EK, O’Neill LA. Negative regulation of toll-like receptor-mediated immune responses. Nat Rev Immunol. 2005;5:446–458. doi: 10.1038/nri1630. [DOI] [PubMed] [Google Scholar]
- 8.Wang J, Hu Y, Deng WW, Sun B. Negative regulation of Toll-like receptor signaling pathway. Microbes Infect. 2009;11:321–327. doi: 10.1016/j.micinf.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 9.Kawai T, Adachi O, Ogawa T, Takeda K, Akira S. Unresponsiveness of MyD88-deficient mice to endotoxin. Immunity. 1999;11:115–122. doi: 10.1016/s1074-7613(00)80086-2. [DOI] [PubMed] [Google Scholar]
- 10.Fitzgerald KA, Palsson-McDermott EM, Bowie AG, Jefferies CA, Mansell AS, Brady G, Brint E, Dunne A, Gray P, Harte MT, McMurray D, Smith DE, Sims JE, Bird TA, O’Neill LA. Mal (MyD88-adapter-like) is required for Toll-like receptor-4 signal transduction. Nature. 2001;413:78–83. doi: 10.1038/35092578. [DOI] [PubMed] [Google Scholar]
- 11.Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, Takeuchi O, Sugiyama M, Okabe M, Takeda K, Akira S. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, Chen T, Han C, He D, Liu H, An H, Cai Z, Cao X. Lysosome-associated small Rab GTPase Rab7b negatively regulates TLR4 signaling in macrophages by promoting lysosomal degradation of TLR4. Blood. 2007;110:962–971. doi: 10.1182/blood-2007-01-066027. [DOI] [PubMed] [Google Scholar]
- 13.Wang D, Lou J, Ouyang C, Chen W, Liu Y, Liu X, Cao X, Wang J, Lu L. Ras-related protein Rab10 facilitates TLR4 signaling by promoting replenishment of TLR4 onto the plasma membrane. Proc Natl Acad Sci USA. 2010;107:13806–13811. doi: 10.1073/pnas.1009428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Husebye H, Aune MH, Stenvik J, Samstad E, Skjeldal F, Halaas O, Nilsen NJ, Stenmark H, Latz E, Lien E, Mollnes TE, Bakke O, Espevik T. The Rab11a GTPase controls Toll-like receptor 4-induced activation of interferon regulatory factor-3 on phagosomes. Immunity. 2010;33:583–596. doi: 10.1016/j.immuni.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ikeda K, Kundu RK, Ikeda S, Kobara M, Matsubara H, Quertermous T. Glia maturation factor-gamma is preferentially expressed in microvascular endothelial and inflammatory cells and modulates actin cytoskeleton reorganization. Circ Res. 2006;99:424–433. doi: 10.1161/01.RES.0000237662.23539.0b. [DOI] [PubMed] [Google Scholar]
- 16.Aerbajinai W, Liu L, Chin K, Zhu J, Parent CA, Rodgers GP. Glia maturation factor-gamma mediates neutrophil chemotaxis. J Leukoc Biol. 2011;90:529–538. doi: 10.1189/jlb.0710424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lippert DN, Wilkins JA. Glia maturation factor gamma regulates the migration and adherence of human T lymphocytes. BMC Immunol. 2012;13:21. doi: 10.1186/1471-2172-13-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gandhi M, Smith BA, Bovellan M, Paavilainen V, Daugherty-Clarke K, Gelles J, Lappalainen P, Goode BL. GMF is a cofilin homolog that binds Arp2/3 complex to stimulate filament debranching and inhibit actin nucleation. Curr Biol. 2010;20:861–867. doi: 10.1016/j.cub.2010.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toshima J, Toshima JY, Martin AC, Drubin DG. Phosphoregulation of Arp2/3-dependent actin assembly during receptor-mediated endocytosis. Nat Cell Biol. 2005;7:246–254. doi: 10.1038/ncb1229. [DOI] [PubMed] [Google Scholar]
- 20.Llado A, Timpson P, Vila de Muga S, Moreto J, Pol A, Grewal T, Daly RJ, Enrich C, Tebar F. Protein kinase Cdelta and calmodulin regulate epidermal growth factor receptor recycling from early endosomes through Arp2/3 complex and cortactin. Mol Biol Cell. 2008;19:17–29. doi: 10.1091/mbc.E07-05-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.An H, Xu H, Zhang M, Zhou J, Feng T, Qian C, Qi R, Cao X. Src homology 2 domain-containing inositol-5-phosphatase 1 (SHIP1) negatively regulates TLR4-mediated LPS response primarily through a phosphatase activity-and PI-3K-independent mechanism. Blood. 2005;105:4685–4692. doi: 10.1182/blood-2005-01-0191. [DOI] [PubMed] [Google Scholar]
- 22.An H, Zhao W, Hou J, Zhang Y, Xie Y, Zheng Y, Xu H, Qian C, Zhou J, Yu Y, Liu S, Feng G, Cao X. SHP-2 phosphatase negatively regulates the TRIF adaptor protein-dependent type I interferon and proinflammatory cytokine production. Immunity. 2006;25:919–928. doi: 10.1016/j.immuni.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 23.Wakabayashi Y, Kobayashi M, Akashi-Takamura S, Tanimura N, Konno K, Takahashi K, Ishii T, Mizutani T, Iba H, Kouro T, Takaki S, Takatsu K, Oda Y, Ishihama Y, Saitoh S, Miyake K. A protein associated with toll-like receptor 4 (PRAT4A) regulates cell surface expression of TLR4. J Immunol. 2006;177:1772–1779. doi: 10.4049/jimmunol.177.3.1772. [DOI] [PubMed] [Google Scholar]
- 24.Sweet MJ, Leung BP, Kang D, Sogaard M, Schulz K, Trajkovic V, Campbell CC, Xu D, Liew FY. A novel pathway regulating lipopolysaccharide-induced shock by ST2/T1 via inhibition of Toll-like receptor 4 expression. J Immunol. 2001;166:6633–6639. doi: 10.4049/jimmunol.166.11.6633. [DOI] [PubMed] [Google Scholar]
- 25.Husebye H, Halaas O, Stenmark H, Tunheim G, Sandanger O, Bogen B, Brech A, Latz E, Espevik T. Endocytic pathways regulate Toll-like receptor 4 signaling and link innate and adaptive immunity. EMBO J. 2006;25:683–692. doi: 10.1038/sj.emboj.7600991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boone DL, Turer EE, Lee EG, Ahmad RC, Wheeler MT, Tsui C, Hurley P, Chien M, Chai S, Hitotsumatsu O, McNally E, Pickart C, Ma A. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol. 2004;5:1052–1060. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- 27.Chuang TH, Ulevitch RJ. Triad3A, an E3 ubiquitin-protein ligase regulating Toll-like receptors. Nat Immunol. 2004;5:495–502. doi: 10.1038/ni1066. [DOI] [PubMed] [Google Scholar]
- 28.Kobayashi K, Hernandez LD, Galan JE, Janeway CA, Jr, Medzhitov R, Flavell RA. IRAK-M is a negative regulator of Toll-like receptor signaling. Cell. 2002;110:191–202. doi: 10.1016/s0092-8674(02)00827-9. [DOI] [PubMed] [Google Scholar]
- 29.Divanovic S, Trompette A, Atabani SF, Madan R, Golenbock DT, Visintin A, Finberg RW, Tarakhovsky A, Vogel SN, Belkaid Y, Kurt-Jones EA, Karp CL. Negative regulation of Toll-like receptor 4 signaling by the Toll-like receptor homolog RP105. Nat Immunol. 2005;6:571–578. doi: 10.1038/ni1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mansell A, Smith R, Doyle SL, Gray P, Fenner JE, Crack PJ, Nicholson SE, Hilton DJ, O’Neill LA, Hertzog PJ. Suppressor of cytokine signaling 1 negatively regulates Toll-like receptor signaling by mediating Mal degradation. Nat Immunol. 2006;7:148–155. doi: 10.1038/ni1299. [DOI] [PubMed] [Google Scholar]
- 31.Kaksonen M, Toret CP, Drubin DG. Harnessing actin dynamics for clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2006;7:404–414. doi: 10.1038/nrm1940. [DOI] [PubMed] [Google Scholar]
- 32.Smythe E, Ayscough KR. Actin regulation in endocytosis. J Cell Sci. 2006;119:4589–4598. doi: 10.1242/jcs.03247. [DOI] [PubMed] [Google Scholar]
- 33.Okreglak V, Drubin DG. Cofilin recruitment and function during actin-mediated endocytosis dictated by actin nucleotide state. J Cell Biol. 2007;178:1251–1264. doi: 10.1083/jcb.200703092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin MC, Galletta BJ, Sept D, Cooper JA. Overlapping and distinct functions for cofilin, coronin and Aip1 in actin dynamics in vivo. J Cell Sci. 2010;123:1329–1342. doi: 10.1242/jcs.065698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rocca DL, Martin S, Jenkins EL, Hanley JG. Inhibition of Arp2/3-mediated actin polymerization by PICK1 regulates neuronal morphology and AMPA receptor endocytosis. Nat Cell Biol. 2008;10:259–271. doi: 10.1038/ncb1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao B, Wang Q, Du J, Luo S, Xia J, Chen YG. PICK1 promotes caveolin-dependent degradation of TGF-beta type I receptor. Cell Res. 2012;22:1467–1478. doi: 10.1038/cr.2012.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakano K, Kuwayama H, Kawasaki M, Numata O, Takaine M. GMF is an evolutionarily developed Adf/cofilin-super family protein involved in the Arp2/3 complex-mediated organization of the actin cytoskeleton. Cytoskeleton (Hoboken) 2010;67:373–382. doi: 10.1002/cm.20451. [DOI] [PubMed] [Google Scholar]
- 38.Kagan JC, Su T, Horng T, Chow A, Akira S, Medzhitov R. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-beta. Nat Immunol. 2008;9:361–368. doi: 10.1038/ni1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zanoni I, Ostuni R, Marek LR, Barresi S, Barbalat R, Barton GM, Granucci F, Kagan JC. CD14 controls the LPS-induced endocytosis of Toll-like receptor 4. Cell. 2011;147:868–880. doi: 10.1016/j.cell.2011.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakahira K, Kim HP, Geng XH, Nakao A, Wang X, Murase N, Drain PF, Wang X, Sasidhar M, Nabel EG, Takahashi T, Lukacs NW, Ryter SW, Morita K, Choi AM. Carbon monoxide differentially inhibits TLR signaling pathways by regulating ROS-induced trafficking of TLRs to lipid rafts. J Exp Med. 2006;203:2377–2389. doi: 10.1084/jem.20060845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Triantafilou M, Miyake K, Golenbock DT, Triantafilou K. Mediators of innate immune recognition of bacteria concentrate in lipid rafts and facilitate lipopolysaccharide-induced cell activation. J Cell Sci. 2002;115:2603–2611. doi: 10.1242/jcs.115.12.2603. [DOI] [PubMed] [Google Scholar]
- 42.Zhu X, Owen JS, Wilson MD, Li H, Griffiths GL, Thomas MJ, Hiltbold EM, Fessler MB, Parks JS. Macrophage ABCA1 reduces MyD88-dependent Toll-like receptor trafficking to lipid rafts by reduction of lipid raft cholesterol. J Lipid Res. 2010;51:3196–3206. doi: 10.1194/jlr.M006486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thieblemont N, Wright SD. Transport of bacterial lipopolysaccharide to the golgi apparatus. J Exp Med. 1999;190:523–534. doi: 10.1084/jem.190.4.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bihl F, Salez L, Beaubier M, Torres D, Lariviere L, Laroche L, Benedetto A, Martel D, Lapointe JM, Ryffel B, Malo D. Overexpression of Toll-like receptor 4 amplifies the host response to lipopolysaccharide and provides a survival advantage in transgenic mice. J Immunol. 2003;170:6141–6150. doi: 10.4049/jimmunol.170.12.6141. [DOI] [PubMed] [Google Scholar]
- 45.Bruscia EM, Zhang PX, Satoh A, Caputo C, Medzhitov R, Shenoy A, Egan ME, Krause DS. Abnormal trafficking and degradation of TLR4 underlie the elevated inflammatory response in cystic fibrosis. J Immunol. 2011;186:6990–6998. doi: 10.4049/jimmunol.1100396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzuki M, Sugimoto Y, Ohsaki Y, Ueno M, Kato S, Kitamura Y, Hosokawa H, Davies JP, Ioannou YA, Vanier MT, Ohno K, Ninomiya H. Endosomal accumulation of Toll-like receptor 4 causes constitutive secretion of cytokines and activation of signal transducers and activators of transcription in Niemann-Pick disease type C (NPC) fibroblasts: a potential basis for glial cell activation in the NPC brain. J Neurosci. 2007;27:1879–1891. doi: 10.1523/JNEUROSCI.5282-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schlee M, Krug T, Gires O, Zeidler R, Hammerschmidt W, Mailhammer R, Laux G, Sauer G, Lovric J, Bornkamm GW. Identification of Epstein-Barr virus (EBV) nuclear antigen 2 (EBNA2) target proteins by proteome analysis: activation of EBNA2 in conditionally immortalized B cells reflects early events after infection of primary B cells by EBV. J Virol. 2004;78:3941–3952. doi: 10.1128/JVI.78.8.3941-3952.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]